Abstract
MicroRNAs (miRNAs or miRs) have been advocated as potentially robust and highly stable biomarkers of diverse disease conditions including cancer. The primary aim of this study was two-fold: i) to profile the expression levels of selected mature miRNA signature genes, such as miR-145, miR-195, miR-29 and miR-92, in a paired-study design of 20 colorectal cancer (CRC) tissues from patients versus adjacent neoplasm-free mucosal tissues employing reverse transcription-quantitative polymerase chain reaction; and ii) to examine their expression level in the plasma of the same CRC patients in relation to the age-matched plasma of healthy controls. Statistically significant (P<0.01) increases in miR-29 (2.5) and miR-92 (2.6) were observed in CRC tissues compared with adjacent neoplasm-free mucosal tissues. Profiling of CRC plasma samples showed that the expression levels of circulating miR-29 and miR-92 were significantly higher (P<0.01) than in the age-matched normal plasma. By contrast, miR-145 and miR-195 exhibited significant (P<0.05) decreases in their mean expression levels in CRC tissue samples in relation to the normal tissues. The mean expression levels of miR-145 and miR-195 were significantly lower (P<0.05) in CRC plasma than the healthy controls. Distinct stage-dependent changes in the expression level of the four miRNA gene profiles were observed between stages II and IV plasma of CRC patients relative to the control plasma. Taken together, the results clearly reflect a similar trend for the four miRNA expression levels in tissue and plasma as well as the positive correlation in the levels of miRNAs in tissues and plasma. These findings may be useful to clarify the molecular mechanisms underlying colorectal carcinogenesis and to underscore the potential of the investigated miRNAs as novel early diagnostic biomarkers of CRC.
Keywords: microRNA, colorectal cancer, plasma, biomarkers, real-time polymerase chain reaction
Introduction
Worldwide, colorectal cancer (CRC) is ranked the third most commonly diagnosed cancer types of in both genders after lung and breast cancers, accounting for almost 10% of the cancer mortalities each year. Approximately 1.4 million new cases are diagnosed annually with CRC of which there is an estimated mortality rate of 50% (1). In the year 2005, the population of the Kingdom of Saudi Arabia (KSA) was estimated at 16,945,484, comprising mostly of native Saudis (62%). In that same year, the Saudi Cancer Registry reported that CRC was the second most common malignancy among Saudis for all ages (10.3%) and the number one malignancy in males (11.8%) (2,3). Additionally, between 1994 and 2003 age-standardized rates for CRC in KSA almost doubled (2). Between 2001 and 2003, while the annual percent change (APC) of CRC incidence in Saudi females exhibited an insignificant increase of 6%, a profoundly rising incidence among Saudi males was observed, with an APC of 20.5% (4). It has been predicted that by the year 2030, the CRC incidence in KSA may increase four-fold in the two genders (2). This suggests a foreseeable increase in cancer burden mainly attributed to population growth, adoption of unhealthy life styles and its associated risk factors and aging of the population.
Early diagnosis and treatment of CRC positively correlates with higher patient survival rates (5). In the U.S., for example, the 5-year survival rate is as high as 93.2% for stage I as compared to only 8.1% for stage IV (5). Consequently, early diagnosis is a vital goal for any healthcare system to achieve a reliable treatment and successful clinical outcome of CRC patients. Several clinical tests are currently employed for CRC screening, including fecal occult blood tests (FOBT), radiologic tests, colonoscopy and stool DNA test. However, none of these approaches have been regarded as the method of choice due to their invasiveness, low sensitivity or high cost (6,7). Thus, it is fundamentally important to search for novel, non-invasive, specific and sensitive biomarkers for the early diagnosis of CRC.
Recently, microRNAs (miRNAs or miRs), which represent a newly-discovered class of short (19–25 nucleotides) non-coding RNAs, have acquired considerable interest due the roles they play in a variety of cell processes including development, cell cycle progression, cell differentiation, proliferation and apoptosis (8). Bioinformatics and cloning studies have indicated that miRNAs may post-transcriptionally regulate almost 60% of all human genes and control hundreds of cognate gene targets through their oncogenic or tumor-suppressive activity (9). Aberrant miRNA expressions have been associated with several types of hematological and solid malignant tumors (10,11) including CRC (12–17), highlighting their potential for diagnostic and prognostic applications, and classification of human malignancies (18,19). While the discovery of malignancy-linked RNAs in plasma/serum were first described almost 10 years ago (20), scientific reports on the existence of miRNAs in body fluids such as blood plasma/serum, saliva, urine and semen of cancer patients have only been presented more recently (21–24). Notably, various studies have recently confirmed the potential utility of circulating miRNAs as stable biomarkers for the multi-stage process of carcinogenesis in solid tumors and several other malignancies (21,23).
The investigation of cancer-specific circulating miRNA biomarkers is an emerging and promising field of research. Thus, we aimed in the current study to conduct targeted transcriptional profiling of a panel of four mature miRNA signatures (miR-145, miR-195, miR-29 and miR-92) in the plasma and colorectal tissues of Saudi CRC patients in relation to healthy controls to assess their potential diagnostic value using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). These miRNA genes were particularly selected based on a comprehensive review of relevant literatures and previously published data on CRC (15,22,25–28). In addition, the association between the aberrant expression of these circulating miRNA gene profiles and CRC clinical stages was also evaluated.
Materials and methods
Patients
The subjects recruited in this study (CRC patients, n=20; healthy neoplasm-free controls, n=20) were subjected to rigorous eligibility criteria for the selection (Table I). Patients were excluded if they had been undergoing chemotherapy or radiotherapy prior to blood sampling, or clinically diagnosed with familial adenomatous polyposis or hereditary non-polyposis CRC. The tumor-node-metastasis classification system accepted by the Union for International Cancer Control (29) has been employed for staging of malignant tumors. Signed written and informed consent forms were collected from all the subjects participating in this study for the use of their blood and tissue samples. Research protocols conducted in this project were approved by the Institutional Medical Ethics Review board at the College of Applied Medical Sciences Affiliated to King Saud University (Riyadh, Kingdom of Saudi Arabia). All samples were obtained from the Saudi CRC Biobank at King Khalid University Hospital.
Table I.
Eligibility criteria for the selection and exclusion of study subjects.
| Clinicopathological characteristics |
|---|
| General inclusion criteria |
| 1. Age ≥18 years and ≤80 years |
| 2. Not currently residing in an institution, such as a nursing home or shelter |
| 3. Not severely ill in the intensive care unit |
| 4. With the capability to give informed consent |
| 5. Encountered between September 2013 and March 2015 |
| Healthy individuals (plasma control group) |
| 1. Underwent the medical check-up in King Khalid University Hospital |
| 2. Asymptomatic and apparently healthy without a previous history of cancer |
| 3. Confirmed healthy condition without malignancy in the physical examinations |
| 4. No system infection (lung, gastrointestinal or urinary tract) |
| Colorectal cancer patients (CRC group) |
| 1. Underwent colonoscopy biopsy and colorectal surgical resections |
| 2. Diagnosed by 2 experienced pathologists |
| 4. No pre-operative chemotherapy and radiotherapy |
Tissue isolation and RNA extraction
Paired fresh CRC tissue specimens and their adjacent non-cancerous normal mucosa were collected from 20 patients who underwent surgical resection of tumors by surgeons and subsequently examined by pathologists. The CRC tissues were histologically confirmed to be an adenocarcinoma of the colon. Tissue specimens were collected in RNAlater® RNA Stabilization Reagent (Qiagen, Hilden, Germany) tubes, snap-frozen in liquid nitrogen, and stored at −80°C until further analysis. Total RNA including miRNAs was extracted from 50–100 mg of cryo-preserved tissues by use of TRIzol® reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) as described in the manufacturer's protocol. In order to maximize RNA yield, a homogenization step was carried out by use of a TissueLyser LT with 5-mm stainless-steel beads (Qiagen).
Plasma preparation and circulating RNA extraction
Blood was drawn from the 20 CRC patients recruited in this study as well as 20 age-matched neoplasm-free healthy subjects. The cancer-free status of blood-donating healthy control subjects was confirmed a priori based on their negative health examination results including: blood test, chest X-ray, abdominal ultrasound examination, FOBT, rectal examination, computed tomography scan and colonoscopy. None of the controls had been previously diagnosed with any types of malignancy. Peripheral whole blood (8–10 ml) was collected from each individual into BD Vacutainer® blood collection tubes (BD Biosciences, Franklin Lakes, NJ, USA) (EDTA spray-coated). Plasma was fractionated from whole blood samples according to the procedure described by Duttagupta et al (30). Freshly drawn whole blood was processed for plasma fractionation and the obtained plasma was frozen at −80°C within 4 h from blood draw. The plasma samples were spectrophotometrically analyzed to be free from haemoglobin (31). Hemolyzed plasma samples were excluded from further analysis. Whole blood was centrifuged at 1,700 × g for 10 min. The obtained cloudy supernatant was transferred to a fresh tub and centrifuged at 2,000 × g for 10 min. Subsequently, the obtained supernatant was centrifuged at 12,000 × g for 10 min to pellet any remaining cellular debris. Circulating RNA extraction was essentially conducted on 1 ml plasma volume using the Plasma/Serum Circulating and Exosomal RNA Purification kit (Slurry Format; Norgen Biotek Corp., Thorold, ON, Canada) following the manufacturer's instructions. The resulting eluate was subjected to an additional concentration step by the use of the RNA Clean-Up and Concentration kit (Norgen Biotek Corp., Thorold, ON, Canada) using 20 µl elution buffer to collect the RNA.
RNA quality
The concentration of the isolated RNA from tissue specimens was assessed by measuring the optical density at 260 nm (OD260) and 280 nm (OD280) using a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and a NanoDrop 1000 spectrophotometer (Peqlab Biotechnologie GmbH, Erlangen, Germany), whereas the quality of the RNA purified from plasma was assessed by PCR amplification curves and efficiencies, due to the absence of ribosomal RNA. RNA purity was evaluated by the ratio of the absorbance at OD260/OD280. Analysis of RNA integrity (RIN) was conducted using the Agilent 2100 Bioanalyzer (Agilent Technologies GmbH, Waldbronn, Germany) with the RNA 6000 series II Nano LabChip analysis kit. The 2100 Bioanalyzer generates numerical RIN values. RIN is an incremental scale spanning 0–10, where increasing RIN correlates with increasing RIN value. Total RNA samples extracted from CRC and adjacent neoplasm-free mucosal tissues were of high integrity as judged by the obtained RIN values of ≥8.0.
miRNA quantification by real-time (RT)-qPCR
Total RNA including miRNAs, from a fixed plasma volume of 1 ml or 1 µg RNA from colorectal tissues, was polyadenylated and reverse transcribed to cDNA in a final volume of 20 µl using the miScript Reverse Transcription kit (Qiagen) and the 5X miScript HiSpec buffer. Prior to qPCR, cDNA was diluted by adding 200 µl RNase-free water to the 20 µl RT reaction. qPCR assays were performed in triplicate measurements using the miScript SYBR-Green PCR kit (Qiagen, Germany) on the Rotor-Gene Q 5-Plex HRM (Qiagen) according to the manufacturer's instructions. The miRNAs utilized in this study were purchased from Qiagen.
The amplification profile was denatured at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min, in which fluorescence was obtained. After the above PCR cycling program, melting curve analyses and agarose gel electrophoresis (3.0%) on the amplicons were conducted to validate the specificity of the expected PCR amplicon. Raw cycle threshold (Ct) values, defined as the number of cycles required for the fluorescent signal to cross the threshold in an RT-qPCR experiment, were calculated using the Rotor-Gene® Q software 2.1 (Qiagen), and implementing automatic baseline settings and a threshold of 0.1. The ΔCt was then calculated by subtracting the Ct values of the control [housekeeping (HK)] gene from the Ct values of the gene of interest. ΔΔCt was then calculated by subtracting ΔCt of the appropriate controls (plasma, miR-191; tissues, RNUB-6) from the ΔCt of the CRC plasma or tissue sample. The expression level of each miRNA gene was represented by fold change, which was calculated using the equation 2−ΔΔCt. The efficiency of each miRNA assay was determined by constructing a standard curve using a series of total RNA dilutions. Assays exhibited good linearity (R2>0.96) across the obtained Ct values and the log of the initial total RNA quantity of each dilution (data not shown).
Statistical analysis
Data were presented as mean ± standard deviation. The Mann-Whitney U test was employed to evaluate the differential expression level of miRNAs between CRC patients and healthy controls using SPSS Data Analysis Software version 19 (SPSS, Inc., Chicago, IL, USA).
Results and Discussion
Clinicopathological characteristics of CRC patients
Inclusion criteria for the 20 CRC patients and 20 healthy individuals (plasma control group) are detailed in Table I. The age of the participants was within the range of ≥18 and ≤80 years. Clinicopathological parameters of the 20 participants (9 men and 11 women) recruited in the study are shown in Table II. The mean age of the subjects was 61 years (±10.6 standard deviation). None of the participants exhibited any evidence of other disease complications. In addition, 5 participants (25%) had stage I CRC, 3 (15%) had stage II, 7 (25%) had stage III, while the remaining 5 (35%) were diagnosed with stage IV of the disease (Table II). In terms of tumor location, 7 patients had tumors which were localized to the rectum, 5 to the distal colon and 8 to the proximal colon. Histologic examination revealed that the diagnosed CRC tumors were of the adenocarcinoma type: 16 adenocarcinoma, 2 mucous adenocarcinoma and 2 signet ring cell (Table II).
Table II.
Clinicopathological characteristics of CRC patients.
| Variable | Frequency, n=20 |
|---|---|
| Gender | |
| Male | 9 |
| Female | 11 |
| Age at diagnosis | |
| Mean ± standard deviation | 61±10.6 |
| Median range | 61 (26–84) |
| TNM stage | |
| I | 5 |
| II | 3 |
| III | 7 |
| IV | 5 |
| Nodal stage | |
| Positive | 12 |
| Negative | 8 |
| T stage | |
| T1 | 3 |
| T2 | 4 |
| T3 | 9 |
| T4 | 4 |
| Tumor location | |
| Rectum | 7 |
| Distal colon | 5 |
| Proximal colon | 8 |
| Histology | |
| Adenocarcinoma | 16 |
| Mucous adenocarcinoma | 2 |
| Signet ring cell | 2 |
CRC, colorectal cancer; TNM, tumor-node-metastasis.
qPCR analyses of mature miRNAs
Mounting evidence suggests miRNAs function in carcinogenesis as oncogenes or tumor suppressors (8,19,27). Thus, it was fundamentally important to identify CRC-related miRNA signatures by comprehensive quantitative techniques such as qPCR to broaden our understanding of their functional roles in CRC biology and pathogenesis. In this context, we selected four mature miRNAs genes, i.e., miR-145, miR-195, miR-29 and miR-92, to profile their expression levels in the plasma and tissue samples of CRC patients in relation to healthy normal specimens. Previously, the most frequently used method for the quantification of miRNAs was Northern blotting. Alsmost a decade ago, several assays including cDNA arrays, modified invader and flow cytometry were introduced for the quantitative determination of miRNAs (18,32–34). However, these assays often suffer from a serious limitation of low sensitivity due to the fact that they lack a miRNA amplification step (13). Thus, being more sensitive and RT-qPCR technology was employed to overcome the relatively low sensitivity issues of these assays (35). Traditionally, miRNA discovery workflows using global profiling are firstly conducted with either cDNA microarrays or deep sequencing (next generation sequencing). Subsequently, the RT-qPCR assay is employed as the method of choice to evaluate the diagnostic potential on a panel of selected and defined miRNAs of interest (36).
Determination of the most invariantly-expressed housekeeping miRNA gene in plasma and tissues
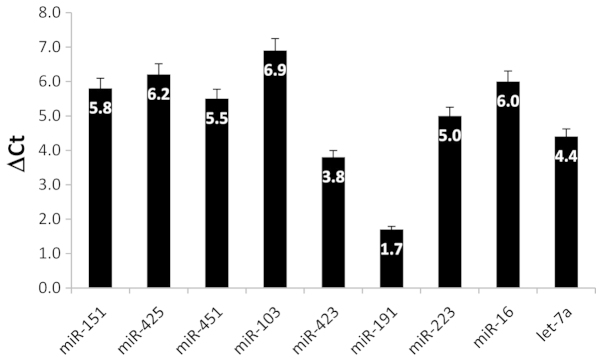
A critical prerequisite for any RT-qPCR assay to robustly profile circulating miRNA species in body fluids including plasma is the ability to isolate and accurately measure those miRNAs under study (30). In order to be able to accurately profile the expression of miRNA biomarkers in CRC plasma samples in relation to normal controls we first identified the most invariantly-expressed miRNA gene to utilize as a reference (HK) gene for RT-qPCR. Thus, we examined the expression of eight different miRNA genes and their differential expression between CRC and cancer-free plasma samples which was calculated by subtracting their obtained Ct values (ΔCt=Cttumor-Ctcontrol). The results shown in Fig. 1 indicated that the least variable gene among the studied miRNA genes is miR-191 with a ΔCt value of only 1.7. Therefore, it was employed as a reliable HK gene for fold change calculations. Previous reports have recommended using miR-16, miR-223 or let-7a as normalizers for RT-qPCR data as they maintain a high and constantly expressed level of expression in plasma/serum samples (37). However, based on our obtained results the abovementioned candidate normalizer miRNAs exhibited the most variable ΔCt values among the 9 genes examined (Fig. 1) and were therefore disregarded as HK genes in miRNA gene expression profiling in plasma. Nevertheless, more empirical validations remain to be systematically conducted in order for a consensus on robust and accurate normalization controls to be identified. As for colorectal tissues, we established RNU6B (a small nucleolar RNA gene) as an ideal internal control HK gene, as it exhibited a consistent, stable and high expression across all the tested tissue samples
Figure 1.
Identification of the most invariantly-expressed housekeeping gene in plasma samples. Cycle threshold (Ct) values of each gene in plasma colorectal cancer samples was subtracted from Ct value in control plasma to obtain the ΔCt value.
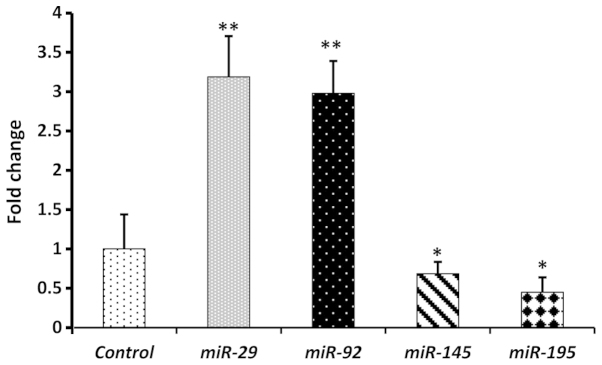
Results of qPCR profiling of 20 clinical CRC plasma samples showed that the expression levels of circulating miR-29 and miR-92 were significantly higher (P<0.01) than in the age-matched normal plasma, with mean fold-change values of (3.2) and (2.9), respectively (Fig. 2). We then investigated the expression levels of miR-145 and miR-195. The mean expression levels of miR-145 and miR-195 were significantly lower (P<0.05) in CRC plasma than in the healthy control plasma, with means of 0.69 and 0.45, respectively (Fig. 2). In a paired-study design employing RT-qPCR, statistically significant (P<0.01) increases in miR-29 (2.5) and miR-92 (2.6) were observed in the 20 CRC tissue samples compared with adjacent non-cancerous colorectal mucosa (Fig. 3). By contrast, miR-145 (0.75) and miR-195 (0.53) exhibited statistically significant (P<0.05) decreases in their mean expression levels in CRC tissue samples in relation to normal tissues (Fig. 3). Taken together, our data clearly reflect similar trends in the expression levels of the four miRNAs in CRC tissues and plasma, as well as the positive correlation between tissue and circulating miRNAs. Moreover, the obtained results are in strong agreement with previous reports on the expression level of the four miRNAs under investigation in CRC tissues and plasma (15,22,25–28), thereby confirming their potential as diagnostic biomarkers.
Figure 2.
Fold change of miR-29, miR-92, miR-145 and miR-195 in colorectal cancer plasma in relation to neoplasm-free plasma controls. Bars show the fold change presented as mean ± standard deviation. Experiments were conducted in triplicate (*P<0.05, **P<0.01).
Figure 3.
Fold change of miR-29, miR-92, miR-145 and miR-195 in colorectal cancer tissues in relation to adjacent neoplasm-free mucsal tissue controls. Bars show the fold change presented as mean ± standard deviation. Experiments were conducted in triplicate (*P<0.05, **P<0.01).
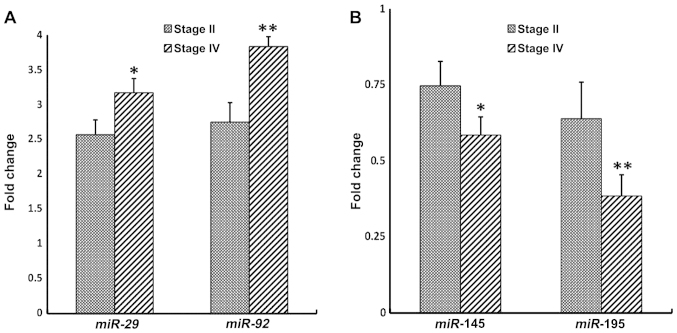
The ideal biomarker should possess a number of essential criteria including non-invasiveness, specificity, reliability in indicating the presence of disease prior to the manifestation of any clinical symptoms, and more importantly sensitivity to changes in the pathology (disease progression or therapeutic response) (38). Therefore, we examined whether there is an association between the aberrant expression of these circulating miRNAs genes in the plasma and CRC clinical stages (Fig. 4). Distinct stage-dependent changes in the expression level of the four miRNA gene profiles were observed between stages II and IV plasma of CRC patients relative to control plasma (Fig. 4). This observation clearly indicates that the four miRNAs under study exhibit some dynamic changes in their levels of expression as a function of the clinical cancer stage.
Figure 4.
Fold change of miR-29, miR-92, miR-145 and miR-195 in colorectal cancer (CRC) plasma at different clinical stages. Expression levels of the four genes varied in different clinical stages. Bars show the fold change represented as mean ± standard deviation in the plasma from 20 CRC patients versus non-neoplastic plasma controls. All experiments were conducted in triplicate (*P<0.05, **P<0.01).
The identification of circulating miRNAs in plasma/serum has led to scientific interest. Therefore, we investigated the circulating levels of two overexpressed and two downregulated miRNAs previously identified in plasma samples of CRC patients compared to healthy individuals. Colorectal adenomas are a precursor stage of adenocarinoma. Notably, it has been previously reported that the aberrantly expressed plasma miR-29a and miR-92a were equally efficient in discriminating advanced adenoma patients from neoplasm-free individuals, albeit with a lesser discriminatory power than for CRC patients (22,39). Similar findings have been recently described in matched CRC and normal tissue samples for miR-145 at different clinical stages, emphasizing its potential as an early diagnostic biomarker for CRC (15). Furthermore, it has been reported that miR-145 acts as a powerful tumor suppressor that regulates multiple cell pathways, making it an ideal biomarker for the diagnosis of various carcinomas (27,28). However, further studies with larger numbers of patients and simultaneous measurements of miRNA expression in plasma and tissue are required to clarify this issue.
There are certain limitations that need to be considered when interpreting the findings of the current study. Firstly, the sample size of this study remains critically small, particularly in the context of miRNA-based biomarker identification and validation. Secondly, numerous miRNA signatures in plasma samples are generally too low to be accurately detected and quantified by RT-qPCR, a well-known limitation of this technique. Therefore, some potentially relevant markers may not be efficiently identified. In addition, the fact that no consensus endogenous control has been established yet in circulating miRNA studies complicates its reliable investigation. Some reports have advocated the use of miR-16 as an endogenous and reliable control because of its relative stability and abundance in plasma/serum (22,40). However, other reports have challenged this suggestion by showing that miR-16 exhibits an aberrantly expression level in plasma/serum which was associated with the risk of prostate cancer and lymphoma (41,42). We haveinitiated a high throughput miRNome profiling strategy based on next generation sequencing technology to aid with the identification of certain novel miRNA biomarkers that are differentially expressed in CRC patients (unpublished data). Thirdly, it is not clear whether the observed mis-expression levels of the four miRNAs signatures, which were examined in the current study, are CRC-specific in a Saudi population context. Additional functional dissection is required to confirm the role of the selected miRNA signatures in CRC and to validate their potential diagnostic and prognostic value.
In conclusion, the qPCR results identified alterations of miRNA expression in CRC plasma and tissues with two downregulated miRNAs (miR-145 and miR-195), and two upregulated miRNAs (miR-29 and miR-92), which are potential candidate biomarkers for CRC. Plasma miRNAs appear to be potentially useful biomarkers for the early detection and diagnosis of CRC. While research on plasma-based miRNA profiling remains at its infancy compared to investigations on tissue-based miRNA profiling, it holds great potential for the development of novel non-invasive blood-based CRC screening approaches. Nevertheless, the diagnostic value of the identified panel of miRNA biomarker signatures may not yet be capable of competing with the performance of other traditional non-invasive tests, such as immunochemical FOBTs (39). Additional investigations on a multi-marker blood-based test, potentially including the panel of miRNAs profiled in this study among those proposed by other study groups may constitute a promising strategy to enhance the repertoire for non-invasive cancer screening programs. A panel of plasma miRNA multi-markers assays may possess a superior sensitivity and specificity as compared with other single-marker ones for CRC screening. Therefore, patients exhibiting elevated plasma miRNAs, as in the case with miR-29 and miR-92 for example, may initiate more targeted and accurate clinical examinations such as colonoscopy.
Acknowledgements
This study was financially supported by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, award no. (11-MED1770-02).
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.1. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 (Internet). Lyon, France: International Agency for Research on Cancer. http://globocan.iarc.fr. [Jan 16;2015 ];2014 Accessed. [Google Scholar]
- 2.Saudi Cancer Registry: Cancer Incidence Report Saudi Arabia 2005. http://www.scr.org.sa/reports/SCR2005.pdf. [Oct 12;2015 ];Kingdom of Saudi Arabia, Ministry of Health, Saudi Cancer Registry. Accessed. [Google Scholar]
- 3.Mosli MH, Al-Ahwal MS. Colorectal cancer in the Kingdom of Saudi Arabia: need for screening. Asian Pac J Cancer Prev. 2012;13:3809–3813. doi: 10.7314/APJCP.2012.13.8.3809. [DOI] [PubMed] [Google Scholar]
- 4.Ibrahim EM, Zeeneldin AA, El-Khodary TR, Al-Gahmi AM, Sadiq Bin BM. Past, present and future of colorectal cancer in the Kingdom of Saudi Arabia. Saudi J Gastroenterol. 2008;14:178–182. doi: 10.4103/1319-3767.43275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 6.Levin B, Lieberman DA, McFarland B, Smith RA, Brooks D, Andrews KS, Dash C, Giardiello FM, Glick S, Levin TR, et al. American Cancer Society Colorectal Cancer Advisory Group; US Multi-Society Task Force; American College of Radiology Colon Cancer Committee: Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–160. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 7.Mandel JS. Screening for colorectal cancer. Gastroenterol Clin North Am. 2008;37(vii):97–115. doi: 10.1016/j.gtc.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannafon BN, Sebastiani P, de las Morenas A, Lu J, Rosenberg P. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 2011;13:R24. doi: 10.1186/bcr2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miko E, Czimmerer Z, Csánky E, Boros G, Buslig J, Dezso B, Scholtz B. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res. 2009;35:646–664. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 12.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and −145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 13.Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29–38. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James P. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 15.Peng J, Xie Z, Cheng L, Zhang Y, Chen J, Yu H, Li Z, Kang H. Paired design study by real-time PCR: miR-378* and miR-145 are potent early diagnostic biomarkers of human colorectal cancer. BMC Cancer. 2015;15:158–165. doi: 10.1186/s12885-015-1123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 17.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 19.Waldman SA, Terzic A. MicroRNA signatures as diagnostic and therapeutic targets. Clin Chem. 2008;54:943–944. doi: 10.1373/clinchem.2008.105353. [DOI] [PubMed] [Google Scholar]
- 20.Tsang JC, Lo YM. Circulating nucleic acids in plasma/serum. Pathology. 2007;39:197–207. doi: 10.1080/00313020701230831. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 22.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Wang Q, Zheng Y, Lv S, Ning S, Sun J, Huang T, Zheng Q, Ren H, Xu J, et al. Prioritizing human cancer microRNAs based on genes' functional consistency between microRNA and cancer. Nucleic Acids Res. 2011;39:e153. doi: 10.1093/nar/gkr770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui SY, Wang R, Chen LB. MicroRNA-145: A potent tumour suppressor that regulates multiple cellular pathways. J Cell Mol Med. 2014;18:1913–1926. doi: 10.1111/jcmm.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou Y, Wang X, Chen Y, Mu S. MicroRNA-145 as an ideal biomaker for the diagnosis of various carcinomas. Tumour Biol. 2015;36:2641–2649. doi: 10.1007/s13277-014-2886-9. [DOI] [PubMed] [Google Scholar]
- 29.Sobin LH, Wittekind Ch, editors. UICC: TNM Classification of Malignant Tumours. 6th. Hoboken, NJ: John Wiley & Sons; 2002. [Google Scholar]
- 30.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP, Takova T, Neri BP, Lyamichev VI. Quantitation of microRNAs using a modified Invader assay. RNA. 2004;10:1153–1161. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 34.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrews WJ, Brown ED, Dellett M, Hogg RE, Simpson DA. Rapid quantification of microRNAs in plasma using a fast real-time PCR system. Biotechniques. 2015;58:244–252. doi: 10.2144/000114287. [DOI] [PubMed] [Google Scholar]
- 37.Zubakov D, Boersma AW, Choi Y, van Kuijk PF, Wiemer EA, Kayser P. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int J Legal Med. 2010;124:217–226. doi: 10.1007/s00414-009-0402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One. 2013;8:e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 41.Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH, Liu L, Fan L, Miao KR, Liu P, Xu W, et al. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol. 2012;91:553–559. doi: 10.1007/s00277-011-1350-9. [DOI] [PubMed] [Google Scholar]
- 42.Watahiki A, Wang Y, Morris J, Dennis K, O'Dwyer HM, Gleave M, Gout PW, Wang Y. MicroRNAs associated with metastatic prostate cancer. PLoS One. 2011;6:e24950. doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]