Abstract
Plant varieties expressing the Bt (Bacillus thuringiensis) insecticidal proteins Cry1Ah and Cry2Ab have potential commercialization prospects in China. However, their potential effects on non-target arthropods (NTAs) remain uncharacterized. The cotton aphid Aphis gossypii is a worldwide pest that damages various important crops. The ladybeetle Propylea japonica is a common and abundant natural enemy in many cropping systems in East Asia. In the present study, the effects of Cry1Ah and Cry2Ab proteins on A. gossypii and P. japonica were assessed from three aspects. First, neither of the Cry proteins affected the growth or developmental characteristics of the two test insects. Second, the expression levels of the detoxification-related genes of the two test insects did not change significantly in either Cry protein treatment. Third, neither of the Cry proteins had a favourable effect on the expression of genes associated with the amino acid metabolism of A. gossypii and the nutrition utilization of P. japonica. In conclusion, the Cry1Ah and Cry2Ab proteins do not appear to affect the cotton aphid A. gossypii or the ladybeetle P. japonica.
In the past two decades, vast plantings of insect-resistant genetically modified (IRGM) crops producing insecticidal proteins from the bacterium Bacillus thuringiensis (Bt) have contributed to the management of several major insect pests and reduced the use of insecticide sprays1,2,3. However, one of the risks associated with the growth of IRGM crops is their potential adverse effect on non-target arthropods (NTAs)4,5. Non-target effects assessment is one of the most important components of environmental risk assessment (ERA), which is required to authorize the IRGM crops to be released into the environment4,5.
The rapid evolution of resistance in several target pests has reduced the benefits of Bt crops6. There are usually two ways to cope with pest resistance, one is the discovery of novel cry genes, and another option is to pyramid different genes in one plant. The discovery of novel cry genes with new or broader activity spectra or higher toxicity is important for the development of new varieties and the management of resistance7. The cry1Ah gene is a novel insecticidal gene that was cloned from B. thuringiensis isolate BT8 and encodes a protein with a molecular weight of 134 kDa7. Although its activity spectrum and activity level are similar to those of other typical Cry1A toxins, the Cry1Ah protein is more toxic to Helicoverpa armigera, Chilo suppressalis, and Ostrinia furnacalis than Cry1Ac and is more toxic to O. furnacalis than Cry1Ab and Cry1Ie7. For Plutella xylostella, Cry1Ah exhibits toxicity similar to that of Cry1Ac, one of the most potent toxins against this pest7. Moreover, cry1Ah transgenic corn exhibited strong resistance to O. furnacalis larvae both in the laboratory and field8. Thus, high toxicity against a range of pest species makes this novel toxin a potential candidate for new Bt varieties. Thus a non-target effects assessment of this novel toxin should be conducted.
Another strategy to delay the evolution of pest resistance to Bt crops is the "pyramid" strategy, which uses plants that produce two or more toxins that kill the same pest6. Currently, second-generation transgenic cotton producing the Bt toxins Cry1Ac and Cry2Ab is the only type of Bt cotton grown in Australia and the predominant type of Bt cotton grown in India and the United States9. However, only first-generation Bt cotton producing one toxin (Cry1Ac) is grown in China, the world’s leading producer of cotton9. To cope with pest resistance, a new cotton cultivar (producing Cry1Ac and Cry2Ab) has been developed in China and will be commercially available in the foreseeable future10,11. The Cry2Ab protein targets various Lepidoptera pests, such as Spodoptera exigua, Spodoptera frugiperda and Pectinophora gossypiella12. Thus, non-target effect assessment before its commercial release is required.
A few studies have evaluated the effects of Cry1Ah or Cry2A proteins on non-target insects. For example, Cry1Ah corn does not appear to affect the survival, development, colony performance or behaviour of the honeybee Apis mellifera13. No significant differences were noted in the survival of A. mellifera or Apis cerana that were fed sugar syrup with Cry1Ah protein14. The risk assessment of Cry2A proteins have also been tested with honey bees and natural enemies. For example, the Cry2A have no acute toxicity to A. mellifera larvae at concentrations >10 times higher than those detected in pollen from Bt plants15. Even at concentrations higher than those expected in real-life situations, Cry2Ab does not have a detrimental effect on the green lacewing Chrysoperla carnea when ingested either directly or through prey16. When Coleomegilla maculata larvae were fed an artificial diet incorporated with Cry2Ab at >10-fold higher concentrations than in cotton tissue, no differences were observed in any life-table parameters in the Cry protein-containing diet treatment17. In addition, Li et al. reported that the life-table parameters of P. japonica were not affected when fed a rapeseed pollen-based diet containing purified Cry2A at concentrations that were >10-fold higher than in pollen18.
However, the potential effects of Cry1Ah and Cry2Ab on cotton aphid A. gossypii and ladybeetle P. japonica remain uncharacterized. The cotton aphid Aphis gossypii Glover (Homoptera: Aphididae) is a worldwide pest that damages various crops, including melon, cotton, potato, chili pepper, sweet pepper, and eggplant19. This pest sucks plant sap throughout the developmental period of cotton, causing seedling death, leaf curling and withering, and serious yield loss20. Throughout East Asia, the ladybird beetle Propylea japonica (Thunberg) (Coleoptera: Coccinellidae) is a common and abundant natural enemy in many cropping systems, including maize, cotton, rice, vegetables, and fruit trees21,22,23. Both the larvae and adults feed on aphids, thrips, spider mites, and the eggs and young larvae of Lepidoptera24. Furthermore, P. japonica can be easily reared and is amenable for testing in the laboratory, making it a suitable surrogate species for evaluating the potential effects of Bt proteins on predacious Coccinellidae18,25,26.
Most studies regarding the risk assessment of aphids and ladybird beetles have focused on biology and ecology20,27,28. However, the molecular response of insects to Bt proteins has not been elucidated. Insects have the ability to protect themselves from secondary plant metabolic and toxic chemicals and pathogens by regulating the expression of genes encoding detoxification responses29. Cytochrome P450 monooxygenases (P450s), glutathione S-transferases (GSTs) and esterases (ESTs) are three major detoxifying enzyme families that are involved in the xenobiotic metabolism of insects30,31,32. Mannakkara et al. reported that transgenic Bt rice expressing Cry1Ab/Cry1Ac, Cry2Aa and Cry1Ca had no marked effects on detoxification responses in Nilaparvata lugens33.
Furthermore, many study designs aimed to investigated the effects (including positive and negative effects) of Bt proteins on aphids and ladybeetles20,27,28,34, and here, we focused on if the effects were positive. Several key genes are involved in amino acid metabolism in pea aphids35, and we are interested in whether A. gossypii could use Bt proteins for amino acid metabolism. Moreover, many researchers have used the quantitative nutritional approach to investigate how food is utilized by organisms36,37. The quantitative nutritional approach consists of measuring the amount of food that is consumed, digested and assimilated, excreted, metabolized, and converted into biomass36. An analysis of these measurements reveals how organisms respond to different foods and which food components exert the greatest effects on growth37. This method has also been used to investigate how Bt proteins influence insects38,39,40.
In the present study, we investigated the effects of Cry1Ah and Cry2Ab proteins on cotton aphid A. gossypii and ladybeetle P. japonica from three aspects: (1) the toxicity of Cry proteins on the growth and developmental characteristics of A. gossypii and P. japonica; (2) the detoxification response of A. gossypii and P. japonica to Cry proteins; and (3) the effects of Cry proteins on the expression of genes that are associated with the amino acid metabolism of A. gossypii and the nutrition utilization of P. japonica. The first two aspects aim to determine whether the Cry1Ah and Cry2Ab proteins affect these two NTAs, whereas the third aspect focuses on whether the effect is positive.
Results
Experiments with A. gossypii
Toxicity of Cry proteins to A. gossypii
More than 90% of A. gossypii nymphs reached the adult stage when fed diets supplemented with Cry1Ah, Cry2Ab or pure diet, and the survival rates did not significantly differ between each Cry protein treatment and the control (χ2 test; Cry1Ah: χ2 = 0.429, P = 0.513; Cry2Ab: χ2 = 0.429, P = 0.513) (Table 1). In contrast, no nymphs developed to adults in the E-64 treatment. Moreover, no differences were noted in the nymphal development time (Mann-Whitney U-test; Cry1Ah: P = 0.963; Cry2Ab: P = 0.794) and adult fresh weight (Dunnett’s test; Cry1Ah: P = 0.077; Cry2Ab: P = 0.119) between each Cry protein treatment and the control (Table 1).
Table 1. Life-table parameters of Aphis gossypii when fed artificial diet containing Cry1Ah, Cry2Ab or E-64.
| Treatment | Percent nymphs developed to adultsa | Days to adultb | Adult fresh weight (mg)c |
|---|---|---|---|
| Control: diet only | 94.67 ± 2.49 | 5.57 ± 0.06 | 0.62 ± 0.02 |
| Cry1Ah 500 μg/ml diet | 93.33 ± 2.98 | 5.58 ± 0.06 | 0.57 ± 0.02 |
| Cry2Ab 500 μg/ml diet | 92.00 ± 2.49 | 5.60 ± 0.06 | 0.57 ± 0.02 |
| E-64 600 μg/ml diet | 0 | – | – |
Data are means ± SE. Each toxin treatment was compared to the control. Each treatment has five replicates. Fifteen 2nd instar nymphs (within 12 h) were tested for each replicate. "-" indicates no individual reached the adult stage in E-64 treatment.
aχ2 test.
bMann–Whitney U test.
cDunnett’s test.
Expression of genes that are associated with detoxification responses and amino acid metabolism
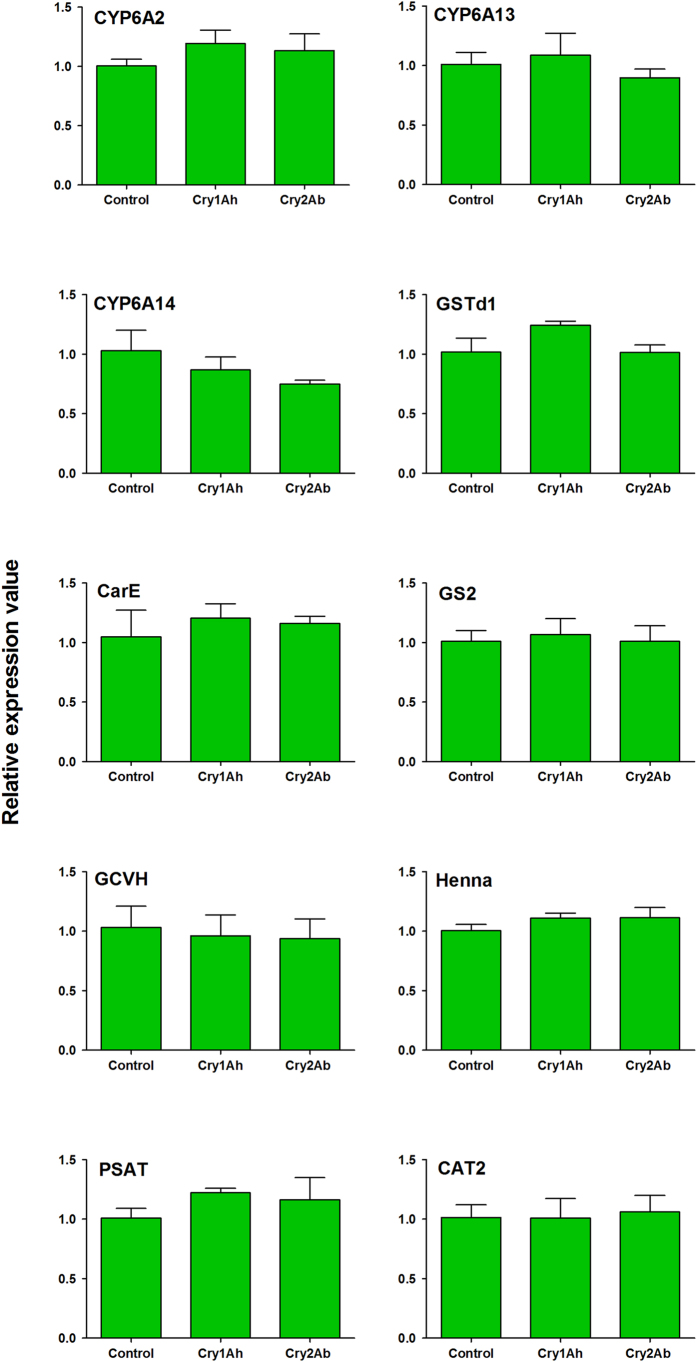
In the Cry1Ah treatment, genes that are associated with A. gossypii detoxification responses [i.e., CYP6A2, CYP6A13, glutathione S-transferase delta 1 (GSTd1), carboxylesterases (CarE)] were slightly up-regulated (regulation factors did not exceed 1.3-fold) compared with the control treatment, whereas only the gene CYP6A14 was slightly down-regulated (Fig. 1). The expression levels of genes that are related to detoxification responses also exhibited minimal change between the Cry2Ab treatment and the control (regulation factors did not exceed 1.3-fold) (Fig. 1). Similarly, the expression of genes relevant to the utilization of essential amino acids [i.e., cationic amino acid transporter 2 (CAT2) and phenylalanine hydroxylase (Henna)], genes relevant to the synthesis of nonessential amino acids [i.e., glutamine synthetase 2 (GS2) and phosphoserine aminotransferase (PSAT)], and a gene that is involved in the catabolism of a nonessential amino acid glycine [i.e., glycine cleavage system h protein (GCVH)], was very similar to the control in both of the Cry protein treatments (Fig. 1). In addition, no significant differences were detected in the expression levels of genes that were associated with detoxification responses and amino acid metabolism between each Cry protein treatment and the control.
Figure 1. Relative expression level of detoxification and amino acid metabolism related genes of Aphis gossypii when fed artificial diet containing Cry1Ah or Cry2Ab.
Data are means ± SE. Three replicates were tested in each treatment. Artificial diet with no Bt protein was served as control treatment. Means were analyzed by Tukey HSD and data yielded no significant differences at P < 0.05 levels.
Experiments with P. japonica
Toxicity of Cry proteins to P. japonica larvae
The larval (Mann-Whitney U-test; Cry1Ah: P = 0.128; Cry2Ab: P = 0.660) and pupal (Cry1Ah: P = 0.469; Cry2Ab: P = 0.413) development time did not significantly differ when P. japonica larvae were fed a sucrose solution-based diet that contained Cry1Ah or Cry2Ab protein (Table 2). Similarly, the pupation (χ2 test; Cry1Ah: χ2 = 0.061, P = 0.806; Cry2Ab: χ2 = 0.015, P = 0.901) and eclosion (Cry1Ah: χ2 = 0.250, P = 0.617; Cry2Ab: χ2 = 0.143, P = 0.705) rates did not significantly differ between each Cry protein treatment and the control (Table 2). Moreover, no differences were found in adult fresh weight (Dunnett’s test; Cry1Ah: P = 0.885; Cry2Ab: P = 0.909) between each Cry protein treatment and the control (Table 2). In contrast, P. japonica in the E-64 treatment exhibited a significantly decreased larval development time (P < 0.001) and pupation rate (χ2 = 19.703, P < 0.001) compared with the control (Table 2).
Table 2. Life-table parameters of Propylea japonica when fed a sucrose solution-based diet containing Cry1Ah, Cry2Ab or E-64.
| Treatment | Larval development time (d)a | Pupation rate (%)b | Pupal development time (d)a | Eclosion rate (%)b | Adult fresh weight (mg)c | |
|---|---|---|---|---|---|---|
| Female | Male | |||||
| Control: pure diet | 11.63 ± 0.14 | 91.43 | 4.25 ± 0.04 | 93.75 | 5.1 ± 0.18 | 4.42 ± 0.15 |
| Cry1Ah 500 μg/ml diet | 11.24 ± 0.18 | 97.14 | 4.15 ± 0.13 | 100 | 5.25 ± 0.18 | 4.25 ± 0.16 |
| Cry2Ab 500 μg/ml diet | 11.55 ± 0.20 | 94.29 | 4.11 ± 0.11 | 100 | 5.24 ± 0.15 | 4.52 ± 0.14 |
| E-64 400 μg/ml diet | 15.9 ± 0.51* | 14.29* | 4.4 ± 0.19 | 100 | 4.87 ± 0.26 | 4.18 |
Data are means ± SE. Each toxin treatment was compared to the control. An asterisk denotes a significant difference between a toxin treatment and the control. Thirty-five larvae were tested for each treatment. Sample of male fresh weight in E-64 treatment was not enough for data analyze.
aMann–Whitney U test.
bχ2 test.
cDunnett’s test.
Expression of genes that are associated with detoxification responses
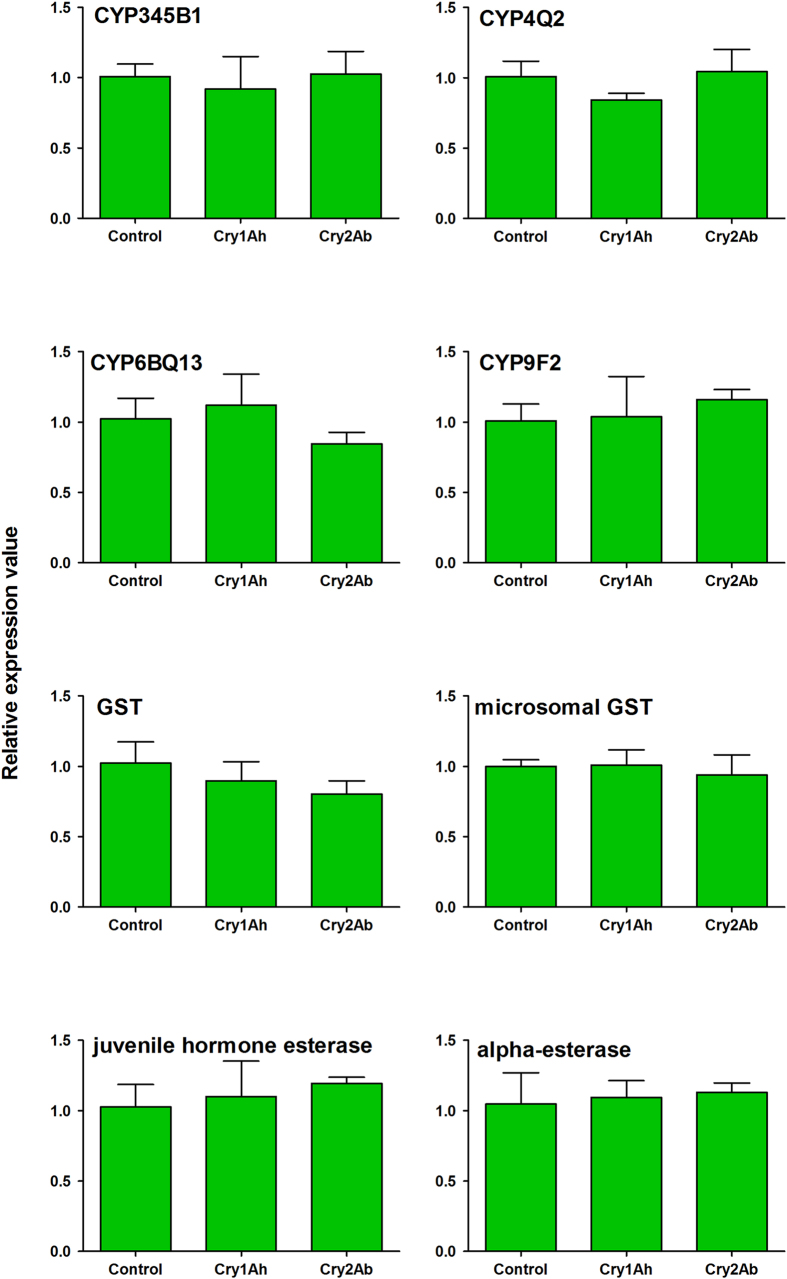
The expression of genes that are associated with P. japonica detoxification responses [i.e., CYP345B1, CYP4Q2, CYP6BQ13, CYP9F2, GST, microsomal GST, juvenile hormone esterase, alpha-esterase] were slightly altered (regulation factors did not exceed 1.2-fold) in each Cry protein treatment compared with the control (Fig. 2), and the alterations were not significantly different.
Figure 2. Relative expression level of detoxification related genes of Propylea japonica when fed sucrose solution containing Cry1Ah or Cry2Ab.
Data are means ± SE. Three replicates were tested in each treatment. Sucrose solution with no Bt protein was served as control treatment. Means were analyzed by Tukey HSD and data yielded no significant differences at P < 0.05 levels.
Nutrition utilization of P. japonica
The ELISA results showed that the concentrations of Cry2Ab was 458.04 ± 12.43 μg/g fresh weight (FW) in the fresh diets and 436.71 ± 13.48 μg/g FW in the diets that had been stored at 4 °C for 24h. The difference was not significant between this two diets (t = 1.163, df = 4, P = 0.309). The relative growth rate (RGR) (least significant difference (LSD) test; Cry1Ah: P = 0.816; Cry2Ab: P = 0.149), efficiency of conversion of ingested materials (ECI) (Cry1Ah: P = 0.554; Cry2Ab: P = 0.072), efficiency of conversion of digested food (ECD) (Cry1Ah: P = 0.748; Cry2Ab: P = 0.085) and efficiency of approximate digestibility (EAD) (Cry1Ah: P = 0.857; Cry2Ab: P = 0.388) did not significantly differ when P. japonica larvae were fed a pork-liver-based artificial diet that contained Cry1Ah or Cry2Ab protein (Table 3). No significant difference was detected in relative consumption rates (RCR) between the Cry1Ah treatment and the control (P = 0.333), but the RCR was significantly lower in the Cry2Ab treatment than in the control treatment (P = 0.038) (Table 3).
Table 3. Nutrition utilization of Propylea japonica when fed artificial diet containing Cry1Ah or Cry2Ab protein.
| Treatment | Nutrition utilization indices | ||||
|---|---|---|---|---|---|
| RGR(mg/mg/day)a | RCR(mg/mg/day)a | ECI(%)a | ECD(%)a | EAD(%)a | |
| Control: pure diet | 0.12 | 0.66 ± 0.03 | 18.95 ± 1.05 | 22.14 ± 1.34 | 86.42 ± 0.81 |
| Cry1Ah 500 μg/ml diet | 0.11 | 0.59 ± 0.03 | 20.02 ± 0.91 | 23.51 ± 1.17 | 85.78 ± 0.85 |
| Cry2Ab 500 μg/ml diet | 0.13 ± 0.01 | 0.61 ± 0.02* | 21.19 ± 0.77 | 25.03 ± 1.02 | 85.34 ± 1.08 |
Data are means ± SE. Each toxin treatment was compared to the control. An asterisk denotes a significant difference between a toxin treatment and the control. Thirty-five larvae were tested for each treatment.
aANCOVA followed by LSD test.
Discussion
Non-target risk assessment for transgenic crops should be case specific, depending on the plant, transgene, and intended release environment4. In the present study, we focused on the novel insecticidal protein Cry1Ah and a toxin with prospects for commercial release in China, Cry2Ab. The concentrations of Cry1Ah and Cry2Ab used in our toxicity bioassays were at least 10 times higher than that measured in Bt cotton pollen11. This high concentration represents a worst-case exposure scenario and could increase the certainty of the hazard assessment4. The bioassay results revealed that neither of the Cry proteins affected the growth or developmental characteristics of the two tested insects. Moreover, the expression of the detoxification-related genes of the two tested insects was not significantly altered in either of the Cry proteins treatments. Our results indicate that neither Cry protein has a favourable effect on the expression of genes that are associated with the amino acid metabolism of A. gossypii and the nutrition utilization of P. japonica.
A number of studies aimed to quantify the Cry protein concentration in aphids that have fed Bt-transgenic plants. In the majority of studies, Cry proteins were either absent or detected at very low levels that might have been due to contamination41. However, Burgio et al. repeated experiments with a pre-flowering stage of Cry1Ac-expressing oilseed rape in a growth chamber and again detected Cry1Ac protein uptake by Myzus persicae42. These results confirm the findings of their previous study and highlight the potential for Cry protein uptake by aphids42. In our previous study, a small fraction of Cry1Ac proteins were detected in A. gossypii feeding on Bt cotton cultivar10. Because the aphids were previously checked under a microscope, it is unlikely that the samples were contaminated10. Moreover, Cry1Ac and Cry3Aa bind to the aphid gut epithelium and exhibited low toxicity against the pea aphid Acyrthosiphon pisum43. Thus, we believe that Cry proteins may potentially affect aphids. In present study, the results suggest that Cry1Ah and Cry2Ab have no impact on the performance of A. gossypii. Similarly, Bt (Cry1Ac) cotton did not have any influence on the performance of A. gossypii27. In contrast, A. gossypii on Bt (Cry1A) + CpTI cotton exhibited reduced survival rates and an earlier occurrence of peak daily mortality in the first generation20.
In a previous study, we evaluated the effects of Bt (Cry1Ac + Cry2Ab) cotton on P. japonica through its prey A. gossypii. Development was delayed, and the pre-oviposition period was significantly longer when P. japonica was fed A. gossypii reared on Bt cotton10. Poor prey quality may account for the negative effects because other studies have indicated that plant allelochemicals can affect the nutritional suitability of herbivores, with potential effects on the performance of predators at the third trophic level of the food chain44. In the present study, a 2 M sucrose solution was used as carrier to deliver Cry proteins to P. japonica because this method worked in previous studies28,45, and this technique may avoid the problems discussed above. Our data revealed no distinct differences in any of the P. japonica life-table parameters between the Cry protein treatments and the control. The results are consistent with those of numerous other studies that were conducted to assess the effect of Cry proteins on P. japonica. The life-table parameters of P. japonica were not affected when fed a rapeseed pollen-based diet containing purified Cry1C or Cry2A at concentrations that were >10-fold higher than in pollen18. Cry proteins Cry1Ab, Cry1Ac, and Cry1F have no direct toxicity to P. japonica46. A tritrophic study also confirmed that the Cry1Ac protein is unlikely to have detrimental effects on this representative species24.
To date, little is known about the detoxification response of non-target insects to Bt proteins. In our study, five detoxification-related genes of A. gossypii were obtained from the transcriptome data47, and eight genes that are involved in the P. japonica detoxification response were obtained from Tang et al.48. Tang et al. analysed the transcriptome of an insecticide-resistant P. japonica strain and identified potential candidate genes for conferring insecticide resistance, including P450s, GSTs and ESTs48. Previous studies have suggested that CYP6s are associated with insecticide resistance49,50. For example, the overexpression of CYP6BQ9 in Tribolium castaneum brain tissue was partly responsible for the deltamethrin resistance49, and the overexpression of CYP6G1 in Drosophila melanogaster caused resistance to dithiothreitol (DDT) and imidacloprid50. CYP340s with a relatively high expression in the midgut probably contribute to the detoxification of insecticides or plant toxins in P. xylostella51. CYP9s also have been indicated in relation to the response to plant allelochemicals and xenobiotics52. In our study, CYP6A2, CYP6A13, and CYP6A14 of A. gossypii and CYP345B1, CYP4Q2, CYP6BQ13, and CYP9F2 of P. japonica did not significantly respond to Cry1Ah and Cry2Ab. Similar results were noted for CYP6AY1 and CYP4CE1 of N. lugens reared on transgenic rice33. Another study found that the expression of CYP6AE14, CYP6B2 and CYP9A12 was suppressed in H. armigera larvae that fed the Cry1Ab toxin, which might be related to the low susceptibility of the species to Cry1Ab toxin53.
Similarly to P450s, GSTs and ESTs can function broadly in xenobiotic detoxification31,32. The delta GST classes were uniquely identified in insects and have been implicated in insecticide resistance31. CarE is involved in organophosphorus insecticide resistance and the metabolism of xenobiotic compounds in numerous insect species54. Our data revealed that GSTd1 and CarE of A. gossypii and GST, microsomal GST, juvenile hormone esterase and alpha-esterase of P. japonica were not significantly regulated by Cry1Ah and Cry2Ab. Likewise, GST did not exhibit any significant difference when N. lugens was reared on transgenic rice33. In total, our results suggest that the Cry1Ah and Cry2Ab proteins have no distinct effects on the detoxification-related gene regulation of A. gossypii and P. japonica.
Our results demonstrate that the genes of A. gossypii that are relevant to the utilization of essential amino acids (CAT2 and Henna) and the synthesis of nonessential amino acids (GS2 and PSAT) as well as a gene that is involved in the catabolism of a nonessential amino acid (GCVH) did not distinctly respond to the Cry1Ah and Cry2Ab proteins. Elevated CO2 up-regulated the expression of genes that are relevant to amino acid metabolism in the aphid A. pisum, indicating that pea aphids manipulate their amino acid metabolism to favour the population growth of the aphid under elevated CO255. However, similar results were not observed in our study, indicating that the amino acid metabolism of A. gossypii was not altered by either of the Cry proteins.
Nutritional ecology is central to proper interpretations of life history phenomena (e.g., manner of feeding, habitat selection, defence, and reproduction), both in ecological and evolutionary time37. Many studies of nutritional ecology have focused on target pests38,39,40. When H. armigera were fed transgenic Bt (Cry1Ac) cotton, the RGR, ECI and ECD were significantly reduced38. This is not surprising, given that H. armigera is sensitive to Cry1Ac. Similarly, the Bt (Cry1Ac) cotton significantly reduced the relative growth, consumption, metabolic rates and other nutritional indices of S. frugiperda39. In contrast, no significant difference was detected for any nutritional indices of O. furnacalis and H. armigera when fed phytase transgenic maize40. In our study, the RGR, ECI, ECD and EAD indices of P. japonica were not significantly altered in the Bt treatment. However, the RCR of the Cry1Ah treatment was reduced by 7%, and a marginally significant difference (P = 0.038) was observed compared with the control. We believe that this decrease is minor and is unlikely to affect this species in the field given that the Bt concentration that was used in our study is considerably higher than that noted under field conditions.
In summary, the current study suggests that Cry1Ah and Cry2Ab proteins have no adverse or beneficial effect on the cotton aphid A. gossypii or the ladybeetle P. japonica. Because these two Bt toxins have not been promoted in China, the present study is valuable for decision-making in the commercialization of new Bt varieties and in the establishment of appropriate integrated pest management strategies in the field.
Methods
Insects
Ladybeetles P. japonica were collected in April 2014, and cotton aphids A. gossypii were collected in July 2013 at the experimental field station of the Institute of Cotton Research, Chinese Academy of Agricultural Sciences (CAAS). P. japonica was maintained on pea aphids Acyrthosiphon pisum over 4 generations, and a colony of A. gossypii was maintained on a non-transgenic cotton cultivar. Both insects were reared at 25 ± 1 °C and 75 ± 5% relative humidity (RH) under a16:8 h light:dark (L:D) cycle.
Insecticidal compounds
The insecticidal compounds that were used in this study include the protease inhibitor E-64 [N-[N-(L-3-trans-carboxyoxirane-2-carbonyl)-L-leucyl]-agmatine)] and the Bt proteins Cry1Ah and Cry2Ab. E-64 was purchased from Sigma-Aldrich (St. Louis, MO, USA). Activated Cry1Ah and Cry2Ab proteins were supplied by the Biotechnology Research Laboratory, Institute of Plant Protection, Chinese Academy of Agricultural Sciences. The Cry1Ah and Cry2Ab protoxins from Bacillus thuringiensis had been expressed as single-gene products in Escherichia coli. The E. coli-expressed protoxin inclusion bodies then were dissolved and trypsinized. Subsequently they were isolated and purified by ion exchange HPLC followed by the desalting and lyophilization of the pure fractions. A bioassay showed that the LC50 (concentration resulting 50% mortality) of our batch of Cry1Ah and Cry2Ab proteins were 7 μg/ml and 10 μg/ml diet when neonates of H. armigera were fed a Bt-containing artificial diet for one week.
Experiments with A. gossypii
Toxicity of Bt proteins to A. gossypii
Membrane feeding assays43 were used to assess the toxicity of Cry1Ah and Cry2Ab against A. gossypii. The artificial diet was the diet A5 reported by Febvay et al.56, with a sucrose content changed to 25%. Glass tubes (cylindrical glass tubes, 25 mm diameter, and 50 mm height) with openings at both ends were used as feeding containers. One end of the glass tube was covered with a stretched Parafilm sachet containing 200 μl of artificial diet. Aphids were introduced into the tube from the other end, which was then covered with a fine mesh.
Fifteen 2nd instar (1st instar aphids were not able to penetrate through the stretched Parafilm) aphids that were grown on non-transgenic cotton were transferred to the artificial diet device, with five replicates for each treatment. The aphids were fed the artificial diet containing (1) 500 μg/ml Cry1Ah; (2) 500 μg/ml Cry2Ab; (3) 600 μg/ml E-64 (a highly specific cysteine proteinase inhibitor57; positive control); or (4) no added toxin (negative control). The diet was replaced every two days, and assays were conducted for a period of 7 days. The assays were conducted in a growth chamber at 24 ± 1 °C and 75 ± 5% RH under a 16:8 L:D cycle. Aphids were observed twice per day (9:00 am and 9:00 pm), and their development and mortality were recorded. When the adults emerged, ten randomly selected aphids were weighed (within 12 h) for each replicate.
Expression of genes that are associated with detoxification responses and amino acid metabolism
A. gossypii were reared on artificial diets that did or did not include Bt proteins as described above. When the adults emerged, aphids were frozen in liquid nitrogen and stored at –80 °C for further use. The entire bodies of three pools of ten individuals from each sample were used for total RNA extraction. Total RNA was extracted by the SV Total Isolation System (Promega, Madison, WI, USA) following the manufacturer’s instructions. The concentration and quality of total RNA were determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, USA). The first-strand cDNA of each sample was synthesized from 1 μg of total RNA using a PrimeScript RT reagent kit with gDNA eraser (Perfect Real Time) (TaKaRa Biotechnology (Dalian) Co., Ltd.).
Gene functions were determined by searching BLASTX and Kyoto Encyclopedia of Genes and Genomes with the cotton aphid transcriptome data47. To confirm the transcriptome assemblies, the PCR products of all of the putative genes were sequenced. Quantitative real-time PCR (qPCR) was performed using the Mastercycler ep realplex system (Eppendorf, Hamburg, Germany). Gene-specific primers (Supplementary Table S1) were designed using Beacon Designer 7.6 and synthesized by GENEWIZ Co. Ltd. (Beijing, China). Dimethyladenosine transferase (GenBank: KF018923) and peptidyl-prolyl cis-trans isomerase (GenBank: KF018924) were used as endogenous controls47. The reaction was performed as follows: 2 min at 95 °C followed by 40 cycles at 95 °C for 15 s, 58 °C for 30 s and 72 °C for 30 s. A melting curve was used to detect a single gene-specific peak and the absence of primer dimer peaks. GoTaq qPCR Master Mix (Promega, Madison, WI, USA) was used to measure the mRNA levels according to the manufacturer’s instructions. A fivefold dilution series was used to construct a relative standard curve to determine the PCR efficiencies and for quantification analysis. Each reaction was performed in triplicate (technical repeat) with three independent biological replicates. The transcript levels were calculated by the comparative 2−△△CT method58.
Experiments for P. japonica
Toxicity of Bt proteins to P. japonica larvae
A bioassay was conducted to test the toxicity of Cry1Ah and Cry2Ab to P. japonica as described by Alvarez-Alfageme et al.45. On the first day of each instar, P. japonica larvae were individually fed with a 2 M sucrose solution containing (1) 500 μg/ml Cry1Ah; (2) 500 μg/ml Cry2Ab; (3) 400 μg/ml E-64 (positive control); or (4) no added toxin (negative control). After 24 h, larvae were individually transferred to a clean 7-ml centrifuge tube that was sealed with cotton gauze and subsequently fed ad libitum with pea aphids until the next moult. Thirty-five individual P. japonica larvae were tested for each treatment. Larval development and mortality were recorded twice per day (9:00 am and 9:00 pm), and emerging adults were sexed and weighed (within 12 h).
Expression of genes that are associated with detoxification responses
Newly hatched P. japonica larvae were reared on A. pisum until the 3rd instar. On the first day of the 4th instar, P. japonica larvae were individually fed the 2 M sucrose solution containing Cry1Ah, Cry2Ab or no added toxins. After 24 h, larvae were frozen in liquid nitrogen and stored at –80 °C for further use. The entire bodies of three pools of five individuals from each sample were used for total RNA extraction. RNA extraction, first-strand cDNA synthesis and qPCR were performed as described above. Gene-specific primers (Supplementary Table S2) were from Tang et al.48, and β-actin was used as a reference gene.
Nutrition utilization of P. japonica
Many previous studies have used P. japonica artificial diets containing saccharides, insect tissue and viscera of livestock59,60. An artificial diet that was optimized previously60 was used in the present study. This artificial diet includes fresh pork liver + pure honey + sucrose (regulated weight ratio of 5:1:1, respectively) and 0.3% (to the total weight of the diet) olive oil. Fresh pork liver was mashed in a blender with pure honey, sucrose and olive oil in the described proportions. Cry1Ah and Cry2Ab were mixed with the artificial diet respectively at final concentrations of 500 μg/ml. An artificial diet with no Bt protein was used as a control. The artificial diet of each treatment was kept in a plastic jar that was covered by Parafilm and stored at 4 °C. Every other day, another batch of pork-liver-based artificial diets (with or without Cry proteins) was prepared to keep the diets fresh and to avoid the degradation of the Cry proteins. The methods of measuring the stability of the Cry2Ab protein in the artificial diets are provided in the Supplementary Information.
The 4th instar larvae (within 12 h) were tested in this assay. Before this assay, samples of artificial diets (n = 50) and 4th instar larvae (n = 50) were oven-dried at 80 °C for 72 h to calculate the proportion of dry matter and water content36,38. Every day, approximately 60 μl of artificial diet was placed on a square of Parafilm (length of a side = 20 mm) and offered to each larva, which was reared in a 7-ml centrifuge tube sealed with cotton gauze. After 24 h, the remaining artificial diets and the frass that was produced by each larva were collected and oven-dried as above. The development time of each 4th instar larva was calculated from the beginning of 4th instar to pre-pupa. The pre-pupa were oven-dried as above, and the dry weights were determined. Thirty-five larvae were reared individually for each treatment.
The nutrition utilization indices, including the RGR, RCR, ECI, ECD, and EAD, were determined gravimetrically following the methods of Waldbauer (1968)36 and Scriber & Slansky (1981)37. The amount of artificial diet (mg) that was ingested, the amount of frass that was produced and the larval body weight were calculated as dry weights.
Data analyses
The details of the data analyses are provided in the Supplementary Information.
Additional Information
How to cite this article: Zhao, Y. et al. Bt proteins Cry1Ah and Cry2Ab do not affect cotton aphid Aphis gossypii and ladybeetle Propylea japonica. Sci. Rep. 6, 20368; doi: 10.1038/srep20368 (2016).
Supplementary Material
Acknowledgments
We acknowledge Fen Zhu and Peng Han for their critical comments on an earlier version of this manuscript. This research was supported by the National Special Transgenic Project of China (2014ZX08011-002).
Footnotes
Author Contributions C.L., J.C. and S.Z. conceived and designed the experiments; Y.Z. performed the experiments; Y.Z., J.L., C.W., L.L. and X.W. analyzed the data and prepared the figures and tables; Y.Z. wrote the manuscript. All authors reviewed the manuscript.
References
- Wu K. M., Lu Y. H., Feng H. Q., Jiang Y. Y. & Zhao J. Z. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321, 1676–1678 (2008). [DOI] [PubMed] [Google Scholar]
- Hutchison W. et al. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science 330, 222–225 (2010). [DOI] [PubMed] [Google Scholar]
- Lu Y., Wu K., Jiang Y., Guo Y. & Desneux N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 487, 362–365 (2012). [DOI] [PubMed] [Google Scholar]
- Romeis J. et al. Assessment of risk of insect-resistant transgenic crops to nontarget arthropods. Nat. Biotechnol. 26, 203–208 (2008). [DOI] [PubMed] [Google Scholar]
- Desneux N. & Bernal J. S. Genetically modified crops deserve greater ecotoxicological scrutiny. Ecotoxicology 19, 1642–1644 (2010). [DOI] [PubMed] [Google Scholar]
- Brévault T. et al. Potential shortfall of pyramided transgenic cotton for insect resistance management. Proc. Natl. Acad. Sci. USA 110, 5806–5811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J. et al. Cloning and characterization of a novel Cry1A toxin from Bacillus thuringiensis with high toxicity to the Asian corn borer and other lepidopteran insects. FEMS Microbiol. Lett. 280, 95–101 (2008). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Ubi1 intron-mediated enhancement of the expression of Bt cry1Ah gene in transgenic maize (Zea mays L.). Chinese Sci. Bull. 53, 3185–3190 (2008). [Google Scholar]
- Tabashnik B. E., Brévault T. & Carrière Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat. Biotechnol. 31, 510–521 (2013). [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. Bt cotton expressing Cry1Ac/Cry2Ab or Cry1Ac/epsps does not harm the predator Propylaea japonica through its prey Aphis gossypii. Agr. Ecosyst. Environ. 179, 163–167 (2013). [Google Scholar]
- Niu L. et al. Impact of single and stacked insect-resistant Bt-cotton on the honey bee and silkworm. PLoS One 8, e72988 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasupramaniam S. et al. Toxicity and characterization of cotton expressing Bacillus thuringiensis Cry1Ac and Cry2Ab2 proteins for control of lepidopteran pests. J. Econ. Entomol. 101, 546–554 (2008). [DOI] [PubMed] [Google Scholar]
- Dai P. L. et al. Field assessment of Bt cry1Ah corn pollen on the survival, development and behavior of Apis mellifera ligustica. Ecotox. Environ. Safe. 79, 232–237 (2012). [DOI] [PubMed] [Google Scholar]
- Dai P. L. et al. The effects of Bt Cry1Ah toxin on worker honeybees (Apis mellifera ligustica and Apis cerana cerana). Apidologie 43, 384–391 (2012). [Google Scholar]
- Wang Y. Y. et al. Toxicological, biochemical, and histopathological analyses demonstrating that Cry1C and Cry2A are not toxic to larvae of the honeybee, Apis mellifera. J. Agric. Food Chem. 63, 6126–6132 (2015). [DOI] [PubMed] [Google Scholar]
- Rodrigo-Simón A. et al. Lack of detrimental effects of Bacillus thuringiensis Cry toxins on the insect predator Chrysoperla carnea: a toxicological, histopathological, and biochemical analysis. Appl. Environ. Microb. 72, 1595–1603 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Romeis J., Wang P., Peng Y. & Shelton A. M. A comprehensive assessment of the effects of Bt cotton on Coleomegilla maculata demonstrates no detrimental effects by Cry1Ac and Cry2Ab. PloS One 6, e22185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Consumption of Bt rice pollen containing Cry1C or Cry2A does not pose a risk to Propylea japonica (Thunberg) (Coleoptera: Coccinellidae). Sci. Rep. 5, 7679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletto J. et al. Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Mol. Ecol. 18, 2198–2212 (2009). [DOI] [PubMed] [Google Scholar]
- Liu X. D., Zhai B. P., Zhang X. X. & Zong J. M. Impact of transgenic cotton plants on a non-target pest, Aphis gossypii Glover. Ecol. Entomol. 30, 307–315 (2005). [Google Scholar]
- Bai Y., Jiang M. & Cheng J. Effects of transgenic cry1Ab rice pollen on fitness of Propylea japonica (Thunberg). J. Pest Sci. 78, 123–128 (2005). [Google Scholar]
- Zhang S. Z., Li J. J., Shan H. W., Zhang F. & Liu T. X. Influence of five aphid species on development and reproduction of Propylaea japonica (Coleoptera: Coccinellidae). Biol. Control 62, 135–139 (2012). [Google Scholar]
- Han P., Niu C. Y. & Desneux N. Identification of top-down forces regulating cotton aphid population growth in transgenic Bt cotton in central China. PloS One 9, e102980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Y., Li D. M., Cui J. & Xie B. Y. Effects of Bt-toxin Cry1Ac on Propylaea japonica Thunberg (Col., Coccinellidae) by feeding on Bt-treated Bt-resistant Helicoverpa armigera (Hübner)(Lep., Noctuidae) larvae. J. Appl. Entomol. 130, 206–212 (2006). [Google Scholar]
- Carstens K. et al. Surrogate species selection for assessing potential adverse environmental impacts of genetically engineered insect-resistant plants on non-target organisms. GM Crops Food 5, 11–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis J. et al. Deriving criteria to select arthropod species for laboratory tests to assess the ecological risks from cultivating arthropod-resistant genetically engineered crops. Chemosphere 90, 901–909 (2013). [DOI] [PubMed] [Google Scholar]
- Lawo N. C., Wäckers F. L. & Romeis J. Indian Bt cotton varieties do not affect the performance of cotton aphids. PLoS One 4, e4804 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Alfageme F., Bigler F. & Romeis J. Laboratory toxicity studies demonstrate no adverse effects of Cry1Ab and Cry3Bb1 to larvae of Adalia bipunctata (Coleoptera: Coccinellidae): the importance of study design. Transgenic Res. 20, 467–479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y. Y. et al. De novo intestine-specific transcriptome of the brown planthopper Nilaparvata lugens revealed potential functions in digestion, detoxification and immune response. Genomics 99, 256–264 (2012). [DOI] [PubMed] [Google Scholar]
- Feyereisen R. Insect P450 enzymes. Annu. Rev. Entomol. 44, 507–533 (1999). [DOI] [PubMed] [Google Scholar]
- Enayati A. A., Ranson H. & Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14, 3–8 (2005). [DOI] [PubMed] [Google Scholar]
- Ramsey J. S. et al. Comparative analysis of detoxification enzymes in Acyrthosiphon pisum and Myzus persicae. Insect Mol. Biol. 19, 155–164 (2010). [DOI] [PubMed] [Google Scholar]
- Mannakkara A., Niu L., Ma W. & Lei C. Zero effect of Bt rice on expression of genes coding for digestion, detoxification and immune responses and developmental performances of brown planthopper Nilaparvata lugens (Stål). J. Insect Physiol. 59, 985–993 (2013). [DOI] [PubMed] [Google Scholar]
- Ramirez-Romero R., Desneux N., Chaufaux J. & Kaiser L. Bt-maize effects on biological parameters of the non-target aphid Sitobion avenae (Homoptera: Aphididae) and Cry1Ab toxin detection. Pestic. Biochem. Physiol. 91, 110–115 (2008). [Google Scholar]
- Nakabachi A. et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc. Natl. Acad. Sci. USA 102, 5477–5482 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldbauer G. The consumption and utilization of food by insects. Adv. Inesct Physiol. 5, 229–288 (1968). [Google Scholar]
- Scriber J. & Slansky F. The nutritional ecology of immature insects. Annu. Rev. Entomol. 26, 183–211 (1981). [Google Scholar]
- Chen F., Wu G., Ge F., Parajulee M. N. & Shrestha R. B. Effects of elevated CO2 and transgenic Bt cotton on plant chemistry, performance, and feeding of an insect herbivore, the cotton bollworm. Entomol. Exp. Appl. 115, 341–350 (2005). [Google Scholar]
- de Sousa Ramalho F. et al. Feeding of fall armyworm, Spodoptera frugiperda, on Bt transgenic cotton and its isoline. Entomol. Exp. Appl. 139, 207–214 (2011). [Google Scholar]
- Zhang Y., Liu C., Li Y. & Wu K. Phytase transgenic maize does not affect the development and nutrition utilization of Ostrinia furnacalis and Helicoverpa armigera. Environ. Entomol. 39, 1051–1057 (2010). [DOI] [PubMed] [Google Scholar]
- Romeis J. & Meissle M. Non-target risk assessment of Bt crops – Cry protein uptake by aphids. J. Appl. Entomol. 135, 1–6 (2011). [Google Scholar]
- Burgio G. et al. Bt-toxin uptake by the non-target herbivore, Myzus persicae (Hemiptera: Aphididae), feeding on transgenic oilseed rape in laboratory conditions. B. Entomol. Res. 101, 241–247 (2011). [DOI] [PubMed] [Google Scholar]
- Li H., Chougule N. P. & Bonning B. C. Interaction of the Bacillus thuringiensis delta endotoxins Cry1Ac and Cry3Aa with the gut of the pea aphid, Acyrthosiphon pisum (Harris). J. Invertebr. Pathol. 107, 69–78 (2011). [DOI] [PubMed] [Google Scholar]
- Du L., Ge F., Zhu S. & Parajulee M. N. Effect of cotton cultivar on development and reproduction of Aphis gossypii (Homoptera: Aphididae) and its predator Propylaea japonica (Coleoptera: Coccinellidae). J. Econ. Entomol. 97, 1278–1283 (2004). [DOI] [PubMed] [Google Scholar]
- Álvarez-Alfageme F., Palinkas Z., Bigler F. & Romeis J. Development of an early-tier laboratory bioassay for assessing the impact of orally-active insecticidal compounds on larvae of Coccinella septempunctata (Coleoptera: Coccinellidae). Environ. Entomol. 41, 1687–93 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Use of a pollen-based diet to expose the ladybird beetle Propylea japonica to insecticidal proteins. PloS One 9, e85395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Q. et al. Ecological adaption analysis of the cotton aphid (Aphis gossypii) in different phenotypes by transcriptome comparison. PloS One 8, e83180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L. D. et al. De novo sequencing-based transcriptome and digital gene expression analysis reveals insecticide resistance-relevant genes in Propylaea japonica (Thunberg) (Coleoptea: Coccinellidae). PLoS One 9, e100946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F. et al. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. USA 107, 8557–8562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn P., Boundy S., Yen J. & Pittendrigh B. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet. Genomics 266, 556–563 (2001). [DOI] [PubMed] [Google Scholar]
- Yu L. et al. Characterization and expression of the cytochrome P450 gene family in diamondback moth, Plutella xylostella (L.). Sci. Rep. 5, 8952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. L., Snyder M. J., Koener J. F. & Feyereisen R. Inducible P450s of the CYP9 family from larval Manduca sexta midgut. Insect Biochem. Molec. Biol. 30, 559–568 (2000). [DOI] [PubMed] [Google Scholar]
- Muñoz P., López C., Moralejo M., Pérez-Hedo M. & Eizaguirre M. Response of last instar Helicoverpa armígera larvae to Bt toxin ingestion: changes in the development and in the CYP6AE14, CYP6B2 and CYP9A12 gene expression. PloS One 9, e99229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y. H., Yu X. R., Shang Q. L., Shi X. Y. & Gao X. W. Oral delivery mediated RNA interference of a carboxylesterase gene results in reduced resistance to organophosphorus insecticides in the cotton aphid, Aphis gossypii Glover. PloS One 9, e102823 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. et al. Pea aphid promotes amino acid metabolism both in Medicago truncatula and bacteriocytes to favor aphid population growth under elevated CO2. Global Change Biol. 19, 3210–3223 (2013). [DOI] [PubMed] [Google Scholar]
- Febvay G., Delobel B. & Rahbé Y. Influence of the amino acid balance on the improvement of an artificial diet for a biotype of Acyrthosiphon pisum (Homoptera: Aphididae). Can. J. Zool. 66, 2449–2453 (1988). [Google Scholar]
- Deraison C. et al. Cloning and characterization of a gut-specific cathepsin L from the aphid Aphis gossypii. Insect Mol. Biol. 13, 165–177 (2004). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Li K., Zhang T., Zhang L., Wang B. & Wang Q. Effects of artificial diets on female Propylea japonica’s feeding and body composition. J. East China Norm. Univ. Nat. Science 6, 97–105 (2007). [Google Scholar]
- Zhang L. L., Li K., Zhang T. S. & Wang B. The effect of the artificial diets on Propylea japonica growth and fecundity. Chinese Bull. Entomol. 44, 871–876 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.