Abstract
Background
The platelet fibrinogen receptor represents the final common pathway of platelet activation, and is formed from two glycoprotein (GP) subunits (GPIIb/IIIa). Carriage of the mutant PlA2 allele of GPIIIa has been shown to confer an increased risk of cardiovascular events, but published studies have disagreed as to the mechanism for this association.
Objectives
To assess whether carriage of the PlA2 allele conforms to Mendelian patterns of expression and to identify whether carriage of the mutant allele modulates platelet function.
Methods
Expression of the PlA2 allele was assessed in both healthy subjects (n = 25) and patients with known coronary artery disease (n = 90) through the development and validation of a liquid chromatography, tandem mass spectrometry (LC-MS/MS) assay. Platelet function was assessed in the patient cohort in response to multiple agonists, and these data were analysed in the context of the proteomic data.
Results
Expression of the wild-type PlA1 allele and mutant PlA2 alleles was readily quantifiable and conformed to Mendelian patterns in both healthy and patient cohorts. Patients who were homozygous for the mutant PlA2 allele had an increased aggregatory response to adenosine diphosphate, collagen, adrenaline, ristocetin, thrombin receptor-activating peptide 6 and U46619, when assessed using agonist-concentration response curves.
Conclusions
These findings support the hypothesis that carriage of the mutant PlA2 allele mediates an increased risk of cardiovascular events through the modulation of platelet reactivity.
Keywords: Platelets, clinical genetics, mass spectrometry, platelet function testing
Introduction
The fibrinogen receptor is the most abundant integrin on the platelet surface, and represents the final common pathway of platelet activation, adhesion and aggregation.1,2 Formed from two glycoprotein (GP) subunits (GPIIb/IIIa), the receptor binds a combination of fibrinogen, von Willebrand factor (vWF) and fibronectin in a process that terminates haemorrhage following vascular injury.3–5 The GPIIIa subunit is polymorphic with single amino acid substitutions resulting in a number of stable allelic variants, the PlA1/A2 diallelic antigen system being one of the more heavily studied due to its association with neonatal alloimmunity and an increased risk of cardiovascular events.6–8 The prevalence of the PlA2 allele varies between ethnic groups, with a frequency of approximately 1% in Oriental populations, rising to 15% in Caucasian populations.9,10
There had been much disagreement as to the extent, and indeed validity, of any association between carriage of the mutant PlA2 allele and cardiovascular disease until recent meta-analyses showed that carriage of the PlA2 allele does indeed confer a moderate increased risk of both myocardial infarction (n = 40,692; OR 1.08, 95% CI 1.02–1.13; p = 0.004) and ischaemic stroke (n = 11,873; OR 1.12, 95% CI 1.03–1.22; p = 0.011).11,12 Significant heterogeneity was observed across these analyses, and it is unclear whether this represents the challenge of identifying the contribution of a single polymorphism to a multifactorial, polygenic pathological process13 or variation in an individual’s expression of the PlA1/A2 proteins in heterozygotes. It is not yet known whether expression of the PlA1/A2 proteins obey Mendelian rules or whether subjects identified as genetically heterozygous may in fact have non-uniform patterns of protein expression. A non-uniform pattern of expression would be represented by a heterozygous individual expressing the PlA1/A2 proteins in a ratio that diverges from the 1:1 ratio proscribed by Mendelian rules. If the ratio of expression was found to vary significantly between heterozygous individuals, the utility of genomic techniques to quantify the cardiovascular risk conferred by heterozygous expression would be significantly impaired. If the allele is to realise any potential as a biomarker of cardiovascular disease, either as a standalone test or as part of broader risk stratification, it is vital to understand its proteomic expression.
Uncertainty also remains as to the aetiology of the increased risk conferred by carriage of the PlA2 allele. The mutant allele encodes a single amino acid substitution of proline for leucine adjacent to the ligand binding site,6 and it has therefore been hypothesised that the polymorphism leads to increased platelet reactivity. Studies in static systems have shown no impact of the polymorphism on ligand binding,14,15 although those performed in cell culture under conditions of shear stress have noted enhanced binding to both fibrinogen and vWF.16,17 The response to platelet agonists has similarly found to be mixed.14,18 It has also been suggested that carriage of the PlA2 allele leads to aspirin resistance, a phenomenon that confers increased cardiovascular risk,19 although a recent meta-analysis does not support this hypothesis.20
In this study, we have developed a liquid chromatography, tandem mass spectrometry (LC-MS/MS) assay to measure the expression of platelet PlA1/A2 peptides in both healthy subjects and patients with cardiovascular disease. The use of a LC-MS/MS assay containing internal reference peptides provides a truly quantitative measure of peptide expression. We hypothesise that the proteomic expression of the PlA1/A2 alleles can be quantified using LC-MS/MS, conforms to Mendelian rules of expression, and that increased cardiovascular risk conferred by carriage of the PlA2 allele is secondary to increased levels of platelet reactivity.
Methods
Subject recruitment
Twenty-five clinically healthy subjects, without personal or family history of a bleeding disorder, and having not consumed any medication for at least 14 days, were recruited as the ‘healthy cohort’ to facilitate the development of the LC-MS/MS assay.
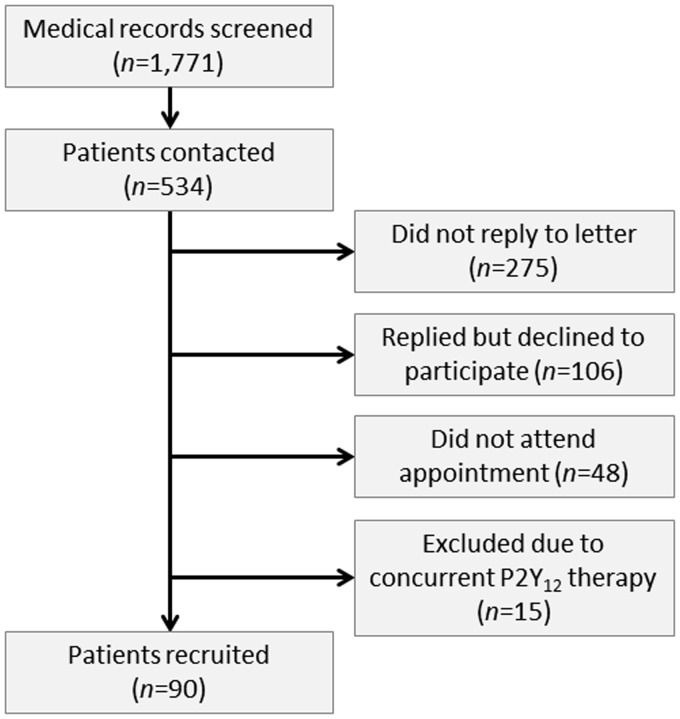
Patients with stable coronary artery disease were recruited from Guy’s and St Thomas’ NHS Foundation Trust following an automated search of the TOMCAT clinical database (Phillips) covering the period 6 January 2012 to 11 January 2013. A summary of the recruitment protocol can be seen in Figure 1. Ninety patients were recruited as the ‘patient cohort’, and satisfied the following inclusion criteria: (i) presence of angiographic coronary artery disease and (ii) prescribed daily 75 mg aspirin therapy as their sole antiplatelet agent. Patients were excluded if they had a clinical history suggestive of unstable angina or acute coronary syndrome (ACS) in the previous four months.21,22 Ethical approval for the study was obtained from London-Bloomsbury Research Ethics Committee (Reference: 11/LO/1371). All participants were recruited from King’s College London and Guy’s and St Thomas’ NHS Trust, provided written informed consent to participate in this study, with the consent procedure approved by the ethics committee.
Figure 1.
Summary of patient recruitment.
Platelet function testing
Blood was drawn by venipuncture into trisodium citrate (final concentration 0.32%) from an antecubital vein, with samples sent for biochemical and haematological analyses. Whole blood was centrifuged (15 min, 200 × g) at room temperature (RT) to obtain platelet-rich plasma (PRP), with platelet-poor plasma (PPP) obtained by further centrifugation of PRP (5 min, 7000 × g, RT). The platelet count of PRP was not adjusted, and this approach is consistent with current recommendations.23
Light transmission aggregometry (LTA) was performed in response to arachidonic acid (1.6 mM; AA, Sigma) and adenosine diphosphate (10 µM; ADP, LabMedics) using a PAP8E aggregometer (Bio/Data Corporation) as previously described.24 Briefly, 225 µL PRP was incubated for 2 min at 37℃, before the addition of 25 µL agonist. Agonist and PRP were continually mixed by a siliconised stirrer bar generating a low-shear vortex (1200 r/min), with measurements of light transmission recorded over 5 min. Light transmission through unstimulated PRP and PPP was calibrated to 0% and 100% aggregation, respectively. Aspirin resistance status in the patient cohort was assigned based on ≥20% aggregation on LTA in response to 1.6 mM AA and/or ≥70% aggregation in response to 10 µM ADP.25,26
Categorisation of platelet function was performed using the Optimul assay, which utilises a similar principle to LTA, but enables multiple agonist concentrations to be assessed simultaneously.27 Briefly, a 96-well flat-bottomed plate was prepared with platelet agonists to achieve the following concentrations on the addition of 100 µL PRP: AA (0.03–1.6 mM), ADP (0.3–30 µM), collagen (0.1–30 µg/mL; LabMedics), adrenaline (0.001–100 µM; LabMedics), ristocetin (0.18–2 mg/mL; Helena Laboratories), thrombin receptor-activating peptide 6 (0.1–30 µM; TRAP-6, Bachem) and U46619 (0.1–30 µM; Cayman Chemical). On addition of PRP, the plate was immediately placed in a 96-well plate reader (Tecan Sunrise) and absorbance determined at 595 nm every 15 s for 16 min, between vigorous linear shaking at 37℃.
Mass spectrometry
An expanded description on the development and validation of the LC-MS/MS assay is available in the Supplemental Material.
A selective reaction monitoring (SRM) assay was developed to enable the absolute quantification of PlA1/A2 peptides through the use of synthetic peptides as internal reference standards.28,29 The assay was validated in accordance with standard criteria,30 and the lower limit of measuring range (LMR) observed to be 20 fmol for both wild-type and mutant peptides. Peptide quantification ≥20 fmol could therefore be regarded as an accurate representation of the amount of peptide within the sample.
Briefly, 10 µg platelet lysates from each subject underwent in-gel enzymic digestion before PlA1/A2 peptides were quantified with reference to a 200 fmol spike of isotopically distinct synthetic peptides.29 Analyses were performed sequentially on a Vantage triple stage quadrupole mass spectrometer (Thermo), with regular quality control samples. Data processing was performed using Pinpoint v1.0 (Thermo), and underwent manual interrogation to ensure the presence of appropriate time-aligned transitions of peptide fragmentation.
Statistical methods
All data are expressed as mean ± standard error (SEM), unless otherwise stated. Analyses of unpaired data were performed using Mann-Whitney test, with analyses of categorical data performed using Fisher’s exact test. The Bonferroni method was used to correct for multiple comparisons. Agonist-concentration response curves were plotted and analysed according to a four parameter logistic equation, where the lower and upper boundaries were set at 0 and 100%, respectively. Area under the curve (AUC) values were calculated using the trapezoid rule from the log-concentration response curve. Analyses of pooled data based on genotype were performed using a Friedman test for paired, non-parametric data. Inter-assay agreement of aspirin resistance status used Cohen’s Kappa (κ).31 All statistical analyses were performed using GraphPad version 4 (Prism), and significance was taken as p < 0.05 (two-tailed).
Results
The proteomics of platelet PlA1/A2 expression
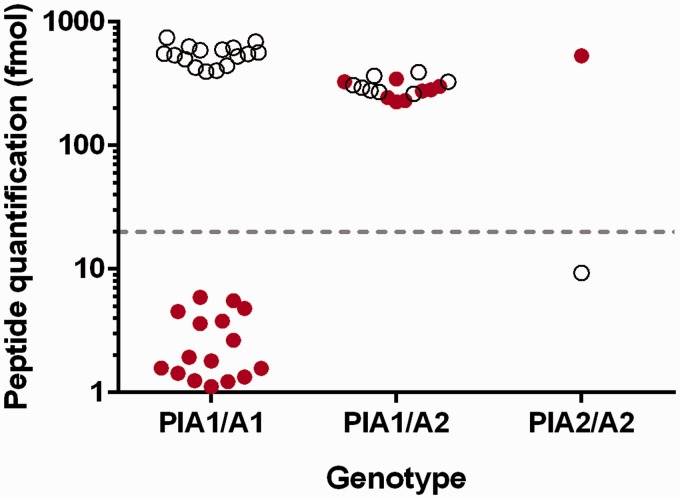
Expression of the PlA1/PlA2 alleles was inferred through the presence of the corresponding prototypic peptides (PlA1 peptide: DEALPLGSPRC; PLA2 peptide: DEALPPGSPRC). The assignment of genotype based on peptide detection above the LMR enabled healthy subjects to be clearly divided into three groups: expression of only the wild-type peptide (PlA1/A1; n = 16), expression of only the mutant peptide (PlA2/A2; n = 1) and expression of both wild-type and mutant peptides (PlA1/A2; n = 8) (Figure 2). There was no significant difference in the measured expression of PlA1/A2 peptides in heterozygous subjects within the healthy cohort.
Figure 2.
Peptide expression in healthy subjects based on inferred genotype.
Scatter plot showing the results from peptide quantification in healthy subjects for PlA1/A1 (n = 16), PlA1/A2 (n = 8) and PlA2/A2 (n = 1) genotypes. Open circles represent the PlA1 peptide and filled circles the PlA2 peptide. Dotted line indicates the lower limit of measuring range for the PlA1/A2 peptides, above which peptide quantification can be reliably determined.
In the patient cohort, genotyping of subjects based on peptide expression was similarly unequivocal, with 69 patients identified as homozygous for wild-type, two patients homozygous for mutant and 19 patients heterozygous (Table 1). The overall prevalence for carriage of the PlA2 allele was 28% (32% in Caucasian individuals) when the cohorts were combined.
Table 1.
Frequency of PlA2 allele expression.
| Ethnicity | Healthy cohort (n = 25) | Patient cohort (n = 90) | Cohorts combined (n = 115) |
|---|---|---|---|
| Caucasian | 18.0 | 12.7 | 13.9 |
| Asian | 2.0 | – | 0.4 |
Patients who carried the PlA2 allele did not generally differ significantly from PlA1 homozygotes in either biological/haematological profiles (Table 2) or general characteristics (Table 3). Carriers of the PlA2 allele were noted to have reduced low density lipoprotein (LDL) concentrations when compared to PlA1 homozygotes, but this was not significant following Bonferroni correction for multiple comparisons. Carriage of the PlA2 allele has been previously identified as a mechanism for neonatal thrombocytopenia.7 Within this adult population, carriers of the PlA2 allele did not have an increased odds ratio for thrombocytopenia (OR 0.82 for platelet count < 150 × 109/L, 95% CI 0.21–3.26; p = 0.284).
Table 2.
Biochemical and haematological profiles based on carriage of PlA2 allele.
| Parameter | PlA1/A1 (n = 69) | PlA2 carriers (n = 21) | Significance (p*) |
|---|---|---|---|
| Sodium (mmol/L) | 139 ± 0.3 | 138 ± 0.7 | 0.372 |
| Potassium (mmol/L) | 4.4 ± 0.0 | 4.4 ± 0.1 | 0.983 |
| Creatinine (µmol/L) | 89 ± 2.7 | 96 ± 7.4 | 0.279 |
| HDL cholesterol (mmol/L) | 1.4 ± 0.1 | 1.5 ± 0.1 | 0.486 |
| LDL cholesterol (mmol/L) | 2.2 ± 0.1 | 1.7 ± 0.1 | 0.029 |
| HbA1c (mmol/mol) | 45 ± 2.3 | 42 ± 3.3 | 0.564 |
| Haemoglobin (g/L) | 140 ± 0.1 | 139 ± 0.3 | 0.809 |
| White blood cells (×109/L) | 7.3 ± 0.2 | 6.6 ± 0.4 | 0.154 |
| Platelets(×109/L) | 199 ± 6.3 | 189 ± 8.6 | 0.405 |
Significant p values following Bonferroni correction for multiple comparisons.
HbA1c: glycated haemoglobin; HDL: high density lipoprotein; LDL: low density lipoprotein.
Table 3.
Patient characteristics based on carriage of PlA2 allele.
| Parameter | PlA1/A1 (n = 69) | PlA2 carriers (n = 21) | Significance (p*) |
|---|---|---|---|
| Sex | |||
| Male | 65 (94%) | 19 (90%) | 0.621 |
| Female | 4 (6%) | 2 (10%) | |
| Age (years) | 67.8 ± 1.2 | 69.0 ± 1.7 | 0.618 |
| Body mass index (kg/m2) | 28.2 ± 0.6 | 26.8 ± 0.8 | 0.217 |
| Ethnicity | |||
| Caucasian | 65 (94%) | 21 (100%) | 0.569 |
| Asian | 4 (6%) | – | |
| Smoking status | |||
| Current smoker | 4 (6%) | 2 (10%) | 0.817 |
| Ex-smoker | 32 (46%) | 9 (43%) | |
| Never smoker | 35 (48%) | 10 (47%) | |
| Blood pressure (mmHg) | |||
| Systolic | 137 ± 2.2 | 140 ± 3.6 | 0.583 |
| Diastolic | 77 ± 1.3 | 80 ± 2.2 | 0.313 |
Significant p values following Bonferroni correction for multiple comparisons.
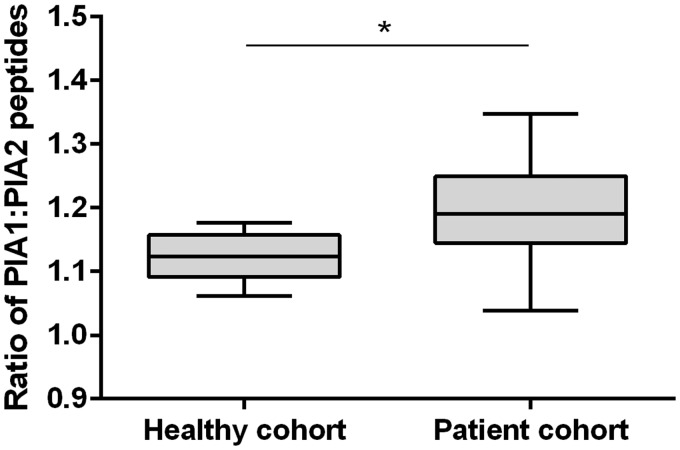
In heterozygous subjects, no significant differences were observed between expression of the PlA1 and PlA2 peptides in healthy subjects (ratio PlA1/PlA2 1.12 ± 0.01; p = 0.245), but a significant difference was observed in the patients cohort (ratio PlA1/PlA2 1.17 ± 0.02, p = 0.023), the ratio of PlA1/A2 expression in heterozygotes being significantly greater in the patient cohort than in the healthy cohort (p = 0.013) (Figure 3).
Figure 3.
Box and whisker plot for PlA1:PlA2 peptide expression in heterozygote subjects.
*p < 0.05
Carriage of the PlA2 allele and platelet function testing in the patient cohort
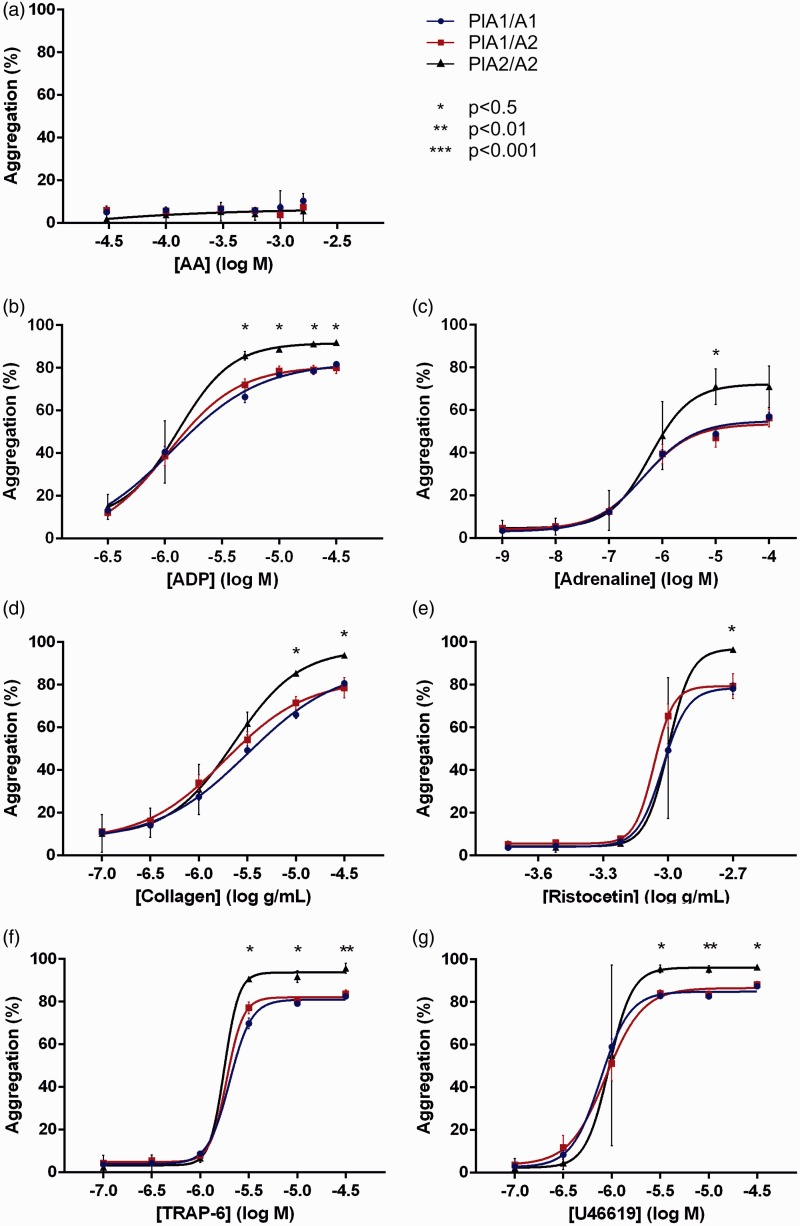
Analysis of platelet function by Optimul revealed significant differences in aggregation between PlA1 homozygotes and PlA2 homozygotes at higher concentrations of ADP, adrenaline, collagen, ristocetin, TRAP-6 and U46619. Optimul agonist-concentration response curves were plotted according to a four parameter logistic equation for all agonists except AA, where inhibition of platelet aggregation by aspirin therapy did generate sufficient data (Figure 4).
Figure 4.
Impact of carriage of the PlA2 allele on platelet function as assessed by Optimul.
A-G) Optimul aggregation in response to agonist. Statistical analyses displayed compare PlA1 homozygotes to PlA2 homozygotes for individual agonist concentrations. Individual data points plotted as mean ± SEM. PlA1/A1 (n = 69), PlA1/A2 (n = 19), PlA2/A2 (n = 2)
AA: arachidonic acid: ADP: adenosine diphosphate; SEM: standard error; TRAP-6: thrombin receptor activating peptide 6.
Analysis of agonist-concentration response curve parameters revealed a significant increase in maximal aggregation for PlA2 homozygotes versus PlA1 homozygotes irrespective of agonist used (all p < 0.05), but significant results were not consistently observed for analyses of AUC, Hill slope and half-maximum effective concentration (EC50). To increase the power of these analyses to identify any differences in global platelet reactivity between the genotypes, data for all agonists (with the exception of AA) were pooled using the mean results for each parameter. The rationale for this pooled approach is that if the mutant allele does indeed modulate platelet reactivity through the activity of GPIIIa, then the affect should be observed irrespective of agonist used as the GPIIb/IIIa complex represents the final common pathway of platelet aggregation. This approach identified significant differences between the genotypes for all parameters except EC50, with PlA2 homozygotes demonstrating an increased AUC and Hill slope in addition to the higher maximum aggregation already observed (Table 4).
Table 4.
Analysis of agonist-concentration response curves.
| Parameter | Genotype |
Significance (p) | ||
|---|---|---|---|---|
| PlA1/A1 | PlA1/A2 | PlA2/A2 | ||
| Maximum aggregation (%) | 78 ± 5 | 77 ± 5 | 91 ± 4 | 0.008* |
| AUC (Arb) | 103 ± 16 | 106 ± 16 | 122 ± 20 | 0.029* |
| Hill slope | 2.88 ± 1.05 | 3.41 ± 1.40 | 3.693 ± 1.14 | 0.012* |
| logEC50 (Arb) | −5.44 ± 0.50 | −5.49 ± 0.49 | −5.42 ± 0.49 | 0.142* |
Note: Pooled data for all agonist-concentration response curves generated by Optimul in response to ADP, collagen, adrenaline, ristocetin, U46619 and TRAP-6. Arbitrary units (Arb) presented for logEC50 due to variation in units between agonists. Data presented as mean ± SEM. *p < 0.05
ADP: adenosine diphosphate; AUC: area under curve; EC50: half-maximum effective concentration; SEM: standard error; TRAP-6: thrombin receptor-activating peptide 6.
Association between carriage of the PlA2 allele and aspirin resistance
Twelve patients were identified as aspirin-resistant by AA-induced LTA (13.3%), of which none were carriers of the PlA2 allele. Fourteen patients were identified as aspirin-resistant by ADP-induced LTA (15.6%), of which two were carriers of the PlA2 allele. Inter-assay agreement on the assignment of aspirin resistance status as identified by ADP-induced LTA was poor (κ = 0.192, 95% CI − 0.064 to 0.447).
Discussion
Meta-analyses have shown that carriage of the PlA2 allele of GPIIIa is a risk factor for myocardial infarction and ischaemic stroke, but there remains uncertainty as to the aetiology of this increased risk.11,12 To date, all published studies have assessed PlA1/A2 status using genetic techniques, with the underlying assumption that subjects labelled as heterozygous had equal expression of the corresponding proteins within their platelets in accordance with Mendelian rules of inheritance. However, with the identification of X-chromosome inactivation in females, imprinting of some autosomal genes and preferential expression of certain single nucleotide polymorphisms, it is clear that Mendelian gene expression is not absolute and that carriage of heterozygous alleles does not always result in equal expression of the proteins they encode.32–34 Here, we have developed an LC-MS/MS assay to enable the absolute quantification of platelet PlA1/A2 peptides and to assess whether Mendelian rules of expression do indeed apply to this important polymorphism.
Exploration of the performance characteristics of the assay demonstrated that it was ‘fit for purpose’ to accurately quantify the PlA1/A2 peptides, as measures of both reproducibility and precision conformed to accepted standards.30 The LMR for both peptides was 20 fmol and, as shown in Figure 2, this facilitated a clear division of genotypes based on the presence of the corresponding peptides. In the healthy cohort, there was no significant difference in the expression of PlA1/A2 peptides in heterozygous subjects clearly indicating that expression does indeed obey Mendelian rules within this cohort.
The data for the patient cohort were less clear in regard to obeying Mendelian expression patterns, as there was a reduced expression of the mutant peptide when compared to the wild-type in heterozygous individuals. It has previously been observed that both cardiovascular disease and aspirin therapy can modulate gene expression. It is therefore possible that either, or both, of these mechanisms may be contributing to a down regulation of the mutant allele and a consequent reduction in cardiovascular risk.35–37 Alternatively, in patients with stable cardiovascular disease their underlying inflammatory phenotype may lead to a preferential activation and subsequent degradation of mutant fibrinogen receptors in heterozygous subjects.38 However, the wider interquartile range observed in the patient cohort combined with a relatively small difference in the PlA1/PlA2 ratio suggests that these data are more likely to represent variation in sample storage, preparation and analysis between the cohorts. Further investigation in a larger cohort is required to explore the PlA1/PlA2 ratio further.
The prevalence of the PlA2 allele has been previously suggested to be approximately 0.15 in Caucasian populations, based on a study of 200 random Dutch blood donors.10 Here, we have observed a similar prevalence of the mutant allele in Caucasian patients (0.13), with none of the four Asian patients identified as carriers.
Carriage of the PlA2 allele in the patient cohort gave rise to a demonstrable difference in platelet function as assessed by the Optimul assay, which enabled us to produce a comprehensive assessment of how carriage of the PlA2 allele impacts on platelet function in response to a broad range of agonists. The increase in platelet aggregation for PlA2 homozygotes was shown to be not restricted to a single pathway of platelet activation, but rather was observed all agonists under investigation (other than AA). The demonstrable aspirin-medicated inhibition of platelet COX-1 in all carriers of the PlA2 allele impaired the analysis of how the PlA1/A2 polymorphism may modulate AA-induced aggregation.
The use of Optimul also enabled the plotting of agonist-concentration response curves and the analysis of aggregation responses in their totality rather than solely a comparison of isolated data points. Variation within the patient cohort (including demographics, biochemical/haematological parameters, prescribed medication and co-morbidities) combined with only two PlA2 homozygous subjects meant that analyses of curve parameters were underpowered when considered for each agonist separately. However, the pooling of data for all agonists revealed a significant increase in maximal aggregation, AUC and Hill slope for PlA2 homozygotes.
These findings support the hypothesis that carriage of the PlA2 allele does indeed modulate platelet reactivity through a final common pathway, and the putative mechanism of enhanced ligand binding in the presence of this mutation is compatible with the observed increase in Hill slope. The significant increase in aggregation was only observed for PlA2 homozygotes, but we hypothesise that a moderate increase in aggregation would also be observed for heterozygotes within an appropriately powered data set. The difference in aggregation response cannot be explained by either differing haematological/biological profiles or patient characteristics. However, the identification of only two PlA2 homozygotes is a clear limitation of this study, and a larger dataset would be required to corroborate these findings.
Carriage of the PlA2 allele was not observed to confer an increased risk of aspirin resistance, defined by either AA-induced or ADP-induced LTA. This is perhaps unsurprising given the importance of shear forces to the phenotype determined by the polymorphism and indicates substantial issues with most previously published studies investigating the association between aspirin resistance and this polymorphism.20 The poor inter-assay agreement on aspirin resistance status further highlights the controversies surrounding the precise identification of this phenomenon.39
Aside from the findings presented here, this work raises a broader issue about how one should approach the analysis of platelet function within a clinical environment. LTA is the current ‘gold standard’, but in previously published work appears to provide an inaccurate assessment of platelet function for carriers of a common genetic polymorphism. These findings may be the result of differences in assay mechanics when compared to Optimul or possibly represent the limitations of a technique that precludes plotting agonist-concentration response curves for multiple agonists. Further investigation in a healthy population should further categorise of how the choice of platelet function assay may impact on the observed effect of the polymorphism.
Conclusions
Platelet PlA1/A2 peptides are readily quantifiable by LC-MS/MS and appear to obey the rules of Mendelian expression, with homozygosity for the mutant allele appearing to confer a global increase in platelet reactivity. The pairing of proteomic and platelet function data as we have demonstrated here has potential for the future understanding of platelet aggregation responses.
Acknowledgements
The authors would like to acknowledge the technical assistance for the mass spectrometry provided by Proteome Sciences plc.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The work presented here was funded by Guy’s and St Thomas’ Charity (Registered Charity no. 251983).
Ethical approval
Ethical approval for the study was obtained from London-Bloomsbury Research Ethics Committee (Reference: 11/LO/1371).
Guarantor
AF is the guarantor for all the content presented in this paper.
Contributorship
All authors conceived the study question, and CNF was responsible for the data collection. All authors were involved in the design phase, the analysis and the interpretation of the results. CNF drafted the manuscript, and all authors approved the final version.
References
- 1.Wagner CL, Mascelli MA, Neblock DS, et al. Analysis of GPIIb/IIIa receptor number by quantification of 7E3 binding to human platelets. Blood 1996; 88: 907–914. [PubMed] [Google Scholar]
- 2.Floyd CN, Ferro A. The platelet fibrinogen receptor: from megakaryocyte to the mortuary. JRSM Cardiovas Dis 2012, pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marguerie GA, Plow EF, Edgington TS. Human platelets possess an inducible and saturable receptor specific for fibrinogen. J Biol Chem 1979; 254: 5357–5363. [PubMed] [Google Scholar]
- 4.Reininger AJ. Function of von Willebrand factor in haemostasis and thrombosis. Haemophilia 2008; 14(Suppl 5): 11–26. [DOI] [PubMed] [Google Scholar]
- 5.Pinniger JL, Prunty FT. Some observations on the blood-clotting mechanism; the role of fibrinogen and platelets, with reference to a case of congenital afibrinogenaemia. Br J Exp Pathol 1946; 27: 200–210. [PMC free article] [PubMed] [Google Scholar]
- 6.Newman PJ, Derbes RS, Aster RH. The human platelet alloantigens, PlA1 and PlA2, are associated with a leucine33/proline33 amino acid polymorphism in membrane glycoprotein IIIa, and are distinguishable by DNA typing. J Clin Invest 1989; 83: 1778–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman PJ, Valentin N. Human platelet alloantigens: recent findings, new perspectives. Thromb Haemost 1995; 74: 234–239. [PubMed] [Google Scholar]
- 8.Weiss EJ, Bray PF, Tayback M, et al. A polymorphism of a platelet glycoprotein receptor as an inherited risk factor for coronary thrombosis. N Engl J Med 1996; 334: 1090–1094. [DOI] [PubMed] [Google Scholar]
- 9.Lim J, Lal S, Ng KC, et al. Variation of the platelet glycoprotein IIIa PI(A1/A2) allele frequencies in the three ethnic groups of Singapore. Int J Cardiol 2003; 90: 269–273. [DOI] [PubMed] [Google Scholar]
- 10.Simsek S, Faber NM, Bleeker PM, et al. Determination of human platelet antigen frequencies in the Dutch population by immunophenotyping and DNA (allele-specific restriction enzyme) analysis. Blood 1993; 81: 835–840. [PubMed] [Google Scholar]
- 11.Floyd CN, Mustafa A, Ferro A. The PlA1/A2 polymorphism of glycoprotein IIIa as a risk factor for myocardial infarction: a meta-analysis. PloS One 2014; 9: e101518–e101518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Floyd CN, Ellis BH, Ferro A. The PlA1/A2 polymorphism of glycoprotein IIIa as a risk factor for stroke: a systematic review and meta-analysis. PloS One 2014; 9: e100239–e100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lander ES, Schork NJ. Genetic dissection of complex traits. Science 1994; 265: 2037–2048. [DOI] [PubMed] [Google Scholar]
- 14.Corral J, Gonzalez-Conejero R, Rivera J, et al. HPA-1 genotype in arterial thrombosis–role of HPA-1b polymorphism in platelet function. Blood Coagul Fibrinolysis 1997; 8: 284–290. [DOI] [PubMed] [Google Scholar]
- 15.Bennett JS, Catella-Lawson F, Rut AR, et al. Effect of the Pl(A2) alloantigen on the function of beta(3)-integrins in platelets. Blood 2001; 97: 3093–3099. [DOI] [PubMed] [Google Scholar]
- 16.Vijayan KV, Huang TC, Liu Y, et al. Shear stress augments the enhanced adhesive phenotype of cells expressing the Pro33 isoform of integrin beta3. FEBS Lett 2003; 540: 41–46. [DOI] [PubMed] [Google Scholar]
- 17.Vijayan KV, Liu Y, Sun W, et al. The Pro33 isoform of integrin beta3 enhances outside-in signaling in human platelets by regulating the activation of serine/threonine phosphatases. J Biol Chem 2005; 280: 21756–21762. [DOI] [PubMed] [Google Scholar]
- 18.Michelson AD, Furman MI, Goldschmidt-Clermont P, et al. Platelet GP IIIa Pl(A) polymorphisms display different sensitivities to agonists. Circulation 2000; 101: 1013–1018. [DOI] [PubMed] [Google Scholar]
- 19.Krasopoulos G, Brister SJ, Beattie WS, et al. Aspirin “resistance” and risk of cardiovascular morbidity: systematic review and meta-analysis. BMJ 2008; 336: 195–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Floyd CN, Ferro A. The PlA1/A2 polymorphism of glycoprotein IIIa in relation to efficacy of antiplatelet drugs: a systematic review and meta-analysis. Br J Clin Pharmacol 2013; 77: 446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trip MD, Cats VM, van Capelle FJ, et al. Platelet hyperreactivity and prognosis in survivors of myocardial infarction. N Engl J Med 1990; 322: 1549–1554. [DOI] [PubMed] [Google Scholar]
- 22.Ault KA, Cannon CP, Mitchell J, et al. Platelet activation in patients after an acute coronary syndrome: results from the TIMI-12 trial. Thrombolysis in myocardial infarction. J Am Coll Cardiol 1999; 33: 634–639. [DOI] [PubMed] [Google Scholar]
- 23.Cattaneo M, Lecchi A, Zighetti ML, et al. Platelet aggregation studies: autologous platelet-poor plasma inhibits platelet aggregation when added to platelet-rich plasma to normalize platelet count. Haematologica 2007; 92: 694–697. [DOI] [PubMed] [Google Scholar]
- 24.Born GV, Cross MJ. The Aggregation of blood platelets. J Physiol 1963; 168: 178–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gum PA, Kottke-Marchant K, Poggio ED, et al. Profile and prevalence of aspirin resistance in patients with cardiovascular disease. Am J Cardiol 2001; 88: 230–235. [DOI] [PubMed] [Google Scholar]
- 26.Lordkipanidze M, Pharand C, Schampaert E, et al. A comparison of six major platelet function tests to determine the prevalence of aspirin resistance in patients with stable coronary artery disease. Eur Heart J 2007; 28: 1702–1708. [DOI] [PubMed] [Google Scholar]
- 27.Chan MV, Warner TD. Standardised optical multichannel (optimul) platelet aggregometry using high-speed shaking and fixed time point readings. Platelets 2012; 23: 404–408. [DOI] [PubMed] [Google Scholar]
- 28.Stahl-Zeng J, Lange V, Ossola R, et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics 2007; 6: 1809–1817. [DOI] [PubMed] [Google Scholar]
- 29.Gerber SA, Rush J, Stemman O, et al. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA 2003; 100: 6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Protocols for determination of limits of detection and limits of quantification. Approved guidelines. NCCLS Document EP-17-A. Pennsylvania, USA: The National Committee for Clinical Laboratory Standards.
- 31.McHugh ML. Interrater reliability: the kappa statistic. Biochem Medi 2012; 22: 276–282. [PMC free article] [PubMed] [Google Scholar]
- 32.Gartler SM, Goldman MA. Biology of the X chromosome. Curr Opin Pediatr 2001; 13: 340–345. [DOI] [PubMed] [Google Scholar]
- 33.Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol 2002; 192: 245–258. [DOI] [PubMed] [Google Scholar]
- 34.Lo HS, Wang Z, Hu Y, et al. Allelic variation in gene expression is common in the human genome. Genome Res 2003; 13: 1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flescher E, Rotem R, Kwon P, et al. Aspirin enhances multidrug resistance gene 1 expression in human Molt-4 T lymphoma cells. Anticancer Res 2000; 20: 4441–4444. [PubMed] [Google Scholar]
- 36.Massimi I, Guerriero R, Lotti LV, et al. Aspirin influences megakaryocytic gene expression leading to upregulation of multidrug resistance protein-4 in human platelets. Br J Clin Pharmacol 2014; 78: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McManus DD, Beaulieu LM, Mick E, et al. Relationship among circulating inflammatory proteins, platelet gene expression, and cardiovascular risk. Arterioscler Thromb Vasc Biol 2013; 33: 2666–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med 2011; 17: 1410–1422. [DOI] [PubMed] [Google Scholar]
- 39.Floyd CN, Ferro A. Mechanisms of aspirin resistance. Pharmacol Ther 2014; 141: 69–78. [DOI] [PubMed] [Google Scholar]