Abstract
Purpose
To develop a PCR gel analysis method for assessing the bacterial bioburden in orthokeratology contact lens (OK) case fluid determined by culture.
Methods
A prospective study with the participation of 41 OK wearers (20 girls, 21 boys) was performed. The mean OK-wearing experience was 3.5±1.9 years. PCR was used to assess the bacterial bioburden (colony-forming units per milliliter) of OK after removal and soaking in the storage case for 6 h. The signal intensity of the PCR bands was analyzed after grayscale image transformation. The difference (cPCR-d) and ratio (cPCR-r) between a PCR signal and its background were used as two standardized indices of PCR signals. The association between the two indices of the PCR signals and the bacterial bioburden determined by culture were analyzed with Pearson’s correlation coefficient (r) and receiver operating characteristic (ROC) plots.
Results
At least one microbe was isolated from the OK lens case from 38 of the 41 subjects. Both cPCR-d and cPCR-r showed strong correlations with the bacterial bioburden (r>0.7, p<0.0001). ROC analysis enabled good determination of the cutoff values for the two PCR indices with acceptable sensitivity and specificity (78−89%) to assess the degree of bacterial contamination.
Conclusions
The high microbial contamination rate of the OK lens cases revealed the general inappropriate lens care by OK wearers. PCR analysis provides an alternative and rapid method for assessing the bacterial bioburden of OK lens cases, and these results should serve as a warning to OK wearers to follow appropriate lens care procedures to prevent infection.
Introduction
Contact lenses (CL) are common and convenient optical aids for correcting visual acuity. The U.S. Centers for Disease Control and Prevention (CDC, Atlanta, GA) recently stated that, regardless of type, CL wear is the largest single risk factor for developing infection [1-3]. According to the CDC’s statement, wearing CL overnight and not cleaning and replacing storage cases frequently are some of the key behaviors that increase the risk for keratitis [3-5]. Orthokeratology contact lenses (OK) are designed using reverse-geometry rigid gas-permeable CL for temporarily flattening the central cornea after overnight wear, thus effectively molding the cornea to reduce myopia and astigmatism [6,7]. OK have become increasingly popular for pediatric myopic control in areas with a high prevalence of myopia [8,9]; however, safety concerns related to their use remain. Several studies have reported an association between OK wear and microbial keratitis (MK) [10-13]. The microbial contamination (bioburden) of the CL/OK system has been proposed to be an important causal factor of MK [14-19] and is generally high in the OK system [20]. The lens storage case is the most frequently contaminated item in the lens care system [14,18,21]. Furthermore, an association between cultures of corneal scrapings and CL storage case fluids has been observed in cases of CL-related MK, suggesting that case fluids may be useful for identifying pathogens in patients with negative corneal scraping results [22,23]. Therefore, assessing lens case contamination may provide information about microbes with the potential for transfer to the lens surface and ultimately to the ocular surface [24,25].
Quantification of the overall level of bacterial contamination may be more valuable than identification of individual bacteria in the assessment of lens care quality. For example, Pseudomonas aeruginosa, the most common pathogen causing CL-related MK, has been recognized in the normal flora of CL wearers [26]. Therefore, we previously proposed that the total bioburden, determined as the colony-forming units per milliliter (CFU/ml) of all isolated bacteria, in a lens care system could be used as a key indicator of lens care quality [20].
Culture-based assessment of the bioburden is a time-consuming process and may fail in the presence of fastidious microbes. To solve this problem, our study group previously proposed a dot hybridization model for assessment of the bacterial bioburden. This model showed good correlation with the bacterial bioburden determined by culture and could effectively classify the contamination level into different grades with high sensitivity and specificity [27]. However, some laboratories may not be able to adopt this technique given that it requires use of a spotter and is relatively costly. Therefore, we here evaluated the application of PCR gel electrophoresis, a common and popular laboratory technique, as an alternative method for determining the bacterial bioburden. This method is easy to implement and may be suitable for the rapid assessment of lens care quality in combination with digital image analysis. The aim of this study was to elucidate the efficacy of PCR gel electrophoresis combined with standardized digital image analysis as an alternative approach to culture-based methods for assessing the bioburden of lens case fluid in determining pediatric OK care quality.
Methods
Subjects
Subjects were included in this prospective study if they had undergone OK treatment (Euclid Systems Orthokeratology, Euclid Systems Corporation, Herndone, VA) for 1 year or more at Kaohsiung Chang Gung Memorial Hospital (CGMH), a tertiary hospital center in southern Taiwan [20]. Subjects who were younger than 8 or older than 18 years of age, had renewed their lens cases within 3 days before sample collection, and were unable to complete the scheduled follow-up were excluded from the study. The aims and procedures were explained to the subjects and their parents/guardians before the study, and informed consent was obtained from all subjects [20]. All procedures involving human subjects adhered to the principles set forth in the Declaration of Helsinki. This study also adhered to the ARVO statement on human subjects. Institutional Review Board (IRB)/Ethics Committee approval was obtained from the Committee of Medical Ethics and Human Experiments of CGMH.
Sample collection
Subjects were asked to remove their OK after overnight wearing and then to rinse, clean, and soak the OK in their own lens cases for approximately 6 h according to their usual habits as described in our previous study [20]. Briefly, the subjects and/or their responsible parents/guardians brought the entire OK case, including the OK and fluids contained within the case, to our outpatient department on the same day, and the OK care system was transferred to the laboratory for assessment. After the OK was removed from the lens case, the inner surface of the case was rubbed with an aseptic cotton swab. The OK case fluids were collected by micropipettes and transferred into microcentrifuge tubes. Part of the case fluid was immediately inoculated on the culture media (see below), and the other part was stored at 0 °C for subsequent DNA extraction.
Microbiological investigation: Culture-based methods
Blood agar, chocolate agar, and eosin methylene blue agar (all from BBLTM, Becton Dickinson, Sparks, MD) were used as the standard culture system [20]. Ten microliters of the case fluid was then homogenously spread on each culture plate at room temperature. After incubation at 35 °C in an atmosphere containing 5% CO2 for 72 h, the bacteria obtained from the three culture media were identified with Gram staining, followed by standard biochemical tests for identification of bacteria [20]. The bioburden (CFU/ml) of each isolated strain was recorded for each case, which was used to characterize the degree of bacterial contamination in each lens case.
DNA extraction and PCR amplification
The case fluid sample (1 ml) was centrifuged at 13,200 ×g for 10 min in a microcentrifuge tube. DNA in the precipitates was extracted using a commercial kit (DNeasy Blood & Tissue Kit, Qiagen, Valencia, CA), and then stored at −70 °C. Bacterial universal primers, designed for targeting the 16S rRNA gene of bacteria, with two forward primers (Ba2-f and Ba3-f) and one reverse primer (Ba2-r), were used (Table 1).
Table 1. Primers used in this study.
| Primer | Orientation | Primer sequence (5′ to 3′)a | Pseudomonas aeruginosa Accession number KC211291 |
|---|---|---|---|
| Ba2-f |
Forward |
GCTCAACCTGGGAACTGC |
604~621 bp |
| Ba3-f |
Forward |
GAGGGTCATTGGAAACTG |
346~363 bp |
| Ba2-r | Reverse | AACCCAACATCTCACGACAC | 1059~1078 bp |
aThe target gene of the primer pairs is the 16S rRNA gene of bacteria.
After thawing, each PCR mixture (25 µl) was prepared with 2.5 μl template DNA, 0.2 μM of each forward primer, 0.4 μM of reverse primer, and other reagents obtained from the PCR kit (JMR-THS5, JMR Holdings, Inc., St. Augustine, FL). PCR amplification was performed in an automated thermocycler (GeneAmp PCR system 2700, Applied Biosystems, Foster City, CA) under the following cycling conditions: initial denaturation (95 °C for 3 min), 35 cycles of denaturation (95 °C for 30 s), annealing (55 °C for 30 s), extension (72 °C for 20 s), and final extension (72 °C for 10 min). The amplicons were examined through electrophoresis on a 2% agarose gel containing DNA binding fluorescent dye (SMOBIO Technology Inc., Hsinchu, Taiwan). Two negative controls were also run simultaneously by replacing the template DNA with human DNA (extracted from the corneal epithelium of a noninfectious keratitis patient) and a new opened multipurpose solution [Bioclean (OPHTECS Corporation, Toyooka, Japan) multipurpose solution routinely used by these OK wearers for lens care] in the PCR mixture. Successful amplification was indicated if a clear band of 400–600 bp appeared on the agarose gel.
PCR gel analysis
The signal intensities of the PCR products were quantified with ImageJ software (developed by Wanye Rasband, National Institutes of Health, Bethesda, MD; available at RSB). Briefly, each captured image of the PCR gel was transformed to grayscale for subsequent single-band quantification. The background for a specific PCR band was defined as the hollow rectangle area extending outward from the PCR band by about 1 PCR band width (Figure 1). The sampling procedure was performed in triplicate for each PCR band and its background, and the mean pixel intensities were obtained by averaging the sampled PCR band and background gray levels, respectively. Two indices of the signal intensity of the PCR band were used: the corrected difference in the intensity of the PCR band and its background (cPCR-d) and the corrected ratio of the PCR band intensity and background intensity (cPCR-r).
Figure 1.
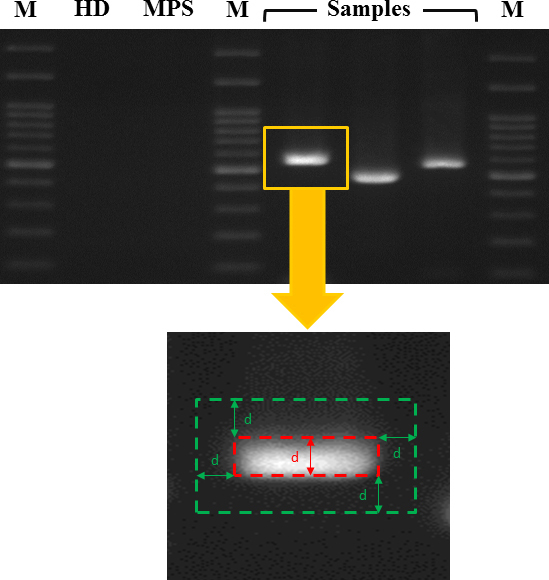
The PCR gel electrophoresis signal was analyzed with ImageJ software from digital images of the gel. A PCR band was recognized and defined visually. The background of a PCR band was defined as the hollow rectangular area extending outward by about 1 PCR band width. After grayscale image transformation, the signals of a PCR band and its background were each recorded in triplicate to determine their mean intensities. M=marker lane. HD=human DNA. MPS=multipurpose solution.
Statistical analyses
A general linear model was applied to determine the correlation between the signal intensities of the PCR products and the culture-based bacterial bioburden. Pearson’s correlation coefficient was calculated using Excel 2010 (Microsoft Corporation, Redmond, WA). Receiver operating characteristic (ROC) analysis was used to determine the threshold PCR signal intensity at the predetermined cutoff values of the bacterial bioburden determined by culture. The predetermined cutoff values were used to classify the levels of bacterial contamination as rare (≤200 CFU/ml), light (201–2,000 CFU/ml), moderate (2,001–12,000 CFU/ml), and heavy (>12,000 CFU/ml) contamination [20]. Statistical significance was accepted at p<0.05.
Results
Participants
The demographic data were collected based on related studies for the same project [20,27]. A total of 41 experienced OK wearers, including 20 girls and 21 boys, participated in this study. The mean age and OK-wearing experience were 12.7±2.6 (range 8–18) years and 3.5±1.9 (range 1–8) years, respectively.
Bacterial bioburden of the OK lens cases determined by culture
The culture results were summarized according to previous related studies of the same project [20,27]. Of the 41 samples collected, 38 (93%) were contaminated with at least one microbe. The median bacterial bioburden of each case estimated by culture was 2,600 CFU/ml (25th and 75th percentile, 350 and 11,000 CFU/ml, respectively). Subjects with three isolated strains (n=19; median=10,900 CFU/ ml; 25th and 75th percentile, 3,300 and 22,500 CFU/ml, respectively) had a significantly higher bacterial bioburden (p=0.00028) than those with fewer isolated strains (≤2 strains, n=22, median=600 CFU/ ml; 25th and 75th percentile, 100 and 2,225 CFU/ml, respectively). Male subjects (n=21, median=6,500 CFU/ ml; 25th and 75th percentile, 250 and 21,250 CFU/ml, respectively) had a significantly higher (p=0.018) bacterial bioburden than female subjects (n=20, median=1,600 CFU/ ml; 25th and 75th percentile, 400 and 4,625 CFU/ml, respectively). This result might be confounded by tap water–rinsing behavior because male subjects showed an increased tendency to use tap water for lens care (p=0.08). No significant difference in bacterial bioburden was found when the subjects were stratified by age.
Association between culture-based bacterial bioburden and PCR analysis
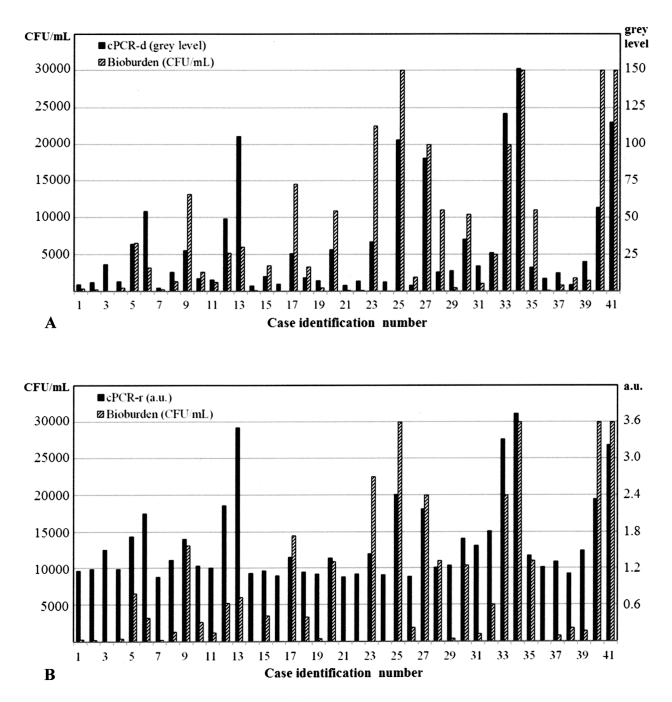
The comparison of the bacterial bioburden determined by culture and the PCR signals from each subject is shown in Figure 2. Three culture-negative subjects showed positive PCR signal intensities.
Figure 2.
Comparison of the bacterial bioburden (colony-forming units per milliliter [CFU/ml]) and two indices of polymerase chain reaction (PCR) analysis for each case. A: Bacterial bioburden versus cPCR-d (the corrected difference in PCR intensity, defined as the difference in the mean intensities of a PCR band and its background). B: Bacterial bioburden versus cPCR-r (the corrected ratio of the PCR intensity, defined as the ratio of the mean intensities of a PCR band and its background). a.u., arbitrary unit.
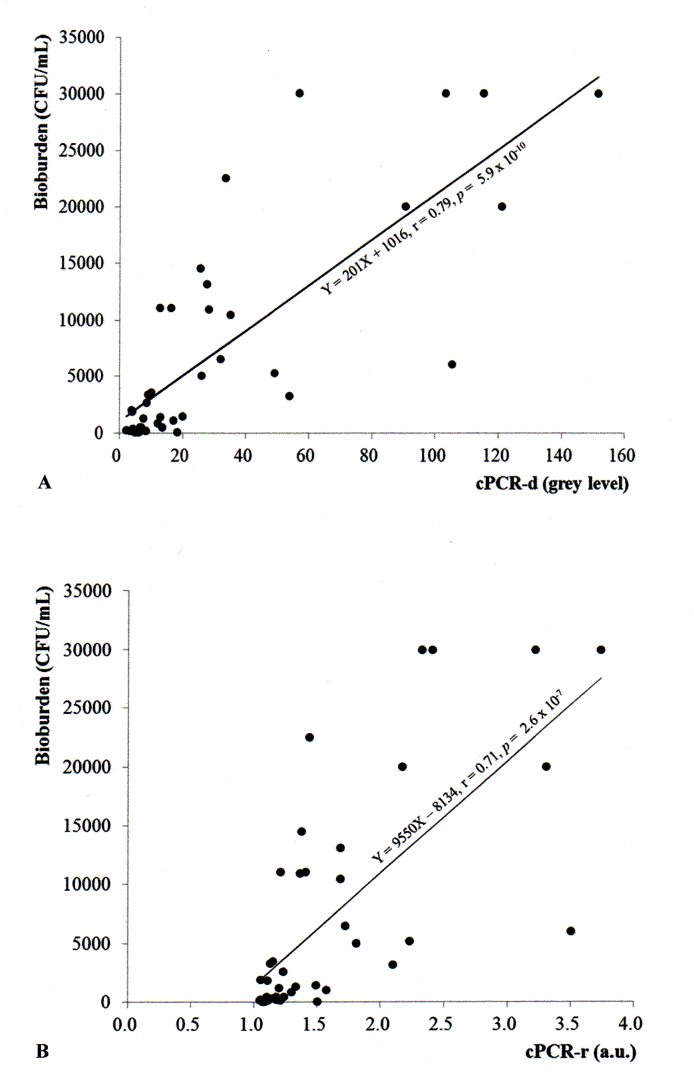
Both indices of the PCR analysis (cPCR-d and cPCR-r) were significantly correlated with the bacterial bioburden determined by culture (Figure 3), according to the following linear models: [bacterial bioburden=201 × (cPCR-d) + 1016] and [bacterial bioburden=9550 × (cPCR-r) – 8134]. The difference-based index cPCR-d showed a slightly stronger correlation with bacterial bioburden than the ratio-based index cPCR-r (r=0.79, p=5.9 × 10−10 and r=0.71, p=2.6 × 10−7, respectively).
Figure 3.
Correlation analysis between signal indices of polymerase chain reaction (PCR) bands and bacterial bioburden (colony-forming units per milliliter [CFU/ml]). A: Correlation between cPCR-d (corrected difference in PCR band intensity and background intensity) and the bacterial bioburden. B: Correlation between cPCR-r (corrected ratio of the PCR intensity and background intensity) and the bacterial bioburden.
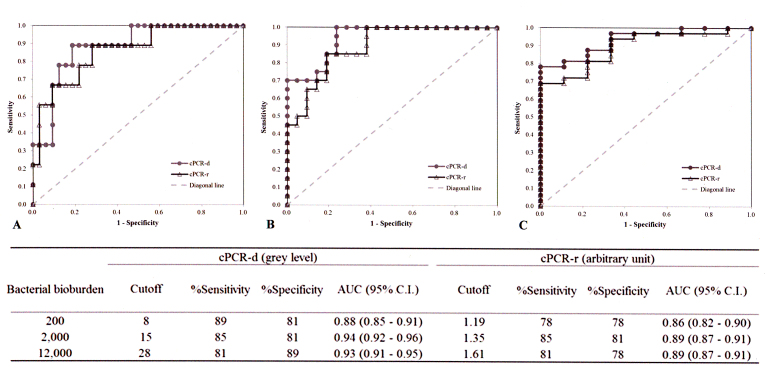
ROC analysis was applied to determine the cutoff values of the two indices of PCR analysis (cPCR-d and cPCR-r) based on three predetermined bacterial bioburden levels that are commonly used in clinical practice to grade bacterial contamination (Figure 4). According to the area under the curves of ROC plots, cPCR-d showed slightly better grading performance than cPCR-r (0.88−0.93 versus 0.86−0.89, respectively) and had higher sensitivity and specificity than cPCR-r (81−89% versus 78−85% and 81−89% versus 78−81%, respectively). The interpretation of the bioburden using the two indices of PCR analysis is summarized in Table 2.
Figure 4.
Determination of cutoff values of the signal indices of PCR analysis based on bacterial bioburden determined by culture. The cutoff values corresponding to the predetermined bacterial bioburden, 200 colony-forming units (CFU)/ml (A), 2,000 CFU/ml (B), and 12,000 CFU/ml (C) of bacteria, were determined with receiver operating characteristic plots. The performances of the cutoff values of PCR indices are summarized in the inserted table. AUC=area under curve. C.I.=confidence interval.
Table 2. Interpretation of bioburden using the signal indices of polymerase chain reaction (PCR) analysis.
| Gradea | Culture |
Indices of PCRb analysis |
|
|---|---|---|---|
| Bioburden (CFU/ml) | cPCR-dc (gray level) | cPCR-rd (a.u.e) | |
| Rare |
≤200 |
≤8 |
≤1.19 |
| Light |
201–2000 |
9–15 |
1.20–1.35 |
| Moderate |
2001–12000 |
16–28 |
1.36–1.61 |
| Heavy | ≥12,001 | ≥29 | ≥1.62 |
aGrade of bacterial contamination. bPCR, polymerase chain reaction gel electrophoresis. ccPCR-d, corrected difference in the PCR band intensity from the background. dcPCR-r, corrected ratio in the PCR band intensity from the background. ea.u., arbitrary unit.
Discussion
The issue of lens care quality is of great importance, and rapid assessment of bacterial contamination in the OK care system is a useful method for identifying and warning wearers who practice improper lens care to prevent ocular infections. Conventional culture procedures are time-consuming and may fail because of the use of inappropriate media or due to the presence of fastidious or nonviable microorganisms. In this study, we evaluated the feasibility of PCR analysis for the rapid assessment of lens care quality by comparing the results obtained with this method to the bacterial bioburden determined by culture. Our results showed that PCR analysis is a rapid and useful alternative method for assessing bioburden, based on two novel indices: cPCR-d and cPCR-r.
PCR is a technology developed in the 1980s and is now widely applied in molecular biology to amplify a small amount of DNA across several orders of magnitude through thermal cycling [28]. Although real-time PCR [29,30] is a faster and more quantitative method [31], it requires more sophisticated equipment and may not be cost-effective for broad assessments of the bacterial bioburden in the lens care system. We previously proposed the dot hybridization assay as a valuable tool for preventing CL-related MK by assessing bacterial bioburden of an OK lens storage case [27]. However, this assay requires a spotter, which limits its application for several laboratories. Therefore, the PCR analysis proposed herein, in combination with free digital image software to objectively quantify the signal intensities of the PCR gel, should serve as an inexpensive and easy method for all laboratories to assess the bacterial bioburden. Compared with the universal bacteria probes PB2 and PB3 of the dot hybridization model [27], the indices c-PCR-d and cPCR-r showed higher correlations with the culture results and higher sensitivity and specificity for classifying the degree of bacterial contamination into different grades of severity.
Nonetheless, there are some limitations to this study. The PCR efficiency might not be equal for different microorganisms due to differences in the DNA extraction efficiency. This factor may create a selection bias. Three subjects (case no. 3, 16, and 24) were culture-negative but PCR-positive (Figure 2), indicating that PCR gel electrophoresis detected some nonviable bacteria instead of only culture-positive bacteria. However, the PCR analysis indices (cPCR-d and cPCR-r) showed high correlations with the bioburden determined by culture (Figure 3A,B). Moreover, ROC analysis enabled the establishment of cutoff signals for the PCR analysis indices (cPCR-d and cPCR-r) to grade the bacterial bioburden as rare, light, moderate, or heavy (Table 2) with high sensitivity and specificity.
In conclusion, we developed a PCR-based analysis method for assessing the bacterial bioburden of OK lens cases, which is easy to implement, rapid, and effective for identification of improper contact lens care to promote improved lens care quality. This method should be useful to prevent OK-related MK by enabling rapid assessment and feedback for the growing population of OK wearers.
Acknowledgments
The authors acknowledge the Genomic & Proteomic Core Laboratory, Department of Medical Research, Chang Gung Memorial Hospital at Kaohsiung for supplying a high-resolution image scanner. This work was supported by Chang Gung Research Proposal (CMRPG8B1181, CMRPG8C0761), and from the Ministry of Science and Technology (Grant No. MOST 103–2314-B-182A-044). The funding organizations had no role in the design or conduct of this research. There are no potential conflicts of interest to declare. Some of the results of this study were presented at the 50th mid-year Taiwan academic congress in May 2015.
References
- 1.Dart JK, Stapleton F, Minassian D. Contact lenses and other risk factors in microbial keratitis. Lancet. 1991;338:650–3. doi: 10.1016/0140-6736(91)91231-i. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=1679472&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 2.Green M, Apel A, Stapleton F. Risk factors and causative organisms in microbial keratitis. Cornea. 2008;27:22–7. doi: 10.1097/ICO.0b013e318156caf2. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18245962&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 3.Keay L, Stapleton F, Schein O. Epidemiology of contact lens-related inflammation and microbial keratitis: a 20-year perspective. Eye Contact Lens. 2007;33:346–53. doi: 10.1097/ICL.0b013e318157c49d. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17975418&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 4.Schein OD, Glynn RJ, Poggio EC, Seddon JM, Kenyon KR. The relative risk of ulcerative keratitis among users of daily-wear and extended-wear soft contact lenses. A case-control study. Microbial Keratitis Study Group. N Engl J Med. 1989;321:773–8. doi: 10.1056/NEJM198909213211201. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=2671733&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 5.Poggio EC, Glynn RJ, Schein OD, Seddon JM, Shannon MJ, Scardino VA, Kenyon KR. The incidence of ulcerative keratitis among users of daily-wear and extended-wear soft contact lenses. N Engl J Med. 1989;321:779–83. doi: 10.1056/NEJM198909213211202. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=2770809&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 6.Woo GC, Wilson MA. Current methods of treating and preventing myopia. Optom Vis Sci. 1990;67:719–27. doi: 10.1097/00006324-199009000-00012. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=2234833&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 7.Nichols JJ, Marsich MM, Nguyen M, Barr JT, Bullimore MA. Overnight orthokeratology. Optom Vis Sci. 2000;77:252–9. doi: 10.1097/00006324-200005000-00012. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10831215&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 8.Cho P, Cheung SW, Edwards M. The longitudinal orthokeratology research in children (LORIC) in Hong Kong: a pilot study on refractive changes and myopic control. Curr Eye Res. 2005;30:71–80. doi: 10.1080/02713680590907256. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15875367&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 9.Chan B, Cho P, Cheung SW. Orthokeratology practice in children in a university clinic in Hong Kong. Clin Exp Optom. 2008;91:453–60. doi: 10.1111/j.1444-0938.2008.00259.x. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18355342&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 10.Tseng CH, Fong CF, Chen WL, Hou YC, Wang IJ, Hu FR. Overnight orthokeratology-associated microbial keratitis. Cornea. 2005;24:778–82. doi: 10.1097/01.ico.0000153101.81657.0b. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16160491&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 11.Hsiao CH, Yeung L, Ma DH, Chen YF, Lin HC, Tan HY, Huang SC, Lin KK. Pediatric microbial keratitis in Taiwanese children: a review of hospital cases. Arch Ophthalmol. 2007;125:603–9. doi: 10.1001/archopht.125.5.603. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17502497&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 12.Young AL, Leung KS, Tsim N, Hui M, Jhanji V. Risk factors, microbiological profile, and treatment outcomes of pediatric microbial keratitis in a tertiary care hospital in Hong Kong. Am J Ophthalmol. 2013;156:1040–4. doi: 10.1016/j.ajo.2013.06.019. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23972308&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 13.Lee YS, Tan HY, Yeh LK, Lin HC, Ma DH, Chen HC, Chen SY, Chen PY, Hsiao CH. Pediatric microbial keratitis in Taiwan: clinical and microbiological profiles, 1998–2002 versus 2008–2012. Am J Ophthalmol. 2014;157:1090–6. doi: 10.1016/j.ajo.2014.01.013. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24487048&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 14.Boost MV, Cho P. Microbial flora of tears of orthokeratology patients, and microbial contamination of contact lenses and contact lens accessories. Optom Vis Sci. 2005;82:451–8. doi: 10.1097/01.opx.0000168587.72893.ec. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15976581&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 15.Wu YT, Zhu H, Harmis NY, Iskandar SY, Willcox M, Stapleton F. Profile and frequency of microbial contamination of contact lens cases. Optom Vis Sci. 2010;87:E152–8. doi: 10.1097/OPX.0b013e3181cf86ee. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20101194&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 16.Devonshire P, Munro FA, Abernethy C, Clark BJ. Microbial contamination of contact lens cases in the west of Scotland. Br J Ophthalmol. 1993;77:41–5. doi: 10.1136/bjo.77.1.41. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8435399&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boost MV, Shi G, Cho P. Comparison of contamination rates of designs of rigid contact lens cases. Optom Vis Sci. 2012;89:E1030–4. doi: 10.1097/OPX.0b013e31825da44a. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22729171&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 18.Cho P, Boost M, Cheng R. Non-compliance and microbial contamination in orthokeratology. Optom Vis Sci. 2009;86:1227–34. doi: 10.1097/OPX.0b013e3181bbc55d. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19786928&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 19.Szczotka-Flynn LB, Pearlman E, Ghannoum M. Microbial contamination of contact lenses, lens care solutions, and their accessories: a literature review. Eye Contact Lens. 2010;36:116–29. doi: 10.1097/ICL.0b013e3181d20cae. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20168237&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo J, Kuo MT, Chien CC, Tseng SL, Lai YH, Fang PC. Microbial Bioburden of Orthokeratology Contact Lens Care System. Eye Contact Lens. 2016;42:61–7. doi: 10.1097/ICL.0000000000000130. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25723564&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 21.Kuzman T, Kutija MB, Juri J, Jandroković S, Skegro I, Olujić SM, Kordić R, Cerovski B. Lens wearers non-compliance - is there an association with lens case contamination? Cont Lens Anterior Eye. 2014;37:99–105. doi: 10.1016/j.clae.2013.08.004. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24012202&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 22.Das S, Sheorey H, Taylor HR, Vajpayee RB. Association between cultures of contact lens and corneal scraping in contact lens related microbial keratitis. Arch Ophthalmol. 2007;125:1182–5. doi: 10.1001/archopht.125.9.1182. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17846356&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 23.Chan TC, Li EY, Wong VW, Jhanji V. Orthokeratology-Associated Infectious Keratitis in a Tertiary Care Eye Hospital in Hong Kong. Am J Ophthalmol. 2014;158:1130–5. doi: 10.1016/j.ajo.2014.08.026. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25158307&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 24.Mayo MS, Schlitzer RL, Ward MA, Wilson LA, Ahearn DG. Association of Pseudomonas and Serratia corneal ulcers with use of contaminated solutions. J Clin Microbiol. 1987;25:1398–400. doi: 10.1128/jcm.25.8.1398-1400.1987. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=3114318&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keay LJ, Gower EW, Iovieno A, Oechsler RA, Alfonso EC, Matoba A, Colby K, Tuli SS, Hammersmith K, Cavanagh D, Lee SM, Irvine J, Stulting RD, Mauger TF, Schein OD. Clinical and microbiological characteristics of fungal keratitis in the United States, 2001–2007: a multicenter study. Ophthalmology. 2011;118:920–6. doi: 10.1016/j.ophtha.2010.09.011. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21295857&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen EP, Tsay RY, Chia JS, Wu S, Lee JW, Hu FR. The role of type III secretion system and lens material on adhesion of Pseudomonas aeruginosa to contact lenses. Invest Ophthalmol Vis Sci. 2012;53:6416–26. doi: 10.1167/iovs.11-8184. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22918630&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 27.Kuo MT, Chien CC, Lo J, Hsiao CC, Tseng SL, Lai YH, Fang PC, Chang TC. A DNA dot hybridization model for assessment of bacterial bioburden in orthokeratology lens storage cases. Invest Ophthalmol Vis Sci. 2015;56:445–50. doi: 10.1167/iovs.14-15920. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25537200&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 28.Bartlett JM, Stirling D. A short history of the polymerase chain reaction. Methods Mol Biol. 2003;226:3–6. doi: 10.1385/1-59259-384-4:3. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12958470&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 29.Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology. 1992;10:413–7. doi: 10.1038/nbt0492-413. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=1368485&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 30.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16460794&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 31.Taravati P, Lam D, Van Gelder RN. Role of molecular diagnostics in ocular microbiology. Curr Ophthalmol Rep. 2013;1:181–9. doi: 10.1007/s40135-013-0025-1. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24416712&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]