Abstract
The prognosis of oral squamous cell carcinoma (OSCC) patients is affected by tumor recurrence and metastasis, and cancer stem cells are hypothesized to be involved in these processes. Thus, the aim of the present study was to determine whether the expression levels of five stem cell-related transcription factors, sex determining region Y-box 2 (SOX2), octamer-binding transcription factor 4 (Oct4), avian myelocytomatosis viral oncogene homolog (c-Myc), Krüppel-like factor 4 (KLF4) and brachyury, are associated with metastasis and survival in OSCC. Immunohistochemistry was performed to analyze the expression of these proteins in biopsy specimens obtained from 108 OSCC patients. The results revealed that the expression of SOX2, Oct4, KLF4 and brachyury were significantly associated with lymph node metastasis (P=0.002, P=0.031, P=0.003 and P=0.007, respectively). In addition, the expression of KLF4 and brachyury were significantly associated with distant metastasis (P=0.014 and P=0.012, respectively). Furthermore, multivariate analysis revealed that SOX2 and KLF4 are predictive factors for lymph node metastasis [odds ratios (ORs), 4.526 and 4.851, respectively], and KLF4 is also a predictive factor for distant metastasis (OR, 9.607). In addition, OSCC patients with low co-expression of SOX2, KLF4 and brachyury exhibited a significantly lower disease-specific survival rate (78.6 vs. 100%; P=0.025; χ2=5.033) and disease-free survival rate (60.7 vs. 90.9%; P=0.015; χ2=5.897) when compared with OSCC patients with high co-expression of these factors. The results indicate that SOX2, KLF4 and brachyury serve important roles in tumor progression, and these transcription factors may thus represent clinically useful prognostic markers for OSCC.
Keywords: SOX2, KLF4, brachyury, squamous cell carcinoma, metastasis, lymph nodes
Introduction
Oral squamous cell carcinoma (OSCC) accounts for 90% of all malignant head and neck tumors worldwide (1). Furthermore, metastasis to regional lymph nodes and distant sites, which occurs in 40 and 10% of all OSCC cases, respectively, is associated with poor prognosis (2). Although the underlying mechanisms of metastasis remain unclear, recent studies have demonstrated that a small subset of tumor cells known as cancer stem cells (CSCs), which exhibit similar characteristics to normal stem cells (including self-renewal and pluripotency), may be involved in cancer invasion and metastasis (3).
Previous studies have revealed that the expression of four transcription factors [octamer-binding transcription factor 4 (Oct4), sex determining region Y-box 2 (SOX2), avian myelocytomatosis viral oncogene homolog (c-Myc) and Krüppel-like factor 4 (KLF4)] is sufficient to reprogram differentiated cells to pluripotency (4,5). SOX2 and Oct4 are important for maintaining self-renewal and pluripotency in pluripotent stem cells (6). KLF4, which is involved in tissue development, differentiation and maintenance of homeostasis, may act as either an oncogene or a tumor suppressor in certain types of cancer, including gastric adenocarcinoma and colon cancer (7–9). c-Myc is an oncogenic transcription factor that is involved in cell proliferation, differentiation and apoptosis (10). In addition, the expression of these transcription factors is associated with several types of malignant cancer, including oesophageal (11) breast (12), bladder (13) and lung cancer (14,15). However, the role of these genes in CSCs remains unclear.
Recently, the T-box transcription factor brachyury, which is essential for mesoderm formation during early development (16,17), has been found to regulate the epithelial-mesenchymal transition (EMT) and CSC potential in human salivary carcinoma cells (18–21). In addition, brachyury expression was found to correlate with lymph node metastasis in OSCC (22). However, to date, the association between SOX2, Oct4, KLF4, c-Myc and brachyury expression in OSCC has not been investigated. Therefore, the aim of the present study was to determine whether these transcription factors may represent potential CSC markers and prognostic factors for OSCC.
Materials and methods
Patients and tumor specimens
A total of 108 OSCC patients who were treated at the Department of Oral and Maxillofacial Surgery, Kyushu University Hospital (Fukuoka, Japan) between March 2001 and December 2006 were retrospectively enrolled in the present study. Pretreatment biopsies were obtained from 108 patients. Clinicopathological information, including age, gender, tumor size and location, nodal status, treatments and the presence or absence of disease recurrence and metastasis, was obtained from patient records. The protocol for this study was approved by the Ethics Committee of Kyushu University.
Histopathology and immunohistochemistry
Consecutive 4-µm sections were cut from formalin-fixed paraffin-embedded (FFPE) biopsy samples and deparaffinized with xylene, rehydrated in a graded alcohol series, and heat-treated with Target Retrieval Solution (Dako, Carpinteria, CA, USA) prior to histopathological and immunohistochemical analyses. Tumors were staged according to the International Union for Cancer Control tumor-node-metastasis classification system (7th edition) (23). In addition, tumors were graded using World Health Organization criteria (24) and Anneroth's multifactorial classification system (25,26).
Immunohistochemistry was performed to analyze the expression patterns of SOX2, Oct4, c-Myc, KLF4 and brachyury in OSCC samples. FFPE sections were treated with 3% H2O2 and serum-free protein in phosphate-buffered saline with 0.015 M sodium azide to block endogenous peroxide activity and nonspecific antibody binding. The sections were then incubated overnight at 4°C with the following primary antibodies: Monoclonal rabbit anti-human SOX2 (clone D6D9; #3579; 1:50; Cell Signaling Technology, Inc., Danvers, MA, USA), polyclonal rabbit anti-human Oct4 (clone POU5F1; #2750; 1:100; Cell Signaling Technology, Inc.), polyclonal rabbit anti-brachyury (clone H-210; #sc-20109; 1:200; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), monoclonal mouse anti-human c-Myc (clone 9E10; #sc-40; 1:200; Santa Cruz Biotechnology, Inc.) and monoclonal mouse anti-human KLF4 (clone AT4E6; #NBP1-50367; 1:100; Novus Biologicals, LLC, Littleton, CO, USA). Subsequently, immunostaining was visualized with the CSA II Biotin-Free Tyramide Signal Amplification System (Dako), CSA II Rabbit Link amplification reagent (Dako) and 3,3-diaminobenzidine according to the manufacturer's instructions. Briefly, the sections were incubated with horseradish-peroxidase conjugated anti-mouse or rabbit IgG secondary antibodies (CSA II Biotin-Free Tyramide Signal Amplification System; Dako) for 15 min at room temperature, followed by incubation with CSA II amplification reagent (Dako) and 3,3′-diaminobenzidine. Finally, the sections were counterstained with 0.5% hematoxylin.
The staining pattern was evaluated at three randomly selected locations along the invasive edge of OSCC tumors using an optical microscope equipped with a charge-coupled device camera (BZ-9000; Keyence Corporation, Osaka, Japan). Specifically, the intensity of staining was quantified as the difference between the mean pixel density in 10 randomly selected stained carcinoma cells and that of the background using the BZ-II Analyzer (Keyence Corporation). To account for staining heterogeneity, the expression intensity (EI) of a protein was defined as the ratio of the immunostain density in the nuclei of tumor cells to that of normal basal epithelial cells in the same OSCC sample (Table IA; Fig. 1), according to the following formula: EI = (mean density of positive signal in OSCC cells - mean density of background staining) / (mean density of positive signal in normal cells - mean density of background staining). The results were classified into two groups (high or low expression) for each protein according to the mean value, as shown in Table IA.
Table I.
Classification of EI and positive ER.
| A, EI classification | |||
|---|---|---|---|
| Relative mean pixel densitya | |||
| Factor | Low | Cut-off | High |
| SOX2, Oct4, KLF4, c-Myc, brachyury | < | 1 | ≤ |
| B, Positive ER classification | |||
| Positively stained nuclei, % | |||
| Factor | Low | Cut-off (median) | High |
| SOX2 | < | 66.57 | ≤ |
| Oct4 | < | 54.74 | ≤ |
| KLF4 | < | 66.72 | ≤ |
| c-Myc | < | 71.92 | ≤ |
| Brachyury | < | 71.86 | ≤ |
Density ratio of immunostained OSCC cells to normal epithelium. EI, expression intensity; ER, expression ratio; SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
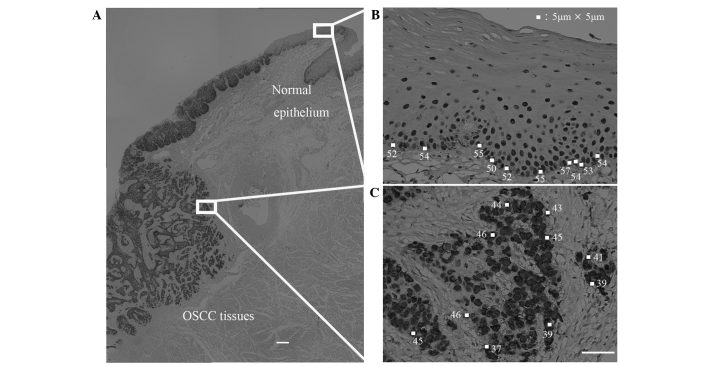
Figure 1.
Determination of immunostain density in OSCC tissue. Photomicrographs show the procedure used to determine SOX2 immunostain density in an OSCC biopsy section that includes both tumor cells and normal epithelial cells. (A) OSCC tissue immunostained with anti-SOX2 antibody. Scale bar, 300 µm. (B and C) The staining density was quantified as the mean pixel density of 10 randomly selected normal basal epithelial cells and OSCC cells along the invasive edge of the tumor [white boxes in (B) and (C), respectively; the pixel densities of each selected area are also shown]. Scale bar, 50 µm. OSCC, oral squamous cell carcinoma; SOX2, sex determining region Y-box 2; KLF4, Krüppel-like factor 4.
The positive expression ratio (ER) was calculated as the ratio of positively stained nuclei to total number of carcinoma cells in each field. The results were classified into two groups (high or low expression) for each protein according to the median value, as shown in Table IB. All samples were scored by two independent pathologists who were blinded to the patient's clinical information and diagnosis.
Statistical analysis
The associations between protein expression and clinicopathological factors were assessed using the χ2 test and Fisher's exact test. Univariate and multivariate logistic regression analyses were performed to identify independent risk factors for lymph node and distant metastasis. Overall survival, disease-specific survival and disease-free survival were analyzed with the Kaplan-Meier method and the log-rank test. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using SPSS 22.0 statistical software (SPSS, Inc., Chicago, IL, USA).
Results
Patient characteristics
The patient cohort included 69 males and 39 females, with a median age of 62 years (range, 24–85 years). Primary OSCC tumors were most frequently identified on the tongue (55/108; 50.9%). Lymph node metastasis occurred in 40/108 patients (37.0%) and distant metastasis occurred in 9/108 patients (8.3%). The median follow-up period was 60 months (range, 5–60 months). Further patient characteristics are shown in Table II.
Table II.
Association between SOX2, Oct4, KLF4, c-Myc and brachyury expression intensity and clinicopathological factors in 108 oral squamous cell carcinoma patients.
| SOX2 expression, n | Oct4 expression, n | KLF4 expression, n | c-Myc expression, n | Brachyury expression, n | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinicopathological parameter | Cases, n | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value |
| Age, years | 0.870 | 0.341 | 0.055 | 0.610 | 0.190 | |||||||||||
| <65 | 61 | 25 | 36 | 38 | 23 | 36 | 25 | 40 | 21 | 35 | 26 | |||||
| ≥65 | 47 | 20 | 27 | 25 | 22 | 19 | 28 | 33 | 14 | 21 | 26 | |||||
| Gender | 0.919 | 0.611 | 0.956 | 0.784 | 0.130 | |||||||||||
| Male | 69 | 29 | 40 | 39 | 30 | 35 | 34 | 46 | 23 | 32 | 37 | |||||
| Female | 39 | 16 | 23 | 24 | 15 | 20 | 19 | 27 | 12 | 24 | 15 | |||||
| Clinical stage | 0.242 | 0.407 | 0.097 | 0.003a | 0.158 | |||||||||||
| T1 | 18 | 8 | 10 | 13 | 5 | 12 | 6 | 16 | 2 | 11 | 7 | |||||
| T2 | 46 | 23 | 23 | 28 | 18 | 26 | 20 | 36 | 10 | 28 | 18 | |||||
| T3 | 21 | 5 | 16 | 11 | 10 | 10 | 11 | 10 | 11 | 8 | 13 | |||||
| T4 | 23 | 9 | 14 | 11 | 12 | 7 | 16 | 11 | 12 | 9 | 14 | |||||
| Primary tumor site | 0.217 | 0.407 | 0.001a | 0.481 | 0.390 | |||||||||||
| Buccal mucosa | 8 | 6 | 2 | 5 | 3 | 1 | 7 | 6 | 2 | 4 | 4 | |||||
| Upper gingiva | 12 | 5 | 7 | 5 | 7 | 4 | 8 | 9 | 3 | 7 | 5 | |||||
| Lower gingiva | 22 | 7 | 15 | 9 | 13 | 8 | 14 | 11 | 11 | 7 | 15 | |||||
| Tongue | 55 | 22 | 33 | 37 | 18 | 34 | 21 | 41 | 14 | 34 | 21 | |||||
| Oral floor | 10 | 4 | 6 | 6 | 4 | 8 | 2 | 5 | 5 | 3 | 7 | |||||
| Palate | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | |||||
| Lymph node metastasis | 0.002a | 0.031a | 0.003a | 0.386 | 0.007a | |||||||||||
| Positive | 40 | 9 | 31 | 18 | 22 | 13 | 27 | 25 | 15 | 14 | 26 | |||||
| Negative | 68 | 36 | 32 | 45 | 23 | 42 | 26 | 48 | 20 | 42 | 26 | |||||
| Distant metastasis | 0.051 | 0.109 | 0.014a | 0.322 | 0.012a | |||||||||||
| Positive | 9 | 1 | 8 | 3 | 6 | 1 | 8 | 5 | 4 | 1 | 8 | |||||
| Negative | 99 | 44 | 55 | 60 | 39 | 54 | 45 | 68 | 31 | 55 | 44 | |||||
| Tumor differentiation | 0.263 | 0.232 | 0.213 | 0.057 | 0.660 | |||||||||||
| Well | 83 | 37 | 46 | 51 | 32 | 45 | 38 | 60 | 23 | 44 | 39 | |||||
| Moderate | 25 | 8 | 17 | 12 | 13 | 10 | 15 | 13 | 12 | 12 | 13 | |||||
| Poor | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Anneroth score | <0.001a | 0.007a | 0.496 | 0.115 | <0.001a | |||||||||||
| 1 | 7 | 4 | 3 | 6 | 1 | 6 | 1 | 7 | 0 | 5 | 2 | |||||
| 2 | 14 | 9 | 5 | 8 | 6 | 2 | 12 | 8 | 6 | 10 | 4 | |||||
| 3 | 54 | 29 | 25 | 37 | 17 | 31 | 23 | 38 | 16 | 34 | 20 | |||||
| 4 | 33 | 3 | 30 | 12 | 21 | 16 | 17 | 20 | 13 | 7 | 26 | |||||
Significant. SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
Subcellular localization of SOX2, Oct4, KLF4, c-Myc and brachyury expression
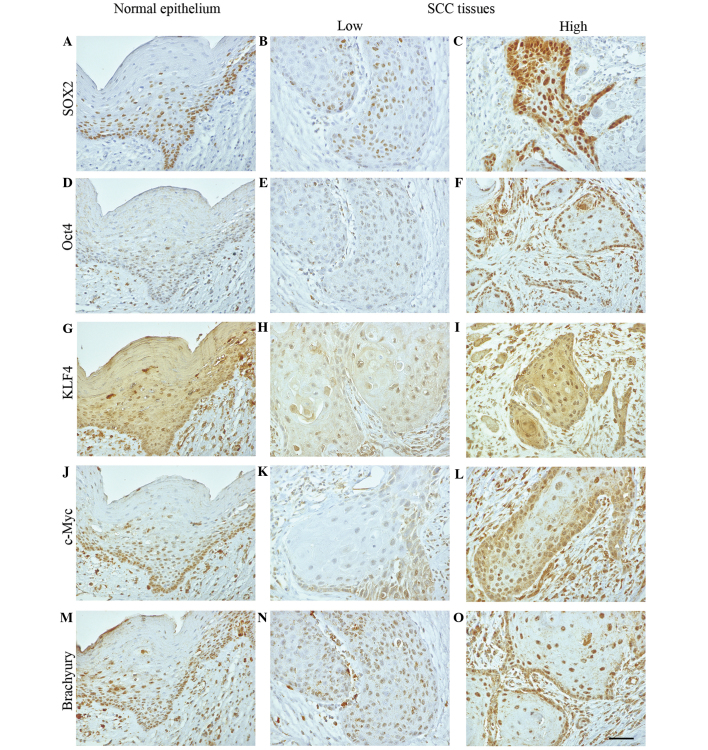
SOX2, Oct4, c-Myc and brachyury were predominantly localized to the nucleus of OSCC cells; however, in certain cases, they were localized to the cytoplasm and nucleus (Fig. 2). KLF4 was primarily localized to the cytoplasm and nucleus of OSCC cells. All proteins were also detected in the nucleus of normal basal epithelial cells.
Figure 2.
EI of SOX2, Oct4, KLF4, c-Myc and brachyury in oral squamous cell carcinoma tissue. Photomicrographs show representative examples of normal epithelium (left column) and low (middle column) or high (right column) EI of (A-C) SOX2, (D-E) Oct4, (G-I) KLF4, (J-L) c-Myc and (M-O) brachyury. Scale bar, 50 µm. EI, expression intensity; SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
Association between SOX2, Oct4, KLF4, c-Myc and brachyury expression and clinicopathological factors
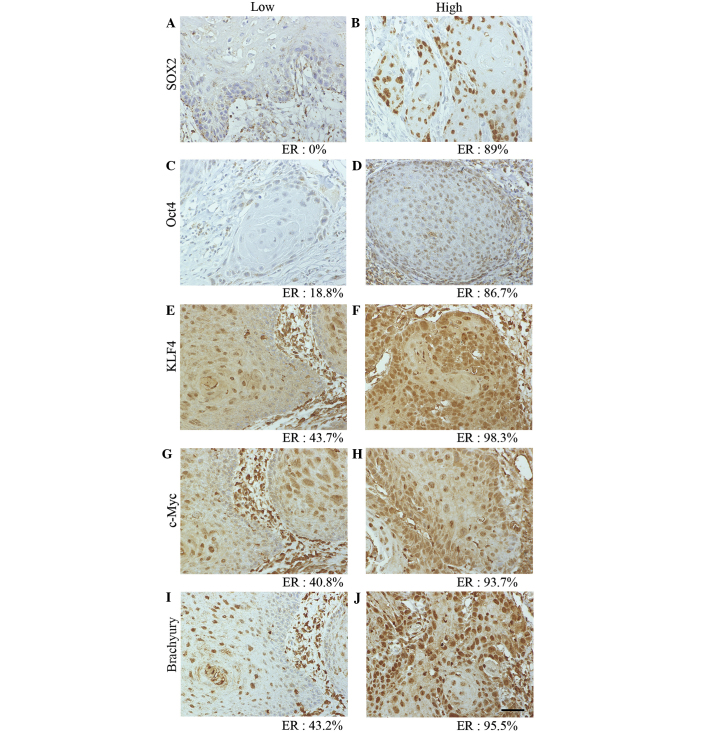
The median ERs of SOX2, Oct4, KLF4, c-Myc and brachyury, which were used as the cut-off values for low or high expression, were 66.6, 54.7, 66.7, 71.9 and 71.9%, respectively (Fig. 3). The EIs and ERs of these transcription factors were found to be significantly associated with several clinicopathological factors (Tables II–IV). For example, c-Myc EI was significantly associated with clinical tumor stage (P=0.003), while SOX2, Oct4, KLF4 and brachyury EIs were significantly associated with lymph node metastasis (P=0.002, P=0.031, P=0.003 and P=0.007, respectively) (Table II). KLF4 and brachyury EIs were also significantly associated with distant metastasis (P=0.014 and P=0.012, respectively). However, no significant differences were identified between the EIs of these proteins and the degree of tumor differentiation. Notably, the EIs of SOX2, Oct4 and brachyury were significantly associated with Anneroth scores (P<0.001, P=0.007 and P<0.001, respectively). χ2 tests revealed that the EIs of SOX2, KLF4 and brachyury in tumors with an Anneroth score of 3 were significantly associated with lymph node metastasis (P=0.015, P=0.005 and P=0.025, respectively) (Table V). However, no significant differences were identified between EIs of SOX2, KLF4 and brachyury in tumors with Anneroth scores of 1, 2 or 4.
Figure 3.
Positive ER of SOX2, Oct4, KLF4, c-Myc and brachyury in oral squamous cell carcinoma tissue. Photomicrographs show representative examples of low (left column) or high (right column) positive ER for (A and B) SOX2, (C and D) Oct4, (E and F) KLF4, (G and H) c-Myc, and (I and J) brachyury. Scale bar, 50 µm. ER, expression ratio; SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
Table IV.
Predictive factors for lymph node and distant metastasis in oral squamous cell carcinoma patients.
| Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|
| Type of metastasis | Comparison | OR | P-value | 95% CI | OR | P-value | 95% CI |
| Lymph node | |||||||
| SOX2 | Low vs. high EI | 3.875 | 0.003 | 1.604–9.359 | 4.526 | 0.011 | 1.404–14.588 |
| Oct4 | Low vs. high EI | 2.391 | 0.033 | 1.074–5.323 | 1.148 | 0.795 | 0.405–3.255 |
| KLF4 | Low vs. high EI | 3.355 | 0.004 | 1.474–7.639 | 4.851 | 0.004 | 1.667–14.116 |
| c-Myc | Low vs. high EI | 1.440 | 0.387 | 0.631–3.288 | 0.559 | 0.284 | 0.193–1.622 |
| Brachyury | Low vs. high EI | 3.000 | 0.008 | 1.330–6.766 | 0.999 | 0.998 | 0.312–3.193 |
| Distant | |||||||
| SOX2 | Low vs. high EI | 6.400 | 0.086 | 0.771–53.123 | 3.766 | 0.314 | 0.285–49.820 |
| Oct4 | Low vs. high EI | 3.077 | 0.127 | 0.727–13.030 | 1.003 | 0.997 | 0.188–5.359 |
| KLF4 | Low vs. high EI | 9.600 | 0.036 | 1.157–79.673 | 9.607 | 0.053 | 0.974–94.804 |
| c-Myc | Low vs. high EI | 1.755 | 0.425 | 0.441–6.987 | 0.579 | 0.494 | 0.121–2.775 |
| Brachyury | Low vs. high EI | 10.000 | 0.033 | 1.205–83.005 | 3.301 | 0.360 | 0.256–42.542 |
OR, odds ratio; CI, confidence interval; EI, expression intensity. SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
Table V.
Association between SOX2, Oct4, KLF4, c-Myc and brachyru protein expression intensity and metastasis in oral squamous cell carcinoma patients according to Anneroth score.
| SOX2 expression, n | Oct4 expression, n | KLF4 expression, n | c-Myc expression, n | Brachyury expression, n | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Cases, n | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value |
| Anneroth score=1 | ||||||||||||||||
| Lymph node metastasis | – | – | – | – | – | |||||||||||
| Positive | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Negative | 7 | 4 | 3 | 6 | 1 | 6 | 1 | 7 | 0 | 5 | 2 | |||||
| Distant metastasis | – | – | – | – | – | |||||||||||
| Positive | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Negative | 7 | 4 | 3 | 6 | 1 | 6 | 1 | 7 | 0 | 5 | 2 | |||||
| Anneroth score=2 | ||||||||||||||||
| Lymph node metastasis | 0.545 | 0.594 | 0.495 | 0.594 | 0.689 | |||||||||||
| Negative | 10 | 6 | 4 | 6 | 4 | 2 | 8 | 6 | 4 | 7 | 3 | |||||
| Distant metastasis | 0.110 | 0.165 | 0.725 | 0.165 | 0.066 | |||||||||||
| Positive | 2 | 0 | 2 | 0 | 2 | 0 | 2 | 0 | 2 | 0 | 2 | |||||
| Negative | 12 | 9 | 3 | 8 | 4 | 2 | 10 | 8 | 4 | 10 | 2 | |||||
| Anneroth score=3 | ||||||||||||||||
| Lymph node metastasis | 0.015a | 0.095 | 0.005a | 0.537 | 0.025a | |||||||||||
| Positive | 17 | 5 | 12 | 9 | 8 | 5 | 12 | 11 | 6 | 7 | 10 | |||||
| Negative | 37 | 24 | 13 | 28 | 9 | 26 | 11 | 27 | 10 | 27 | 10 | |||||
| Distant metastasis | 0.716 | 0.535 | 0.177 | 0.509 | 0.608 | |||||||||||
| Positive | 2 | 1 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | |||||
| Negative | 52 | 28 | 24 | 36 | 16 | 31 | 21 | 37 | 15 | 33 | 19 | |||||
| Anneroth score=4 | ||||||||||||||||
| Lymph node metastasis | 0.384 | 0.947 | 0.393 | 0.727 | 0.652 | |||||||||||
| Positive | 19 | 1 | 18 | 7 | 12 | 8 | 11 | 12 | 7 | 4 | 15 | |||||
| Negative | 14 | 2 | 12 | 5 | 9 | 8 | 6 | 8 | 6 | 3 | 11 | |||||
| Distant metastasis | 0.600 | 0.612 | 0.187 | 0.331 | 0.277 | |||||||||||
| Positive | 5 | 0 | 5 | 2 | 3 | 1 | 4 | 4 | 1 | 0 | 5 | |||||
| Negative | 28 | 3 | 25 | 10 | 18 | 15 | 13 | 16 | 12 | 7 | 21 | |||||
Statistically significant (P<0.05). SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
In addition, clinical tumor stage was significantly associated with Oct4 and KLF4 ERs (P=0.048 and P=0.028, respectively) (Table III). Lymph node metastasis was significantly associated with Oct4 ER (P=0.046) and distant metastasis was significantly associated with SOX2 (P=0.016). Anneroth scores were significantly associated with SOX2, Oct4, KLF4, c-Myc and brachyury ERs (P=0.005, P=0.019, P=0.003, P=0.019 and P=0.010, respectively); however, only Oct4 expression was significantly associated with tumor differentiation (P=0.012).
Table III.
Association between SOX2, Oct4, KLF4, c-Myc and brachyury positive expression ratio and clinicopathological factors in 108 oral squamous cell carcinoma patients.
| SOX2 expression, n | Oct4 expression, n | KLF4 expression, n | c-Myc expression, n | Brachyury expression, n | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinicopathological parameter | Cases, n | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value |
| Age, years | 0.56 | 0.846 | 0.846 | 0.332 | 0.560 | |||||||||||
| <65 | 61 | 29 | 32 | 31 | 30 | 31 | 30 | 33 | 28 | 32 | 29 | |||||
| ≥65 | 47 | 25 | 22 | 23 | 24 | 23 | 24 | 21 | 26 | 22 | 25 | |||||
| Gender | 0.317 | 0.161 | 0.317 | 0.548 | 0.071 | |||||||||||
| Male | 69 | 32 | 37 | 31 | 38 | 32 | 37 | 33 | 36 | 30 | 39 | |||||
| Female | 39 | 22 | 17 | 23 | 16 | 22 | 17 | 21 | 18 | 24 | 15 | |||||
| Clinical stage | 0.550 | 0.048a | 0.028a | 0.845 | 0.420 | |||||||||||
| T1 | 18 | 11 | 7 | 13 | 5 | 12 | 6 | 9 | 9 | 11 | 7 | |||||
| T2 | 46 | 24 | 22 | 22 | 24 | 27 | 19 | 25 | 21 | 25 | 21 | |||||
| T3 | 21 | 10 | 1 | 6 | 15 | 9 | 12 | 9 | 12 | 8 | 13 | |||||
| T4 | 23 | 9 | 14 | 13 | 10 | 6 | 17 | 11 | 12 | 10 | 13 | |||||
| Primary tumor site | 0.030a | 0.465 | 0.329 | 0.091 | 0.045a | |||||||||||
| Buccal mucosa | 8 | 6 | 2 | 4 | 4 | 6 | 2 | 6 | 2 | 5 | 3 | |||||
| Upper gingiva | 12 | 8 | 4 | 5 | 7 | 6 | 6 | 5 | 7 | 8 | 4 | |||||
| Lower gingiva | 22 | 9 | 13 | 11 | 11 | 8 | 14 | 12 | 10 | 10 | 12 | |||||
| Tongue | 55 | 29 | 26 | 30 | 25 | 28 | 27 | 29 | 26 | 29 | 26 | |||||
| Oral floor | 10 | 1 | 9 | 3 | 7 | 6 | 4 | 1 | 9 | 2 | 8 | |||||
| Palate | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | |||||
| Lymph node metastasis | 1.000 | 0.046a | 0.425 | 0.111 | 0.425 | |||||||||||
| Positive | 40 | 20 | 20 | 15 | 25 | 18 | 22 | 16 | 24 | 18 | 22 | |||||
| Negative | 68 | 34 | 34 | 39 | 29 | 36 | 32 | 38 | 30 | 36 | 32 | |||||
| Distant metastasis | 0.016a | 0.244 | 0.081 | 0.500 | 0.081 | |||||||||||
| Positive | 9 | 1 | 8 | 3 | 6 | 2 | 7 | 4 | 5 | 2 | 7 | |||||
| Negative | 99 | 53 | 46 | 51 | 48 | 52 | 47 | 50 | 49 | 52 | 47 | |||||
| Tumor differentiation | 0.254 | 0.012a | 0.820 | 0.110 | 0.110 | |||||||||||
| Well | 83 | 44 | 39 | 47 | 36 | 42 | 41 | 45 | 38 | 45 | 38 | |||||
| Moderate | 25 | 10 | 15 | 7 | 18 | 12 | 13 | 9 | 16 | 9 | 16 | |||||
| Poor | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Anneroth score | 0.005a | 0.019a | 0.003a | 0.019a | 0.010a | |||||||||||
| 1 | 7 | 3 | 4 | 6 | 1 | 5 | 2 | 4 | 3 | 5 | 2 | |||||
| 2 | 14 | 12 | 2 | 6 | 8 | 9 | 5 | 10 | 4 | 10 | 4 | |||||
| 3 | 54 | 30 | 24 | 31 | 23 | 31 | 23 | 29 | 25 | 27 | 27 | |||||
| 4 | 33 | 9 | 24 | 11 | 22 | 9 | 24 | 11 | 22 | 12 | 21 | |||||
Significant. SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
Predictive factors for lymph node and distant metastasis
As the results of the present study indicated that lymph node and distant metastases were more significantly associated with EI than ER, whether SOX2, Oct4, KLF4, c-Myc and brachyury EIs are significant predictive factors for lymph node and distant metastases was investigated. Univariate analyses revealed that high SOX2, Oct4, KLF4 and brachyury EIs were significantly associated with lymph node metastasis [odds ratios (ORs), 3.875, 2.391, 3.355 and 3.000, respectively], and high KLF4 and brachyury EIs were associated with distant metastasis (ORs, 9.600 and 10.000, respectively) (Table IV). Multivariate analysis also revealed that high SOX2 and KLF4 EIs were significantly associated with lymph node metastasis (ORs, 4.526 and 4.851, respectively).
Correlation between SOX2, Oct4, KLF4, c-Myc and brachyury expression and survival in OSCC patients
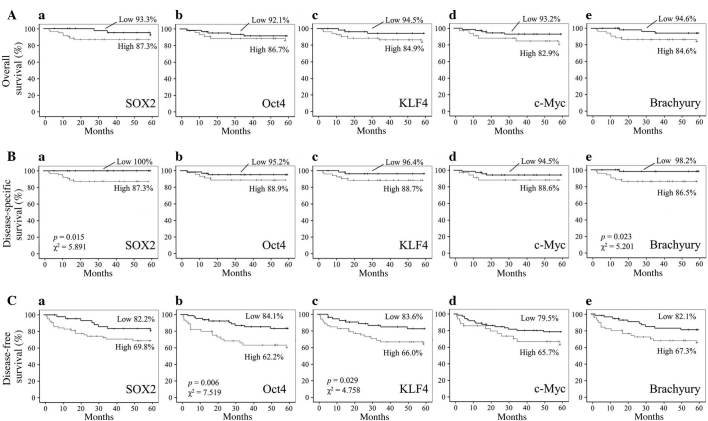
No significant associations between the five-year overall survival rates of OSCC patients and the EIs of SOX2, Oct4, KLF4, c-Myc or brachyury were identified (Fig. 4A). However, the five-year disease-specific survival rates of OSCC patients with high SOX2 and brachyury expression were significantly decreased when compared with those exhibiting low expression [SOX2, 87.3% vs. 100%, respectively (P=0.015; χ2=5.891); brachyury, 86.5% vs. 98.2%, respectively (P=0.023; χ2=5.201); Fig. 4B]. In addition, the five-year disease-free survival rates of OSCC patients with high Oct4 and KLF4 expression were significantly decreased when compared with those exhibiting low expression [Oct4, 62.2% vs. 84.1%, respectively (P=0.006; χ2=7.519); KLF4, 66.0% vs. 83.6%, respectively (P=0.029; χ2=4.758); Fig. 4C].
Figure 4.
Correlation between SOX2, Oct4, KLF4, c-Myc and brachyury expression and survival in OSCC patients. (A) Overall, (B) disease-specific and (C) disease-free survival of OSCC patients with high or low expression intensity of (a) SOX2, (b) Oct4, (c) KLF4, (d) c-Myc and (e) brachyury. P-values and χ2 statistics are shown in plots with statistically significant differences. OSCC, oral squamous cell carcinoma; SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
As SOX2, KLF4 and brachyury EIs were found to be associated with lymph node and distant metastasis in this study, the association between patient survival and the co-expression of SOX2, KLF4 and brachyury was also investigated. The results revealed that the co-expression of SOX2, KLF4 and brachyury was not significantly associated with overall survival (Fig. 5A). However, the five-year disease-specific survival rate of patients with high co-expression of these proteins was decreased when compared with that of patients exhibiting low co-expression (78.6% vs. 100%, respectively; P=0.025; χ2=5.033) (Fig. 5B). Similarly, the five-year disease-free survival rate of patients with high co-expression was decreased compared with that of patients exhibiting low co-expression (60.7% vs. 90.9%, respectively; P=0.015; χ2=5.897) (Fig. 5C).
Figure 5.
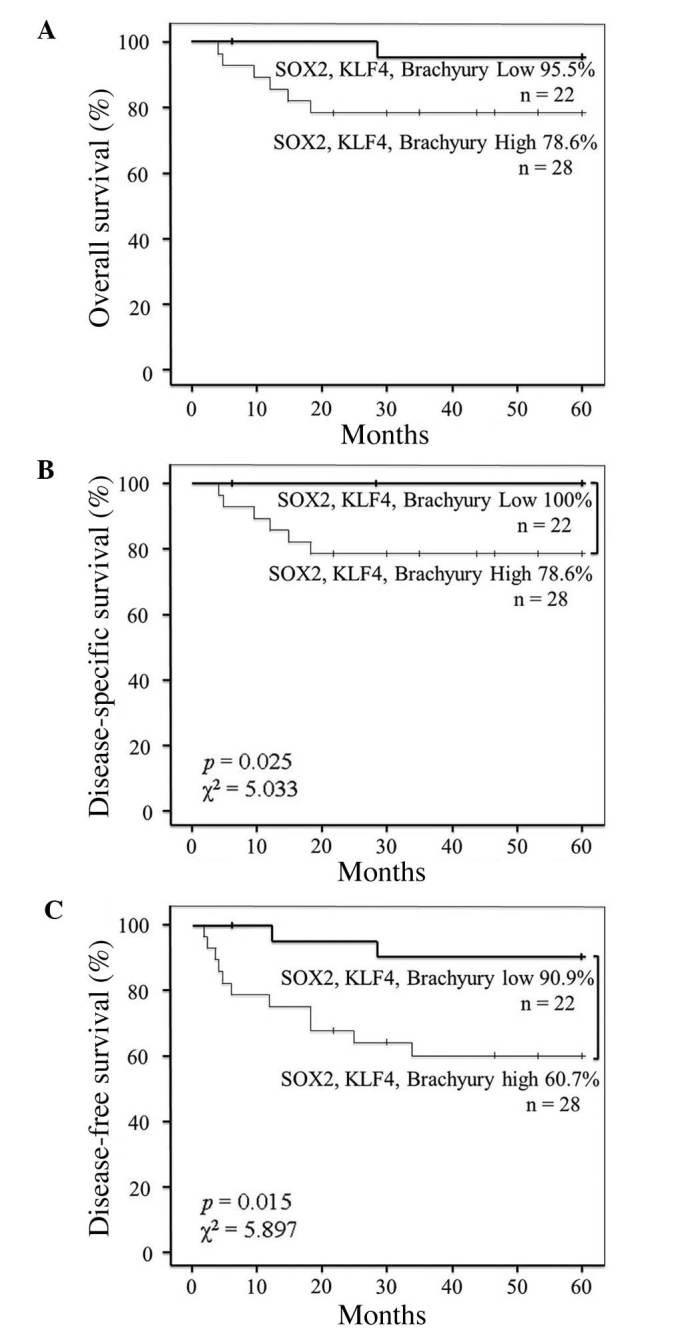
Correlation between co-expression of SOX2, KLF4 and brachyury and survival in OSCC patients. (A) Overall, (B) disease-specific and (C) disease-free survival of OSCC patients with tumors with indicated high or low co-expression intensity status of SOX2, KLF4 and brachyury. P-values and χ2 statistics are shown in plots with statistically significant differences. OSCC, oral squamous cell carcinoma; SOX2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; c-Myc, avian myelocytomatosis viral oncogene homolog; KLF4, Krüppel-like factor 4.
Discussion
The self-renewal and pluripotent properties of CSCs, which are hypothesized to enable primary tumors to metastasize (27,28), indicate that their identification in tumor samples may be important for cancer diagnosis and treatment. However, to date, few CSC markers in OSCC have been identified (29,30). The results of the current study indicate that SOX2, KLF4 and brachyury may present clinically useful CSC markers, and their expression levels may be prognostic factors for OSCC. The expression levels of these transcription factors were quantified in terms of EI and ER, which reflect the level of protein expression and the number of cells expressing a protein, respectively. As high expression levels of CSC-related transcription factors may promote tumorigenesis (31,32), EI may also be a measure of tumor invasiveness and local metastasis. Similarly, as large numbers of CSCs increase the chance that some will maintain stemness when they disseminate to other sites (33), ER may be a measure of the likelihood of distant metastasis. Thus, the EI and ER of CSC-related transcription factors may be associated with survival outcomes in cancer patients.
The results of the present study revealed that SOX2 EI and ER were significantly associated with lymph node metastasis and distant metastasis, respectively, indicating that SOX2 expression is involved in OSCC metastasis. In addition, the significant association between high SOX2 expression and reduced five-year disease-specific survival rate indicates that SOX2 may be a prognostic factor in OSCC patients. These results are consistent with those of previous reports, which have revealed that SOX2 is associated with poor prognosis in several types of cancer (12,13,15,34,35). Furthermore, SOX2 regulates stemness (36) and upregulates CSC-related gene expression in skin squamous cell carcinoma, and helps maintain neural stem cells (37).
Similarly, a significant association between KLF4 EI and lymph node metastasis was identified in the present study; however, this association was not observed between KLF4 ER and distant metastasis, which indicates that KLF4 may be less important for metastasis than SOX2. Furthermore, the association identified between high KLF4 EI and decreased five-year disease-free survival rate in OSCC patients is consistent with the association between increased nuclear expression of KLF4 and poor prognosis in breast cancer and head and neck cancer patients, which has been reported in previous studies (38,39).
In the present study brachyury EI was found to significantly correlate with lymph node metastasis, distant metastasis and Anneroth scores, which indicates that brachyury is also involved in OSCC metastasis. These results are consistent with those of previous studies, which revealed that silencing brachyury expression inhibits tumor formation and metastasis in human adenoid cystic carcinoma cells (19,20). In addition, a previous study revealed that brachyury expression is associated with EMT and lymph node metastasis in OSCC patients (22).
The results of the present study found that c-Myc EI and ER were not associated with lymph node or distant metastasis. By contrast, Oct4 EI and ER were significantly associated with lymph node metastasis and Anneroth scores, which suggests that Oct4 may be involved in tumor metastasis. c-Myc EI was only significantly associated with clinical tumor stage, which is consistent with its reported association with tumorigenesis and sustained tumor growth (40).
Two conclusions may be drawn from the results of the present study. Firstly, the EI of CSC markers is a better indicator of metastasis and survival than ER in OSCC patients. This may be due to the relative uniformity of ER in normal and tumor cells in biopsy specimens (data not shown). In addition, the normal expression level of the transcription factors examined in this study was higher in OSCC tissue samples than in noncancerous tissue samples. Secondly, high SOX2, KLF4 and brachyury expression is significantly associated with tumor invasion and metastasis, as well as decreased disease-specific survival and disease-free survival, in OSCC patients. Thus, these transcription factors may be involved in tumor progression, and may represent clinically useful prognostic markers in OSCC.
In conclusion, the expression of SOX2, KLF4 and brachyury may present novel prognostic factors in OSCC and thus, the combined use of these factors and classical prognostic factors, such as Anneroth score, may improve the accuracy of metastasis prediction. Therefore, future prospective studies investigating clinical intervention in OSCC patients with positive SOX2, KLF4 and brachyury expression are required.
Acknowledgements
This study was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (no. 23390465 to Professor Tsuyoshi Sugiura; no. 25861958 to Dr Yosuke Kobayashi; and no. 25893174 to Dr Satomi Chigita).
References
- 1.Slootweg PJ, Eveson JW. Tumours of the oral cavity and oropharynx. World Health Organization Classification of Tumours. In: Barnes L, Eveson JW, Reichart P, Sidransky D, editors. Pathology & Genetics of Head and Neck Tumours. Lyon: IARC Press; 2005. pp. 166–167. [Google Scholar]
- 2.Genden EM, Ferlito A, Bradley PJ, Rinaldo A, Scully C. Neck disease and distant metastases. Oral Oncol. 2003;39:207–212. doi: 10.1016/S1368-8375(02)00049-0. [DOI] [PubMed] [Google Scholar]
- 3.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Kaichi S, Hasegawa K, Takaya T, et al. Cell line-dependent differentiation of induced pluripotent stem cells into cardiomyocytes in mice. Cardiovasc Res. 2010;88:314–323. doi: 10.1093/cvr/cvq189. [DOI] [PubMed] [Google Scholar]
- 6.Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136:2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans PM, Liu C. Roles of Krüppel-like factor 4 in normal homeostasis cancer and stem cells. Acta Biochim Biophys Sin (Shanghai) 2008;40:554–564. doi: 10.1111/j.1745-7270.2008.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu LS, Chan CP, Chen CJ, Lin SH, Lai MT, Hsu JD, Yeh KT, Soon MS. Decreased Krüppel-like factor 4 (KLF4) expression may correlate with poor survival in gastric adenocarcinoma. Med Oncol. 2013;30:632. doi: 10.1007/s12032-013-0632-6. [DOI] [PubMed] [Google Scholar]
- 9.Patel NV, Ghaleb AM, Nandan MO, Yang VW. Expression of the tumor suppressor Krüppel-like factor 4 as a prognostic predictor for colon cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:2631–2638. doi: 10.1158/1055-9965.EPI-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Bio. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 11.He W, Li K, Wang F, Qin YR, Fan QX. Expression of OCT4 in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. World J Gastroenterol. 2012;18:712–719. doi: 10.3748/wjg.v18.i7.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann J, Bahr F, Horst D, Kriegl L, Engel J, Luque RM, Gerhard M, Kirchner T, Jung A. SOX2 expression correlates with lymph-node metastases and distant spread in right-sided colon cancer. BMC Cancer. 2011;11:518. doi: 10.1186/1471-2407-11-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruan J, Wei B, Xu Z, Yang S, Zhou Y, Yu M, Liang J, Jin K, Huang X, Lu P, Cheng H. Predictive value of SOX2 expression in transurethral resection specimens in patients with T1 bladder cancer. Med Oncol. 2013;30:445. doi: 10.1007/s12032-012-0445-z. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Han B, Huang J, Zheng B, Geng Q, Aziz F, Dong Q. Prognostic significance of OCT4 expression in adenocarcinoma of the lung. Jpn J Clin Oncol. 2010;40:961–966. doi: 10.1093/jjco/hyq066. [DOI] [PubMed] [Google Scholar]
- 15.Sholl LM, Barletta JA, Yeap BY, Chirieac LR, Hornick JL. SOX2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am J Surg Pathol. 2010;34:1193–1198. doi: 10.1097/PAS.0b013e3181e5e024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidricaire G, Jardine K, McBurney MW. Expression of the brachyury gene during mesoderm development in differentiating embryonal carcinoma cell cultures. Development. 1994;120:115–122. doi: 10.1242/dev.120.1.115. [DOI] [PubMed] [Google Scholar]
- 17.Kispert A, Herrmann BG, Leptin M, Reuter R. Homologs of the mouse brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium and Locusta. Genes Dev. 1994;8:2137–2150. doi: 10.1101/gad.8.18.2137. [DOI] [PubMed] [Google Scholar]
- 18.Ishii K, Shimoda M, Sugiura T, Seki K, Takahashi M, Abe M, Matsuki R, Inoue Y, Shirasuna K. Involvement of epithelial-mesenchymal transition in adenoid cystic carcinoma metastasis. Int J Oncol. 2011;38:921–931. doi: 10.3892/ijo.2011.917. [DOI] [PubMed] [Google Scholar]
- 19.Shimoda M, Sugiura T, Imajyo I, Ishii K, Chigita S, Seki K, Kobayashi Y, Shirasuna K. The T-box transcription factor brachyury regulates epithelial-mesenchymal transition in association with cancer stem-like cells in adenoid cystic carcinoma cells. BMC Cancer. 2012;12:377. doi: 10.1186/1471-2407-12-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi Y, Sugiura T, Imajyo I, Shimoda M, Ishii K, Akimoto N, Yoshihama N, Mori Y. Knockdown of the T-box transcription factor brachyury increases sensitivity of adenoid cystic carcinoma cells to chemotherapy and radiation in vitro, Implications for a new therapeutic principle. Int J Oncol. 2014;44:1107–1117. doi: 10.3892/ijo.2014.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C. The T-box transcription factor brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest. 2010;120:533–544. doi: 10.1172/JCI38379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imajyo I, Sugiura T, Kobayashi Y, Shimoda M, Ishii K, Akimoto N, Yoshihama N, Kobayashi I, Mori Y. T-box transcription factor brachyury expression is correlated with epithelial-mesenchymal transition and lymph node metastasis in oral squamous cell carcinoma. Int J Oncol. 2012;41:1985–1995. doi: 10.3892/ijo.2012.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobin LH, Gospodarowicz MK, Wittekind C, editors. TNM Classification of Malignant Tumours. 7th. Hoboken, NJ: Wiley-Blackwell; 2009. [Google Scholar]
- 24.Pindborg JJ, Reichart PA, Smith CJ, van der Waal I. Histological Typing of Cancer and Precancer of the Oral Mucosa. Berlin: Springer-Verlag; 1997. World Health Organization International Histological Classification of Tumours. [DOI] [Google Scholar]
- 25.Anneroth G, Hansen LS, Silverman S., Jr Malignancy grading in oral squamous cell carcinoma. I. Squamous cell carcinoma of the tongue and floor of mouth Histologic grading in the clinical evaluation. J Oral Pathol. 1986;15:162–168. doi: 10.1111/j.1600-0714.1986.tb00599.x. [DOI] [PubMed] [Google Scholar]
- 26.Anneroth G, Batsakis J, Luna M. Review of the literature and a recommended system of malignancy grading in oral squamous cell carcinomas. Scand J Dent Res. 1987;95:229–249. doi: 10.1111/j.1600-0722.1987.tb01836.x. [DOI] [PubMed] [Google Scholar]
- 27.Nomura A, Banerjee S, Chugh R, Dudeja V, Yamamoto M, Vickers SM, Saluja AK. CD133 initiates tumors, induces epithelial-mesenchymal transition and increases metastasis in pancreatic cancer. Oncotarget. 2015;6:8313–8322. doi: 10.18632/oncotarget.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González-Moles MA, Scully C, Ruiz-Ávila I, Plaza-Campillo JJ. The cancer stem cell hypothesis applied to oral carcinoma. Oral Oncol. 2013;49:738–746. doi: 10.1016/j.oraloncology.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Yu CC, Hu FW, Yu CH, Chou MY. Targeting CD133 in the enhancement of chemosensitivity in oral squamous cell carcinoma-derived side population cancer stem cells. Head Neck Dec. 2014;24 doi: 10.1002/hed.23975. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 30.Patel SS, Shah KA, Shah MJ, Kothari KC, Rawal RM. Cancer stem cells and stemness markers in oral squamous cell carcinomas. Asian Pac J Cancer Prev. 2014;15:8549–8556. doi: 10.7314/APJCP.2014.15.20.8549. [DOI] [PubMed] [Google Scholar]
- 31.Liu K, Lin B, Zhao M, Yang X, Chen M, Gao A, Liu F, Que J, Lan X. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25:1264–1271. doi: 10.1016/j.cellsig.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S, Xu Y, Chen Y, Li X, Mou W, Wang L, Liu Y, Reisfield RA, Xiang R, Xiang R, Lv D, Li N. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PLoS One. 2012;7:e36326. doi: 10.1371/journal.pone.0036326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forghanifard MM, Khales Ardalan S, Javdani-Mallak A, Rad A, Farshchian M, Abbaszadegan MR. Stemness state regulators SALL4 and SOX2 are involved in progression and invasiveness of esophageal squamous cell carcinoma. Med Oncol. 2014;31:922. doi: 10.1007/s12032-014-0922-7. [DOI] [PubMed] [Google Scholar]
- 34.Lengerke C, Fehm T, Kurth R, Neubauer H, Scheble V, Müller F, Schneider F, Petersen K, Wallwiener D, Kanz L, et al. Expression of the embryonic stem cell marker SOX2 in early-stage breast carcinoma. BMC Cancer. 2011;11:42. doi: 10.1186/1471-2407-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitamura H, Torigoe T, Hirohashi Y, Asanuma H, Inoue R, Nishida S, Tanaka T, Fukuta F, Masumori N, Sato N, Tsukamoto T. Prognostic impact of the expression of ALDH1 and SOX2 in urothelial cancer of the upper urinary tract. Mod Pathol. 2013;26:117–124. doi: 10.1038/modpathol.2012.139. [DOI] [PubMed] [Google Scholar]
- 36.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 37.Episkopou V. SOX2 functions in adult neural stem cells. Trends Neurosci. 2005;28:219–221. doi: 10.1016/j.tins.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Tai SK, Yang MH, Chang SY, Chang YC, Li WY, Tsai TL, Wang YF, Chu PY, Hsieh SL. Persistent Krüppel-like factor 4 expression predicts progression and poor prognosis of head and neck squamous cell carcinoma. Cancer Sci. 2011;102:895–902. doi: 10.1111/j.1349-7006.2011.01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.CCR-03-0484. [DOI] [PubMed] [Google Scholar]
- 40.Chen BJ, Wu YL, Tanaka Y, Zhang W. Small molecules targeting c-Myc oncogene, Promising anti-cancer therapeutics. Int J Biol Sci. 2014;10:1084–1096. doi: 10.7150/ijbs.10190. [DOI] [PMC free article] [PubMed] [Google Scholar]