This report describes the process of radiomics, its challenges, and its potential power to facilitate better clinical decision making, particularly in the care of patients with cancer.
Abstract
In the past decade, the field of medical image analysis has grown exponentially, with an increased number of pattern recognition tools and an increase in data set sizes. These advances have facilitated the development of processes for high-throughput extraction of quantitative features that result in the conversion of images into mineable data and the subsequent analysis of these data for decision support; this practice is termed radiomics. This is in contrast to the traditional practice of treating medical images as pictures intended solely for visual interpretation. Radiomic data contain first-, second-, and higher-order statistics. These data are combined with other patient data and are mined with sophisticated bioinformatics tools to develop models that may potentially improve diagnostic, prognostic, and predictive accuracy. Because radiomics analyses are intended to be conducted with standard of care images, it is conceivable that conversion of digital images to mineable data will eventually become routine practice. This report describes the process of radiomics, its challenges, and its potential power to facilitate better clinical decision making, particularly in the care of patients with cancer.
Introduction
With high-throughput computing, it is now possible to rapidly extract innumerable quantitative features from tomographic images (computed tomography [CT], magnetic resonance [MR], or positron emission tomography [PET] images). The conversion of digital medical images into mineable high-dimensional data, a process that is known as radiomics, is motivated by the concept that biomedical images contain information that reflects underlying pathophysiology and that these relationships can be revealed via quantitative image analyses. Although radiomics is a natural extension of computer-aided diagnosis and detection (CAD) systems, it is significantly different from them. CAD systems are usually standalone systems that are designated by the Food and Drug Administration for use in either the detection or diagnosis of disease (1). Early successes of CAD have been greatest in breast cancer imaging (2,3). Unlike CAD systems, which are directed toward delivering a single answer (ie, presence of a lesion or cancer), radiomics is explicitly a process designed to extract a large number of quantitative features from digital images, place these data in shared databases, and subsequently mine the data for hypothesis generation, testing, or both. Radiomics is designed to develop decision support tools; therefore, it involves combining radiomic data with other patient characteristics, as available, to increase the power of the decision support models. As radiomics is intended to extract maximal information from standard of care images, the creation of databases that combine vast quantities of radiomics data (and ideally other complementary data) from millions of patients is foreseeable.
Although radiomics can be applied to a large number of conditions, it is most well developed in oncology because of support from the National Cancer Institute (NCI) Quantitative Imaging Network (QIN) and other initiatives from the NCI Cancer Imaging Program. As described in subsequent sections of this article, the potential of radiomics to contribute to decision support in oncology has grown as knowledge and analytic tools have evolved. Quantitative image features based on intensity, shape, size or volume, and texture offer information on tumor phenotype and microenvironment (or habitat) that is distinct from that provided by clinical reports, laboratory test results, and genomic or proteomic assays. These features, in conjunction with the other information, can be correlated with clinical outcomes data and used for evidence-based clinical decision support (Fig 1). Radiomics appears to offer a nearly limitless supply of imaging biomarkers that could potentially aid cancer detection, diagnosis, assessment of prognosis, prediction of response to treatment, and monitoring of disease status.
Figure 1:
Flowchart shows the process of radiomics and the use of radiomics in decision support. Patient work-up requires information from disparate sources to be combined into a coherent model to describe where the lesion is, what it is, and what it is doing. Radiomics begins with acquisition of high-quality images. From these images, a region of interest (ROI) that contains either the whole tumor or subregions (ie, habitats) within the tumor can be identified. These are segmented with operator edits and are eventually rendered in three dimensions (3D). Quantitative features are extracted from these rendered volumes to generate a report, which is placed in a database along with other data, such as clinical and genomic data. These data are then mined to develop diagnostic, predictive, or prognostic models for outcomes of interest.
The mining of radiomic data to detect correlations with genomic patterns is known as radiogenomics, and it has elicited especially great interest in the research community. To avoid confusion, it should be noted that the term radiogenomics is also used in the field of radiation oncology to describe whole-genome analyses aimed at determining the genetic causes of variations in radiosensitivity (4,5). Henceforward in this article, we will refer to radiogenomics only as the combination of radiomic features with genomic data for the purpose of enabling decision support. The value of radiogenomics stems from the fact that while virtually all patients with cancer undergo imaging at some point and often multiple times during their care, not all of them have their disease genomically profiled. Furthermore, when genomic profiling is performed, it is done one time at one location and is susceptible to sampling error. Thus, radiogenomics has two potential uses, which will be described in detail in the Examples of Radiomics Results section. First, a subset of the radiomic data can be used to suggest gene expression or mutation status that potentially warrants further testing. This is important because the radiomic data are derived from the entire tumor (or tumors) rather than from just a sample. Thus, radiomics can provide important information regarding the sample genomics and can be used for cross-validation. Second, a subset of radiomic features is not significantly related to gene expression or mutational data and, hence, has the potential to provide additional, independent information. The combination of this subset of radiomic features with genomic data may increase diagnostic, prognostic, and predictive power.
While radiomics primarily grew out of basic research, lately it has also elicited interest from those in clinical research, as well as those in daily clinical practice. For a clinical radiologist, radiomics has the potential to help with the diagnosis of both common and rare tumors. Visualization of tumor heterogeneity may prove critical in the assessment of tumor aggressiveness and prognosis. For example, research has already shown the capacity of radiomics analyses to help distinguish prostate cancer from benign prostate tissue or add information about prostate cancer aggressiveness (6). In the evaluation of lung cancer and in the evaluation of glioblastoma multiforme, radiomics has been shown to be a tool with which to assess patient prognosis (7). The tools developed for radiomics can help in daily clinical work, and radiologists can play a pivotal role in continuously building the databases that are to be used for future decision support.
The suffix -omics is a term that originated in molecular biology disciplines to describe the detailed characterization of biologic molecules such as DNA (genomics), RNA (transcriptomics), proteins (proteomics), and metabolites (metabolomics). Now, the term is also being used in other medical research fields that generate complex high-dimensional data from single objects or samples (8). One desirable characteristic of -omics data is that these data are mineable and, as such, can be used for exploration and hypothesis generation. The -omics concept readily applies to quantitative tomographic imaging on multiple levels: One multisection or three-dimensional image from one patient may easily contain millions of voxels. Also, one tumor (or other abnormal entity) may contain hundreds of measurable features describing size, shape, and texture.
Radiomics analyses epitomize the pursuit of precision medicine, in which molecular and other biomarkers are used to predict the right treatment for the right patient at the right time. The availability of robust and validated biomarkers is essential to move precision medicine forward (9). Around the world, efforts are underway to improve the availability of such biomarkers, and in the United States, the effort is most notably through The Precision Medicine Initiative (10,11). This initiative will provide funding for a new model of patient-powered research that promises to accelerate biomedical discoveries and provide clinicians with new tools, knowledge, and therapies that enable more precise personalized care.
A major strength of a radiomics approach for cancer is that digital radiologic images are obtained for almost every patient with cancer, and all of these images are potential sources for radiomics databases (Table 1). In the United States alone, there are approximately 1.6 million new cancer cases every year (12). Most of these patients will undergo multiple CT, MR imaging, and PET examinations. In the future, it is possible that image interpretation for all these studies will be augmented by using radiomics, building an unprecedented source of big data that will expand the potential for discovering helpful correlations. While radiomics will allow better characterization of patients and their diseases through new applications of genomics and improved methods of phenotyping, it will also add to the challenges of data management, as we will discuss later in this article.
Table 1.
Important Aspects of Radiomics

Radiomics offers important advantages for assessment of tumor biology. It is now appreciated that most clinically relevant solid tumors are highly heterogeneous at the phenotypic, physiologic, and genomic levels (13–15) and that they continue to evolve over time. In this emerging era of targeted therapies, it is notable that most responses are not durable and that benefit is generally measured in months, not years. For example, this is the case with (a) gefitinib in patients with epidermal growth factor receptor–mutated lung cancer (16), (b) trastuzamab in those with human epidermal growth factor receptor 2 (or HER2) overexpressing breast cancer (17), and (c) vemurafenib in those with B-Raf–mutated melanoma (18). Genomic heterogeneity within tumors and across metastatic tumor sites in the same patient is the major cause of treatment failure and emergence of therapy resistance (19). Thus, precision medicine requires not only in vitro biomarkers and companion diagnostics but also spatially and temporally resolved in vivo biomarkers of tumor biology. A central hypothesis driving radiomics research is that radiomics has the potential to enable quantitative measurement of intra- and intertumoral heterogeneity. Moreover, radiomics offers the possibility of longitudinal use in treatment monitoring and optimization or in active surveillance. Although such applications of radiomics have yet to be explored in depth, they may provide the most value going forward.
It should be emphasized that radiomic and radiogenomic analyses can be used to identify correlations, not causes; thus, they are not expected to enable definitive assessment of genetic or other contents of tissue through imaging alone. However, correlation of radiomic data with genomic or other -omic data could inform not only the decision about whether to test for certain gene alterations in biopsy samples but also the choice of biopsy sites. It also could provide confirmatory information to support histopathologic findings. This is important, as it is estimated that the error rate of cancer histopathology can be as high as 23% (20–23). Errors in histopathology are due to both sampling errors and observer variability; thus, there is a great need for additional quantitative diagnostic information.
We believe that radiomics is rapidly expanding beyond a boutique research area and is emerging as a translational technology. Hence, this is an appropriate time to begin to establish benchmarks for data extraction, analysis, and presentation. The goal of this report is to introduce the practice of radiomics to a wide audience of practicing clinicians, including radiologists, to engage a broader community in establishing benchmarks. In doing so, we will describe the processes involved in radiomics and the unique information it offers, as well as its challenges and their potential solutions. We will also highlight some of the more recent findings of importance and, finally, offer a vision for radiomics of the future.
Process of Radiomics
While conceptually simple, the practice of radiomics involves discrete steps, each with its own challenges (24,25). These steps are shown in Figure 1 and include: (a) acquiring the images, (b) identifying the volumes of interest (ie, those that may contain prognostic value), (c) segmenting the volumes (ie, delineating the borders of the volume with computer-assisted contouring), (d) extracting and qualifying descriptive features from the volume, (e) using these to populate a searchable database, and (f) mining these data to develop classifier models to predict outcomes either alone or in combination with additional information, such as demographic, clinical, comorbidity, or genomic data. We will discuss these processes in turn.
Image Acquisition
Modern CT, MR imaging, and combined PET/CT units allow for wide variations in acquisition and image reconstruction protocols, and standardization of these protocols across medical imaging centers is typically lacking. This is generally not a problem in the routine identification of radiologic features used in clinical practice. However, when images are analyzed numerically to extract meaningful data, variations in acquisition and image reconstruction parameters can introduce changes that are not due to underlying biologic effects. This has been well recognized in the emerging field of quantitative imaging, in which the intent is to generate medical images with describable limits of bias and variance. In other words, it is not sufficient to report a number or a set of numbers derived from images; instead, we must also be able to provide error bars, as is done with every other credible laboratory measurement.
There have been multiple efforts to advance quantitative imaging, including definition of acquisition and reconstruction standards, over the past 15 years (26,27). The QIN is a cooperative network initiated by the NCI with the goal of developing quantitative imaging methods that improve the effectiveness of clinical trials of new cancer therapies (28). The QIN is a major initiative from the NCI and can be regarded as the leading edge of new imaging methods, including radiomics. Also, the Radiological Society of North America (RSNA) and the National Institute for Biomedical Imaging and Bioengineering have sponsored the Quantitative Imaging Biomarkers Alliance (QIBA), which is a major effort in quantitative imaging (29). The goal of QIBA is to industrialize quantitative imaging by bringing together the entire spectrum of groups involved in its development and implementation. The main product of QIBA is a new type of a document termed a profile that provides a consensus on the measurement accuracy of a quantitative imaging biomarker for a specific use and the requirements and procedures needed to achieve this level of measurement accuracy. More than 100 participants were involved in the creation of the initial QIBA fluorodeoxyglucose PET/CT profile, which was released in 2014 (30,31). The American Association of Physicists in Medicine is providing technical guidelines in quantitative imaging in the form of modality-dependent reports on imager operation and testing. Finally, relevant professional societies, such as the American College of Radiology, RSNA, the Society of Nuclear Medicine and Molecular Imaging, the International Society of Magnetic Resonance in Medicine, and the World Molecular Imaging Society, are increasingly including aspects of the bedrock of quantitative imaging in their guidelines.
Volume of Interest Identification
Identification of tissue volumes of prognostic value is the core of the practice of radiology in oncology. Although at the time of diagnosis cancer can be detected at one tumor site or multiple tumor sites, most patients with cancer metastasis have multiple lesions. In either scenario, we need to identify tumors and suspected tumors as volumes of interest. However, detailed analysis of subvolumes within tumors (the manifestations of tumor heterogeneity) that may have prognostic value generally are not captured in a radiology report because of the spatial and contrast limitations of digital images. While heterogeneity is not included in Response Evaluation Criteria in Solid Tumors, version 1.1 (32), a few texture descriptors have been incorporated in more complex diagnostic imaging reporting and data systems, such as the Breast Imaging Reporting and Data System (BI-RADS) (33), the Prostate Imaging Reporting and Data System (PI-RADS) (34), and the Lung Imaging Reporting and Data System (Lung-RADS) (35). In the practice of radiomics, so-called subvolumes of interest can be captured and added to the analyses. The basic philosophy, which has its foundation in process engineering, is to capture as much data as possible at the front end and use downstream database mining to identify the features with the highest prognostic value. This is driven by the knowledge that attempting to filter the data at input would be inefficient and would presuppose knowledge regarding the value of the features in classifier models before they were tested.
Recently, the concept of using image data to identify physiologically distinct regions within lesions has been described (36). In this approach, images with different acquisition parameters (eg, contrast material–enhanced T1-weighted MR imaging, diffusion-weighted, and fluid attenuation sequences) can be combined to yield regions with specific combinations of quantitative image data. Notably, when this is performed, the combinations reside in spatially explicit regions of the tumors (Fig 2). We have termed these regions habitats because they represent physiologically distinct volumes, each with a specific combination of blood flow, cell density, necrosis, and edema. Additional radiomic features can be extracted from each of these habitats to obtain highly granular descriptions of cancer lesions. The distribution of these habitats in patients with glioblastoma multiforme, for example, can enable us to discriminate between cancers that progress quickly (<400 days of survival) and those that are more indolent (37). Furthermore, these habitats change after treatment (eg, treatment with radiation and temazolamide), and the pattern of change has been observed to be predictive of response.
Figure 2:
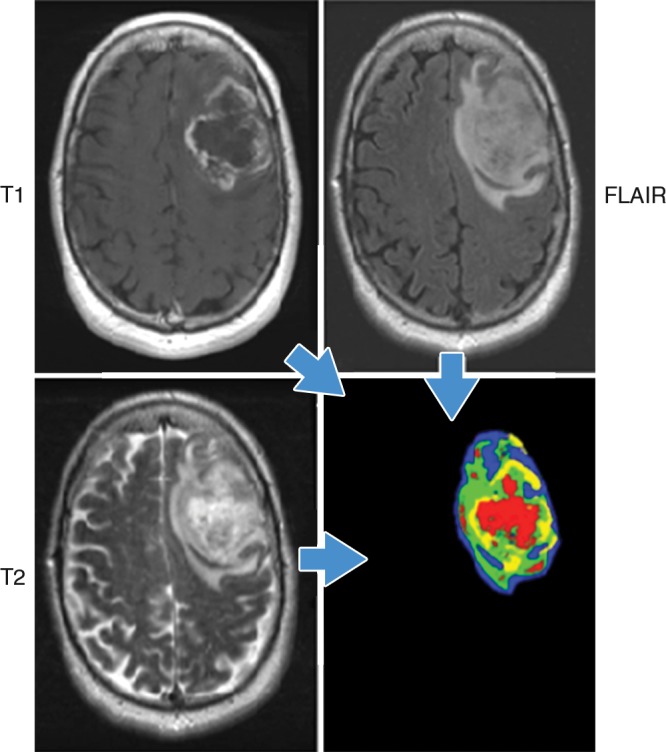
Habitats in a patient with glioblastoma multiforme. Habitats were defined by combining unenhanced and contrast-enhanced T1-weighted, 120-msec echo time T2-weighted, and fluid-attenuated inversion recovery (FLAIR) images. Data from each acquisition were sorted into low and high values with automated histogram analyses, yielding a potential for eight different combinations. In practice, only four distinct combinations were observed. They correspond to the red (low T1, high T2 and FLAIR), yellow (low T1 and T2, high FLAIR), blue (High T1 and FLAIR, low T2), and green (high T1, low FLAIR and T2) areas. Notably, while the identities of individual voxels were determined mathematically, they spatially clustered into contiguous regions reflecting different physiologic microenvironments. (Image courtesy of R. A. Gatenby.)
Segmentation
Segmentation is the most critical, challenging, and contentious component of radiomics. It is critical because the subsequent feature data are generated from the segmented volumes. It is challenging because many tumors have indistinct borders. It is contentious because there are ongoing debates over whether to seek ground truth or reproducibility and how much to rely on manual or automatic segmentation. However, a consensus is emerging that truth is elusive and that optimum reproducible segmentation is achievable with computer-aided edge detection followed by manual curation. It is well recognized that interoperator variability of manually contoured tumors is high (38,39). Segmentation of normal structures, such as skeletal elements and organs, can now be achieved with full automation. However, any disease, especially cancer, requires operator input because of inter- and intrasubject morphologic and contrast heterogeneity at the initial examination.
Feature Extraction and Qualification
The heart of radiomics is the extraction of high-dimension feature data to quantitatively describe attributes of volumes of interest. In practice, “semantic” and “agnostic” features are the two types of features extracted in radiomics. (Table 2). Semantic features are those that are commonly used in the radiology lexicon to describe regions of interest, while agnostic features are those that attempt to capture lesion heterogeneity through quantitative descriptors.
Table 2.
Examples of Semantic and Agnostic Features of Radiomics

Semantic features.—Although semantic features are commonly used by radiologists to describe lesions, in this article we refer to their quantification with computer assistance. With the foreknowledge that semantic features are of prognostic value, early investigations in radiomics developed radiology lexicons, much the same as BI-RADS, PI-RADS, and Lung-RADS attempt to do. A watershed article in this regard came from Segal et al, who, in an early example of radiogenomics, used a finite series of radiologist-scored quantitative features to predict gene expression patterns in hepatocellular carcinoma (40). This approach continues to have high value, and there is a movement to capture such semantic data with the aid of computers to achieve higher interreader agreement, faster throughput, and lower variance.
Agnostic features.—Agnostic radiomic features on an image are mathematically extracted quantitative descriptors, which are generally not part of the radiologists’ lexicon. These can be divided into first-, second-, or higher-order statistical outputs. First-order statistics describe the distribution of values of individual voxels without concern for spatial relationships. These are generally histogram-based methods and reduce a region of interest to single values for mean, median, maximum, minimum, and uniformity or randomness (entropy) of the intensities on the image, as well as the skewness (asymmetry) and kurtosis (flatness) of the histogram of values. Second-order statistical descriptors generally are described as “texture” features; they describe statistical interrelationships between voxels with similar (or dissimilar) contrast values. Texture analysis of images was first introduced in 1973 by Haralick et al (41). In radiomics, texture analyses can readily provide a measure of intratumoral heterogeneity. In practice, there are dozens of methods and multiple variables that can be used to extract texture features, resulting in hundreds of values—far too many to elaborate on in this article. Readers are referred to some excellent reviews on the subject, with specific reference to intratumoral heterogeneity (42,43). Higher-order statistical methods impose filter grids on the image to extract repetitive or nonrepetitive patterns. These include fractal analyses, wherein patterns are imposed on the image and the number of grid elements containing voxels of a specified value is computed (44); Minkowski functionals, which assess patterns of voxels whose intensity is above a threshold (45); wavelets, which are filter transforms that multiply an image by a matrix of complex linear or radial “waves”; and Laplacian transforms of Gaussian bandpass filters that can extract areas with increasingly coarse texture patterns from the image (46).
There has been a sustained effort to identify, define, and extract more agnostic features. The first such study used 182 texture features in combination with 22 semantic features to describe CT images of lung cancer (24). This was followed by a 442-member feature set that also contained wavelets (47). More recently, this has been expanded to 662 features that also contain Laplace transforms of Gaussian fits (46) and 522 features that include texture and fractal dimension features (48). These features potentially can be extracted from individual habitats, thereby yielding thousands of data elements with which to describe each volume of interest, with many volumes of interest available in each patient.
Thus, it is readily apparent that the number of descriptive image features can approach the complexity of data obtained with gene expression profiling, which commonly yields information on more than 30 000 different sequences. With such large complexity, there is a danger of overfitting analyses, and hence, dimensionality must be reduced by prioritizing the features (49,50). The most systematic approach is to first identify features that may be redundant (ie, those that are highly correlated with one another). Figure 3 is a covariance matrix of 219 features extracted from CT scans in 143 patients with non–small cell lung cancer. Those features that are highly correlated (r2 > 0.95) with each other are shown as red off-diagonal elements. Clusters of highly correlated features can be collapsed into one representative feature, usually the one with the largest intersubject variability or highest dynamic range. Figure 3 also provides a conceptual bridge to the other -omics fields, where the data content of the images is indicated by the false-color map. If available, test-retest data are also extremely helpful, as they can help prioritize features on the basis of their reproducibility (51,52). A further level of prioritization, described by Aerts et al (47), is to rank order features within separate categories representing different agnostic and semantic classes of features (eg, size, shape, and first-, second-, and higher-order textures). The classifier models can then be built with the two or three highest-priority features in each class. In the final analysis, the value of feature sets is determined by their contribution to classifier models created through database mining.
Figure 3:
Covariance matrix of radiomic features. A total of 219 features were extracted from each non–small cell lung cancer tumor in 235 patients. Across all tumors, each feature was individually compared with all other features by using regression analysis, thereby generating correlation coefficients (R2). Individual features were then clustered and plotted along both axes, and R2 is shown as a heat map, with areas of high correlation (R2 > 0.95) shown in red. Thus, each of the red squares along the diagonal contains a group of features that are highly correlated with one another and are thus redundant. For data analysis, one feature was chosen to be representative of each of these groups. The representative feature chosen was the one that had the highest natural biologic range (interpatient variability) across the entire patient data set, with the explicit assumption that features that show the highest interpatient variability will be the most informative. (Image courtesy of Y. Balagurunathan.)
Building Databases: Numbers Are King, Quality Is Queen
In radiomics and elsewhere, the power of the predictive classifier model is dependent on having sufficient data. It has been our experience that a reasonable rule of thumb is that 10 samples (patients) are needed for each feature in a model based on binary classifiers. Furthermore, the best models are those that can accommodate additional clinical or genomic covariates, and this increases the need for large high-quality data sets. Radiomics can be performed with as few as 100 patients, although larger data sets provide more power. It is time consuming to capture and curate large high-quality sets from retrospective data. For example, in a recent study, we curated a data set of patients with non–small cell lung cancer adenocarcinoma who had gene expression profiles (46). Within a local database, 285 such patients were readily identified as potential candidates for such a cohort study. The need to validate these via chart and pathology review required 188 hours and resulted in the loss of 50 patients from the study cohort because of missing data or equivocal histopathologic findings. When histopathologic findings were equivocal, a pathologist reviewed the slides; this only added to the curation time. Further validation via picture archiving and communication system review of images captured with standardized acquisition and reconstruction parameters required 94 hours and resulted in the attrition of an additional 92 patients. Segmentation and extraction of features into the database required an additional 145 hours. Thus, the curation of a data set of 143 patients required an initial cohort of 285 patients (approximately 50% attrition) and required 430 hours of processing, or approximately 3 hours of processing per patient. As these patients were not filtered for medical or demographic issues, there was no selection bias. In the future, capturing images and other data prospectively and with higher quality and standards should reduce data attrition and make the process more efficient.
Classifier Modeling and Data Sharing
Once large high-quality and well-curated data sets are available, they can be used for data mining, which refers to the process of discovering patterns in large data sets. This process can use artificial intelligence, machine learning, or statistical approaches. At one end, these include both supervised and nonsupervised machine learning approaches, such as neural networks, support vector machines, or Bayesian networks. Although these approaches use a priori knowledge through training sets, they are agnostic in that they make no assumptions about the meaning of the individual features. Hence, all features are treated with equal weight at the initiation of learning. At the other end of the data-mining spectrum are hypothesis-driven approaches that cluster features according to predefined information content. While both of these approaches have merit, the best models are those that are tailored to a specific medical context and, hence, start out with a well-defined endpoint.
Ideally, robust models accommodate patient features beyond imaging. Covariates include genomic profiles (expression, mutation, polymorphisms), histology, serum markers, patient histories, and biomarkers that are qualified for the specific-use case (Fig 1). In practice, not all information is available for all patients; hence, models should also be designed to accommodate sparse data. As mentioned previously, the power of the model is entirely dependent on the size and quality of the data within the database. Quality depends not only on the image acquisition conditions but also on the availability and reliability of covariates. For example, overall survival is a common endpoint for many studies, but this includes death from all causes, which may not be related to the disease being studied. More exact endpoints include progression-free survival and disease-free survival or recurrence; however, these data are not readily available and require a dedicated abstraction effort with chart review. Hence, there is a pressing need to capture such data and to share data across institutions to accumulate sufficient numbers for statistical power. Such data sharing is a major initiative of the QIN, whose members are committed to depositing well-curated data sets into The Cancer Imaging Archive for public and private data mining efforts.
Examples of Radiomics Results
In the past 10 years, radiomics and radiogenomics research in tomographic imaging (CT, MR imaging, and PET) has increased dramatically. Two well-written and relatively recent reviews describe some of the advances through 2014 (42,43). In the subsequent section, we will highlight selected findings, some of which are very recent, that show the potential of radiomics to substantially aid clinical care in several areas.
Enabling Diagnosis
In a study of 147 men with biopsy-proven prostate cancer, Wibmer et al (6) showed that Haralick texture analysis has the potential to enable differentiation of cancerous from noncancerous prostate tissue on both T2-weighted MR images and apparent diffusion coefficient (ADC) maps derived from diffusion-weighted MR images (Fig 4). In the peripheral zone of the prostate, all five features assessed (entropy, inertia, energy, correlation, and homogeneity) differed significantly between benign and cancerous tissue on both types of images; however, in the transition zone, significant differences were found for all five features on ADC maps and for two features (inertia and correlation) on T2-weighted images. In a follow-up study, these features were used to automatically compute Gleason grade and were found to enable discrimination between cancers with a Gleason score of 6 (3+3) and those with a Gleason score of 7 of more with 93% accuracy. Furthermore, these analyses could be used to distinguish between two different forms of Gleason score 7 disease (4+3 vs 3+4) with 92% accuracy (53).
Figure 4:
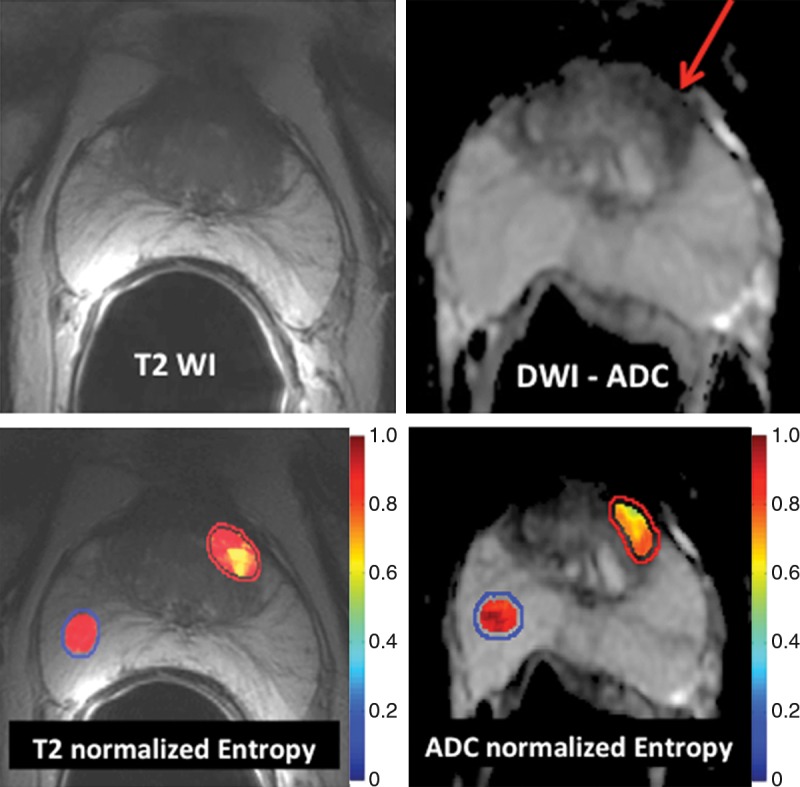
Application of texture analysis to T2-weighted MR images and ADC maps of prostate cancer. A lesion in the transition zone is barely discernible on the T2-weighted image (top left) and has higher conspicuity on the ADC map (top right). Texture features were computed on a per-voxel basis (using a 5 × 5 × 1 pixel window) from manually segmented regions of interest identifying the normal peripheral zone (outlined in blue) and cancer (outlined in red). From the computed texture features, a machine learning method was applied to distinguish between normal and cancerous structures and to stratify the Gleason patterns. Heat map images show clear differences between healthy tissue and cancer and depict intratumoral heterogeneity that may be useful in assessing tumor aggressiveness and informing fused MR imaging–ultrasonography biopsy.
Tumor Prognosis
Seminal radiogenomic studies were the first to show a relationship between quantitative image features and gene expression patterns in patients with cancer (40,54,55). In the first of these studies, the investigators compared semantic radiologist-defined features extracted from contrast-enhanced CT images in patients with hepatocellular carcinoma to gene expression patterns by using machine learning with a neural network. They found that combinations of 28 imaging traits could be used to reconstruct 78% of the global gene expression profiles, which in turn were linked to cell proliferation, liver synthetic function, and patient prognosis (40). In a subsequent study and with a similar approach, the investigators compared image features extracted from MR images to predict global gene expression patterns in patients with glioblastoma multiforme (54). They found that an “infiltrative” imaging phenotype was associated with significantly shorter survival (54).
In patients with lung cancer, there is incontrovertible evidence for intratumoral heterogeneity on lung CT images (Fig 5). These heterogeneities can be captured with features such as spiculation or entropy gradients. Grove et al found these measures to be strong prognostic indicators in patients with early-stage lung cancer (P < .01) (56). A study by Aerts et al (47) showed that a radiomic signature could be used to predict outcome in completely independent cohorts of patients with lung cancer from two separate institutions. Further, this same signature could be applied to cohorts of patients with head and neck cancer with equivalent prognostic power. Notably, the signature was comprised of the top features from four classes (size, shape, texture, and wavelets) that were prioritized from a database of 442 features by using test-retest reproducibility and intersubject range. This study and others like it demonstrate the potential of radiomics for the identification of a general prognostic imaging phenotype existing in several forms of cancer.
Figure 5:
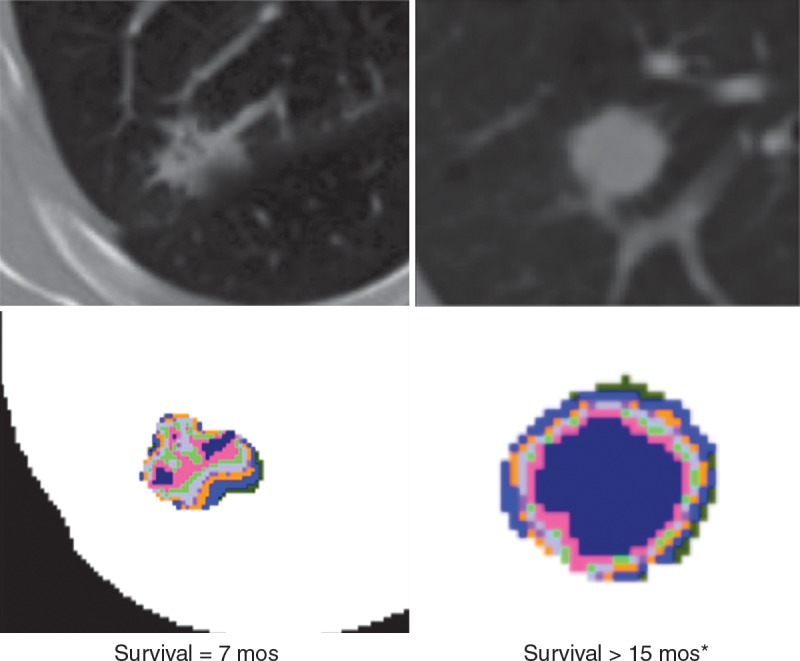
Attenuation gradients of lung CT images. Data are representative of patients with (left) and without (right) recurrence after lobectomy. Although differences in texture were visible on the CT images (top), the color-coded attenuation maps (bottom) more dramatically show intratumoral complexity. Maps were generated by separating the attenuation into quartiles, with hotter colors representing higher attenuation.
It is well known in the radiology community that contrast enhancement at MR imaging is often heterogeneous, with complex patterns. In a landmark article, Rose et al (44) analyzed the pattern of enhancement on dynamic contrast-enhanced MR images in simulations, phantoms, and 23 patients with glioma by using second-order and higher statistical measures to represent enhancement heterogeneity. They convincingly showed that complex measures of texture heterogeneity in transfer coefficient maps from dynamic contrast-enhanced MR images could be used to distinguish high- and low-grade gliomas with much higher statistical power (P < .00005) than could median transfer coefficient maps alone (P = .005). In a more recent study, Gevaert et al extracted a large number of semantic and agnostic features in 55 patients with glioma who had undergone gene expression profiling (57). The feature set was then filtered for reproducibility, yielding 18 features that were assessed in three distinct habitats. Of the agnostic features, most could be correlated with the semantic features; three of 54 were related to survival, and seven were related to gene expression. When gene expression was assessed via pathways, approximately half of the imaging features showed strong correlation to genomics. These analyses show that power for predicting gene expression patterns, outcomes, and staging of gliomas can be significantly increased with radiomics-based approaches.
Recently, Vignati et al performed a thorough prospective radiomic analysis of diffusion- and T2-weighted MR imaging examinations in 49 patients with prostate cancer (58). Agnostic features extracted from T2-weighted images and ADC maps were compared with more traditional ADC cutoff metrics to test the hypothesis that textures could help differentiate between men with a pathologic Gleason score of 6 and those with a pathologic Gleason score of 7 or higher. This is an important cutoff, as men with a pathologic Gleason score of 6 may be candidates for active surveillance. For standard ADC cutoff metrics, the area under the receiver operator characteristic curve ranged from 0.82 to 0.85. When ADC and T2 maps were analyzed for heterogeneity, the area under the curve improved to impressive values of 0.92 and 0.96, respectively. Although this study may have been underpowered, it shows the potential value of quantitative analysis of tumor heterogeneity in assessing tumor aggressiveness and informing major clinical decisions, such as whether to treat the cancer at all. Of note, other investigators have also found entropy determined from ADC maps to be significantly associated with the pathologic Gleason score, even after controlling for the median ADC (6,53).
Treatment Selection
In a seminal study, Kuo et al identified hepatocellular carcinoma imaging phenotypes that correlated with a doxorubicin drug response gene expression program (55). Their results suggested that radiogenomic analyses could be used to guide the selection of therapy for individual tumors. More recently, a study of 58 women who underwent treatment for locally advanced breast cancer suggested that texture analysis of dynamic contrast-enhanced MR imaging could help predict response to neoadjuvant chemotherapy before its initiation (59).
Deciding Where to Biopsy or Resect
It is axiomatic that images can be used to guide biopsy. It is our opinion that quantitative analyses of regionally distinct radiomic features can also precisely inform biopsy; that is, they can be used to identify a priori those locations within complex tumors that are most likely to contain important diagnostic, prognostic, or predictive information. This has already been shown with the use of PET to overlay functional information on CT or MR images to better guide biopsies in the abdomen and in patients with bone disease (60,61). Figures 4, 6, and 7 show recent examples of applications of radiomics to MR imaging, CT, and PET/CT in patients with prostate, bladder, and metastatic breast cancer, respectively, and show the potential of radiomics to enable better-informed decisions about where to biopsy.
Figure 6:
Application of texture analysis to CT images of bladder cancer. On original contrast-enhanced CT image of bladder cancer (top left), a high-attenuation lesion is clearly visible, and there is some evidence of intratumoral heterogeneity. However, when the texture features of energy (top right), entropy (bottom left), and homogeneity (bottom right) are displayed over the source image, intratumoral heterogeneity can be readily appreciated. Other studies have shown that higher intratumoral heterogeneity is associated with a worse prognosis.
Figure 7:
Application of radiomics to FDG-avid lymph nodes on PET and CT images in a patient with metastatic breast cancer. Left: Standard PET image shows there is little evidence of intranodal heterogeneity. Right: CT image shows calculation and display of the Haralick co-occurrence statistics with a 9 × 9 × 9 voxel matrix and clearly reveals some areas with lower co-occurrence (red), which have higher regional heterogeneity and would therefore be considered more suspicious for cancer. The results were used to select lymph nodes for image-informed biopsy.
Challenges for Radiomics
In this article, we have already discussed technical challenges to the individual steps in the process of radiomics. We will now we present broader concerns that arise from radiomics as a whole.
Reproducibility
Radiomics is a young discipline. As with therapies motivated by molecular biology, radiomics offers great potential to accelerate precision medicine. However, it is also possible that radiomics will undergo the same slow progress already experienced with molecular biology–based systemic diagnostic techniques and therapies. That slow progress can be attributed to a number of causes, including technical complexity, poor study design (in particular, mixing hypothesis generation with hypothesis testing) and overfitting of data, lack of standards for validating results, incomplete reporting of results, and unrecognized confounding variables in the databases used, particularly if data are derived retrospectively. Hence, as with any biomarker study, a retrospective radiomics investigation must be validated against a completely independent data set, preferably from another institution. Furthermore, the most rigorous biomarker qualification requires a prospective multicenter trial wherein the biomarker is one of the primary endpoints (62,63).
While standardized tools for genomic profiling (GenomeDx for prostate cancer [GenomeDx Biosciences, San Diego, Calif], Oncotype Dx for breast cancer [Genomic Health, Redwood City, Calif]) have been developed, they are not universally agreed upon or applied across medical centers, hampering efforts to share data and reproduce results. Studies have documented these problems in biomedical research generally and in molecular-targeted drug development specifically. A 2009 analysis of biomedical research reports found that at least 50% of studies were too poor, insufficient, or incomplete to be usable (64). Scientists at Bayer HealthCare (Leverkusen, Germany) reported that they were able to successfully reproduce the published results from only a quarter of 67 seminal studies (65,66). Furthermore, when scientists at Amgen (Thousand Oaks, Calif) tried to replicate 53 landmark studies in the basic science of cancer, they were able to reproduce the original results of just six (67). These issues have become serious enough that editors from more than 30 high-impact-factor biomedical journals have united to impose common standards for statistical testing and to improve access to raw data (68,69). The standards have been adopted by the National Institutes of Health (8,70). Although these standards were generated to address preclinical data, they can be applied across all areas of research and can provide a roadmap for navigating the complex issues associated with acquisition and analysis of high-dimensional data inherent in radiomics. Superb reporting guidelines for clinical studies have been developed by many organizations. An excellent overview is provided by the Equator network, which promotes the quality and transparency of health research (65). Challenges with study design were also identified in the 2012 report Omics from the Institute of Medicine (8). A clear solution to these challenges is to establish benchmarks for the conduct of radiomics studies and for their reporting in the literature.
Big Data
In the era of precision medicine, gigabytes of data are collected for each patient, and radiomics data can provide a significant component of this. The exponential growth in the numbers of patients and the data elements being harvested from each is known colloquially as “big data”. Big data initiatives are aimed at drawing inferences from large data sets that are not derived from carefully controlled experiments. Although correlations among observations can be vast in number and easy to obtain, causality is much harder to assess and establish, partly because it is a vague and poorly specified construct for complex systems. Across big data disciplines there are basic questions: Will access to massive data be a key to understanding the fundamental questions of basic and applied science? Or, does the vast increase in data confound analysis, produce computational bottlenecks, and decrease the ability to draw valid causal inferences? As in radiomics, the field of big data is in its early phases. The aforementioned questions were addressed in a meeting on big data that was sponsored by the National Academy of Sciences (71), and the radiomics field will benefit from this effort.
Data Sharing
The biggest challenge to establishing radiomics-based models as biomarkers to use in decision support is the sharing of image data and metadata across multiple sites. Multisite trials are required to interrogate separate cohorts of patients and to create databases with sufficient size for statistical power. Data sharing is a common challenge in all biomedical research, and it must overcome cultural, administrative, regulatory, and personal issues (72). Notably, communities like the Children’s Oncology Group have established a history and culture of data sharing (73) and therefore are in a prime position to expand their efforts to include radiomic image analysis. Data sharing in radiomics is especially daunting because shared data must include images and sharing must be in compliance with the Health Insurance Portability and Accountability Act, as a substantial amount of personal health information is needed to build models of sufficient complexity. Solutions to this challenge are many and can include: (a) large centralized data repositories, such as The Cancer Imaging Archive and The Cancer Genome Atlas, wherein access can be limited to institutional review board–approved users or data can be stripped of personal health information; (b) federated approaches, wherein each institution maintains their individual data, and query models are sent to extract the relevant metadata; or (c) federated approaches, wherein the institutions are all accessible by an honest broker (ie, a superuser with multisite institutional review board–approved access). No matter which solution is applied, the infrastructure costs can be substantial.
Standards
While standards exist or are being developed in many of the areas already mentioned in this article, there are still gaps. For example, while the value of test-retest subject or patient image studies is well recognized, many of the published studies have small sample sizes. Ideally, these studies should be combined to provide a meta-analysis; however, as noted earlier, there are often problems with how results are reported. While there are guidelines for reporting, none yet exist for the reporting of quantitative imaging results, let alone for the reporting of much more complex radiomics results. Testing of the core technology of texture analysis also is among the areas in which standards are lacking. It will be necessary to provide standards for all aspects of radiomics if the field is to realize its potential.
Radiomics: The Next Frontier in Clinical Decision Making
Our vision for radiomics is optimistic and clear. In the foreseeable future, we expect that data gleaned from radiologic examinations throughout the world will be converted into quantitative feature data and that these data will be interfaced with knowledge bases to improve diagnostic accuracy and predictive power for decision support. For this to have high penetrance in clinical settings, practitioners must be given an incentive to participate in the process. How do we get there from here? Clearly, part of the solution involves addressing the aforementioned challenges of standardization and data sharing. In addition, the data must be collected prospectively. There are central and critical roles for radiologists to play in identifying and curating data at the front end and in applying classifier models at the user end to improve diagnostic and prognostic accuracy. In between, this will be a multidisciplinary effort involving information technologists, bioinformaticists, statisticians, and treating physicians. Here, we outline what needs to be done at the local and transnational levels in short- and intermediate-term time frames.
Curation of High-Quality Data by Radiologists
In current general practice, radiologic examinations are qualitatively evaluated, and the generated reports often do not use a standard lexicon, despite a number of efforts to embrace a uniform lexicon, such as RadLex® (74). (If used routinely, annotations with RadLex®-type image features could greatly contribute to mineable databases [75,76].) Furthermore, once images are archived, they are rarely reaccessed. Although massive image repositories exist, they are virtually inaccessible for curation because of the limitations described previously. The only viable solution is to capture data prospectively at the point of care. Hence, we envision a transition from classic radiology to a new paradigm in which the radiologist actively participates in the curation of quantitative image databases. Collection of high-quality image data requires sophisticated content expertise to identity and circumscribe (with computer assistance) and annotate (with a standardized and mineable lexicon) the volumes of interest. To make high-quality data curation a reality, we must first convince the imaging practitioners of its value, and we must streamline the process so it can occur within the limitations of a clinical practice. By playing a crucial role in data curation and analysis of big data, radiologists and physicians alike will be able to make radiomics an important, valuable new dimension of their field.
Health Informatics
To be of maximal value, the various kinds of high-quality data that are obtained during the work-up and monitoring of individual patients must interface with each other. This is well recognized, and most large medical centers are now investing in appropriate electronic medical record systems to make patient data accessible in a mineable form. Currently, radiomic data are typically not incorporated as part of this data stream; however, this is changing with the adoption of structured radiology reporting. The challenge going forward will be to capture radiomic data as part of the structured report.
Data Sharing
As discussed previously, the quality of classifier models is limited by the size of the data sets used to create them. Even if one institution were to capture all of its radiomic data prospectively, it would be years before sufficient power could be generated. Additionally, these data are a moving target, as there are continuous improvements in medical image acquisition. Differences in image acquisition and reconstruction are covariates that must be incorporated in the mining of quantitative image data and therefore increase the amount of data required for model building by an order of magnitude. The solution is for multi-institutional, national, or international consortia to agree to share data either through centralized or distributed (federated) networks. These challenges have been met and solved in the basic sciences of gene expression, sequencing, and protein structure databases. They are beginning to be solved through sharing of oncology metadata (www.oriencancer.org or the Children’s Oncology Group), and some sharing of image files has been enabled by The Cancer Imaging Archive (77).
To date, collaborative efforts to develop quantitative image-driven biomarkers have been remarkable. Radiomics has tremendous potential to further enrich image interpretations and to expand the horizons of imaging toward greater precision and extraction of in vivo biologic information. To fully exploit the potential of radiomics, we need to embrace an interdisciplinary shared vision and make a joint commitment.
The Radiology Reading Room of the Future
The aforementioned scenario will entail a reading room wherein practicing radiologists interact with picture archiving and communication system software to identify, segment, and extract features from regions of interest. If prior studies obtained in the same patient are available, the previous regions of interest will be automatically identified by the reading software. As part of the reading, the extracted size, shape, location, and textural features will be automatically uploaded to a shared database and algorithmically compared with prior images to enable more precise diagnoses. Such capabilities are nearly at hand, as most picture archiving and communication systems have the capability to coregister current images with prior images and perform user-interactive segmentation. For the foreseeable future, the field of radiomics research will be concentrated on improving classifier models to provide the most accurate possible diagnoses and, hence, better patient care and outcomes.
Radiomics Resources
Readers may find the following resources helpful: QIN, http://imaging.cancer.gov/programsandresources/specializedinitiatives/qin; QIBA, http://rsna.org/QIBA.aspx; The Cancer Imaging Archive, http://www.cancerimagingarchive.net; Food and Drug Administration CAD Guidance, http://www.fda.gov/RegulatoryInformation/Guidances/ucm187249.htm; and National Institutes of Health Principles and Standards of Research Reporting, http://www.nih.gov/about/reporting-preclinical-research.htm.
Advances in Knowledge
■ Radiomics is defined as the conversion of images to higher-dimensional data and the subsequent mining of these data for improved decision support.
■ Radiomics has been initiated in oncology studies, but it is potentially applicable to all diseases.
■ Radiomics can be performed with tomographic images from CT, MR imaging, and PET studies.
■ Image features are extracted from volumes of interest, which can be either entire tumors or defined subvolumes within tumors, known as habitats.
■ Radiomics is a new field, and there are substantial challenges to its implementation in a clinical setting.
Implications for Patient Care
■ Radiomics is designed to be used in decision support of precision medicine.
■ Although relationships between radiomics and outcome are defined with populations, they can be applied to individual patients.
■ Radiomic analysis promises to increase precision in diagnosis, assessment of prognosis, and prediction of therapy response.
Acknowledgments
Acknowledgments
This article was an outgrowth of the 2014 Radiological Society of North America/American Association of Physicists in Medicine Plenary Session. The authors thank the following colleagues for their substantial input: Daniel Seeburg, MD (Johns Hopkins University), for his timely review of an early draft of the manuscript; Olya Stringfield, PhD (Moffitt Cancer Center), for production of images; Robert A. Gatenby, MD (Moffitt Cancer Center), for providing images, inspiration. and edits; Yoganand Balagurunathan, PhD (Moffitt Cancer Center), for his review of the manuscript; Sandy Napel, PhD (Stanford University), for providing a comprehensive list of relevant references; and Ada Muellner, MS (Memorial Sloan-Kettering Cancer Center), for her spectacular editing. We also acknowledge funding and support from RSNA’s sponsorship of QIBA and the NCI’s sponsorship of QIN.
Received May 29, 2015; revision requested July 6; revision received August 14; accepted September 14; final version accepted October 16.
Funding: This research was supported by the National Institutes of Health (grants U54CA143970, U01CA143062, R01CA190105, and R01CA187532).
Disclosures of Conflicts of Interest: R.J.G. Activities related to the present article: none to disclose. Activities not related to the present article: is on the advisory board of and is an investor in Health Myne. Other relationships: has a patent pending for Radiology Reading Room of the Future. P.E.K. Activities related to the present article: none to disclose. Activities not related to the present article: received a grant from GE Healthcare, is the cofounder of PET/X. Other relationships: none to disclose. H.H. disclosed no relevant relationships.
Abbreviations:
- ADC
- apparent diffusion coefficient
- BI-RADS
- Breast Imaging Reporting and Data System
- CAD
- computer-aided diagnosis and detection
- Lung-RADS
- Lung Imaging Reporting and Data System
- NCI
- National Cancer Institute
- PI-RADS
- Prostate Imaging Reporting and Data System
- QIBA
- Quantitative Imaging Biomarkers Alliance
- QIN
- Quantitative Imaging Network
- RSNA
- Radiological Society of North America
References
- 1.Institute of Medicine I . Evolution of Translational Omics. Lessons learned and the path forward. Washington, DC: National Academies Press, 2012. [PubMed] [Google Scholar]
- 2.Giger ML. Update on the potential of computer-aided diagnosis for breast cancer. Future Oncol 2010;6(1):1–4. [DOI] [PubMed] [Google Scholar]
- 3.Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph 2007;31(4-5):198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Z, Shu Y, Zhou H, Zhang W, Wang H. Radiogenomics helps to achieve personalized therapy by evaluating patient responses to radiation treatment. Carcinogenesis 2015;36(3):307–317. [DOI] [PubMed] [Google Scholar]
- 5.West CM, Barnett GC. Genetics and genomics of radiotherapy toxicity: towards prediction. Genome Med 2011;3(8):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wibmer A, Hricak H, Gondo T, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol 2015;25(10):2840–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015;114(3):345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials, Board on Health Care Services, Board on Health Sciences Policy, Institute of Medicine . Evolution of Translational Omics: Lessons Learned and the Path Forward. Micheel CM, Nass SJ, Omenn GS, eds. Washington, DC: National Academies Press. [PubMed] [Google Scholar]
- 9.Bates SE. It’s all about the test: the complexity of companion diagnostic co-development in personalized medicine. Clin Cancer Res 2014;20(6):1418. [DOI] [PubMed] [Google Scholar]
- 10.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372(9):793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Office of the Press Secretary . Fact sheet: President Obama’s precision medicine initiative. The White House Web site. http://www.whitehouse.gov/the-press-office/2015/01/30/fact-sheet-president-obama-s-precision-medicine-initiative. Updated January 30, 2015. Accessed April 4, 2015. [Google Scholar]
- 12.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014;64(1):9–29. [DOI] [PubMed] [Google Scholar]
- 13.Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010;467(7319):1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol 2008;26(15):2442–2449. [DOI] [PubMed] [Google Scholar]
- 17.Burstein HJ, Harris LN, Marcom PK, et al. Trastuzumab and vinorelbine as first-line therapy for HER2-overexpressing metastatic breast cancer: multicenter phase II trial with clinical outcomes, analysis of serum tumor markers as predictive factors, and cardiac surveillance algorithm. J Clin Oncol 2003;21(15):2889–2895. [DOI] [PubMed] [Google Scholar]
- 18.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 2011;29(22):3085–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3(75):75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clauson J, Hsieh YC, Acharya S, Rademaker AW, Morrow M. Results of the Lynn Sage Second-Opinion Program for local therapy in patients with breast carcinoma: changes in management and determinants of where care is delivered. Cancer 2002;94(4):889–894. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen PL, Schultz D, Renshaw AA, et al. The impact of pathology review on treatment recommendations for patients with adenocarcinoma of the prostate. Urol Oncol 2004;22(4):295–299. [DOI] [PubMed] [Google Scholar]
- 22.Staradub VL, Messenger KA, Hao N, Wiley EL, Morrow M. Changes in breast cancer therapy because of pathology second opinions. Ann Surg Oncol 2002;9(10):982–987. [DOI] [PubMed] [Google Scholar]
- 23.Kronz JD, Westra WH, Epstein JI. Mandatory second opinion surgical pathology at a large referral hospital. Cancer 1999;86(11):2426–2435. [PubMed] [Google Scholar]
- 24.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30(9):1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48(4):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke LP, Sriram RD, Schilling LB. Imaging as a Biomarker: Standards for Change Measurements in Therapy workshop summary. Acad Radiol 2008;15(4):501–530. [DOI] [PubMed] [Google Scholar]
- 27.Shankar LK, Hoffman JM, Bacharach S, et al. Consensus recommendations for the use of 18F-FDG PET as an indicator of therapeutic response in patients in National Cancer Institute Trials. J Nucl Med 2006;47(6):1059–1066. [PubMed] [Google Scholar]
- 28.Clarke LP, Nordstrom RJ, Zhang H, et al. The Quantitative Imaging Network: NCI’s Historical Perspective and Planned Goals. Transl Oncol 2014;7(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC; for the Group. A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology 2011;258(3):906–914. [DOI] [PubMed] [Google Scholar]
- 30.Kinahan P, Wahl R, Shao L, Frank R, Perlman E. The QIBA profile for quantitative FDG-PET/CT oncology imaging [abstr]. J Nucl Med 2014;55(Suppl 1):1520. [Google Scholar]
- 31.FDG-PET/CT Technical Committee . FDG-PET/CT as an Imaging Biomarker Measuring Response to Cancer Therapy, Quantitative Imaging Biomarkers Alliance, Version 1.05, Publicly Reviewed Version. RSNA Web site. http://www.rsna.org/qiba/. Published December 11, 2013. Accessed May 20, 2015.
- 32.van Persijn van Meerten EL, Gelderblom H, Bloem JL. RECIST revised: implications for the radiologist: a review article on the modified RECIST guideline. Eur Radiol 2010;20(6):1456–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burnside ES, Sickles EA, Bassett LW, et al. The ACR BI-RADS experience: learning from history. J Am Coll Radiol 2009;6(12):851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bomers JG, Barentsz JO. Standardization of multiparametric prostate MR imaging using PI-RADS. BioMed Res Int 2014;2014:431680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kazerooni EA, Armstrong MR, Amorosa JK, et al. ACR CT accreditation program and the lung cancer screening program designation. J Am Coll Radiol 2015;12(1):38–42. [DOI] [PubMed] [Google Scholar]
- 36.Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology 2013;269(1):8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou M, Hall L, Goldgof D, et al. Radiologically defined ecological dynamics and clinical outcomes in glioblastoma multiforme: preliminary results. Transl Oncol 2014;7(1):5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rios Velazquez E, Aerts HJ, Gu Y, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol 2012;105(2):167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dam IE, van Sörnsen de Koste JR, Hanna GG, Muirhead R, Slotman BJ, Senan S. Improving target delineation on 4-dimensional CT scans in stage I NSCLC using a deformable registration tool. Radiother Oncol 2010;96(1):67–72. [DOI] [PubMed] [Google Scholar]
- 40.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007;25(6):675–680. [DOI] [PubMed] [Google Scholar]
- 41.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern 1973;SMC-3(6):610–621. [Google Scholar]
- 42.Davnall F, Yip CS, Ljungqvist G, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 2012;3(6):573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res 2015;21(2):249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose CJ, Mills SJ, O’Connor JP, et al. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magn Reson Med 2009;62(2):488–499. [DOI] [PubMed] [Google Scholar]
- 45.Larkin TJ, Canuto HC, Kettunen MI, et al. Analysis of image heterogeneity using 2D Minkowski functionals detects tumor responses to treatment. Magn Reson Med 2014;71(1):402–410. [DOI] [PubMed] [Google Scholar]
- 46.Grossmann P, Grove O, El-Hachem N, et al. Identification of molecular phenotypes in lung cancer by integrating radiomics and genomics. Sci Transl Med (in press). [Google Scholar]
- 47.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickup L, Talwar A, Stalin S, et al. Lung nodule classification using learnt texture features on a single patient population [abstr]. In: Radiological Society of North America Scientific Assembly and Annual Meeting Program. Oak Brook, Ill: Radiological Society of North America, 2015; SSM06. [Google Scholar]
- 49.Schabath MB, Massion PP, Thompson ZJ, et al. Behavior of incidence lung cancers in the CT arm of the National Lung Screening Trial. JAMA (in press). [DOI] [PMC free article] [PubMed]
- 50.Balagurunathan Y, Kumar V, Gu Y, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging 2014;27(6):805–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989;45(1):255–268. [PubMed] [Google Scholar]
- 52.Carrasco JL, King TS, Chinchilli VM. The concordance correlation coefficient for repeated measures estimated by variance components. J Biopharm Stat 2009;19(1):90–105. [DOI] [PubMed] [Google Scholar]
- 53.Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multi-parametric magnetic resonance images. Proc Natl Acad Sci USA (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008;105(13):5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol 2007;18(7):821–831. [DOI] [PubMed] [Google Scholar]
- 56.Grove O, Berglund AE, Schabath MB, et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS One 2015;10(3):e0118261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 2014;273(1):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vignati A, Mazzetti S, Giannini V, et al. Texture features on T2-weighted magnetic resonance imaging: new potential biomarkers for prostate cancer aggressiveness. Phys Med Biol 2015;60(7):2685–2701. [DOI] [PubMed] [Google Scholar]
- 59.Teruel JRHM, Heldahl MG, Goa PE, et al. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed 2014;27(8):887–896. [DOI] [PubMed] [Google Scholar]
- 60.Klaeser B, Wiskirchen J, Wartenberg J, et al. PET/CT-guided biopsies of metabolically active bone lesions: applications and clinical impact. Eur J Nucl Med Mol Imaging 2010;37(11):2027–2036. [DOI] [PubMed] [Google Scholar]
- 61.Tatli S, Gerbaudo VH, Mamede M, Tuncali K, Shyn PB, Silverman SG. Abdominal masses sampled at PET/CT-guided percutaneous biopsy: initial experience with registration of prior PET/CT images. Radiology 2010;256(1):305–311. [DOI] [PubMed] [Google Scholar]
- 62.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009;101(21):1446–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayes DF, Bast RC, Desch CE, et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J Natl Cancer Inst 1996;88(20):1456–1466. [DOI] [PubMed] [Google Scholar]
- 64.Chalmers I, Glasziou P. Avoidable waste in the production and reporting of research evidence. Lancet 2009;374(9683):86–89. [DOI] [PubMed] [Google Scholar]
- 65.Simera I, Moher D, Hirst A, Hoey J, Schulz KF, Altman DG. Transparent and accurate reporting increases reliability, utility, and impact of your research: reporting guidelines and the EQUATOR Network. BMC Med 2010;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov 2011;10(9):712. [DOI] [PubMed] [Google Scholar]
- 67.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature 2012;483(7391):531–533. [DOI] [PubMed] [Google Scholar]
- 68.Journals unite for reproducibility. Nature 2014;515(7525):7. [DOI] [PubMed] [Google Scholar]
- 69.McNutt M. Journals unite for reproducibility. Science 2014;346(6210):679. [DOI] [PubMed] [Google Scholar]
- 70.Landis SC, Amara SG, Asadullah K, et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature 2012;490(7419):187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drawing Causal Inference from Big Data . Arthur M. Sackler Colloquia of the National Academy of Sciences. https://www.youtube.com/playlist?list=PLGJm1x3XQeK0NgFOX2Z7Wt-P5RU5Zv0Hv. Published April 2, 2015. Accessed May 5, 2015. [DOI] [PMC free article] [PubMed]
- 72.Nelson B. Data sharing: empty archives. Nature 2009;461(7261):160–163. [DOI] [PubMed] [Google Scholar]
- 73.Devidas M, London WB, Anderson JR. The use of central laboratories and remote electronic data capture to risk-adjust therapy for pediatric acute lymphoblastic leukemia and neuroblastoma. Semin Oncol 2010;37(1):53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heilbrun ME. Evaluating RadLex and real world radiology reporting: are we there yet? Acad Radiol 2013;20(11):1327–1328. [DOI] [PubMed] [Google Scholar]
- 75.Kahn CE, Jr. Annotation of figures from the biomedical imaging literature: a comparative analysis of RadLex and other standardized vocabularies. Acad Radiol 2014;21(3):384–392. [DOI] [PubMed] [Google Scholar]
- 76.Mabotuwana T, Lee MC, Cohen-Solal EV, Chang P. Mapping institution-specific study descriptions to RadLex Playbook entries. J Digit Imaging 2014;27(3):321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]






