Abstract
MicroRNAs (miRNAs) are involved in tumorigenesis. However, little is known about their role in renal cell carcinoma (RCC). In the present study, the function of the miRNA miR-877 in RCC was investigated, and its expression levels in blood and paired RCC tissues were measured by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Bioinformatics analysis predicted eukaryotic elongation factor-2 kinase (eEF2K) to be the potential mRNA target of miR-877, which was verified by luciferase assay. The expression levels of eEF2K in RCC tissues were evaluated by western blot analysis and qPCR. The proliferation and migration abilities of RCC cells were measured by MTT and in vitro wound healing assays, respectively. The present results indicated that the expression levels of miR-877 were downregulated in blood and paired RCC tissues, whereas the expression levels of eEF2K were upregulated in RCC tissues. In addition, overexpression of miR-877 and knockdown of eEF2K significantly reduced the proliferation and migration abilities of RCC cells in vitro. Furthermore, miR-877 affected the eEF2K/eEF2 signaling pathway in these cells. In conclusion, the present study has demonstrated that miR-877 suppresses the proliferation and migration abilities of RCC cells by modulating the eEF2K/eEF2 signaling cascade. Therefore, miR-877 may be considered a potential biomarker for the diagnosis of RCC.
Keywords: miR-877, renal cell carcinoma, eukaryotic elongation factor-2 kinase
Introduction
Renal cell carcinoma (RCC) originates in the renal cortex, and is a highly metastatic urinary tumor that accounts for ~2% of adult malignancies and 3% of childhood malignancies (1–3). The incidence of RCC increases with age, and is higher in males than females, with a male-to-female ratio of 2:1 (4,5). In addition, the incidence of RCC is significantly higher in European and American countries compared with Asian countries, and the incidence is particularly low in Japan and India (6). Due to the resistance to chemotherapy and radical nephrectomy demonstrated by RCC, the use of microRNAs (miRNAs) as potential biomarkers for the diagnosis, prognosis and therapeutic treatment of RCC has increased in recent years (7–10).
miRNAs are a group of small RNAs ~22 nucleotides in length that negatively regulate gene expression by binding to the 3′-untranslated region (3′-UTR) of the target mRNAs (11,12). miRNAs are involved in the regulation of cell proliferation, development, differentiation and apoptosis (13–15). It is estimated that ~30% of human genes are regulated by miRNAs, and there is increasing evidence supporting the participation of miRNAs in human cancer (16,17).
Eukaryotic elongation factor-2 kinase (eEF2K) is a calmodulin (CaM)-dependent protein kinase that phosphorylates and inhibits eEF2, thus preventing the elongation phase of protein translation (18). eEF2K consists of several domains, including an N-terminal catalytic domain, a C-terminal α-helical region and a linker containing several regulatory phosphorylation sites. Previous studies have identified high expression levels of eEF2K in several types of cancer, and suggested that eEF2K may promote cell proliferation during nutrient starvation in these tumors (19–21). eEF2K has been considered as a potential cancer-therapeutic target, and the use of miRNA delivery agents for the treatment of different types of cancer has been investigated in previous clinical trials (1,22–25). Therefore, the identification of novel anticancer miRNAs capable of targeting eEF2K is of importance.
Previous studies have reported that miRNA-877 acts as a tumor suppressor in human cancer (26). However, the role of miRNA-877 in RCC has not been investigated at present. In the present study, the expression levels of miRNA-877 were observed to be downregulated in the serum and tissues derived from patients with RCC. Furthermore, miRNA-877 was demonstrated to be directly bound to eEF2K, thus inhibiting its transcription and preventing the proliferation of RCC cells. In summary, the results of the present study suggest that miRNA-877 may be a potential biomarker for the diagnosis and therapeutic treatment of RCC.
Materials and methods
Human blood and tissues
Human plasma and surgical specimens, consisting of 100 RCC tissues and 100 paired adjacent normal tissues, were collected from The Third Affiliated Hospital of Soochow University (Changzhou, Jiangsu, China) between January 2012 and December 2014. The total cohort included 55 men and 45 women, with an age range of 45–73 years. The tissues were freshly frozen in liquid nitrogen and subsequently stored at −80°C. Written informed consent for participation in the present study was obtained from all patients and the study was approved by the ethics committee of the Third Affiliated Hospital of Soochow University.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
miRNA was extracted from the tissue specimens using the miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions. The miScript II RT kit (Qiagen GmbH) was used for the reverse transcription of miRNA and mRNA in these tissues. The levels of miRNA-877 in the samples were quantified by RT-qPCR, using the miScript SYBR Green PCR kit (Guangzhou Funeng Gene Co., Ltd., Guangzhou, Guangdong, China), according to the manufacturer's instructions. The primer for U6, used as an internal control, was provided in the kit. The relative expression levels of miRNA-877 in the samples were calculated by the 2−∆∆Ct method, and normalized to the levels of U6.
Cell culture and transfection
The human RCC ACHN, Caki-2, 769-p, 786-O and HEK293 cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA), and cultured in Dulbecco's modified Eagle medium containing 5% or 10% (HEK293 cells) heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific Inc., Waltham, MA, USA), at 37°C in a 5% CO2 tissue culture incubator. The cells were then subcultured at 90% confluency, and maintained in DMEM supplemented with 10% FBS, at 37°C in a 5% CO2 tissue culture incubator. The human RCC cells (5×105 cells/well) were seeded into 6-well plates for in vitro wound healing assays and transfection. The HEK293 cells (1×104 cells/well) were seeded into 24-well plates for the luciferase assay. In order to investigate the role of miR-877 in RCC, miR-877 mimic or inhibitor (Guangzhou Funeng Gene Co., Ltd.) were transfected into the cells using Lipofectamine 2000 (Thermo Fisher Scientific Inc.), according to the manufacturer's protocol.
Western blot analysis
Total protein content was extracted from the tissue specimens using 250 µl RIPA lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) and 5 mM phenylmethanesulfonyl fluoride (Beyotime Institute of Biotechnology), according to the manufacturer's instructions. Total protein was quantified by Bradford assay (27), and 30–50 µg protein was subsequently used for western blot assay. The proteins were subjected to 8–12.5% gel electrophoresis, and transferred onto nitrocellulose membranes (EMD Millipore, Boston, MA, USA). The membranes were treated for 1 h with Tris-buffered saline containing 0.1% Tween 20 (TBST) and 5% skim milk (Amresco LCC, Solon, OH, USA), and then incubated overnight at 4°C with primary polyclonal or monoclonal antibodies as follows: Anti-eEF2K (polyclonal rabbit anti-human; cat no. 3692; 1:1,000); anti-phosphorylated (p) EF2 (Thr56) (polyclonal rabbit anti-human; cat no. 2331; 1:1,000); anti-pAkt (Ser473) (monoclonal rabbit anti-human; cat no. 4060; 1:1,200); and anti-cyclin D1 (monoclonal rabbit anti-human; cat no. 2922; 1:1,000; Cell Signaling Technology, Inc., Danvers, USA). Next, the membranes were washed three times with TBST for 6 min each, and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (cat no. 7074; 1:2,000; Cell Signaling Technology, Inc.) secondary antibody for 2 h at room temperature. Proteins were visualized by enhanced chemiluminescence. The expression levels of the target proteins were normalized to those of GAPDH.
In vitro wound healing assay
Cells were transfected with miR-877 mimic or scramble control miRNA and cultured in 60-mm tissue culture plates under standard conditions (25). At 24 h post-transfection, the cells were incubated with mitomycin C (10 µg/ml; Sigma-Aldrich, St. Louis, MO, USA) for 2 h to prevent any potential cell migration due to cell proliferation. Subsequently, mitomycin C was removed from the medium, and the cells were washed 3 times with 1X phosphate-buffered saline (PBS; Nanjing Sunshine Biotechnology Co., Ltd., Nanjing, China), prior to being scratched with a Axygen P200 pipette tip (Corning, Inc., Corning, NY, USA), in order to create an artificial wound (width, ~0.5 mm; length, 2 cm). The cells were then washed 3 times with PBS to remove any cellular debris. Next, fresh medium was added to the cells, which were cultured for additional 24 h. To monitor the healing process, the wounds were photographed at 0 and 24 h after the cells had been scratched.
Luciferase assay and constructs
The 3′-UTRs of the wild-type and mutant eEF2K sequences were cloned into the PsiCHECK-2 Vector (Promega Corporation, Madison, WI, USA), as previously described (12). A Fast Mutagenesis System mutation kit (TransGen Biotech, Inc., Beijing, China) was used for mutant construction, and DNA sequences were confirmed by DNA sequencing. A total of 2×105 RCC cells were seeded into 48-well plates and incubated for 24 h prior to transfection. Then, 40 ng of the PsiCHECK-2-eEF2K-3′-UTR plasmid and 10 pmol scramble control miRNA or miR-877 mimic were added to 50 µl Opti-MEM (Thermo Fisher Scientific Inc.), and co-transfection was subsequently performed using 0.5 µl Lipofectamine 2000/50 µl Opti-MEM. The luciferase activity of the cell extracts was measured at 24 h post-transfection, using the Dual-Luciferase Reporter Assay System (Promega Corporation). The transfection efficiency was normalized by detecting the activity of Renilla luciferase, according to the Dual-Luciferase Reporter Assay System manufacturer's instructions.
Bioinformatics
The binding sites for miR-877 on the 3′-UTR of the potential target genes were predicted by miRanda software (available from http://www.microrna.org).
Statistical analysis
The RT-qPCR results were analyzed using the 2−∆∆Ct method. Multiple regression analysis was used to evaluate the association between the expression levels of miR-877 and the clinical features of RCC. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of miR-877 expression in plasma and RCC tissues
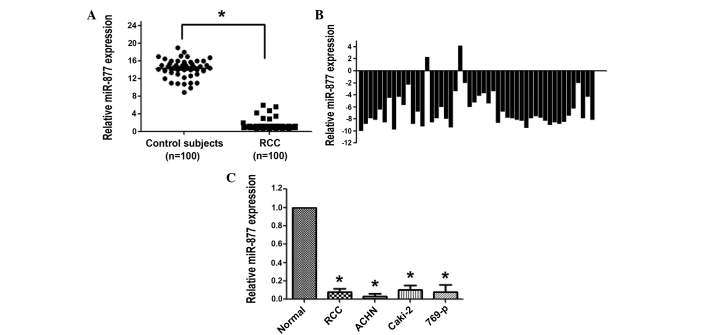
The plasma levels of miR-877 in 100 patients with RCC and 100 healthy control individuals were measured by RT-qPCR. The data indicated that the concentration of miR-877 in the plasma was markedly reduced in patients with RCC compared with the healthy control individuals (P<0.001; Fig. 1A). The clinical significance of miR-877 was further validated by analyzing the expression levels in 50 RCC and paired tissues via RT-qPCR. In agreement with the results obtained in the plasma, RT-qPCR revealed that miR-877 was significantly downregulated in 96% of the RCC samples (P<0.001; Fig. 1B). Notably, miR-877 was barely expressed in the RCC ACHN, Caki-2 and 769-p cell lines (Fig. 1C).
Figure 1.
The expression levels of the miRNA miR-877 were lower in RCC cells, plasma of patients with RCC and renal tumor tissues, compared with normal tissues. (A) The relative expression levels of miR-877 in the plasma of patients with RCC and healthy control subjects were measured by RT-qPCR. Each dot in the graph represents 1 sample. *P<0.05 vs. control subjects. (B) The relative expression levels of miR-877 in renal tumors were presented as fold differences in tumor vs. paired normal tissues. (C) RT-qPCR analysis was conducted to measure the expression levels of miR-877 in normal tissues, RCC tissues and the RCC ACHN, Caki-2 and 769-p cell lines. The levels of miR-877 in normal tissues were set to 1. The data were normalized and expressed as the mean ± standard error. *P<0.05 vs. control (normal tissue). miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; RCC, renal cell carcinoma.
Overexpression of miR-877 suppressed RCC cell proliferation
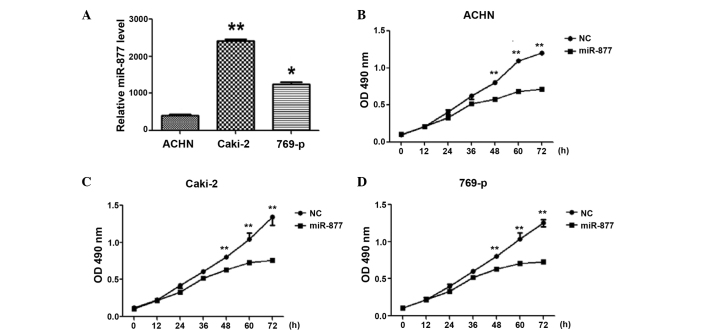
To explore the biological significance of miR-877, miR-877 mimic and control miRNA (Guangzhou Funeng Gene Co., Ltd.) were transfected into the RCC ACHN, Caki-2 and 769-p cell lines. The efficiency of the transfection was assessed by RT-qPCR. The expression of miR-877 was observed to demonstrate a 400–2,400-fold increase in the ACHN, Caki-2 and 769-p cells. There were no evident morphological changes in the cells overexpressing miR-877, but cellular proliferation analyses demonstrated that the overexpression of miR-877 suppressed cell proliferation in the 3 RCC cell lines (P<0.05; Fig. 2).
Figure 2.
Effect of the overexpression of the miRNA miR-877 on the proliferation ability of RCC cells. (A) RT-qPCR analysis was used to evaluate the efficiency of the transfection of miR-877 into different RCC cell lines, including ACHN, Caki-2 and 769-p. *P<0.01, **P<0.001 vs. ACHN. RCC cell lines (B) ACHN, (C) Caki-2 and (D) 769-p, were transfected with miR-877 or control miRNA, and their proliferation ability was investigated by MTT assay.**P<0.01 vs. control. miRNA, microRNA; RCC, renal cell carcinoma, RT-qPCR, reverse transcription-quantitative polymerase chain reaction; OD, optical density; NC, negative control.
Overexpression of miR-877 reduced RCC cell motility during wound healing
To investigate the effect of miR-877 on the migration ability of RCC cells, a scratch wound healing assay was performed. For this purpose, 4 different RCC cell lines, consisting of the ACHN, Caki-2, 769-p and 786-O cell lines, were transfected with miR-877 mimic or scramble control miRNA, and the migration distance was measured at 0 and 24 h after the cells had been scratched. Compared with the scramble control miRNA, all the RCC cell lines that were transfected with miR-877 mimic exhibited enhanced migration at 24 h (P<0.01; Fig. 3).
Figure 3.
The miRNA miR-877 promoted cell migration in RCC (A) ACHN, (B) Caki-2, (C) 769-p and (D) 786-O cells. Scratch wound-healing assays were conducted in RCC cells transfected with miR-877 and scramble control miRNA RCC cells. The migration distance was measured at 0 and 24 h after the cells had been scratched. miRNA, microRNA; RCC, renal cell carcinoma; NC, negative control.
miR-877 directly targeted eEF2K by affecting the eEF2K/EF2 signaling pathway
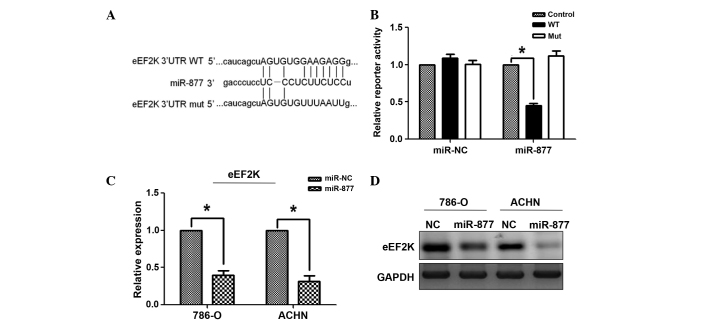
Bioinformatics analysis predicted the 3′-UTR of eEF2K to contain complementary binding sites for miR-877 (Fig. 4A). To test whether eFF2K was a functional target of miR-877, dual luciferase reporter assays were performed, and to assess whether eEF2K was a functional target of miR-877, dual luciferase reporter assays were performed. Therefore, HEK293 cells were transfected with a wild-type or mutant PsiCHECK-2-eEF2K-3′-UTR plasmid, and the cells were co-transfected with miR-877 mimic or scramble control miRNA. Compared with the scramble control miRNA, the miR-877 mimic significantly reduced the luciferase activity of wild-type PsiCHECK-2-eEF2K-3′-UTR, but did not alter the activity of the mutant plasmid (Fig. 4B). In addition, the mRNA and protein levels of eEF2K were significantly reduced in Caki-2 and 786-O cells transfected with miR-877 mimic or scramble control miRNA (Fig. 4C and D). These results suggest that eEF2K mRNA was a direct target of miRNA-877. In order to assess whether miR-877 affects the eEF2K/EF2 signaling pathway, the levels of downstream proteins in the cascade, including Akt, cyclin D1 and pEF2 were measured by western blot analysis. As presented in Fig. 5, the protein levels of cyclin D1, Akt, pAkt and pEF2 were reduced in the miR-877-transfected RCC cells.
Figure 4.
The miRNA miR-877 downregulated the expression of eEF2K by targeting the 3′-UTR of eEF2K. (A) Sequence alignment of the wild-type and mutant eEF2K 3′-UTR, indicating the potential binding sites for miR-877. (B) Luciferase reporter assays revealed a reduction in reporter activity following the transfection of the wild-type eEF2K 3′-UTR reporter construct into the 786-O and ACHN RCC cells overexpressing miR-877. The eEF2K 3′-UTR mutant and control constructs did not exhibit any effect on reporter activity. A Renilla luciferase construct was co-transfected into the cells as an internal control. The normalized luciferase activity of the control construct in each experiment was set to 1. The data represent the mean ± standard error. *P<0.01 vs. control. The (C) mRNA and (D) protein levels of eEF2K were examined by RT-qPCR and western blot analysis, respectively, in RCC 786-O and ACHN cells overexpressing miR-877. The mRNA data were normalized to the levels of GAPDH, which was used as a loading control in western blot analysis. *P<0.01 vs. control miRNA. miR, microRNA; eEF2K, eukaryotic elongation factor-2 kinase; UTR, untranslated region; RT-qPCR, reverse transcription-polymerase quantitative chain reaction; NC, negative control; WT, wild-type; mut, mutant.
Figure 5.
Downstream molecular effects of the overexpression of mimic of the miRNA miR-877 in the RCC ACHN, Caki-2 and 769-p cells. The RCC cells were transfected with miR-877 mimic, and the cell lysates were subjected to western blot analysis. GAPDH was used as a control. Overexpression of miR-877 mimic reduced the protein levels of eEF2K, pEF2, cyclin D1, Akt and pAkt. miRNA, microRNA; RCC, renal cell carcinoma; eEF2K, eukaryotic elongation factor-2 kinase; p, phosphorylated; Akt, protein kinase B; NC, negative control.
eEF2K protein levels are upregulated in RCC tissues
To investigate whether miR-877 was involved in the pathogenesis of human RCC, the protein levels of eEF2K in RCC tissues were evaluated. The mRNA and protein expression levels of eEF2K in 16 pairs of RCC and normal tissues were analyzed by RT-qPCR, western blotting and immunohistochemistry (IHC). The 16 pairs of samples exhibited a significant increase in the levels of eEF2K in the RCC tissues, compared with the control tissues (Fig. 6A). In addition, low levels of miR-877 were associated with increased expression levels of eEF2K mRNA, and vice versa. Notably, compared with the normal tissues, marked overexpression of the eEF2K protein was detected in the 16 RCC tissue specimens by IHC and western blot analysis (Fig. 6B and C, respectively), further supporting that eEF2K is upregulated in RCC tissues.
Figure 6.
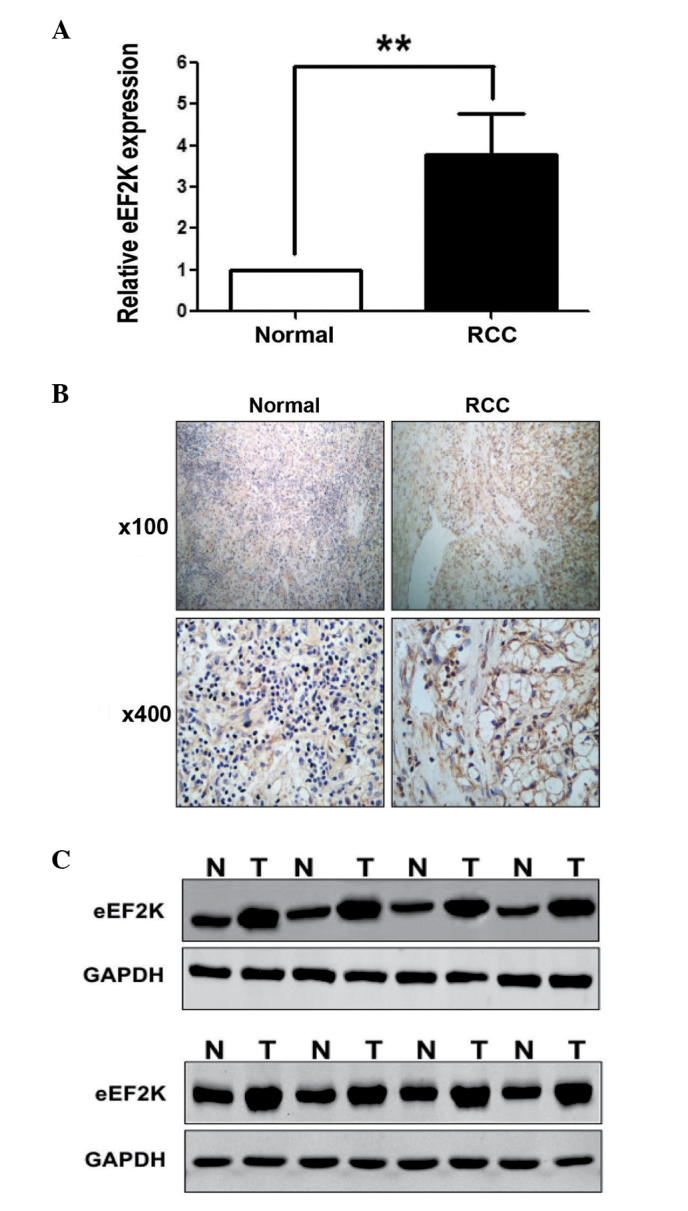
eEF2K was upregulated in RCC tissues compared with the paired normal tissues. (A) The mRNA expression levels of eEF2K were examined by reverse transcription-quantitative polymerase chain reaction in 50 pairs of normal and RCC tissues. **P<0.01 vs. normal tissues. (B) Immunostaining of eEF2K in RCC and paired tissues. (C) The protein expression levels of eEF2K were assessed by western blot analysis in 16 RCC and paired tissues. GAPDH was used as control. eEF2K, eukaryotic elongation factor-2 kinase; RCC, renal cell carcinoma; N, normal tissue; T, tumor tissue.
Discussion
RCC is one of the most common neoplasms of the kidney in adults (28). Metastatic RCC is challenging to treat, and the 5-year survival rate is <10% (3). Therefore, early diagnosis of RCC is of importance. miRNAs are small non-coding RNA molecules containing ~22 nucleotides. miRNAs are well conserved in plants and animals, and are considered to be a vital and evolutionarily ancient component of genetic regulation (13). In animals, miRNAs are able to recognize target mRNAs through specific binding sites located at the 3′ end of the target RNA molecule (14). The binding of the miRNA to its target mRNA induces the cleavage of the targeted mRNAs, which constitutes a post-transcriptional regulatory mechanism of gene expression (14).
Previously, miRNAs have been revealed to perform an important role in cancer metastasis, and miRNAs are considered to be prognostic biomarkers in cancer (13). It has been demonstrated that miRNAs are stable and resistant to extreme pH and temperature in body fluids (29,30), and serum miRNAs have been reported as the best biomarker for cancer diagnosis (31,32). Thus, the use of serum as a diagnostic tool for patients with RCC is of relevance. In the present study, a miRNA signature was identified in plasma and tissues of patients with RCC. The expression levels of miR-877 were observed to be downregulated in the RCC specimens, whereas the expression levels of its target gene eEF2K were upregulated in these samples. This trend has also been previously reported in other types of cancer, which suggests that miRNAs may be involved in the development of metastatic RCC (33,34).
The results of the present study indicated that miR-877 was downregulated in blood and tissues of patients with RCC. These findings suggest that miR-877 may be a potential biomarker for RCC. During the process of tumor development, miRNAs may act as oncogenes or tumor suppressors, depending on the target genes (35). Therefore, gene regulation by miRNAs is a complex process, since miRNAs may possess direct and indirect targets (36). The direct targets of miRNAs may be affected at the mRNA level by mRNA degradation, or at the protein level by translation inhibition (37). The indirect targets of miRNAs may be downstream pathway molecules of the miRNA target genes (13).
In the present study, miR-877 was predicted to target sites on the 3′-UTR of eEF2K, according to bioinformatics analysis. eEF2K is a CaM-dependent protein kinase III that is activated by Ca2+/CaM. The activation of eEF2K triggers the phosphorylation of eEF2, which leads to an increase in nuclear transcription (20). It has previously been demonstrated that the activity of eEF2K was associated with proliferation of tumor cells, and high expression levels of eEF2K have been detected in several types of malignancies (18). eEF2K has been found to modulate the activity of certain apoptotic proteins, which resulted in the inhibition of cancer apoptosis (20). eEF2K has also been reported to perform a regulatory role in autophagy (21). eEF2K has been previously been considered a potential cancer target, but therapeutic interventions based on eEF2K have not been developed at present (38).
In the present study, eEF2K was upregulated in RCC tissues. In order to investigate the role of miR-877 in RCC, the RCC cells were transfected with miR-877 mimic and western blot analysis was performed. The results demonstrated a significant downregulation of eEF2K in the cells transfected with miR-877 mimic compared with the control group. In addition, certain cellular phenotypes, including cell proliferation and migration, were demonstrated to be regulated by miR-877 via the eEF2K/eEF2 signaling pathway.
In conclusion, the results of the present study may aid the understanding of the mechanism behind RCC metastasis. Furthermore, the specific expression pattern of miR-877 observed in the present study and the effects exerted by miR-877 on multiple signaling pathways in RCC cells suggest that miR-877 may be a potential therapeutic intervention for the treatment of cancer.
References
- 1.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: A new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 2.Yu Z, Jian Z, Shen SH, Purisima E, Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steffens S, Roos FC, Janssen M, Becker F, Steinestel J, Abbas M, Steinestel K, Wegener G, Siemer S, Thüroff JW, et al. German Renal Cell Cancer Network: Clinical behavior of chromophobe renal cell carcinoma is less aggressive than that of clear cell renal cell carcinoma, independent of Fuhrman grade or tumor size. Virchows Arch. 2014;465:439–444. doi: 10.1007/s00428-014-1648-9. [DOI] [PubMed] [Google Scholar]
- 4.Doehn C, Witzsch U, Siebels M. Active surveillance for renal cell carcinoma. Aktuelle Urol. 2012;43:243–249. doi: 10.1055/s-0032-1321828. (In German) [DOI] [PubMed] [Google Scholar]
- 5.Chen T, Fallah M, Sundquist K, Liu H, Hemminki K. Risk of subsequent cancers in renal cell carcinoma survivors with a family history. Eur J Cancer. 2014;50:2108–2118. doi: 10.1016/j.ejca.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Hew MN, Zonneveld R, Kümmerlin IP, Opondo D, de la Rosette JJ, Laguna MP. Age and gender related differences in renal cell carcinoma in a European cohort. J Urol. 2012;188:33–38. doi: 10.1016/j.juro.2012.02.2573. [DOI] [PubMed] [Google Scholar]
- 7.Cairns P. Renal cell carcinoma. Cancer Biomark. 2010;9:461–473. doi: 10.3233/CBM-2011-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olshan AF, Kuo TM, Meyer AM, Nielsen ME, Purdue MP, Rathmell WK. Racial difference in histologic subtype of renal cell carcinoma. Cancer Med. 2013;2:744–749. doi: 10.1002/cam4.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banyra O, Tarchynets M, Shulyak A. Renal cell carcinoma: How to hit the targets? Cent European J Urol. 2014;66:394–404. doi: 10.5173/ceju.2013.04.art2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ngo TC, Wood CG, Karam JA. Biomarkers of renal cell carcinoma. Urol Oncol. 2014;32:243–251. doi: 10.1016/j.urolonc.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 11.White NM, Khella HW, Grigull J, Adzovic S, Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA, et al. miRNA profiling in metastatic renal cell carcinoma reveals a tumor-suppressor effect for miR-215. Br J Cancer. 2011;105:1741–1749. doi: 10.1038/bjc.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beresneva EV, Rykov SV, Hodyrev DS, Pronina IV, Ermilova VD, Kazubskaia TP, Braga EA, Loginov VI. Methylation profile of group of miRNA genes in clear cell renal cell carcinoma; involvement in cancer progression. Genetika. 2013;49:366–375. doi: 10.7868/s001667581303003x. (In Russian) [DOI] [PubMed] [Google Scholar]
- 13.Mei Q, Li X, Guo M, Fu X, Han W. The miRNA network: Micro-regulator of cell signaling in cancer. Expert Rev Anticancer Ther. 2014;14:1515–1527. doi: 10.1586/14737140.2014.953935. [DOI] [PubMed] [Google Scholar]
- 14.Filip A. MiRNA - new mechanisms of gene expression control. Postepy Biochem. 2007;53:413–419. (In Polish) [PubMed] [Google Scholar]
- 15.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannakakis A, Coukos G, Hatzigeorgiou A, Sandaltzopoulos R, Zhang L. miRNA genetic alterations in human cancers. Expert Opin Biol Ther. 2007;7:1375–1386. doi: 10.1517/14712598.7.9.1375. [DOI] [PubMed] [Google Scholar]
- 17.Mezzanzanica D, Canevari S, Cecco LD, Bagnoli M. miRNA control of apoptotic programs: Focus on ovarian cancer. Expert Rev Mol Diagn. 2011;11:277–286. doi: 10.1586/erm.11.1. [DOI] [PubMed] [Google Scholar]
- 18.Rose AJ, Alsted TJ, Jensen TE, Kobberø JB, Maarbjerg SJ, Jensen J, Richter EA. A Ca(2+)-calmodulin-eEF2K-eEF2 signalling cascade, but not AMPK, contributes to the suppression of skeletal muscle protein synthesis during contractions. J Physiol. 2009;587:1547–1563. doi: 10.1113/jphysiol.2008.167528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu LL, Xie T, Zhang SY, Liu B. Eukaryotic elongation factor-2 kinase (eEF2K): A potential therapeutic target in cancer. Apoptosis. 2014;19:1527–1531. doi: 10.1007/s10495-014-1019-7. [DOI] [PubMed] [Google Scholar]
- 20.Arora S, Yang JM, Kinzy TG, Utsumi R, Okamoto T, Kitayama T, Ortiz PA, Hait WN. Identification and characterization of an inhibitor of eukaryotic elongation factor 2 kinase against human cancer cell lines. Cancer Res. 2003;63:6894–6899. [PubMed] [Google Scholar]
- 21.Xie CM, Liu XY, Sham KW, Lai JM, Cheng CH. Silencing of EEF2K (eukaryotic elongation factor-2 kinase) reveals AMPK-ULK1-dependent autophagy in colon cancer cells. Autophagy. 2014;10:1495–1508. doi: 10.4161/auto.29164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu H, Yang X, Liu J, Zhou L, Zhang C, Xu L, Qin Q, Zhan L, Lu J, Cheng H, Sun X. Eukaryotic elongation factor 2 kinase confers tolerance to stress conditions in cancer cells. Cell Stress Chaperones. 2014;20:217–220. doi: 10.1007/s12192-014-0545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Cheng Y, Liu Q, Bao JK, Yang M. Autophagic pathways as new targets for cancer drug development. Acta Pharmacol Sin. 2010;31:1154–1164. doi: 10.1038/aps.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tekedereli I, Alpay SN, Tavares CD, Cobanoglu ZE, Kaoud TS, Sahin I, Sood AK, Lopez-Berestein G, Dalby KN, Ozpolat B. Targeted silencing of elongation factor 2 kinase suppresses growth and sensitizes tumors to doxorubicin in an orthotopic model of breast cancer. PLoS One. 2012;7:e41171. doi: 10.1371/journal.pone.0041171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi M, Huang X, Zhou L, Zhang J. Identification of differentially expressed microRNAs in metastatic melanoma using next-generation sequencing technology. Int J Mol Med. 2014;33:1117–1121. doi: 10.3892/ijmm.2014.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruger NJ. The Bradford method for protein quantitation. In: Walker JM, editor. Basic Protein and Peptide Protocols. Vol. 32. Totowa, NJ: Humana Press; 1994. pp. 9–15. [DOI] [Google Scholar]
- 28.Creel PA. Optimizing patient adherence to targeted therapies in renal cell carcinoma. Clin J Oncol Nurs. 2014;18:694–700. doi: 10.1188/14.CJON.694-700. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Ai H, Ren J, Li W, Li P, Qiao R, Ouyang J, Yang M, Ma J, Huang L. A global view of porcine transcriptome in three tissues from a full-sib pair with extreme phenotypes in growth and fat deposition by paired-end RNA sequencing. BMC Genomics. 2011;12:448. doi: 10.1186/1471-2164-12-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyons PJ, Lang-Ouellette D, Morin P., Jr CryomiRs: Towards the identification of a cold-associated family of microRNAs. Comp Biochem Physiol Part D Genomics Proteomics. 2013;8:358–364. doi: 10.1016/j.cbd.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Dai Y, Yang J, Chen T, Yin Y, Tang M, Hu C, Zhang L. Microarray analysis of microRNA expression in renal clear cell carcinoma. Eur J Surg Oncol. 2009;35:1119–1123. doi: 10.1016/j.ejso.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Guo Y, Shang C, Song Y, Wu B. miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology. 2012;80:1298–1302. doi: 10.1016/j.urology.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Shenouda SK, Alahari SK. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 37.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knebel A, Haydon CE, Morrice N, Cohen P. Stress-induced regulation of eukaryotic elongation factor 2 kinase by SB 203580-sensitive and -insensitive pathways. Biochem J. 2002;367:525–532. doi: 10.1042/bj20020916. [DOI] [PMC free article] [PubMed] [Google Scholar]