Abstract
MicroRNA-21 (miR-21) is overexpressed in the serum of patients with colorectal cancer (CRC), suggesting that miR-21 is a promising diagnostic biomarker for CRC. Therefore, a meta-analysis was performed to assess the diagnostic value of serum miR-21 in CRC patients. This meta-analysis included 9 studies with 746 CRC patients and 476 healthy controls. Two independent researchers identified and extracted study characteristics from eligible studies that were searched from literature databases. The quality of the eligible studies was assessed in accordance with the Quality Assessment of Diagnostic Accuracy Studies. Bivariate and hierarchical summary receiver operating characteristic models were used in the meta-analysis. The pooled sensitivity and specificity were 72% [95% confidence interval (CI), 62–80] and 85% (95% CI, 80–88), respectively. The pooled positive- and negative-likelihood ratios were 4.65 (95% CI, 3.42–6.33) and 0.33 (95% CI, 0.24–0.47), respectively. The diagnostic odds ratio value was 14.03 (95% CI, 7.65–25.74), and the area under the summary receiver operator characteristic curve was 0.87. The present results indicated that miR-21 has a potential diagnostic value with moderate sensitivity and good specificity for CRC. Further prospective studies must be conducted to evaluate the diagnostic accuracy of miR-21.
Keywords: colorectal cancer, microRNA-21, serum, diagnosis, biomarker, meta-analysis
Introduction
Colorectal cancer (CRC) is a major cause of cancer-related fatalities worldwide (1). In 2012, the American Cancer Society estimated that 143,460 new cases of CRC would be diagnosed, and 51,690 would succumb to this disease (2). The majority of CRC-related fatalities can be prevented through early diagnosis and surgical removal of early-stage cancer. The five-year survival rates of CRC patients range from <8% at stage IV to >93% at stage I (3). However, the majority of early-stage CRC patients exhibit no symptoms. In addition, the available methodologies for early detection are based on traditional screening methods, such as the fecal occult blood test (FOBT) as the primary screening tool, followed by colonoscopy for FOBT-positive patients. However, these techniques have inherent limitations. Although widely used, FOBT has poor sensitivity for detecting early-stage CRC. Colonoscopy, despite its specificity and sensitivity, is not suitable for the general population due to its high cost, invasiveness, requirement for bowel preparation and sedation, and association with medical complications. A stool DNA test may be a promising alternative in the future, but the widespread application of this technique is limited by labor-intensive handling and high costs (4). Thus far, indicators that precisely predict the presence of early-stage tumors are lacking. Therefore, the development of new markers is urgently required for the rapid, noninvasive, and highly sensitive screening of CRC patients.
MicroRNAs (miRNAs) are small (18–25 nucleotides) noncoding RNAs that regulate the translation of specific genes through sequence-specific binding to the 3′ untranslated region of target mRNAs. MiRNAs reportedly have important roles in various cellular processes that are commonly involved in cancer; these processes include cell growth, differentiation, invasion, angiogenesis and epithelial-mesenchymal transition (5,6). Due to their oncogenic or tumor-suppressive properties, certain miRNAs participate in carcinogenesis (7,8). A previous study revealed the presence of large quantities of miRNAs in serums. Circulating miRNAs can withstand unfavorable physiological conditions, such as extreme variations in pH, temperature, and multiple freeze/thaw cycles (9). Furthermore, the profiles of circulating miRNAs show consistent expression levels across physiologically healthy individuals (10). The diagnostic value of circulating miRNAs for the early detection of cancer has been successfully investigated in numerous malignancies, including CRC. Therefore, miRNAs are potentially useful biomarkers that may be sensitive and specific for the early detection of CRC.
MicroRNA-21 (miR-21) is an oncomiRNA that modulates the expression of multiple cancer-related target genes, such as PTEN, TPM1 and PDCD. miR-21 is overexpressed in various human tumors, particularly in the serum and tissue of CRC patients (11). This finding indicates that miR-21 can serve as a diagnostic marker for CRC.
Recent studies have investigated the diagnostic value of miR-21 in CRC and have raised concerns regarding the biomarker potential of miR-21. However, the findings of these studies are inconsistent. Therefore, a meta-analysis of these studies was conducted to assess the diagnostic value of miR-21 in CRC.
Materials and methods
Search strategy
Several relevant literature databases (PubMed, Embase, OvidSP, The Cochrane Library and Web of Science) and three Chinese databases (Chinese National Knowledge Infrastructure, Wei Pu DATA and Wan Fang DATA) were searched for studies that estimated the diagnostic value of miR-21 in CRC. The key words for the literature retrieval were ‘microRNA-21,’ ‘miR-21,’ ‘miRNA-21’ or ‘hsa-miR-21;’ ‘colorectal,’ ‘large intestine,’ ‘large bowel,’ ‘colon,’ ‘colonic,’ ‘rectal’ or ‘rectum;’ and ‘cancer,’ ‘cancers,’ ‘carcinoma,’ ‘carcinomas,’ ‘tumor,’ ‘tumors,’ ‘neoplasm’ or ‘neoplasms;’ and ‘serum,’ ‘sera,’ ‘serums,’ ‘blood,’ ‘plasma,’ ‘plasmas’ or ‘circulating.’ To acquire additional relevant studies, conference summaries, letters and other types of studies were scanned in the initial search and certain authors were even contacted to obtain additional information when necessary.
Selection criteria
Two reviewers (Y.W. and W.Z.H.) independently assessed the literature extracted by the search strategy. Whenever they had different opinions, the reviewers discussed until a consensus was reached. The inclusion criteria were as follows: i) CRC was diagnosed by pathological confirmation; ii) circulating miR-21 concentration was tested; iii) no treatment, such as radiotherapy or chemotherapy, was performed prior to blood collection for miR-21 testing; iv) the data were sufficient to form two-by-two tables; and v) healthy individuals or patients with benign disease were included in the control group. The exclusion criteria were as follows: i) Duplicate publications; ii) unqualified data; and iii) reviews, letters and meetings. Studies that met the above criteria were considered eligible.
Data extraction and quality assessment
Two investigators (Y.W. and W.Z.H.) independently identified and retrieved data from each study. The data extracted for this systematic review included first author, publication year, number of patients, ethnicity, country, test method, diagnostic results and others.
Two investigators (Y.W. and W.Z.H.) scored the quality of each study independently in accordance with the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) (12) tool (Table I).
Table I.
QUADAS assessment for the eligible studies.
| Ogata-Kawata | Wang and | Basati | Toiyama | Kanaan | Zhang | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Item no. | Description | Liu et al (21) | et al (22) | Zhang (20) | et al (19) | Luo et al (23) | et al (24) | et al (25) | et al (26) | Du et al (27) |
| 1 | Was the spectrum of patients representative of the patients who will receive the test in practice? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 2 | Were selection criteria clearly described? | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 3 | Is the reference standard likely to correctly classify the target condition? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 4 | Is the time period between reference standard and index test short enough to be reasonably sure that the target condition did not change between the two tests? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 5 | Did the whole sample or a random selection of the sample receive verification using a reference standard? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 6 | Did patients receive the same reference standard regardless of the index test result? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 7 | Was the reference standard independent of the index test (i.e., the index test did not form part of the reference standard)? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 8 | Was the execution of the index test described in sufficient detail to permit replication of the test? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 9 | Was the execution of the reference standard described in sufficient detail to permit its replication? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 10 | Were the index test results interpreted without knowledge of the results of the reference standard? | No | No | No | No | No | No | No | No | No |
| 11 | Were the reference standard results interpreted without knowledge of the results of the index test? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 12 | Were the same clinical data available when test results were interpreted as would be available when the test is used in practice? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 13 | Were uninterpretable/intermediate test results reported? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 14 | Were withdrawals from the study explained? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
Statistical analysis
For the diagnostic meta-analysis, true positive, false positive, true negative and false negative were extracted as bivariate data directly or through recalculation on the basis of relative data from each eligible study. Subsequently, the bivariate model was employed to analyze these data (13). A forest plot of sensitivity and specificity was constructed; the positive-likelihood ratio (PLR), negative-likelihood ratio (NLR), and diagnostic odds ratio (DOR) were calculated; and the summary receiver operator characteristic (SROC) curve was generated. The accuracy of the results was further verified by applying the hierarchical summary receiver operating characteristics (HSROC) model and subsequently presented the HSROC curve (14). The HSROC curve overcomes some of the deficiencies of the traditional SROC curve reported by Moses et al (15). Furthermore, the HSROC curve is closely associated with the bivariate random-effect model. Heterogeneity between studies was tested by χ2 and I2 statistics (16). The null hypothesis that the eligible studies are homogeneous was rejected if P<0.05 or I2 >50% (17). Six factors that may cause heterogeneity between studies were incorporated in the bivariate model as covariates to explore the source of heterogeneity by meta-regression analysis. Subgroup and sensitivity analyses were also conducted to explore the source of heterogeneity when necessary. The presence of publication bias was analyzed by Deeks' funnel plot asymmetry test. Statistical significance was considered, i.e., publication bias was present, at P<0.1. To explore the threshold effect, the ROC plane and Spearman correlation coefficient were employed. All the analyses were conducted by Meta-DiSc and Stata SE12.0 software (18).
Results
Included studies
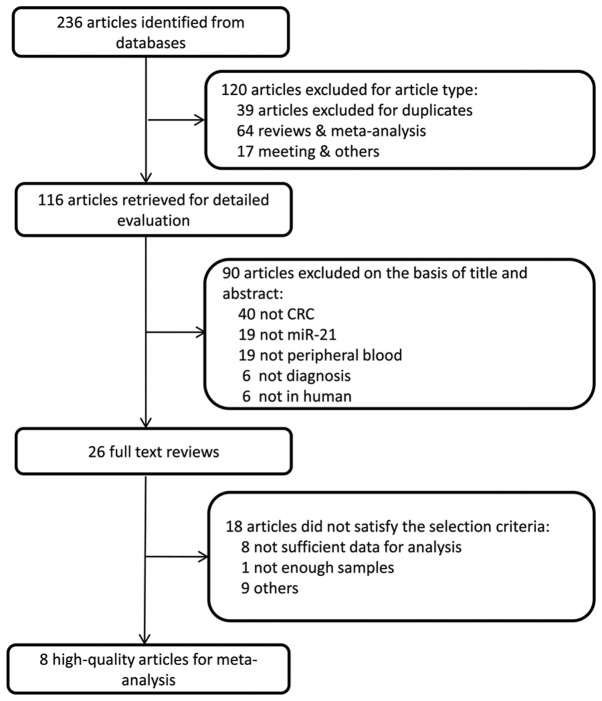
A total of 237 studies were initially retrieved. Considering the study type, we excluded 120, including 39 duplicates, 64 reviews and meta-analyses, and 17 meetings and others. A total of 117 studies were retained for the subsequent evaluation. A total of 90 manuscripts were excluded on the basis of the title and abstract, including 40 studies that did not focus on CRC, 19 that did not focus on miR-21, 19 that did not focus on blood samples, 6 unrelated to diagnosis, and 6 laboratory studies. As a result, 27 adequate studies remained for the full text review. Following reading the whole text, a further 18 studies were excluded, 8 of which contained insufficient information. Finally, 9 studies (19–27) containing 1,222 samples remained for the meta-analysis (Fig. 1).
Figure 1.
Flow chart of the study selection.
Study characteristics and quality assessment
In these selected studies, the 746 CRC patients had been pathologically confirmed. Furthermore, the 476 control individuals were healthy volunteers or patients with benign disease who had never been diagnosed with malignancy. The studies enrolled in the systemic review were conducted in China, Japan, Iran, Germany and the USA. Among the 9 studies included, 6 were conducted in Asian populations and 3 in Caucasian populations. The 9 studies, which were published between 2012 and 2014, investigated the diagnostic value of miR-21 for CRC. Furthermore, 8 of the 9 studies used reverse transcription-quantitative polymerase chain reaction (RT-qPCR) to assess the expression of miR-21. Among them, 6 used TaqMan probe, 2 used the SYBR-Green assay, and 1 used an miRNA microarray. The main characteristics of the eligible studies are presented in Table II. The QUADAS assessment tool was used to assess the quality of the 9 studies. None of the studies conformed to the criteria that the index test results should be interpreted without knowledge of the results of the reference standard (item 10). One of the 9 studies did not clearly describe their selection criteria (item 2) (Table I).
Table II.
Main characteristics of the eligible studies.
| First author, year | Patients, n (controls, n) | Ethnicity | Country | RNA extraction kits | Test method | AUC | QUADAS scores | Se% | Sp% | (Refs.) |
|---|---|---|---|---|---|---|---|---|---|---|
| Liu, 2013 | 200 (80) | Asian | China | Trizol-LS | RT-qPCR (Taqman) | 0.802 | 13 | 65 | 85 | (21) |
| Ogata-Kawata, 2014 | 88 (11) | Asian | Japan | Trizol-LS | miRNA microarray | 0.798 | 12 | 61.4 | 90.9 | (22) |
| Toiyama, 2013 | 186 (53) | Asian | Japan | miRNeasy RNA | RT-qPCR (Taqman) isolation kits | 0.927 | 13 | 82.8 | 90.6 | (24) |
| Kanaan, 2012 | 30 (30) | Caucasian | America | Trizol-LS | RT-qPCR (Taqman) | 0.82 | 13 | 90 | 90 | (25) |
| Wang, 2012 | 32 (39) | Asian | China | Trizol-LS | RT-qPCR (SYBR-Green) | 0.85 | 13 | 87.5 | 74.4 | (20) |
| Basati, 2014 | 40 (40) | Caucasian | Iran | mirVana | RT-qPCR (SYBR-Green) PARIS kit | 0.87 | 13 | 77 | 78 | (19) |
| Luo, 2013 | 80 (144) | Caucasian | Germany | Combination of Trizol LS and miRNeasy mini kit | RT-qPCR (Taqman) | 0.6528 | 13 | 51.7 | 80.7 | (23) |
| Zhang, 2014 | 41 (30) | Asian | China | Trizol-LS | RT-qPCR (Taqman) | 0.657 | 13 | 51.2 | 79 | (26) |
| Du, 2014 | 49 (49) | Asian | China | Trizol-LS | RT-qPCR (Taqman) | 0.877 | 13 | 76.2 | 93.2 | (27) |
AUC, area under the curve; QUADAS, Quality Assessment of Diagnostic Accuracy Studies; Se, sensitivity; Sp, specificity; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Data analysis
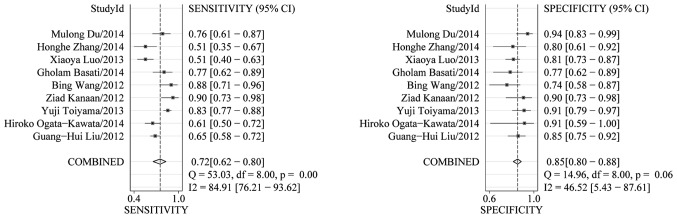
Heterogeneity in sensitivity and specificity was detected in the 9 studies (I2=84.91% and I2=46.52%, respectively), suggesting significant heterogeneity in sensitivity and mild heterogeneity in specificity (Fig. 2). Therefore, the random effects model was employed in this study. The analysis results showed that the pooled sensitivity and specificity of miR-21 for CRC diagnosis were 72% (95% CI, 62–80) and 85% (95% CI, 80–88), respectively.
Figure 2.
Forest plots of sensitivities and specificities for miR-21 test accuracy in the diagnosis of colorectal cancer.
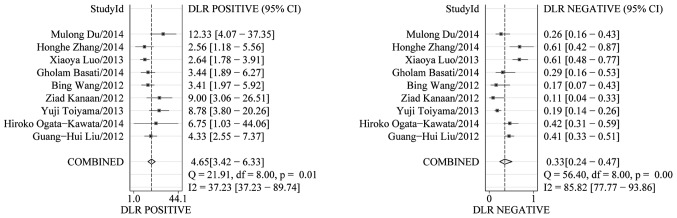
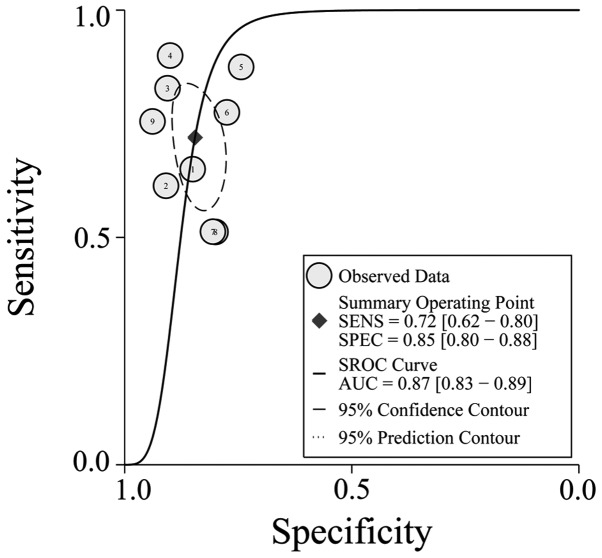
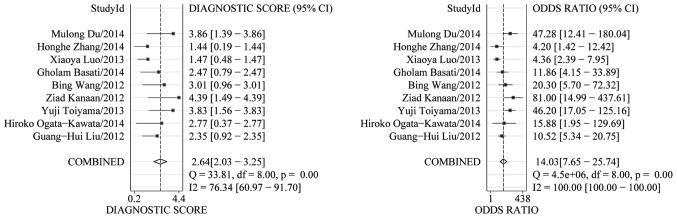
The PLR and NLR of serum miR-21 were calculated for the likelihood ratio; these parameters have been considered more clinically valuable compared to the specificity and sensitivity (28). PLR >10 or NLR <0.1 suggests high diagnostic accuracy. In the present study, the pooled PLR is 4.65 (95% CI, 3.42–6.33; I2=37.23%), indicating that the CRC patients have more than a four-fold probability to express miR-21 in comparison to healthy individuals. The pooled NLR was 0.33 (95% CI, 0.24–0.47; I2=85.82%) (Fig. 3). The SROC curve of the selected studies is shown in Fig. 4. The AUC was 0.87 (95% CI, 0.83–0.89). The DOR value was 14.03 (95% CI, 7.65–25.74), indicating that miR-21 can be used as a good marker for CRC diagnosis (Fig. 5).
Figure 3.
Forest plots of positive- and negative-likelihood ratios for miR-21 test accuracy in the diagnosis of colorectal cancer.
Figure 4.
Summary receiver operating characteristic curves for miR-21 in the diagnosis of colorectal cancer.
Figure 5.
Forest plots of diagnostic odds ratio for miR-21 test accuracy in the diagnosis of colorectal cancer.
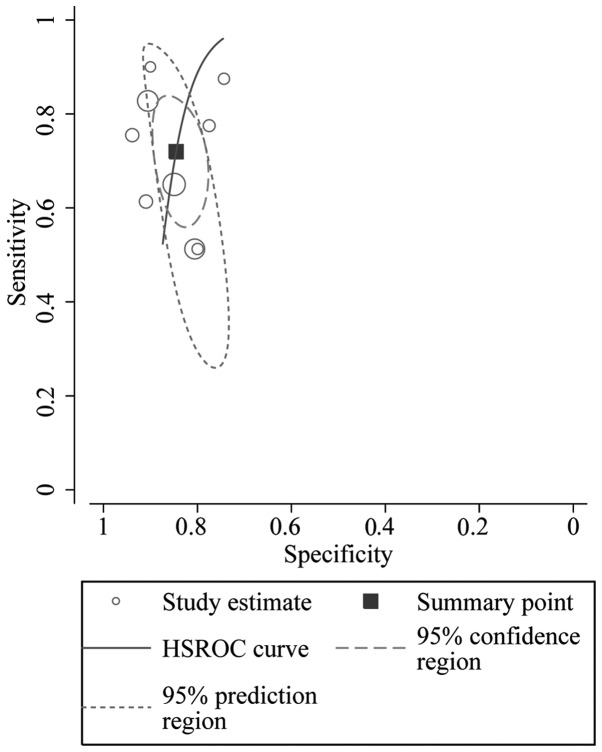
The HSROC curve of the selected studies is shown in Fig. 6, which is consistent with the results from the bivariate model. The summary operating point estimate of sensitivity and specificity is also presented. The 95% prediction and 95% CI are also plotted. The cut-off point was located near the upper left corner of the HSROC curve. The value of β was −1.28, and the P-value was 0.152. This result indicates that the HSROC is symmetrical. The value of γ, which helps distinguish CRC patients from healthy individuals, was 3.941 (2.295–5.588). This result indicates that circulating miR-21 is a relatively accurate diagnostic marker for CRC.
Figure 6.
Hierarchical summary receiver operating characteristics (HSROC) curve for miR-21 in the diagnosis of colorectal cancer.
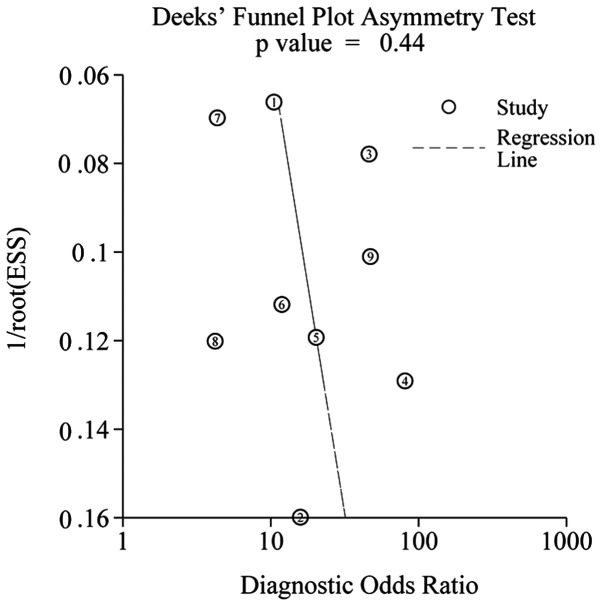
Publication bias
Deeks' funnel plots were used to evaluate the presence of publication bias in this meta-analysis. The funnel plot presents no asymmetry (Fig. 7). The P-value was 0.44, indicating the absence of publication bias in the meta-analysis. However, concluding whether or not publication bias exists is difficult due to the limited number of studies involved in the current meta-analysis.
Figure 7.
Deeks' tests for the assessment of publication bias in miR-21 assays.
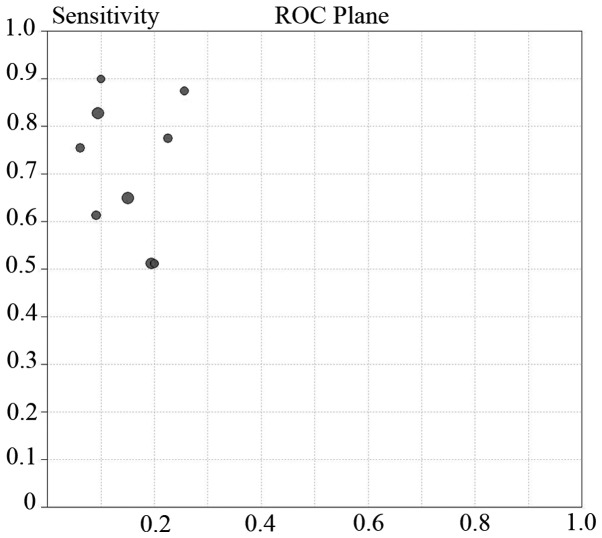
Threshold effect and heterogeneity
Differences in cut-off values cause the threshold effect. The ROC plane and Spearman rank correlation test is a good approach to assess the threshold effect (18). In the present study, the representation of the sensitivity against the specificity of each study is shown in an ROC space (Fig. 8), which can be used to detect the threshold effect. The pattern of the points in this figure does not suggest a ‘shoulder-arm’ shape, indicating the absence of the threshold effect. A Spearman rank correlation was conducted and the absence of heterogeneity was validated from the threshold effect [Spearman correlation coefficient=0.033; P=0.932 (P>0.05)].
Figure 8.
Receiver operating characteristics (ROC) space for the assessment of threshold effect in miR-21 assays.
The I2 of the heterogeneity test is 59%, suggesting moderate heterogeneity. The publication year, test method, RNA extraction kits, ethnicity, patient number and QUADAS score may contribute to heterogeneity. Meta-regression analysis suggests that the above factors are not the sources of heterogeneity in this study. Subsequently, a subgroup analysis was performed on ethnicity, test method and RNA extraction kits; however, the sources of heterogeneity could not be identified. A sensitivity analysis was also performed, but failed to find the sources of heterogeneity.
Discussion
Recent studies have revealed novel diagnostic biomarkers for CRC, including miR-21. miR-21 is an important oncogenic microRNA that participates in tumor initiation, progression and metastasis. Therefore, the present meta-analysis was performed to provide an integrated and up-to-date evaluation of the diagnostic and clinical values of miR-21 as a serum marker for CRC.
In the present meta-analysis, the combined sensitivity and specificity were 72% (95% CI, 62–80) and 85% (95% CI, 80–88), respectively, which imply moderate sensitivity and good specificity. The 2014 NCCN Guidelines for CRC screening highlighted that any positive stool test requires follow-up with a colonoscopy. miR-21 can potentially replace FOBT or serve as a powerful diagnostic tool for CRC. The DOR value reflects the extent of the association between the diagnostic results and the disease. DOR >1 means better discrimination of the index test. DOR <1 means healthy individuals are more likely to be diagnosed as positive by the index test compared to CRC patients. DOR=1 means the index test cannot discriminate healthy individuals and CRC patients. In the present meta-analysis, the DOR value was 14.03, suggesting that miR-21 can serve as a promising diagnostic biomarker for CRC. When the threshold effect exists among studies, the SROC curve appears to be a better method to assess the summary diagnostic accuracy of the test rather than pooled sensitivity, pooled specificity, pooled PLR or pooled DOR. AUC is usually calculated to assess the accuracy of the selected indicator. The closer the value of AUC is to 1, the better the diagnostic accuracy of the test. By contrast, the closer the value of AUC is to 0.5, the worse the diagnostic accuracy of the test. In the present study, the AUC was 0.87, which indicates that miR-21 demonstrates good accuracy for CRC diagnosis. The HSROC model yields similar sensitivity (72.0%) and specificity (84.5%). This result further demonstrates the reliability of the meta-analysis. Overall, miR-21 has a moderate sensitivity and good specificity for CRC diagnosis.
Heterogeneity is a potential problem that can influence the incorporation effect and the interpretation of the meta-analysis results. Although strict inclusion and exclusion criteria were set to gain eligible studies, heterogeneity exists due to the existence of certain potential confounding factors. However, a broad range of sensitivity and specificity was reported. One cause of heterogeneity is the threshold effect, which arises due to different cut-off values used in different studies to define a negative or positive test result. Considering that the 9 studies used different cut-off values, the ROC plane and Spearman rank correlation test were employed to analyze the threshold effect. The ROC plane suggests no ‘shoulder-arm’ shape, and the Spearman correlation coefficient was 0.033 (P=0.932 and >0.05). This result demonstrates that threshold effect is not the source of heterogeneity. The patients involved in the present meta-analysis were from different ethnicities and populations. Not all the detection methods for miR-21 are based on RT-PCR and RT-qPCR. Ogata-Kawata et al (22) used miRNA microarray. Even with RT-qPCR, 6 studies used TaqMan probes and 2 used the SYBR-Green assay. The RNA extraction kits and normalizers also varied. Heterogeneity may result from different laboratories using different methods to quantify miR-21. Therefore, a meta-regression analysis was conducted to evaluate the contribution of the factors above. However, no heterogeneity was detected from these factors. Subsequently, a subgroup analysis was conducted on the ethnicity, test method and RNA extraction kits. In every subgroup, heterogeneity remained, indicating that these factors were not the sources of heterogeneity.
Zhang et al (26) and Du et al (27) also conducted a meta-analysis on the diagnostic value of miR-21 for CTC. Zhang et al (26) reported a pooled sensitivity of 76% and a pooled specificity of 81%. Du et al (27) reported a pooled sensitivity of 76% and a pooled specificity of 82%. The present results were similar to theirs, but not identical. They included only 6 studies, whereas 9 were included in the present meta-analysis. Du et al (27) retained 2 studies of Toiyama et al (24), but 1 of them appears to be a poster presentation. Additionally, the 2 studies were published with an interval of 1 year, which make them potential duplicates or overlapping data. Thus, 1 of these 2 studies was excluded in the present meta-analysis. Zhang et al (26) reported that different cut-off values of miR-21 expression across studies may be a source of heterogeneity. However, they did not demonstrate it further. In the present meta-analyses, ROC plane and Spearman rank correlation were performed, and verified that the threshold effect was not a source of heterogeneity.
miR-21 is overexpressed in numerous other solid types of tumor, indicating that its biology is relevant to numerous types of cancer (29,30). Zeng et al (31) reported that miR-21 had a sensitivity of 66.5% and a specificity of 83.1% in gastric cancer. Yang et al (32) summarized that the sensitivity and specificity of miR-21 in lung cancer were 71% and 84%, respectively. Other studies reported that miR-21 is upregulated in breast cancer, esophageal cancer, pancreatic cancer and glioblastoma (20). miR-21 is unlikely to become a useful blood-based biomarker for diagnosing CRC as an individual miRNA as it is also dysregulated in other diseases (33). Overall, miR-21 is not a special biomarker in CRC; it requires to be combined with another tool for enhanced specificity.
Certain limitations exist in the present study. First, the majority of these studies are retrospective analyses on historical cohorts, which limit the conclusions due to inherent study bias. Therefore, prospective studies that investigate the association between miR-21 and CRC are necessary. Second, evidence that demonstrates the existence of publication bias (P=0.436 for Deeks' test) by funnel plots is lacking. However, the independent patient data of several studies with negative results could not be collected, despite extensively searching other associated references and contacting authors via fax and e-mail. These may have been missed in the meta-analysis, indicating that publication bias may remain. However, current evidence indicates that circulating miR-21 has moderate sensitivity and good specificity as a diagnostic marker for CRC diagnosis. Large-scale prospective studies must be conducted in the future for verification. In addition, improving the diagnostic accuracy of circulating miR-21 and exploring new biomarkers with high diagnostic accuracy in CRC should still be considered in the future. miR-21-related diagnostics have substantial potential for the prevention and treatment of CRC.
Acknowledgements
The present study was funded by the National Natural Science Foundation of China [contract nos. 81071823 and 81201811] and Zhejiang University Research foundation, China [contract no. 11-491020-110].
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American joint committee on cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 4.Collins JF, Lieberman DA, Durbin TE, Weiss DG. Veterans Affairs Cooperative Study #380 Group: Accuracy of screening for fecal occult blood on a single stool sample obtained by digital rectal examination. A comparison with recommended sampling practice. Ann Intern Med. 2005;142:81–85. doi: 10.7326/0003-4819-142-2-200501180-00006. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Wiemer EA. The role of microRNAs in cancer: N o small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PloS one. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schetter AJ, Harris CC. Plasma microRNAs: A potential biomarker for colorectal cancer? Gut. 2009;58:1318–1319. doi: 10.1136/gut.2009.176875. [DOI] [PubMed] [Google Scholar]
- 12.Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS, A tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3:25. doi: 10.1186/1471-2288-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med. 2001;20:2865–2884. doi: 10.1002/sim.942. [DOI] [PubMed] [Google Scholar]
- 15.Moses LE, Shapiro D, Littenberg B. Combining independent studies of a diagnostic test into a summary ROC curve, Data-analytic approaches and some additional considerations. Stat Med. 1993;12:1293–1316. doi: 10.1002/sim.4780121403. [DOI] [PubMed] [Google Scholar]
- 16.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinnes J, Deeks J, Kirby J, Roderick P. A methodological review of how heterogeneity has been examined in systematic reviews of diagnostic test accuracy. Health Technol Assess. 2005;9(iii):1–113. doi: 10.3310/hta9120. [DOI] [PubMed] [Google Scholar]
- 18.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basati G, Razavi Emami A, Abdi S, Mirzaei A. Elevated level of microRNA-21 in the serum of patients with colorectal cancer. Medical Oncol. 2014;31:205. doi: 10.1007/s12032-014-0205-3. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138:1659–1666. doi: 10.1007/s00432-012-1244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, Sun XF. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34:2175–2181. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- 22.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PloS one. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PloS One. 2013;8:e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S. Plasma miR-21: A potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–551. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Li P, Ju H, Pesta M, Kulda V, Jin W, Cai M, Liu C, Wu H, Xu J, et al. Diagnostic and prognostic value of micro RNA-21 in colorectal cancer: An original study and individual participant data meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014;23:2783–2792. doi: 10.1158/1055-9965.EPI-14-0598. [DOI] [PubMed] [Google Scholar]
- 27.Du M, Liu S, Gu D, Wang Q, Zhu L, Kang M, Shi D, Chu H, Tong N, Chen J, et al. Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients. Carcinogenesis. 2014;35:2723–2730. doi: 10.1093/carcin/bgu189. [DOI] [PubMed] [Google Scholar]
- 28.Rosenfeld RM, Shiffman RN. Clinical practice guidelines: A manual for developing evidence-based guidelines to facilitate performance measurement and quality improvement. Otolaryngol Head Neck Surg. 2006;135((Suppl 4)):S1–S28. doi: 10.1016/j.otohns.2006.06.1277. [DOI] [PubMed] [Google Scholar]
- 29.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 31.Zeng Z, Wang J, Zhao L, Hu P, Zhang H, Tang X, He D, Tang S, Zeng Z. Potential role of microRNA-21 in the diagnosis of gastric cancer, A meta-analysis. PloS One. 2013;8:e73278. doi: 10.1371/journal.pone.0073278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X, Guo Y, Du Y, Yang J, Li S, Liu S, Li K, Zhang D. Serum microRNA-21 as a diagnostic marker for lung carcinoma, A systematic review and meta-analysis. PloS One. 2014;9:e97460. doi: 10.1371/journal.pone.0097460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krichevsky AM, Gabriely G. miR-21: A small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]