Abstract
The hepatocyte growth factor (HGF)/c-Met signaling pathway results in cancer cell scattering and invasion, and has been reported to participate in several types of cancer, including prostate and colorectal cancer. The downstream phosphorylation cascade of HGF, particularly the mitogen-activated protein kinase and phosphoinositide 3-kinase/AKT signaling pathway, regulates epithelial-mesenchymal transition (EMT). However, the mechanism by which these signaling pathways govern EMT, and whether certain kinases are able to respond to specific EMT effectors, remains to be elucidated. In the present study, an increase in the levels of vimentin, rather than co-regulation of certain EMT marker proteins, was observed in response to HGF-induced EMT in DU145 prostate cancer cells. In addition, it was observed that curcumin abrogated HGF-induced DU145 cell scattering and invasion. Furthermore, curcumin was able to effectively inhibit the HGF-induced increase in the levels of vimentin by downregulating the expression of phosphorylated c-Met, extracellular signal-regulated kinase and Snail. In conclusion, the results of the present study demonstrated that curcumin was able to reverse HGF-induced EMT, possibly by inhibiting c-Met expression in DU145 prostate cancer cells.
Keywords: epithelial-mesenchymal transition, hepatocyte growth factor, prostate cancer
Introduction
The morbidity and mortality of prostate cancer is increasing every year, with bone metastasis representing the primary cause of disease-associated mortality in patients with prostate cancer (1). Epithelial-mesenchymal transition (EMT) is the primary mechanism of prostate cancer metastasis to bone (2). Therefore, inhibition of this process in prostate cancer cells is an important approach for the suppression of prostate cancer progression.
The occurrence and development of prostate cancer depends on paracrine interactions between matrix and epithelial cells (3). Hepatocyte growth factor (HGF) is a multifunctional growth factor produced by mesenchymal cells, which promotes the migration and growth of epithelial cells (including cancer cells) via EMT (3,4). The levels of c-Met and HGF are significantly correlated, and it has been demonstrated that c-Met expression is a strong prognostic factor for histological tumor grade in patients with prostate cancer (5). In addition, a number of studies have demonstrated overexpression of c-Met in prostate cancer cell models (6,7). Furthermore, inhibition of c-Met expression has been observed to reduce invasion and metastasis in prostate cancer (8).
Curcumin, a plant polyphenol extracted from the turmeric rhizome, possesses antioxidant, anticoagulant, anti-inflammatory, anti-atherosclerotic, lipid-lowering and anti-aging properties (9,10). Previous studies have demonstrated that curcumin is a highly effective anticancer agent (10,11). In addition, curcumin has been revealed to inhibit the progression of prostate cancer and bone metastasis (11). However, the effect of curcumin on EMT in prostate cancer cells remains to be elucidated. In the present study, the effect and mechanism of curcumin on HGF-induced EMT in DU145 prostate cancer cells was investigated, in order to establish a novel approach for the treatment of prostate cancer.
Materials and methods
Materials
Curcumin and dimethyl sulfoxide (DMSO; cat no. CLS3085) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Trypsin and Dulbecco's modified Eagle's medium (DMEM)-high glucose containing trypsin (cat no. SH30022.01B) and fetal bovine serum (FBS; cat no. SH30084.03) were acquired from HyClone (GE Healthcare Life Sciences, Logan, UT, USA). Antibodies against E-cadherin (mouse monoclonal; cat no. sc-59905), vimentin (mouse monoclonal; cat no. sc-373717), AKT (mouse polyclonal; cat no. sc-377457), phosphorylated (p)-AKT (rabbit polyclonal; cat no. sc-33437), extracellular signal-regulated kinase (ERK; mouse monoclonal; cat no. sc-514302), p-ERK (T202/Y204; goat polyclonal; cat no. sc-16982), Snail (goat polyclonal; cat no. sc-10433), c-Met (mouse monoclonal; cat no. sc-8057) and β-actin (mouse monoclonal; cat no. sc-8432) were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). HGF was acquired from Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and dissolved in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4.7 H2O and 1.4 mM KH2PO4, pH 7.4), with 0.1% bovine serum albumin. Rabbit anti-goat immunoglobulin G (IgG; cat no. sc-2768; H+L)-horseradish peroxidase conjugated antibody and rabbit anti-mouse IgG-R (cat no. sc-358922) were purchased from Santa Cruz Biotechnology, Inc. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; cat no. C0009) were obtained from Beyotime Institute of Biotechnology (Shanghai, China). RevertAid™ First Strand cDNA Synthesis kit (cat no. N8080234) and Lipofectamine® 2000 Transfection Reagent (cat no. 11668030) were acquired from Invitrogen (Thermo Fisher Scientific, Inc.).
Cell culture
The DU145 cell line was obtained from the cell repository of the Chinese Academy of Sciences (Shanghai, China). DU145 cells were cultured in DMEM-high glucose supplemented with 10% FBS, 100 U/ml penicillin (cat no. ST488-1; Beyotime Institute of Biotechnology) and 100 U/ml streptomycin (cat no. ST488-2; Beyotime Institute of Biotechnology), and incubated (cat no. 670190; Greiner Bio-One, Stonehouse, UK) in a humidified atmosphere of 5% CO2 and 95% air at 37°C. Cells were cultured without serum for ≥6 h prior to the commencement of the experiments.
Cell proliferation assay
DU145 cells (8×103 cells/ml) were seeded into 96-well culture plates (cat no. CW0543; Beijing ComWin Biotech Co., Ltd., Beijing, China), and incubated overnight in a humidified atmosphere of 5% CO2 and 95% air at 37°C. Following 12 h incubation, the medium was replaced with serum-free DMEM-high glucose, and curcumin was added at various concentrations (0, 5, 10, 15, 20, 25, 30, 35, 40, 45 and 50 µmol/l). Blank (cell-free + medium) and control (cells + medium, without curcumin) groups were additionally evaluated. Following 48 h of incubation, the culture medium was removed, and 5 mg/ml MTT was added to each well. The plates were subsequently incubated for 4 h at 37°C. The culture medium was replaced with 150 µl DMSO/well, and the absorbance (A) at a wavelength of 490 nm was measured using 2104 EnVision® Multilabel Reader (cat no. 2104-0010; PerkinElmer, Inc., Waltham, MA, USA). Cell viability was calculated using the following formula: Cell viability (%) = [(HGF A value - blank group A value)]/[(control group A value - blank group A value)] × 100.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from the tissue samples using TRIzol® reagent, according to the manufacturer's instructions (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Subsequently, cDNA was synthesized using a TaqMan Reverse Transcription Reagents kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The relative expression levels of mRNA were determined using a Power SYBR® Green PCR Master Mix kit (Thermo Fisher Scientific, Inc.) and normalized to GAPDH. RT-PCR was performed using the Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument (cat no. 4425757; Thermo Fisher Scientific, Inc.) and the following gene-specific primers (Sangon Biotech Co., Ltd., Shanghai, China): GAPDH sense, 5′-TGCCATCAACGACCCCTTCA-3′ and antisense, 5′-TGACCTTGCCCACAGCCTTG-3′; c-Met sense, 5′-GAGGCAGTGCAGCATGTAGT-3′ and antisense, 5′-GGTCAGAA-3′; Snail sense, 5′-TTACCTTCCAGCAGCCCTAC-3′ and antisense, 5′-GAG CA-3′; E-cadherin sense, 5′-AGCTATCCTTGCACCTCAGC-3′ and antisense, 5′-CCCAGGAGTTTGAG-3′; and vimentin sense, 5′-CCAAACTTTTCCTCCCTGAACC-3′ and antisense, 5′-GTGATGCTGAGAAGTTTCGTTGA-3′. A control siRNA specific for the red fluorescent protein, 5′-CCACTACCTGAGCACCCAG-3′, was used as negative control (sc-37007; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). All primers were designed using the National Center for Biotechnology Information Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). PCR was performed under the following conditions: Denaturation at 50°C for 2 min, followed by 38 cycles of 95°C for 15 sec and 60°C for 1 min. Gene expression was normalized to internal controls and fold changes were calculated using relative quantification (2−ΔΔCt) (12).
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay buffer (cat no. P0013; Beyotime Institute of Biotechnology) containing 1 mmol/l phenylmethylsulfonyl fluoride (94:6 v/v; cat no. ST506-2; Beyotime Institute of Biotechnology), and subsequently placed on ice for 30 min. The supernatant was collected following centrifugation at 13,600 × g for 10 min at 4°C. Protein concentration was determined using Pierce BCA Protein Assay Kit (cat no. 23227; Thermo Fisher Scientific, Inc.). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (10%; cat no. P0012A; Beyotime Institute of Biotechnology) and subsequently transferred to a polyvinylidene difluoride membrane (cat no. FFP39; Beyotime Institute of Biotechnology) (12). The membrane was immunoblotted with anti-β-actin (1:1,000), anti-c-Met (1:500), anti-ERK1/2 (1:250), anti-p-ERK1/2 (1:250), anti-AKT (1:400), anti-p-AKT (1:200), anti-Snai1 (1:400), anti-E-cadherin (1:500) and anti-vimentin (1:500) at 4°C overnight. All antibodies were diluted with 0.5% bovine serum albumin. Following incubation, the corresponding secondary antibody conjugated with peroxidase (1:1,000) and enhanced chemiluminescence reagents (BeyoECL Plus; cat no. P0018; Beyotime Institute of Biotechnology) were applied, and the blot was visualized (cat no. 121–2550; Beijing Liuyi Biotech Co., Ltd., Beijing, China). The protein content was assessed using LabWorks image acquisition and analysis software (Biomagin Systems Pvt. Ltd., Battaramulla, Sri Lanka).
Scattering assay
DU145 cells (3×105 cells/ml) were seeded into each well of a 24-well plate (cat no. 662102; Greiner Bio-One), and incubated overnight at 37°C in an atmosphere of 5% CO2. DU145 cells were pretreated with 15 µmol/l curcumin for 2 h. HGF was added to each well at a final concentration of 33 ng/ml (12). Cells were subsequently incubated at 37°C for 48 h. Representative images were captured at magnification, ×20 using Eclipse TE2000-U inverted microscope (Nikon, Corporation, Tokyo, Japan).
In vitro invasion assay
The in vitro invasion assay was performed as described previously (13). A Matrigel™ invasion assay was performed in HTS Transwell®-24 Well Permeable Supports (Corning Life Sciences, Tewksbury, MA, USA). Briefly, 25 µl Matrigel Basement Membrane Matrix (BD Biosciences, Franklin Lakes, NJ, USA) was thawed at 4°C overnight and coated on the Transwell insert membrane. The inserts were subsequently incubated at 37°C for 15–30 min to set. Following Matrigel coating, 3×105 DU145 cells/ml in 200 µl minimal essential medium (MEM) with 0.1% serum and 15 µmol/ml curcumin plus 33 ng/ml HGF, 33 ng/ml HGF or 0.1 ml DMSO was added to the upper chamber. Serum-containing MEM (500 µl) was added to the lower chamber. The cells were incubated at 37°C in an atmosphere of 5% CO2 for 48 h. Following incubation, the medium was aspirated, and the non-invading cells were removed with a wet cotton swab. The cells were washed using PBS, and fixed in 4% paraformaldehyde (cat no. P1110; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) at room temperature for 15 min. Fixed cells were washed three times with PBS, and stained with toluidine blue (cat no. 6586-04-5; Sigma-Aldrich) for 3–5 min. Excess stain was removed by washing three times with distilled water for 15 sec (cat no. Z329797; Sigma-Aldrich). The invading cells were visualized under Eclipse TE2000-U inverted microscope at magnification, ×40.
Wound healing assay
DU145 cells (3×105 cells/ml) were seeded into a 6-well plate (cat no. 657160; Greiner Bio-One) in serum-containing medium, and incubated at 37°C in an atmosphere of 5% CO2 in order to form a confluent monolayer. The monolayer was scratched using a sterile plastic pipette tip (cat no. CLS4860; Sigma-Aldrich), and washed with PBS to remove cell debris. Subsequently, fresh medium was added, and 15 µmol/ml curcumin plus 33 ng/ml HGF, 33 ng/ml HGF or 0.1 ml DMSO was added to each well. The scratched monolayer was incubated at 37°C in an atmosphere of 5% CO2 for 48 h. Wound closure was measured in 6 random high-power fields at magnification, ×200, using Image-Pro® Express software version 6 (Media Cybernetics, Inc., Rockville, MD, USA) and Eclipse TE2000-U inverted microscope (13).
Statistical analysis
Experiments were performed at least in triplicate. The data are presented as the mean ± standard deviation. Statistical significance of differences between groups was evaluated by analysis of variance and Student's t-test using SPSS version 11.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism version 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Curcumin blocks HGF-induced scattering of DU145 cells
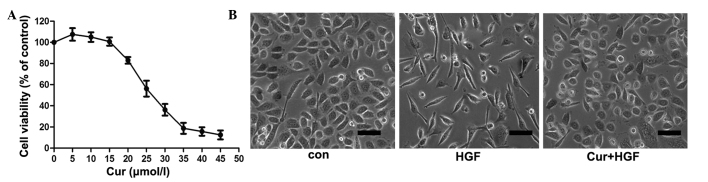
In the present study, the effect of curcumin on DU145 cell viability was examined. As demonstrated in Fig. 1A, curcumin induced DU145 cell death in a concentration-dependent manner, while DU145 cell viability was unaltered at concentrations of curcumin <15 µmol/l. Therefore, 15 µmol/l curcumin was used to treat cancer cells in all subsequent experiments. Activation of HGF/c-Met signaling led to a marked polarized cell morphology in DU145 cells (Fig. 1B). To determine if curcumin caused this HGF-induced phenotypic change, cells were pretreated for 8 h with various concentrations of curcumin (0, 5, 10, 15, 20, 25, 30, 35, 40, 45 and 50 µmol/l), prior to HGF stimulation. As shown in Fig. 1B, curcumin blocked HGF-induced cell scattering, but did not induce cell death under the conditions used in the experiment.
Figure 1.
Effects of curcumin on HGF-induced scattering of DU145 cells. (A) Effect of curcumin on DU145 cell viability. DU145 cells were incubated with various concentrations of curcumin (0, 5, 10, 15, 20, 25, 30, 35, 40, 45 and 50 µmol/l) for 48 h, prior to the detection of cell proliferation by thiazole blue. Experiments were performed in triplicate. *P<0.05 vs. control group. (B) Curcumin inhibited HGF-induced DU145 cell scattering. Cells were pretreated with curcumin for 2 h, and subsequently incubated with HGF for 48 h. Representative images were captured at magnification, ×40 using Eclipse TE2000-U inverted microscope. HGF, hepatocyte growth factor; Con, control; Cur, curcumin. Scale bar, 10 µm.
Curcumin attenuates HGF-induced invasion and migration of DU145 cells
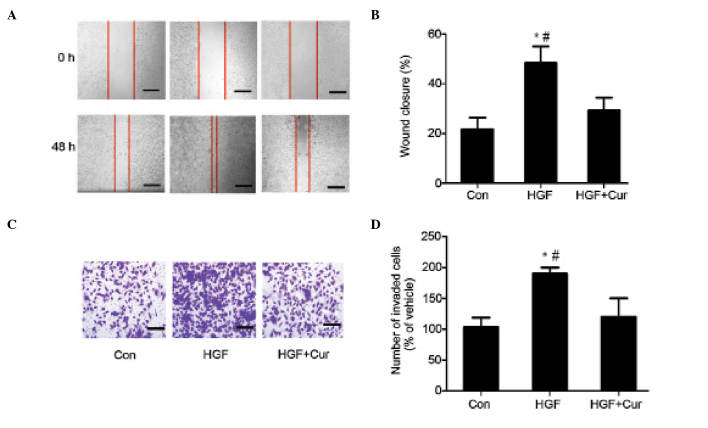
The present study additionally examined the effect of curcumin on HGF-induced cell migration in wound closure assays. Confluent DU145 cells were pretreated with 15 µmol/l curcumin for 2 h, while 0.1 ml DMSO/PBS was used as a control. Cell monolayers were scratched using a pipette tip, and washed to remove any debris. Next, fresh medium containing 0.5% serum with 15 µmol/l curcumin was added. Cells were subsequently incubated with 33 ng/ml HGF for 48 h, and HGF-induced cell migration was determined by measuring wound closure. As shown in Fig. 2A, HGF induced significant cell migration, which was abrogated in the presence of curcumin (Fig. 2A). The present study also determined the effect of curcumin on HGF-induced invasion in Matrigel Transwell chamber invasion assays. Compared with the control, HGF induced an ~2-fold increase in the number of invasive cells. However, prior to treatment with HGF, DU145 cells were pretreated with 15 µmol/l curcumin for 2 h, which significantly reduced the number of HGF-induced invasive cells. Consistent with the results obtained in the cell migration assay (Fig. 2B), curcumin treatment reduced HGF-induced DU145 cell invasion (Fig. 2C and D).
Figure 2.
Curcumin inhibits HGF-induced DU145 cell invasion and migration. (A) In wound closure assays, confluent DU145 cells were pretreated with 15 µmol/l curcumin for 2 h. Dimethyl sulfoxide/phosphate-buffered saline was used as a control. Cell monolayers were scratched using pipette tips and washed to remove any debris, followed by the addition of fresh medium containing 0.5% serum with 15 µmol/l curcumin. Cells were subsequently incubated with 33 ng/ml HGF for 48 h, and HGF-induced cell migration was determined by measuring wound closure. Scale bar, 10 µm. (B) HGF-induced cell motility was determined by measuring the wound closure in six random high-power fields at magnification, ×200 using Image-Pro® Express software. Data are presented as the mean ± standard deviation. *P<0.05 vs. control group, #P<0.05 vs. HGF + curcumin group (C and D) For Matrigel™ Transwell® chamber invasion assays, DU145 cells (3×105 cells/ml) were plated on top of a Matrigel™-coated 24-well Transwell® chamber, and cultured for 48 h in Dulbecco's modified Eagle's medium containing 0.1% serum, 33 ng/ml HGF and 15 µmol/l curcumin. Invading cells were subsequently fixed, stained with toluidine blue and counted in six random high-power fields at magnification, ×200 using Image-Pro® Express software. The experiment was performed in triplicate. *P<0.05 vs. control group, #P<0.05 vs. HGF + curcumin group. HGF, hepatocyte growth factor; Con, control; Cur, curcumin.
Curcumin suppresses HGF-induced EMT in DU145 cells
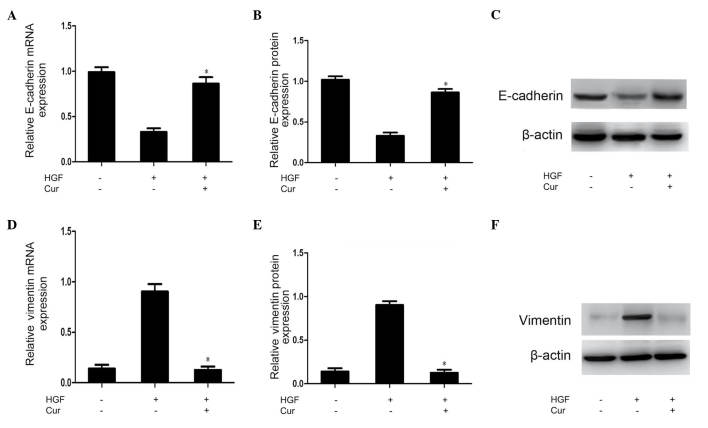
Multiple molecules regarded as epithelial or mesenchymal markers have been reported previously, and downregulation of E-cadherin and upregulation of vimentin are known to be markers of EMT (14). In the present study, RT-qPCR and western blot analysis of the expression of these markers were conducted to investigate whether the abrogation of HGF-induced cell scattering, migration and invasion observed in curcumin-treated DU145 cells was due to alterations in the process of EMT. As shown in Fig. 3, vimentin expression was upregulated, while E-cadherin expression was downregulated in response to HGF activation. These HGF-induced effects were significantly suppressed by pretreatment with curcumin (Fig. 3).
Figure 3.
Curcumin inhibits HGF-induced epithelial-mesenchymal transition in DU145 cells. DU145 cells were pretreated with DMSO (control) or curcumin (15 µmol/l) for 2 h prior to HGF stimulation (33 ng/ml) for 48 h. (A-C) E-cadherin mRNA and protein levels were determined by RT-qPCR and densitometric quantification of western blotting results of whole cell lysates, respectively. (D-F) Vimentin mRNA and protein levels were determined by RT-qPCR and densitometric quantification of western blot results of whole cell lysates, respectively. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05 vs. HGF group. DMSO, dimethyl sulfoxide; HGF, hepatocyte growth factor; mRNA, messenger RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; Cur, curcumin.
Curcumin has an inhibitory effect on HGF/c-Met signaling pathway and Snail expression in DU145 cells
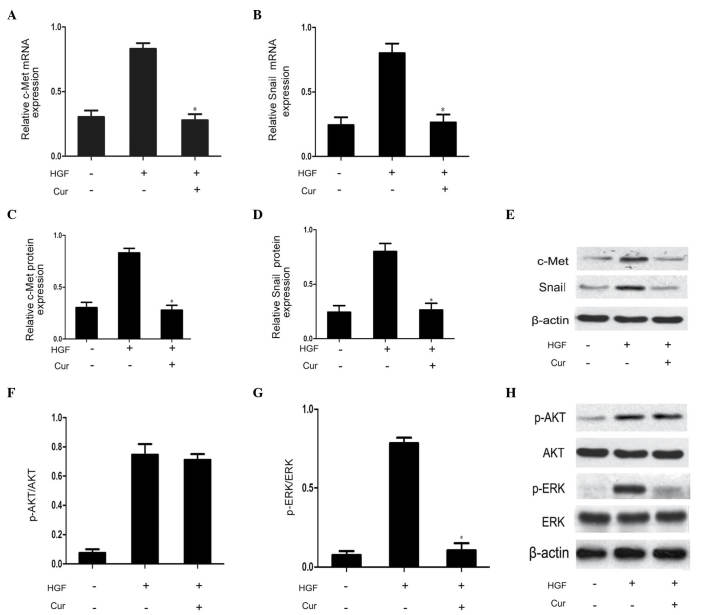
In order to determine the involvement of the HGF/c-Met signaling pathway on the effects of curcumin on DU145 cell scattering and motility, DU145 cells were pretreated with curcumin (10 µmol/l) for 2 h prior to HGF treatment for 48 h. Compared with the HGF group, curcumin significantly suppressed HGF-induced upregulation of c-Met expression, as well as Snail mRNA and protein expression (Fig. 4A–E). This observation indicated that curcumin reduced HGF/c-Met signaling by direct inhibition of c-Met expression. The HGF/c-Met signaling axis is a significant contributor to the promotion of EMT via stimulation of AKT and ERK (3). Therefore, the present study additionally investigated the effect of curcumin on the phosphorylation of AKT and ERK. As demonstrated in Fig. 4F–H, western blot analysis revealed that curcumin pretreatment was able to effectively block HGF-induced ERK phosphorylation. The rapidly accelerated fibrosarcoma/mitogen-activated protein kinase (MAPK) kinase/ERK signaling pathway has been demonstrated to be required for HGF-induced EMT (3). By contrast, HGF-mediated activation of AKT was not affected by curcumin pretreatment in the present study. Therefore, MAPK signaling may be involved in Snail expression. Thus, curcumin was capable of blocking HGF-induced signaling via specific repression of activation of the c-Met/ERK/Snail signaling pathway.
Figure 4.
Curcumin blocks HGF-induced c-Met signaling by downregulating c-Met, Snail and ERK phosphorylation. (A-E) Pretreatment with curcumin inhibited the expression of c-Met and Snail. (F-H) Curcumin slightly decreased phosphorylation of AKT following long-term pretreatment, and significantly decreased phosphorylation of ERK. All experiments were repeated in triplicate. Data are presented as the mean ± standard deviation. *P<0.05 vs. HGF group. HGF, hepatocyte growth factor; ERK, extracellular signal-regulated kinase; p, phosphorylated; mRNA, messenger RNA; Cur, curcumin.
Discussion
EMT is a crucial step in the process of cancer cell invasion and metastasis. During this process, the phenotype of cancer cells undergoes alterations in cellular morphology, with increased expression of mesenchymal markers such as vimentin and decreased expression of epithelial markers such as E-cadherin (15). These changes are associated with decreased cell adhesion capacity, leading to enhanced invasion and metastasis (15,16). Consequently, EMT inhibition represents a significant target for the development of prostate cancer treatments. In the present study, the effects of curcumin on the HGF/c-Met signaling axis were investigated, and it was identified that curcumin was capable of blocking c-Met expression, as well as inhibiting EMT in DU145 cells.
HGF is a multifunctional protein, which has been reported to increase the migration and invasion abilities of prostate cancer cells (17). In the present study, it was demonstrated that HGF induced migration and invasion of DU145 prostate cancer cells, with concomitant downregulation of E-cadherin expression and upregulation of vimentin expression. HGF stimulates c-Met phosphorylation, which activates MAPK signaling and increases Snail expression (18). Snail is a DNA-binding protein that contains a zinc finger-like domain capable of binding to the E-box domain of the E-cadherin promoter, thus inhibiting E-cadherin expression (19). Therefore, it may be speculated that HGF is able to induce EMT in DU145 prostate cancer cells via the c-Met/ERK/Snail signaling pathway. c-Met is known to be associated with prostate cancer cell differentiation (17). Previous studies have revealed that c-Met is overexpressed in poorly differentiated prostate cancer, indicating that reduced expression of c-Met may stimulate prostate cancer cell differentiation (20). Nishimura et al (17) reported that HGF was increased during DU145 cell invasion and metastasis in a c-Met-dependent manner, while this effect of HGF was not observed in LNcaP cell metastasis, and these cells are known to be c-Met-deficient (17). Furthermore, Davies et al (21) additionally indicated that inhibition of c-Met expression by hammerhead ribozyme reduced the invasion and metastasis of prostate cancer cells. Thus, inhibition of prostate cancer cell invasion and metastasis via c-Met blockade may represent a significant approach for the treatment of prostate cancer.
The anticancer effects of curcumin have been widely studied, and it is regarded as a promising drug candidate for the treatment and prevention of cancer (22). It is considered that the mechanism underlying the anticarcinogenic effects of curcumin involves the induction of cancer cell apoptosis via regulation of the expression of anti-oncogenes, oncogenes and the cell cycle (22). However, previous studies have revealed that curcumin is able to inhibit cancer cell invasion and metastasis via signaling changes. Chen et al (23) observed that curcumin was able to inhibit lung cancer cell invasion and metastasis by upregulation of E-cadherin expression, and stimulated the expression of the tumor suppressor DnaJ-like heat shock protein HLJ1 via c-Jun N-terminal kinase/JunD signaling. However, it has additionally been reported that curcumin inhibited post-transcriptional expression of hypoxia-inducible factor 1-α, leading to reversed hypoxia and induction of liver cancer cell proliferation and migration (24). Thus, the role of curcumin in prostate cancer migration remains to be elucidated. In the present study, it was demonstrated that preincubation of DU145 cells for 2 h with curcumin blocked HGF-induced cell scattering with associated alterations in cell morphology (reduction in stress fibers and cell flattening). This is consistent with the effects of the naturally-occurring coumarin compound osthol, which has been observed to inhibit HGF-induced migration and invasion in MCF-7 breast cancer cells (14). Furthermore, the present study demonstrated that HGF stimulated c-Met expression, as well as phosphorylation of AKT and ERK, and additionally increased the protein expression levels of vimentin, while curcumin suppressed HGF-induced EMT, cell migration and invasion via repression of the c-Met/ERK signaling pathway in DU145 prostate cancer cells. However, the AKT signaling pathway was unaffected by curcumin, even following a long preincubation when c-Met was no longer maximally activated by HGF, and the remaining c-Met was likely to be sufficient to induce phosphorylation of AKT. In addition, the present study demonstrated that curcumin was able to inhibit Snail expression, suggesting that ERK may regulate Snail expression. Therefore, it may be speculated that curcumin inhibits Snail expression via suppression of c-Met/ERK signaling, although this remains to be confirmed.
In the present study, curcumin was additionally revealed to suppress c-Met expression, although the exact underlying mechanism remains to be elucidated. Seol et al (25) observed that the transcription factor activator protein-1 (AP-1) binds the c-Met gene promoter and increases c-Met expression. Furthermore, previous studies have indicated that increased fatty acid synthase (FAS) activity maintains lipid rafts, which may aid to stabilize the levels of c-Met (26). These studies suggest that inhibited AP-1 and FAS expression are suppressed by c-Met expression. Thus, it may be hypothesized that curcumin suppresses c-Met expression via inhibition of AP-1 and FAS expression, although this theory requires additional investigation in future studies.
In conclusion, the present study has demonstrated that curcumin may inhibit HGF-induced EMT via reduced c-Met expression, and may attenuate associated signaling pathways. These findings suggest that curcumin may be regarded as an inhibitor of c-Met, the receptor tyrosine kinase for HGF, thus providing a theoretical and experimental basis for the treatment of prostate cancer, and a novel approach for the development of prostate cancer drugs.
Acknowledgements
The present study was supported by the Medical Foundation of Huizhou (Huizhou, China; grant no. 2014Y149) and The Medical Research Foundation of Guangdong Province (Guangzhou, China; grant no. A2014810).
References
- 1.Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: Emerging therapeutic strategies. Nat Rev Clin Oncol. 2011;8:357–368. doi: 10.1038/nrclinonc.2011.127. [DOI] [PubMed] [Google Scholar]
- 2.Josson S, Gururajan M, Hu P, Shao C, Chu GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20:4636–4646. doi: 10.1158/1078-0432.CCR-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parikh RA, Wang P, Beumer JH, Chu E, Appleman LJ. The potential roles of hepatocyte growth factor (HGF)-MET pathway inhibitors in cancer treatment. Onco Targets Ther. 2014;11:969–983. doi: 10.2147/OTT.S40241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott BE, Hung WL, Boag AH, Tuck AB. The role of hepatocyte growth factor (scatter factor) in epithelial-mesenchymal transition and breast cancer. Can J Physiol Pharmacol. 2002;80:91–102. doi: 10.1139/y02-010. [DOI] [PubMed] [Google Scholar]
- 5.Graveel CR, Tolbert D, Van de Woude GF. MET: A critical player in tumorigenesis and therapeutic target. Cold Spring Harb Perspect Biol. 2013;5:a009209. doi: 10.1101/cshperspect.a009209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varkaris A, Corn PG, Gaur S, Dayyani F, Logothetis CJ, Gallick GE. The role of HGF/c-Met signaling in prostate cancer progression and c-Met inhibitors in clinical trials. Expert Opin Investig Drugs. 2011;20:1677–1684. doi: 10.1517/13543784.2011.631523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu P, Chung LW, Berel D, Frierson HF, Yang H, Liu C, Wang R, Li Q, Rogatko A, Zhau HE. Convergent RANK- and c-Met-mediated signaling components predict survival of patients with prostate cancer: An interracial comparative study. PLoS One. 2013;8:e73081. doi: 10.1371/journal.pone.0073081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee RJ, Smith MR. Targeting MET and vascular endothelial growth factor receptor signaling in castration-resistant prostate cancer. Cancer J. 2013;19:90–98. doi: 10.1097/PPO.0b013e318281e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghalandarlaki N, Alizadeh AM, Ashkani-Esfahani S. Nanotechnology-applied curcumin for different diseases therapy. Biomed Res Int. 2014;2014:394264. doi: 10.1155/2014/394264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulbecco P, Savarino V. Therapeutic potential of curcumin in digestive diseases. World J Gastroenterol. 2013;19:9256–9270. doi: 10.3748/wjg.v19.i48.9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorai T, Diouri J, O'Shea O, Doty SB. Curcumin inhibits prostate cancer bone metastasis by up-regulating bone morphogenic protein-7 in vivo. J Cancer Ther. 2014;5:369–386. doi: 10.4236/jct.2014.54044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin XL, He XL, Zeng JF, Zhang H, Zhao Y, Tan JK, Wang Z. FGF21 increases cholesterol efflux by upregulating ABCA1 through the ERK1/2-PPARγ-LXRα pathway in THP1 macrophage-derived foam cells. DNA Cell Biol. 2014;33:514–521. doi: 10.1089/dna.2013.2290. [DOI] [PubMed] [Google Scholar]
- 13.Chang HY, Kao MC, Way TD, Ho CT, Fu E. Diosgenin suppresses hepatocyte growth factor (HGF)-induced epithelial-mesenchymal transition by down-regulation of Mdm2 and vimentin. J Agric Food Chem. 2011;59:5357–5363. doi: 10.1021/jf200598w. [DOI] [PubMed] [Google Scholar]
- 14.Hung CM, Kuo DH, Chou CH, Su YC, Ho CT, Way TD. Osthole suppresses hepatocyte growth factor (HGF)-induced epithelial-mesenchymal transition via repression of the c-Met/Akt/mTOR pathway in human breast cancer cells. J Agric Food Chem. 2011;59:9683–9690. doi: 10.1021/jf2021489. [DOI] [PubMed] [Google Scholar]
- 15.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 16.Książkiewicz M, Markiewicz A, Zaczek AJ. Epithelial-mesenchymal transition: A hallmark in metastasis formation linking circulating tumor cells and cancer stem cells. Pathobiology. 2012;79:195–208. doi: 10.1159/000337106. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura K, Kitamura M, Takada S, Nonomura N, Tsujimura A, Matsumiya K, Miki T, Matsumoto K, Okuyama A. Regulation of invasive potential of human prostate cancer cell lines by hepatocyte growth factor. Int J Urol. 1998;5:276–281. doi: 10.1111/j.1442-2042.1998.tb00603.x. [DOI] [PubMed] [Google Scholar]
- 18.Grotegut S, von Schweinitz D, Christofori G, Lehembre P. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J. 2006;25:3534–3545. doi: 10.1038/sj.emboj.7601213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 20.van Leenders G, van Balken B, Aalders T, de Hulsbergen-van Kaa C, Ruiter D, Schalken P. Intermediate cells in normal and malignant prostate epithelium express c-MET: Implications for prostate cancer invasion. Prostate. 2002;51:98–107. doi: 10.1002/pros.10073. [DOI] [PubMed] [Google Scholar]
- 21.Davies G, Watkins G, Mason MD, Jiang WG. Targeting the HGF/SF receptor c-met using a hammerhead ribozyme transgene reduces in vitro invasion and migration in prostate cancer cells. Prostate. 2004;60:317–324. doi: 10.1002/pros.20068. [DOI] [PubMed] [Google Scholar]
- 22.Park W, Amin AR, Chen ZG, Shin DM. New perspectives of curcumin in cancer prevention. Cancer Prev Res (Phila) 2013;6:387–400. doi: 10.1158/1940-6207.CAPR-12-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HW, Lee JY, Huang JY, Wang CC, Chen WJ, Su SF, Huang CW, Ho CC, Chen JJ, Tsai MF, et al. Curcumin inhibits lung cancer cell invasion and metastasis through the tumor suppressor HLJ1. Cancer Res. 2008;68:7428–7438. doi: 10.1158/0008-5472.CAN-07-6734. [DOI] [PubMed] [Google Scholar]
- 24.Sakulterdkiat T, Srisomsap C, Udomsangpetch R, Svasti J, Lirdprapamongkol K. Curcumin resistance induced by hypoxia in HepG2 cells is mediated by multidrug-resistance-associated proteins. Anticancer Res. 2012;32:5337–5342. [PubMed] [Google Scholar]
- 25.Seol DW, Chen Q, Zarnegar R. Transcriptional activation of the hepatocyte growth factor receptor (c-met) gene by its ligand (hepatocyte growth factor) is mediated through AP-1. Oncogene. 2000;19:1132–1137. doi: 10.1038/sj.onc.1203404. [DOI] [PubMed] [Google Scholar]
- 26.Duhon D, Bigelow RL, Coleman DT, Steffan JJ, Yu C, Langston W, Kevil CG, Cardelli JA. The polyphenol epigallocatechin-3-gallate affects lipid rafts to block activation of the c-Met receptor in prostate cancer cells. Mol Carcinog. 2010;49:739–749. doi: 10.1002/mc.20649. [DOI] [PubMed] [Google Scholar]