Abstract
Di19 (drought-induced protein 19) family is a novel type of Cys2/His2 zinc-finger proteins. In this study, we demonstrated that cotton Di19-1 and Di19-2 (GhDi19-1/-2) proteins could be phosphorylated in vitro by the calcium-dependent protein kinase (CDPK). Mutation of Ser to Ala in N-terminus of GhDi19-1/-2 led to the altered subcellular localization of the two proteins, but the constitutively activated form (Ser was mutated to Asp) of GhDi19-1/-2 still showed the nuclear localization. GhDi19-1/-2 overexpression transgenic Arabidopsis seedlings displayed the hypersensitivity to high salinity and abscisic acid (ABA). However, Ser site-mutated GhDi19-1(S116A) and GhDi19-2(S114A), and Ser and Thr double sites-mutated GhDi19-1(S/T-A/A) and GhDi19-2(S/T-A/A) transgenic Arabidopsis did not show the salt- and ABA-hypersensitive phenotypes. In contrast, overexpression of Thr site-mutated GhDi19-1(T114A) and GhDi19-2(T112A) in Arabidopsis still resulted in salt- and ABA-hypersensitivity phenotypes, like GhDi19-1/-2 transgenic lines. Overexpression of GhDi19-1/-2 and their constitutively activated forms in Atcpk11 background could recover the salt- and ABA-insensitive phenotype of the mutant. Thus, our results demonstrated that Ser phosphorylation (not Thr phosphorylation) is crucial for functionally activating GhDi19-1/-2 in response to salt stress and ABA signaling during early plant development, and GhDi19-1/-2 proteins may be downstream targets of CDPKs in ABA signal pathway.
Drought, high salinity and low temperature are the most common abiotic stresses that limit distribution of plants and affect crop productivity and quality1,2. Plant adaptation to these stresses is dependent on the activation of cascades of molecular networks involving in stress perception, signal transduction and expression of specific stress-related genes3,4. Abscisic acid (ABA) as an important phytohormone is required for plant adaptation to environmental stress by affecting some physiological processes in cells during plant growth and development5. Several signal molecules, such as calcium ion (Ca2+), reactive oxygen species (ROS) and protein kinases [including mitogen-activated protein kinase (MAPK), calcium-dependent protein kinase (CDPK), and calcium/calmodulin-dependent protein kinase (CCaMK)] play important roles in ABA signal transduction for plant response to abiotic stresses6,7,8,9,10. For instance, overexpression of AtCPK4 or AtCPK11 in Arabidopsis enhanced plant ABA- and salt-sensitivity during seed germination and seedling growth, but loss-of-function mutations of CPK4 and CPK11 resulted in plant ABA- and salt-insensitive phenotype11.
Transcriptional modulation is thought to be one of the most important ways for plants responding and adapting to stress conditions, and a number of genes are induced or repressed in plants under abiotic stresses12,13,14,15. Previous studies revealed that plant transcription factors (TFs) (such as bZIP, HD-ZIP, NAC, MYB, MYC and AP2/ERF) are involved in plant stress response3,16. These TFs regulate expression of the stress-related genes by specifically binding to the cis-elements in the target promoters17.
Cys2/His2 (C2H2)-type zinc-finger proteins (ZFPs), also called classical TFIII-types zinc-finger proteins, represent a large family of eukaryotic transcription factors. The C2H2-type zinc finger domain is one of the well-characterized DNA-binding motifs involved in protein-DNA, protein-RNA and protein-protein interactions in plants18,19,20. This domain consists of approximate 30 amino acid residues with the common sequence CX2–4CX3FX5LX2HX3–5H, and a zinc ion is tetrahedrally coordinated by two cysteines and two histidines in order to maintain its stability20. In Arabidopsis and rice, 176 and 189 zinc-finger proteins that contain one or more zinc-finger motifs have been identified, respectively21,22. Some zinc-finger proteins play important roles in plant response to abiotic stresses. For example, Zat10 (STZ) and Zat12 promote plant tolerance to drought, salinity and oxidative stresses22,23,24. ThZF1, a C2H2-type TF from salt cress (Thellungiella halophila), is involved in drought and salt stresses25. Similarly, several C2H2-type ZFPs are related to rice response to drought and salinity stresses4,26,27. Loss of function of a rice DST (drought and salt tolerance) protein leads to the enhanced plant drought- and salt-tolerance28.
Di19 (drought-induced 19) proteins contain two unusual and evolutionarily conserved C2H2 zinc-finger domains29. These proteins may share a common or closely related biological function based on the conserved C2H2 zinc finger-like motif. Arabidopsis Di19 family comprises of seven hydrophilic protein members. AtDi19-1 and AtDi19-3 are rapidly induced by dehydration, while transcript amounts of AtDi19-2 and AtDi19-4 are increased in response to high-salinity stress. However, most of AtDi19 genes are not transcriptionally induced by ABA30. Recently, a study revealed that Arabidopsis Di19-1 as a transcription factor participates in plant response to drought stress by binding to the TACA(A/G)T element within the promoters of PR1 (pathogenesis-related 1), PR2 and PR5 genes31. Subsequently, we reported that AtDi19-3 as a transcriptional activator is involved in plant response to high salinity, drought, ABA and H2O232. In addition, Arabidopsis Di19 proteins could be phosphorylated by CPK11 and CPK3 in vitro in a Ca2+-dependent manner30,33. However, the function of most Di19 proteins in plants remains unknown as yet. Our previous study revealed that two cotton Di19 proteins (GhDi19-1 and GhDi19-2) are involved in plant response to salt stress and ABA signaling. Overexpression of GhDi19-1 and GhDi19-2 in Arabidopsis enhanced plant sensitivity to high salinity and exogenous ABA34. In this study, we further demonstrated that the serine (Ser) site in N-terminus of the GhDi19-1/-2 proteins is crucial for functionally activating GhDi19-1/-2 in response to salt stress and ABA signaling during early seedling development. The calcium-dependent protein kinase (CDPK) is able to phosphorylate these Di19 proteins at Ser116 of GhDi19-1 and at Ser114 of GhDi19-2.
Results
GhDi19-1/-2 is phosphorylated by calcium-dependent protein kinase (CDPK) in a Ca2+-dependent manner
Sequence analysis showed that both GhDi19-1 and GhDi19-2 contain Ser/Thr kinase phosphorylation sites in the conserved nuclear localization signal region next to the two zinc finger domains (GhDi19-1: Thr114 and Ser116; GhDi19-2: Thr112 and Ser114), and the predicted Ser phosphorylation is potentially stronger than the Thr phosphorylation (http://myhits.isb-sib.ch/cgi-bin/motif_scan). Then, we employed an in vitro kinase assay to determine whether GhDi19-1 and GhDi19-2 are phosphorylated by a kinase (such as AtCPK11). As shown in Fig. 1a, the S116 of GhDi19-1 was mutated to A116 [GhDi19-1(S116A)], and the S114 of GhDi19-2 was mutated to A114 [GhDi19-2(S114A)]. The in vitro kinase assay revealed that the recombinant AtCPK11 phosphorylated GhDi19-1 and GhDi19-2 in the presence of Ca2+. Additionally, weaker phosphorylation of GhDi19-1(S116A) was detected in the presence of Ca2+, and no phosphorylation of GhDi19-2(S114A) was found with or without Ca2+ (Fig. 1b). The above results indicated that GhDi19-1/-2 is phosphorylated in vitro by a kinase (such as AtCPK11) in a Ca2+-dependent manner, and the 116th Ser of GhDi19-1 or the 114th Ser of GhDi19-2 is required for the phosphorylation.
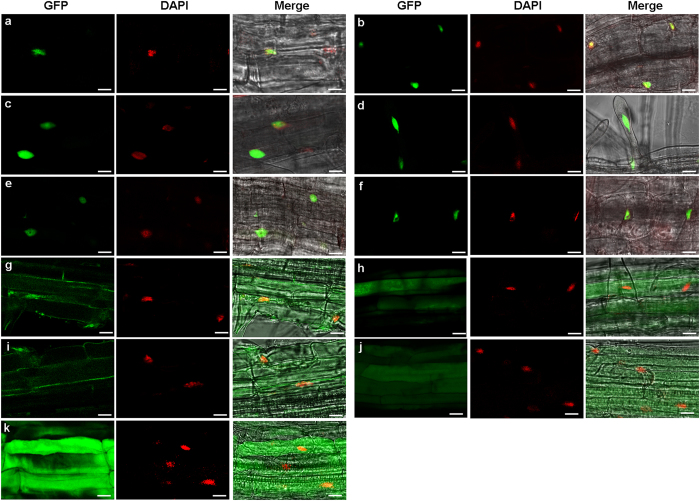
Figure 1. In vitro assay of GhDi19-1 and GhDi19-2 phosphorylation.
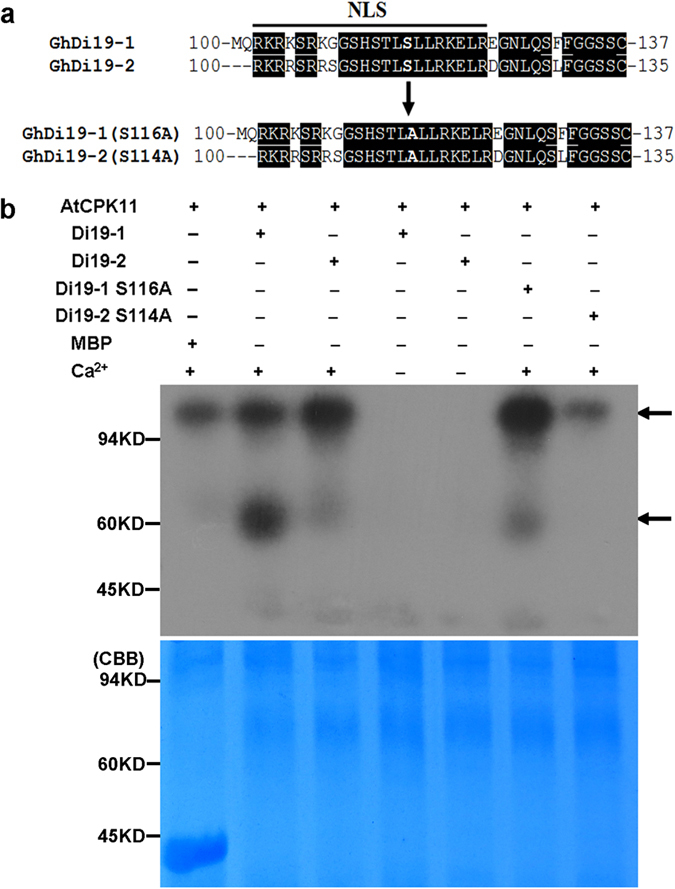
(a) The map of mutation of specific amino acids in the phosphorylation of GhDi19-1 and GhDi19-2. NLS, nuclear localization signal domain; S, serine (Ser); A, alanine (Ala). The arrowhead indicated the mutated amino acids. (b) AtCPK11 phosphorylates GhDi19-1 and GhDi19-2 in vitro. Top: Autoradiograph showing AtCPK11 autophosphorylation and GhDi19-1/-2 phosphorylation; Bottom: the same SDS-PAGE gel with Coomasie Brilliant Blue (CBB)–stained AtCPK11 and GhDi19-1/-2 proteins. GhDi19-1 and GhDi19-2 proteins were purified as a MBP-fusion and incubated with AtCPK11-MBP. A control reaction with the MBP protein was also performed. The upper arrowhead indicated autophosphorylation of AtCPK11-MBP, while the lower arrowhead indicated phosphorylation of GhDi19-1-MBP and GhDi19-2-MBP.
Ser site is crucial for normal subcellular localization of GhDi19-1 and GhDi19-2 proteins
Bioinformatics analysis predicted that both GhDi19-1 and GhDi19-2 contain a conserved nuclear localization signal (NLS) in its sequence. To confirm nuclear localization of the two GhDi19 proteins, the coding regions of the two genes were fused with an enhanced GFP (eGFP) reporter gene under the control of a cauliflower mosaic virus (CaMV) 35S promoter, respectively, and expressed constitutively in Arabidopsis. To determine whether Ser site and/or Thr site play an essential role in subcellular localization of GhDi19-1 and GhDi19-2 proteins, the S116 of GhDi19-1 was mutated to A116 [GhDi19-1(S116A)], the S114 of GhDi19-2 was mutated to A114 [GhDi19-2(S114A)], the T114 of GhDi19-1 was mutated to A114 [GhDi19-1(T114A)], the T112 of GhDi19-2 was mutated to A112 [GhDi19-2(T112A)], both the T114 and S116 of GhDi19-1 were mutated to A [GhDi19-1(S/T-A/A)], and both the T112 and S114 of GhDi19-2 were mutated to A [GhDi19-2(S/T-A/A)]. Furthermore, S116 was mutated to D116 in GhDi19-1 and S114 was mutated to D114 in GhDi19-2 for generating the constitutively activated GhDi19-1 and GhDi19-2 [named GhDi19-1(S116D) and GhDi19-2(S114D), respectively]. The coding sequences of the above mutated GhDi19s were also fused with an enhanced GFP (eGFP) reporter gene under the control of a cauliflower mosaic virus (CaMV) 35S promoter, respectively, and expressed constitutively in Arabidopsis. As shown in Fig. 2a,b, the GhDi19-1:eGFP and GhDi19-2:eGFP fusion proteins were mainly accumulated in the nuclei of root cells of the transgenic seedlings, demonstrating that GhDi19-1 and GhDi19-2 are two nuclear-localized proteins. Likewise, strong GFP fluorescence signals were detected in the nuclei of the transgenic root cells harboring the GhDi19-1(S116D):eGFP (Fig. 2c), GhDi19-2(S114D):eGFP (Fig. 2d), GhDi19-1(T114A):eGFP (Fig. 2e), and GhDi19-2(T112A):eGFP (Fig. 2f), respectively. On the contrary, GFP fluorescence was found in cytoplasm, cytomembrane and nuclei of root cells of the transgenic plants expressing Ser site-mutated GhDi19-1(S116A) (Fig. 2g) and GhDi19-2(S114A) (Fig. 2h), and Ser/Thr double sites-mutated GhDi19-1(S/T-A/A) (Fig. 2i) and GhDi19-2(S/T-A/A) (Fig. 2j), indicating that subcellular localization of both GhDi19-1 and GhDi19-2 proteins was altered as the Ser site (but not Thr site) was mutated to Ala in these proteins, while GFP fluorescence signals were observed in whole cells of the transgenic seedlings harboring only GFP vector (Fig. 2k). The above data implied that the Ser site (but not Thr site) is essential for normal subcellular localization of GhDi19-1 and GhDi19-2 proteins.
Figure 2. Subcellular localization of GhDi19-1/-2 and their mutated proteins in cells.
(a–f) GFP fluorescence signals were mainly detected in nuclei of the transgenic Arabidopsis root cells harboring the GhDi19-1:eGFP, GhDi19-2:eGFP, GhDi19-1(S116D):eGFP, GhDi19-2(S114D):eGFP, GhDi19-1(T114A):eGFP and GhDi19-2(T112A):eGFP, respectively. (a,b) Normal subcellular localization of GhDi19-1 (a) and GhDi19-2 (b). (c,d) Subcellular localization of the Ser site-mutated (constitutively activated) GhDi19-1(S116D) (c) and GhDi19-2(S114D) (d). (e,f) Subcellular localization of the Thr site-mutated GhDi19-1(T114A) (e) and GhDi19-2 (T112A) (f). (g–j) The altered subcellular localization of the Ser site-mutated GhDi19-1(S116A) (g), GhDi19-2(S114A) (h), the Ser and Thr double sites-mutated GhDi19-1(S/T-A/A) (i) and GhDi19-2(S/T-A/A) (j). GFP fluorescence signals were detected in cytoplasm, cytomembrane and nuclei of the transgenic Arabidopsis root cells. (k) pBI121-eGFP construct was used as positive control. (Left) Confocal microsopy of GFP fluorescence. (Midst) Nuclear DAPI staining of the same cell on the left. (Right) Left and midst superimposed over the bright-field image. Bars = 10 μm. S116D, the S116 of GhDi19-1 was mutated to D116; S114D, the S114 of GhDi19-2 was mutated to D114; T114A, the T114 of GhDi19-1 was mutated to A114; T112A, the T112 of GhDi19-2 was mutated to A112; S116A, the S116 of GhDi19-1 was mutated to A116; S114A, the S114 of GhDi19-2 was mutated to A114; S/T-A/A, the S116 and T114 of GhDi19-1 were mutated to A116 and A114, and the S114 and T112 of GhDi19-2 were mutated to A114 and A112, respectively. S, serine (Ser); A, alanine (Ala); T, threonine (Thr); D, aspartic acid (Asp).
Ser site is crucial for activating GhDi19-1 and GhDi19-2 proteins in response to salt stress and ABA signaling
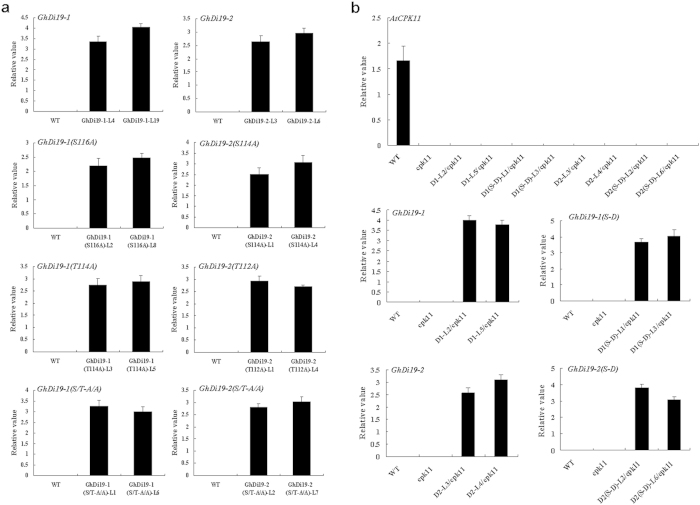
Our previous study indicated that overexpression of the GhDi19-1 and GhDi19-2 genes in Arabidopsis enhances plant sensitivity to salt stress in ABA-dependent manner34. To further investigate the mechanism of GhDi19-1 and GhDi19-2 involving in response to salt stress and ABA signaling, the S116 of GhDi19-1 was mutated to A116, and the S114 of GhDi19-2 was mutated to A114. The coding sequences of GhDi19-1(S116A) and GhDi19-2(S114A) were inserted into plant expression vector pBI121 driven by CaMV 35S promoter, and introduced into Arabidopsis, respectively. Homozygotes of T3 generation of GhDi19-1 overexpression transgenic lines (L4 and L19), GhDi19-1(S116A) overexpression transgenic lines (L2 and L8), GhDi19-2 overexpression transgenic lines (L3 and L6) and GhDi19-2(S114A) overexpression transgenic lines (L1 and L4) were analyzed by PCR to ensure the presence of the respective transgene and by quantitative RT-PCR to confirm transgene expression (Fig. 3a).
Figure 3. Quantitative RT-PCR analysis of expression of the related exogenous GhDi19-1/-2 genes in transgenic Arabidopsis.
(a) Quantitative RT-PCR analysis of expression of the GhDi19-1/-2 and their mutant genes in wild type, and the GhDi19-1/-2 and their site-mutated transgenic lines. 10-day-old seedlings were used for RNA extraction. WT, wild type; Di19-1-L4 and -L19, the GhDi19-1 transgenic line 4 and 19; GhDi19-1(S116A)-L2 and -L8, the site-mutated GhDi19-1(S116A) transgenic line 2 and 8 (S116A, the S116 of GhDi19-1 was mutated to A116). GhDi19-1(T114A)-L3 and -L5, the site-mutated GhDi19-1(T114A) transgenic line 3 and 5 (T114A, the T114 of GhDi19-1 was mutated to A114). GhDi19-1(S/T-A/A)-L1 and -L6, the double sites-mutated GhDi19-1(S/T-A/A) transgenic line 1 and 6 (S/T-A/A, the S116 and T114 of GhDi19-1 were mutated to A116 and A114, respectively). GhDi19-2-L3 and -L6, the GhDi19-2 transgenic line 3 and 6; GhDi19-2(S114A)-L1 and -L4, the site-mutated GhDi19-2(S114A) transgenic line 1 and 4 (S114A, the S114 of GhDi19-2 was mutated to A114); GhDi19-2(T112A)-L1 and -L4, the site-mutated GhDi19-2(T112A) transgenic line 1 and 4 (T112A, the T112 of GhDi19-2 was mutated to A112); GhDi19-2(S/T-A/A)-L2 and -L7, the double sites-mutated GhDi19-2(S/T-A/A) transgenic line 2 and 7 (S/T-A/A, the S114 and T112 of GhDi19-2 were mutated to A114 and A112, respectively). (b) Quantitative RT-PCR analysis of expression of the GhDi19-1/-2 and their mutant genes in wild type, Atcpk11 mutant, and GhDi19-1/2- and their mutated transgenic lines in Atcpk11 mutant. 10-day-old seedlings were used for RNA extraction. WT, wild type; cpk11, Atcpk11-2 mutant; D1-L2/cpk11 and D1-L5/cpk11, the GhDi19-1 overexpression transgenic line 2 and 5 in Atcpk11-2 mutant; D1(S-D)-L1/cpk11 and D1(S-D)-L3/cpk11, the site-mutated GhDi19-1(S-D) overexpression transgenic line 1 and 3 in Atcpk11-2 mutant; D2-L3/cpk11 and D2-L4/cpk11, the GhDi19-2 overexpression transgenic line 3 and 4 in Atcpk11-2 mutant; D2(S-D)-L2/cpk11 and D2(S-D)-L6/cpk11, the site-mutated GhDi19-2(S-D) overexpression transgenic line 2 and 6 in Atcpk11-2 mutant (S-D, the S116 of GhDi19-1 was mutated to D116 and the S114 of GhDi19-2 was mutated to D114). The assays were repeated three times along with three independent repetitions of the biological experiments. S, serine (Ser); A, alanine (Ala); T, threonine (Thr); D, aspartic acid (Asp).
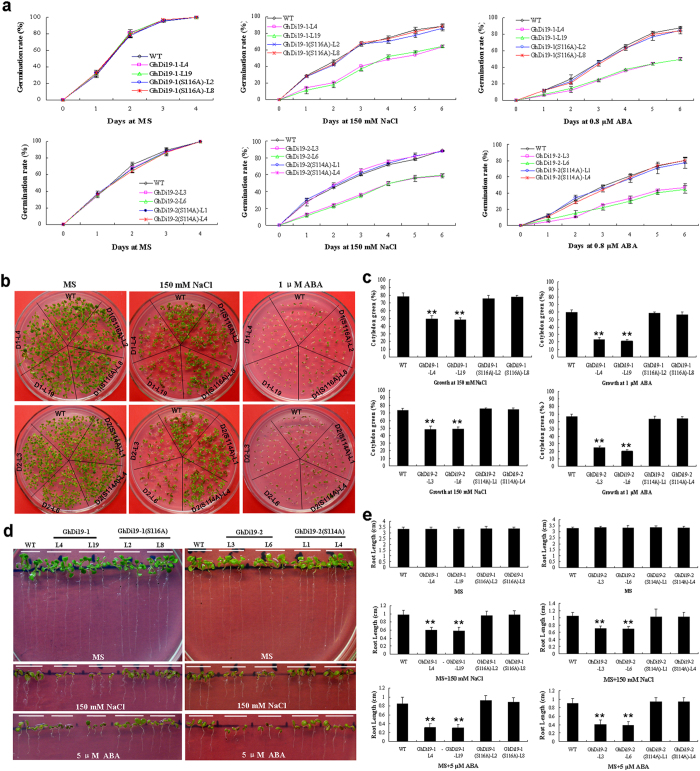
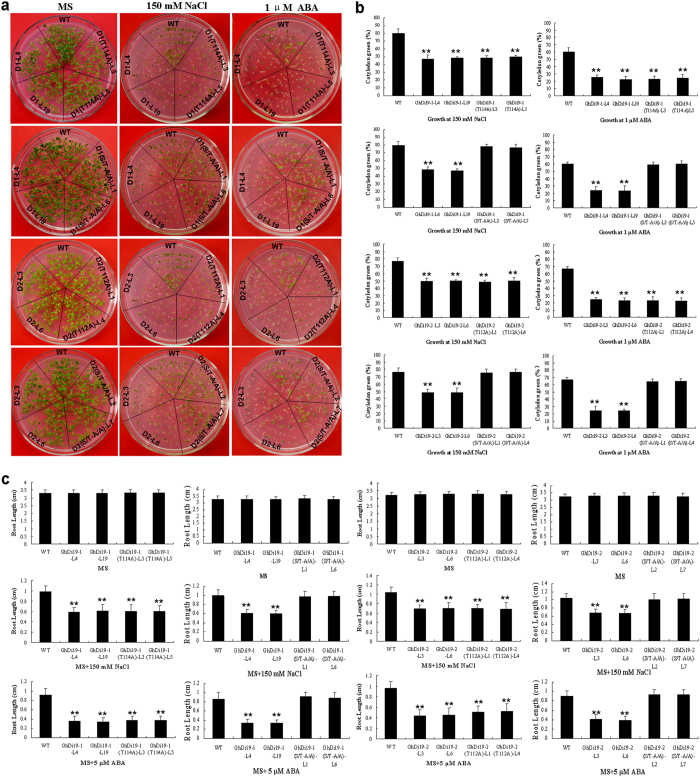
Seeds of GhDi19-1, GhDi19-1(S116A), GhDi19-2 and GhDi19-2(S114A) overexpression transgenic lines and wild type were sowed on MS medium with or without NaCl and ABA, respectively. On MS medium, all the seeds from wild type and GhDi19-1/-2 variants were able to fully germinate (≤ 4 days) after sowing, and there was no significant difference in seed germination rate between each other. In the presence of 150 mM NaCl, however, the GhDi19-1 and GhDi19-2 transgenic seeds germinated much later than those of wild type. After 4 day, approximately 75% of wild type seeds germinated, but only about 35%–40% of GhDi19-1 and GhDi19-2 transgenic seeds germinated. After 6 days, germination rate of wild type seeds reached to about 90%, but both GhDi19-1 and GhDi19-2 transgenic seeds reached to only approximately 60%. In contrast, seed germination rates of GhDi19-1(S116A) and GhDi19-2(S114A) transgenic lines were similar to those of wild type with NaCl treatment. On MS medium supplemented with 0.8 μM ABA, seed germination rate of both GhDi19-1 and GhDi19-2 transgenic lines was significantly lower than that of wild-type. About 60% of wild type seeds, but only 40% of GhDi19-1 and 30% of GhDi19-2 overexpression transgenic seeds germinated after 4 days. On the contrary, seed germination rate of GhDi19-1(S116A) and GhDi19-2(S114A) overexpression transgenic lines was as same as those of wild type under ABA treatment (Fig. 4a).
Figure 4. Effect of serine site-mutation on GhDi19-1 and GhDi19-2 responding to salt and abscisic acid (ABA) during seed germination and early seedling development.
(a) Statistical analysis of seed germination rate. Seeds of the transgenic lines and wild type were germinated on MS medium without (control) or with 150 mM NaCl and 0.8 μM ABA, respectively. (b) Growth status of the transgenic seedlings and wild type grown on MS medium without or with 150 mM NaCl and 1 μM ABA for 10 days, respectively. (c) Statistical analysis of cotyledon expansion and greening of seedlings grown on MS medium with 150 mM NaCl and 1 μM ABA for 10 days, respectively. (d) Phenotypic analysis of GhDi19-1/-2(S-A) transgenic Arabidopsis under NaCl and ABA treatments. 10-day-old seedlings of wild type, GhDi19-1/-2 and GhDi19-1/-2(S-A)-overexpression transgenic lines growing on MS medium with or without 150 mM NaCl and 5 μM ABA, respectively. (e) Statistical analysis of root length of 10-day-old seedlings of wild type and GhDi19-1/2 and GhDi19-1/2(S-A) transgenic lines growing on MS medium without (control) or with 150 mM NaCl and 5 μM ABA, respectively. Mean values and standard errors (bar) were shown from three independent experiments (n > 60 seedlings per each line). Asterisk represents that there was (very) significant difference between the transgenic lines and wild type in independent t-tests (one asterisk: P value<0.05; two asterisks: P value<0.01). WT, wild type; GhDi19-1-L4 (D1-L4) and -L19 (D1-L19), the GhDi19-1 transgenic line 4 and 19; GhDi19-1(S116A)-L2 [D1(S116A)-L2] and -L8 [D1(S116A)-L8], the site-mutated GhDi19-1(S116A) transgenic line 2 and 8 (S116A, the S116 of GhDi19-1 was mutated to A116); Di19-2-L3 (D2-L3) and -L6 (D2-L6), the GhDi19-2 transgenic line 3 and 6. GhDi19-2(S114A)-L1 [D2(S114A)-L1] and -L4 [D2(S114A)-L4], the site-mutated GhDi19-2(S114A) transgenic line 1 and 4 (S114A, the S114 of GhDi19-2 was mutated to A114). S, serine (Ser); A, alanine (Ala).
To assess effects of Ser site on GhDi19-1 and GhDi19-2 involving in response to salt and ABA during early seedling development, we used two approaches in the experiments. One way is that seeds were directly planted on MS medium containing 150 mM NaCl or 1 μM ABA for determining growth status of the seedlings after germination (Fig. 4b). Another is that seeds were germinated on MS medium for 48 h after stratification and then transferred onto MS medium containing 150 mM NaCl and 5 μM ABA in the vertical position (Fig. 4d). The results obtained with these two approaches were similar. There was no significant difference among the GhDi19-1/-2 and their site-mutated transgenic lines and wild type when seedlings grew on MS medium. However, the GhDi19-1 and GhDi19-2 overexpression seedlings were more sensitive to salt and ABA than wild type, but growth status of GhDi19-1(S116A) and GhDi19-2(S114A) transgenic seedlings was almost as same as those of wild type (Fig. 4b). The cotyledon greening rates of GhDi19-1 overexpression lines (L4 and L19) and GhDi19-2 overexpression lines (L3 and L6) were significantly lower than those of wild type under NaCl and ABA treatments, while cotyledon greening rates of GhDi19-1(S116A) and GhDi19-2(S114A) transgenic lines (L2 and L8, L1 and L4, respectively) were similar to those of wild type (Fig. 4c).
When the seedlings were transferred onto MS plates in the vertical position without any stress treatments, root growth of all GhDi19-1/-2 transgenic seedlings and their site-variants was almost as same as those of wild type (Fig. 4d). However, when the seedlings were transferred onto MS medium supplemented with 150 mM NaCl or 5 μM ABA for several days, primary root growth of the GhDi19-1 and GhDi19-2 overexpression transgenic seedlings was significantly inhibited much more than that of wild type, but growth of GhDi19-1(S116A) and GhDi19-2(S114A) transgenic roots was similar to that of wild type (Fig. 4d). Under NaCl and ABA treatments, roots of GhDi19-1 and GhDi19-2 overexpression seedlings were shorter than those of wild type, whereas there was little significant difference in primary root length between GhDi19-1(S116A) and GhDi19-2(S114A) transgenic lines and wild type (Fig. 4e). In addition, we also introduced the “empty vector” (pBI121-eGFP) into Arabidopsis as a negative control in the above experiments, and found that the phenotypes of the transgenic plants harboring the empty vector were similar to those of wild type. Collectively, these data suggested that overexpression of GhDi19-1 and GhDi19-2 in Arabidopsis enhances plant sensitivity to salt stress in ABA-dependent manner, and the Ser site is essential for GhDi19-1 and GhDi19-2 proteins involving in plant response to salt and ABA during seed germination and early seedling development.
Thr site is inessential for GhDi19-1 and GhDi19-2 in response to salt stress and ABA signaling
To understand whether Thr phosphorylation site also play an important role in GhDi19-1 and GhDi19-2 involving in response to salt and ABA signaling, the T114 of GhDi19-1 was mutated to A114, the T112 of GhDi19-2 was mutated to A112, both T114 and S116 of GhDi19-1 were mutated to A, and both T112 and S114 of GhDi19-2 were mutated to A. The coding sequences of GhDi19-1(T114A), GhDi19-1(S/T-A/A), GhDi19-2(T112A) and GhDi19-2(S/T-A/A) were inserted into pBI121 vector, and introduced into Arabidopsis, respectively. Homozygotes of T3 generation of the respective overexpression transgenic lines were analyzed by PCR to ensure confirm the presence of the respective transgene and by quantitative RT-PCR to confirm that the transgene was expressed (Fig. 3a). Two transgenic lines (L3 and L5) with higher GhDi19-1(T114A) expression, two lines (L1 and L6) with higher GhDi19-1(S/T-A/A) expression, two lines (L1 and L4) with higher GhDi19-2(T112A) expression and two lines (L2 and L7) with higher GhDi19-2(S/T-A/A) expression were selected for analyzing their phenotypes under different stresses.
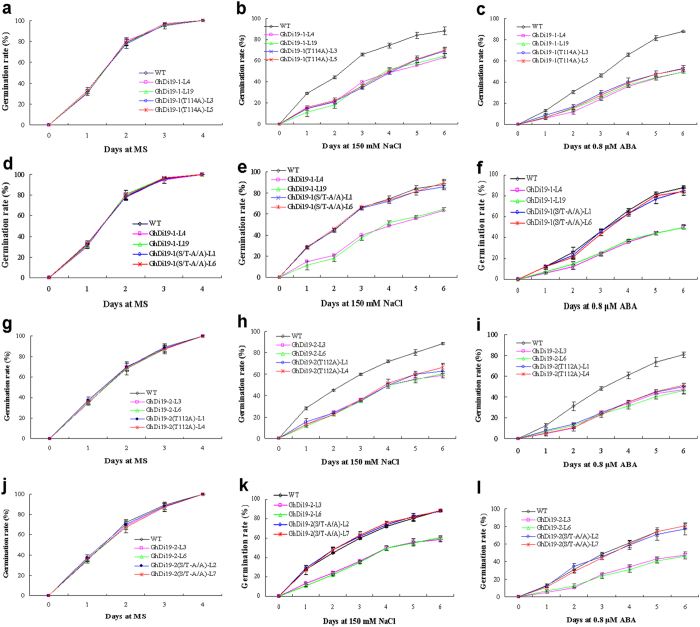
When seeds sowed on MS medium, there was no significant difference in seed germination between all the transgenic lines and wild type under normal conditions (Fig. 5a,d,g, j). When sowing on MS medium containing 150 mM NaCl or 0.8 μM ABA, however, the GhDi19-1(T114A) and GhDi19-2(T112A) transgenic seeds germinated much later than those of wild type, and showed salt- and ABA-sensitivity. In the presence of 150 mM NaCl, only about 35%–40% of the GhDi19-1(T114A) and GhDi19-2(T112A) transgenic seeds germinated, while approximately 65% of wild type germinated after 3 days. And germination rate of the transgenic seeds only reached to 60%, while wild type was about 80% after five days of germination (Fig. 5b,h). In addition, seed germination rate of the GhDi19-1(T114A) and GhDi19-2(T112A) transgenic lines was reduced much more than that of wild type under ABA treatment. When treated with 0.8 μM ABA, the seed germination rates of GhDi19-1, GhDi19-2, GhDi19-1(T114A) and GhDi19-2(T112A) transgenic lines were decreased to approximately 40%, whereas wild type retained 75% seed germination rate after 4 days (Fig. 5c,i). On the other hand, there was no significant difference in seed germination rate between GhDi19-1(S/T-A/A) and GhDi19-2(S/T-A/A) transgenic lines and wild type under NaCl and ABA treatments (Fig. 5e,k,f,l).
Figure 5. Effect of threonine site-mutation on GhDi19-1 and GhDi19-2 responding to salt and abscisic acid (ABA) during seed germination.
Seeds of the transgenic lines and wild type were germinated on MS medium (a,d,g,j), MS medium with 150 mM NaCl (b,e,h,k) and MS medium with 0.8 μM ABA (c,f,i,l), respectively. Statistical analysis of seed germination rate was determined. Mean values and standard errors (bar) were shown from three independent experiments (n > 200 seeds per each line). WT, wild type; Di19-1-L4 and -L19, the GhDi19-1 transgenic line 4 and 19; GhDi19-1(T114A)-L3 and -L5, the site-mutated GhDi19-1(T114A) transgenic line 3 and 5; GhDi19-1(S/T-A/A)-L1 and -L6, the double sites-mutated GhDi19-1(S/T-A/A) transgenic line 1 and 6 (T114A, the T114 of GhDi19-1 was mutated to A114; S/T-A/A, the S116 and T114 of GhDi19-1 were mutated to A116 and A114). Di19-2-L3 and -L6, the GhDi19-2 transgenic line 3 and 6; GhDi19-2(T112A)-L1 and -L4, the site-mutated GhDi19-1(T112A) transgenic line 1 and 4; GhDi19-1(S/T-A/A)-L2 and -L7, the double sites-mutated GhDi19-2(S/T-A/A) transgenic line 2 and 7 (T112A, the T112 of GhDi19-2 was mutated to A112; S/T-A/A, the S114 and T112 of GhDi19-2 were mutated to A114 and A112). The assays were repeated three times along with three independent repetitions of the biological experiments. S, serine (Ser) ; T, threonine (Thr); A, alanine (Ala).
We also calculated cotyledon greening of the transgenic seedlings and wild type under salt stress. As shown in Fig. 6a, both GhDi19-1(T114A) and GhDi19-2(T112A) transgenic seedlings growing on MS containing 150 mM NaCl were more sensitive than wild type. Cotyledon greening of GhDi19-1(T114A) and GhDi19-2(T112A) transgenic seedlings was drastically inhibited by NaCl treatment, compared with that of wild type. Similarly, cotyledon greening of GhDi19-1(T114A) and GhDi19-2(T112A) transgenic plants was also inhibited more than that of wild type. In the presence of 1 μM ABA, approximately 20% and 25% of the GhDi19-1(T114A) and GhDi19-2(T112A) transgenic seedlings with expanded and turned-green cotyledons was observed, respectively, while wild type reached to 60% cotyledon greening rate. In contrast, GhDi19-1(S/T-A/A) and GhDi19-2(S/T-A/A) transgenic plants showed the similar phenotype to wild-type under NaCl and ABA treatments (Fig. 6b).
Figure 6. Effect of threonine site-mutation on GhDi19-1 and GhDi19-2 responding to salt and abscisic acid (ABA) during early seedling development.
(a) Growth status of wild type, and the GhDi19-1/-2, GhDi19-1/-2(T-A) and GhDi19-1/-2(S/T-A/A) overexpression seedlings growing on MS medium with or without 150 mM NaCl and 1 μM ABA, respectively. (b) Statistical analysis of cotyledon expansion and greening of wild type, and the GhDi19-1/-2, GhDi19-1/-2(T-A) and GhDi19-1/-2(S/T-A/A) overexpression seedlings growing on MS medium with 150 mM NaCl and 1 μM ABA, respectively. (c) Statistical analysis of root length of 10-day-old seedlings of wild type, and the GhDi19-1/-2, GhDi19-1/-2(T-A) and GhDi19-1/-2(S/T-A/A) overexpression lines growing on MS medium without (control) or with 150 mM NaCl and 5 μM ABA. Mean values and standard errors (bar) were shown from three independent experiments (n > 60 seedlings per each line). Asterisk represents that there was (very) significant difference between the transgenic lines and wild type in independent t-tests (one asterisk: P value<0.05; two asterisks: P value<0.01). WT, wild type; GhDi19-1-L4 (D1-L4) and -L19 (D1-L19), the GhDi19-1 transgenic line 4 and 19; GhDi19-1(T114A)-L3 and -L5 [D1(T114A)-L3 and -L5], the site-mutated GhDi19-1(T114A) transgenic line 3 and 5; GhDi19-1(S/T-A/A)-L1 and -L6 [D1(S/T-A/A)-L1 and -L6], the double sites-mutated GhDi19-1(S/T-A/A) transgenic line 1 and 6 (T114A indicated the T114 of GhDi19-1 was mutated to A114, and S/T-A/A indicated both S116 and T114 of GhDi19-1 were mutated to A116 and A114). GhDi19-2-L3 (D1-L3) and -L6 (D1-L6), the GhDi19-2 transgenic line 3 and 6; GhDi19-2(T112A)-L1 and -L4 [D2(T112A)-L1 and -L4], the site-mutated GhDi19-2(T112A) transgenic line 1 and 4; GhDi19-2(S/T-A/A)-L2 and -L7 [D2(S/T-A/A)-L2 and -L7], the double sites-mutated GhDi19-2(S/T-A/A) transgenic line 2 and 7 (T112A indicated the T112 of GhDi19-2 was mutated to A112, and S/T-A/A indicated both S114 and T112 of GhDi19-2 were mutated to A114 and A112). S, serine (Ser); T, threonine (Thr); A, alanine (Ala).
Additionally, seeds of all the transgenic Arabidopsis lines and wild type germinated on MS medium for 48 h, and then were transferred onto NaCl- and ABA-containing MS medium for investigating growth of the seedlings responding to high salinity and ABA (Fig. 6c). When transferred onto MS plates, root growth of all the transgenic seedlings was almost as same as that of wild type. However, when the seedlings were transferred onto MS medium supplemented with 150 mM NaCl or 5 μM ABA for several days, primary roots growth of the GhDi19-1(T114A) and GhDi19-2(T112A) transgenic seedlings was more significantly inhibited than that of wild type by NaCl or ABA. The roots of GhDi19-1(T114A) and GhDi19-2(T112A) transgenic seedlings were shorter than those of wild type under NaCl and ABA treatments. However, there was no significant difference in root length between GhDi19-1(S/T-A/A) and GhDi19-2(S/T-A/A) transgenic lines and wild type under NaCl and ABA treatments (Fig. 6c). These results demonstrated that overexpression of the mutated GhDi19-1(T114A) and GhDi19-2(T112A) genes in Arabidopsis still resulted in the transgenic plants salt- and ABA-hypersensitivity, like GhDi19-1/-2 transgenic lines, suggesting that the Thr site is inessential for GhDi19-1 and GhDi19-2 proteins involving in response to salt stress and ABA signaling during seed germination and early seedling development.
Phosphorylation of Ser site by calcium-dependent protein kinase (CDPK) is important for functionally activating GhDi19-1/-2 in response to salt stress and ABA signaling
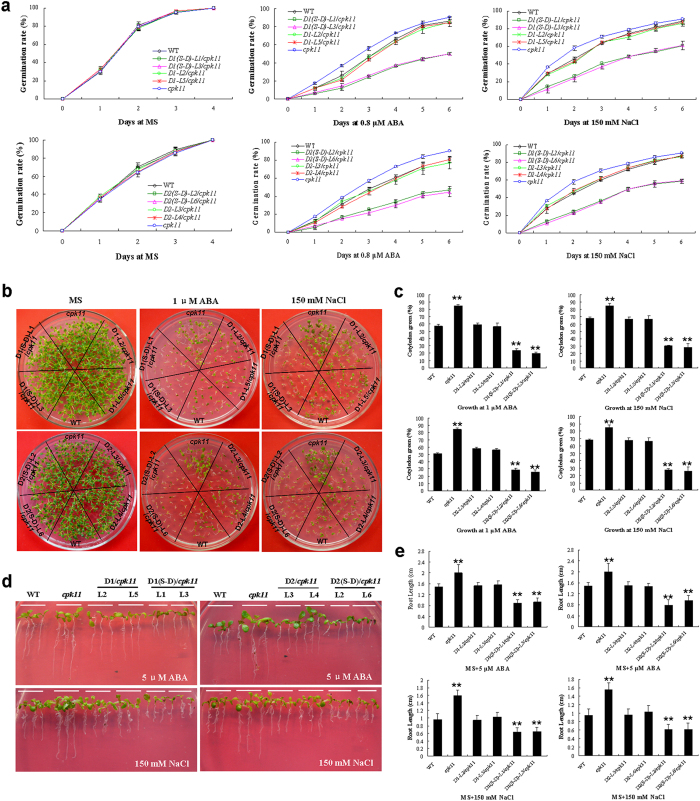
To explore the functional relationship of GhDi19s and CDPK, loss-of-function mutation of CPK11 (Atcpk11-2, SALK_054495) was employed in this study. A single copy of T-DNA was inserted into the genome of the Atcpk11-2 mutant, generating a 39-bp deletion from 320 to 358 bp downstream of the translation start codon11. Quantitative RT-PCR analysis demonstrated that AtCPK11 mRNA was undetectable in Atcpk11-2 mutant. Meanwhile, S116 was mutated to D116 in GhDi19-1 and S114 was mutated to D114 in GhDi19-2 for generating the constitutively activated GhDi19-1 and GhDi19-2 [named GhDi19-1(S116D) and GhDi19-2(S114D), respectively]. The coding sequences of GhDi19-1, GhDi19-2, GhDi19-1(S116D) and GhDi19-2(S114D) under the control of CaMV 35S promoter were introduced into Atcpk11-2 mutant. Homozygotes of T3 generation of these GhDi19 transgenic lines in Atcpk11-2 background were analyzed by quantitative RT-PCR to confirm the transgene expression (Fig. 3b). Seeds of wild type, Atcpk11-2 mutant and GhDi19-1, GhDi19-2, GhDi19-1(S116D) and GhDi19-2(S114D) overexpression transgenic lines in Atcpk11-2 background (GhDi19-1/cpk11, GhDi19-2/cpk11, GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11) were sowed on MS medium with or without ABA and NaCl, respectively. Seed germination rate of wild type and the transgenic lines was monitored under different abiotic stresses. On MS medium, all the seeds from wild type and the transgenic lines were able to fully germinate (≤4 days) after sowing, and there was no significant difference in seed germination rate between the transgenic lines and wild type. However, in the presence of 0.8 μM ABA, seeds of Atcpk11-2 mutant germinated much earlier and faster than those of wild type, and the germination rate of GhDi19-1/cpk11 and GhDi19-2/cpk11 seeds was similar to wild type, whereas seeds of GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 germinated much later than wild type. After 4 days, about 60% of wild type, GhDi19-1/cpk11 and GhDi19-2/cpk11 seeds germinated, and approximately 75% of Atcpk11-2 mutant seeds germinated, but only about 35% of GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 seeds germinated. After 6 days, seed germination rate of Atcpk11-2 mutant reached to about 90%, but those of GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 transgenic lines reached to only approximately 40–50%. In the presence of 150 mM NaCl, seed germination rate of Atcpk11-2 mutant was significantly higher than that of wild type. On the contrary, germination of the GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 transgenic seeds was inhibited much more by NaCl, compared with wild type. Approximately 80% of Atcpk11-2 mutant seeds and 70% of wild type, GhDi19-1/cpk11 and GhDi19-2/cpk11 seeds, but only 50% of GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 transgenic seeds germinated after 4 days (Fig. 7a).
Figure 7. Phenotypic assay of GhDi19-1/-2 and their activated form GhDi19-1/-2 (S-D) transgenic seedlings in Atcpk11 mutant under salt and abscisic acid (ABA) treatments.
(a) Statistical analysis of seed germination rate. Seeds of wild type, Atcpk11 mutant, and GhDi19-1/-2 or GhDi19-1/-2(S-D) overexpression transgenic lines in Atcpk11 mutant were germinated on MS medium with or without 150 mM NaCl and 0.8 μM ABA, respectively. (b) Growth status of seedlings growing on MS medium with or without 1 μM ABA and 150 mM NaCl for 10 days, respectively. (c) Statistical analysis of cotyledon expansion and greening of seedlings growing on MS medium with 1 μM ABA and 150 mM NaCl for 10 days, respectively. (d) Phenotypes of seedlings under NaCl and ABA treatments. After germination on MS medium for 48 h, the wild type, Atcpk11 mutant, and GhDi19-1/-2 or GhDi19-1/-2(S-D) overexpression transgenic seedlings in Atcpk11 mutant were transferred and grew for 10 days on MS medium without or with 150 mM NaCl and 5 μM ABA, respectively, in the vertical position. (e) Statistical analysis of root length of 10-day-old seedlings growing on MS medium with 5 μM ABA and 150 mM NaCl. Mean values and standard errors (bar) were shown from three independent experiments (n > 60 seedlings per each line). Asterisk represents that there was (very) significant difference between the transgenic lines and wild type in independent t-tests (one asterisk: P value<0.05; two asterisks: P value<0.01). WT, wild type; cpk11, the Atcpk11-2 mutant; D1-L2/cpk11 and D1-L5/cpk11, the GhDi19-1 overexpression transgenic line 2 and 5 in Atcpk11-2 mutant; D1(S-D)-L1/cpk11 and D1(S-D)-L3/cpk11, the GhDi19-1(S-D) overexpression transgenic line 1 and 3 in Atcpk11-2 mutant; D2-L3/cpk11 and D2-L4/cpk11, the GhDi19-2 overexpression transgenic line 3 and 4 in Atcpk11-2 mutant; D2(S-D)-L2/cpk11 and D2(S-D)-L6/cpk11, the GhDi19-2(S-D) overexpression transgenic line 2 and 6 in Atcpk11-2 mutant. S-D, the S116 of GhDi19-1 was mutated to D116 and the S114 of GhDi19-2 was mutated to D114. S, serine (Ser); D, aspartic acid (Asp).
During early seedling development, no significant difference was observed among the transgenic variants and wild type when seedlings grew on MS medium. However, the seedlings of Atcpk11-2 mutant grew better than those of wild type on ABA- and NaCl-containing medium, but the GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 transgenic seedlings were more sensitive to ABA and salt than wild type, while growth status of GhDi19-1/cpk11 and GhDi19-2/cpk11 transgenic seedlings was similar to wild type under ABA and NaCl treatments (Fig. 7b). Under ABA and NaCl treatments, cotyledon greening rate of Atcpk11-2 mutant was significantly higher than that of wild type, while cotyledon greening rate of GhDi19-1/cpk11 and GhDi19-2/cpk11 transgenic lines was similar to wild type. In contrast, cotyledon greening of GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 transgenic lines was inhibited much more by ABA and NaCl, compared with wild type (Fig. 7c).
When the seedlings were transferred onto MS plates in the vertical position without any stress treatments, the root growth of all transgenic variants was almost as same as those of wild type. However, when the seedlings were transferred onto MS medium supplemented with 5 μM ABA or 150 mM NaCl for several days, primary root growth of Atcpk11-2 mutant was much better than that of wild type, and root growth of GhDi19-1/cpk11 and GhDi19-2/cpk11 transgenic lines was similar to wild type, whereas root growth of the GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 transgenic seedlings was inhibited much more than that of wild type (Fig. 7d). Roots of Atcpk11-2 mutant were longer than those of wild type, and root length of the GhDi19-1/cpk11 and GhDi19-2/cpk11 transgenic lines was similar to wild type, but roots of GhDi19-1(S116D)/cpk11 and GhDi19-2(S114D)/cpk11 transgenic seedlings were shorter than those of wild type under ABA and NaCl treatments (Fig. 7e). These results indicated that overexpression of GhDi19-1/-2 in Atcpk11 background could recover the salt- and ABA-insensitive phenotype of the mutant, implying the other CDPK (besides CPK11) may also activate GhDi19-1/-2 in the transgenic Arabidopsis plants, and Ser phosphorylation is important for functionally activating GhDi19-1/-2 in response to salt stress and ABA signaling during seed germination and early seedling development.
Discussion
A large number of genes encoding receptors, kinases, transcription factors and other signal molecules in plants are induced after exposure to various abiotic stresses35,36. These genes function in various ways to confer plants stress tolerance37,38,39. Due to much low similarity of the different Di19-related proteins outside the zinc finger domain, Di19 proteins may play diverse roles in plant development and in response to abiotic stresses32. Previous studies revealed that GhDi19-1 and GhDi19-2 are involved in plant response to salt stress and ABA signaling34. In this study, we further investigated the mechanism of GhDi19-1 and GhDi19-2 involving in response to salt stress and ABA signaling, and found the Ser site is essential for GhDi19-1 and GhDi19-2 proteins involving in plant response to salt and ABA during seed germination and early seedling development.
Modification of phosphorylation by kinases and of dephosphorylation by phosphatases is an important physiological process as plants respond to abiotic stresses and ABA signaling. Some ABA/stress signaling regulators are modulated at the post-translational level by changing their phosphorylation states40,41,42,43,44. Calcium-dependent protein kinases (CDPKs) are unique serine/threonine kinases in plants responding in abiotic stresses45. Their multifunctionality and signaling specificity may be conferred by their ability to phosphorylate different substrates. Arabidopsis CPK4 and CPK11 positively regulate ABA signaling via phosphorylating downstream ABA-responsive transcription factors in plants11. Furthermore, previous studies revealed that most of seven Arabidopsis AtDi19s are phosphorylated by CPK3 and CPK11 in vitro in a Ca+2-dependent manner33. Likewise, both GhDi19-1 and GhDi19-2 contain a conserved nuclear localization signal (NLS), in which two putative kinase phosphorylation sites (Thr114 and Ser116 in GhDi19-1, and Thr112 and Ser114 in GhDi19-2, respectively) are located34. In this study, our data indicated that GhDi19-1/-2 could be phosphorylated in vitro by CPK11 in a Ca2+-dependent manner, and the Ser site of GhDi19-1/-2 is required for phosphorylation and normal subcellular localization of GhDi19-1 and GhDi19-2 proteins. Previous study reported that CPK11 phosphorylates Ser104 and Ser107 sites within the AtDi19-1 bipartite NLS probably, but effects of Ser phosphorylation at different sites on Di19 proteins involving in plant response to salt and ABA are unknown so far33. In this study, our data demonstrated the Ser site (but not Thr site) is crucial for functionally activation of cotton Di19-1/-2 in response to salt stress and ABA signaling.
AtCPK11 interacts with AtDi19-1 protein in the cell nucleus, and increases AtDi19-1 transactivation of PR1, PR2, and PR5 expression31. Furthermore, AtCPK4 and AtCPK11 overexpression transgenic plants display the hypersensitivity to ABA during seeds germination and seedlings growth11. Similar phenotype was also observed in the GhDi19-1/-2 overexpression transgenic Arabidopsis34. These findings indicated that there is a specific functional relationship of cotton Di19s and CDPKs. Additionally, overexpression of GhDi19-1/-2 in Atcpk11 could recover the salt- and ABA-insensitive phenotype of the mutant, implying the other kinases (besides CPK11) may also activate GhDi19-1/-2 in the transgenic Arabidopsis plants.
Based on the data presented in this study, we thought that Di19 proteins may be potential downstream targets of CDPKs in ABA signal pathway during early plant growth and development. When plants are exposed to abiotic stress stimuli (such as drought and high salinity etc.), concentrations of intracellular ABA and Ca2+ may be increased in plants for responding the environmental stress signaling. Subsequently, Ser/Thr kinases (e.g. CDPK) are activated by ABA and in turn the activated kinases phosphorylate Di19 proteins at Ser site in the cell nucleus. Finally, the activated Di19 proteins transduce the signals to downstream ABA- and stress-responsive genes, thereby promoting plants response to abiotic stresses.
Methods
Plants materials and growth conditions
A T-DNA insertion mutant (named Atcpk11-2, SALK_054495) of Arabidopsis CPK11 (AT3g05700) was obtained from ABRC (www.arabidopsis.org/abrc). Seeds of Arabidopsis thaliana (Columbia ecotype), including wild type, mutant and transgenic lines, were surface-sterilized with 75% ethanol for 1 min and 10% NaClO for 5 min, followed by washing with sterile water. The sterilized Arabidopsis seeds were plated on Murashige and Skoog (MS) medium. After stratification at 4 °C for 2 days, the plates were transferred to a plant growth incubator (Sanyo, Osaka, Japan) for seed germination (16 h light/8 h dark at 22 °C) ten days later, seedlings were transplanted in soil and grown in a growth room under the conditions of 16 h light/8 h dark cycle at 22–24 °C. Tissues were derived from these seedlings for RNA extraction.
Generation of transgenic plant and mutant lines
The coding sequences of cotton GhDi19-1 and GhDi19-2 genes, amplified from its cDNA by PCR with the proofreading pfu DNA polymerase, were cloned into pBI121 vector with BamH I/Sac I sites to replace the GUS gene, respectively34.
To generate loss-of-phosphorylation GhDi19-1 mutants [GhDi19-1(S116A), GhDi19-1(T114A) and GhDi19-1(S/T-A/A)] and GhDi19-2 mutants [GhDi19-2(S114A), GhDi19-2(T112A) and GhDi19-2(S/T-A/A)], primer-based site-directed mutations were performed, using primer pairs as follows: GhDi19-1(S116A) 5′-TCCTCAGAAGAGCAAGCGTGGAATGAG-3′ and 5′-CTCATTCCACGCTTGCTCTTCTGAGGA-3′; GhDi19-1(T114A) 5′-CAGAAGAGAAAGCGCGGAATGAGATCC-3′ and 5′-GGATCTCATTCCGCGCTTTCTCTTCTG-3′; GhDi19-1(S/T-A/A) 5′-CCTCAGAAGAGCAAGCGCGGAATGA-3′ and 5′-TCATTCCGCGCTTGCTCTTCTGAGG-3′; GhDi19-2(S114A) 5′-GAAGAGAAAGCGCTGAATGAGATCCAC-3′ and 5′-GTGGATCTCATTCAGCGCTTTCTCTTC-3′; GhDi19-2(T112A) 5′-CTTTCCTCAGAAGAGCAAGCGTTGAATGAG-3′ and 5′-CTCATTCAACGCTTGCTCTTCTGAGGAAAG-3′; GhDi19-2(S/T-A/A) 5′-CCTCAGAAGAGCAAGCGCTGAATGAGATC-3′ and 5′-GATCTCATTCAGCGCTTGCTCTTCTGAGG-3′. Mutants were introduced by QuickChange site-directed mutagenesis (Stratagene) and confirmed by sequencing. The coding sequences of GhDi19-1(S116A), GhDi19-1(T114A), GhDi19-1(S/T-A/A) and GhDi19-2(S114A), GhDi19-2(T112A), and GhDi19-2(S/T-A/A) were cloned into pBI121 vector, respectively.
To introduce the Ser116-to-Asp (S116D) mutation of GhDi19-1 or the Ser114-to-Asp (S114D) mutation of GhDi19-2 for constitutively activated GhDi19-1 or GhDi19-2, primer-based site-directed mutations were performed, using primers as follows: GhDi19-1(S116D) 5′-CTCATTCCGACCTTGACCTTCTGAGGA-3′ and 5′- TCCTCAGAAGGTCAAGGTCGGAATGAG-3′; GhDi19-2(S114D) 5′-CATTCAGACCTTGACCTTCTGAGGAAAG-3′ and 5′-CTTTCCTCAGAAGGTCAAGGTCTGAATG-3′. The coding sequences of GhDi19-1(S-D) and GhDi19-2(S-D) were cloned into pCAMBIA1301 vector. Mutant alleles of Atcpk11-2 (SALK_054495) were used for experiments11. Complementation experiments using GhDi19-1/-2 and GhDi19-1/-2(S-D) transgenes were done in Atcpk11-2 background.
All generated binary vectors were transformed into Agrobacterium turmefaciens strain GV3101. Arabidopsis transformation was performed by the floral dip method46, and transformants were identified on selective medium with kanamycin or hygromycin.
Quantitative RT-PCR analysis
Total RNA was extracted from 10-day-old Arabidopsis seedlings of wild type, GhDi19-1-, GhDi19-1(S116A)-, GhDi19-1(T114A)- and GhDi19-1(S/T-A/A)-overexpression transgenic lines, GhDi19-2-, GhDi19-2(S114A)-, GhDi19-2(T112A)- and GhDi19-2(S/T-A/A)-overexpression transgenic lines, Atcpk11 mutant, GhDi19-1(S-D)- and GhDi19-2(S-D)-overexpression transgenic lines in Atcpk11-2 mutant. Real-time quantitative RT-PCR (qRT-PCR) analysis was performed as described as previously47. Primer pairs for qRT-PCR analysis are as follows: GhDi19-1 5′-ATGGATGCTGATTCATGGAGT-3′ and 5′-TTATAAAATTTCATCAGGCAT-3′; GhDi19-1(S116A) 5′-ATGGATGCTGATTCATGGAGT-3′ and 5′-CTCATTCCACGCTTGCTCTTCTGAGGA-3′; GhDi19-1(T114A) 5′-ATGGATGCTGATTCATGGAGT-3′ and 5′-GGATCTCATTCCGCGCTTTCTCTTCTG-3′; GhDi19-1(S/T-A/A) 5′-ATGGATGCTGATTCATGGAGT-3′ and 5′-TCATTCCGCGCTTGCTCTTCTGAGG-3′; GhDi19-1(S116D) 5′-ATGGATGCTGATTCATGGAGT-3′ and 5′-CTCATTCCGACCTTGACCTTCTGAGGA-3′; GhDi19-2 5′-ATGGATGCTGATCCATGGAC-3′ and 5′-TCATAAAACATCATCAAGAAT-3′; GhDi19-2(S114A) 5′-ATGGATGCTGATCCATGGAC-3′ and 5′-CTCATTCAACGCTTGCTCTTCTGAGGAAAG-3′; GhDi19-2(T112A) 5′-ATGGATGCTGATTCATGGAGT-3′ and 5′-GTGGATCTCATTCAGCGCTTTCTCTTC-3′; GhDi19-2(S/T-A/A) 5′-ATGGATGCTGATTCATGGAGT-3′ and 5′-GATCTCATTCAGCGCTTGCTCTTCTGAGG-3′; GhDi19-2(S114D) 5′-ATGGATGCTGATCCATGGAC-3′ and 5′-CATTCAGACCTTGACCTTCTGAGGAAAG-3′; Atcpk11-2 5′- GAGAGAGTCAAAAAAATTGGAGAA-3′ and 5′-AAACCAATTAGGCGATGAACC-3′. Expression level of Arabidopsis ACTIN2 was monitored with forward 5′-GAAATCACAGCACTTGCACC-3′ and reverse 5′-AAGCCTTTGATCTTGAGA GC-3′ primers to serve as an internal control. For all the above qRT-PCR reactions, the assays were repeated three times along with three independent repetitions of the biological experiments, and means of three biological experiments were calculated for estimating gene expression levels.
Phenotypic analysis of the transgenic Arabidopsis plants
Homozygous plants (T3 and T4 generations) of the GhDi19-1/-2 transgenic lines and their site-mutations were used for phenotypic analysis, employing wild type, mutant and the transgenic line harboring the “empty vector” (i.e. pBI121-eGFP) as controls. We first tested a series of ABA and NaCl concentrations in the pre-experiments, and determined which concentration of abscisic acid (ABA) or NaCl is suitable to the respective experiments. Then, seeds of wild type and independent transgenic lines, mutants, or transgenic mutants were germinated on MS medium supplemented without or with 150 mM NaCl, 0.8 and l μM ABA, respectively. The seeds were incubated at 4 °C for 2 days and then transferred into a plant growth incubator at 22 °C conditions (16 h light/8 h dark). Seeds were considered successfully germinated when radicles completely penetrated the seed coats. Germination rate and proportion of seedlings with opened green cotyledons were expressed as a percentage of the total number of seeds plated.
The seedling growth experiments were performed as described previously11. Seeds were germinated after stratification on MS medium for 48 h and then transferred onto MS medium containing 150 mM NaCl and 5 μM ABA in the vertical position. The growth status of seedling was recorded for 10 days and the length of seedling primary roots was measured at tenth day after the transfer. In addition, seedling growth was also assessed by directly planting the seeds on NaCl- or ABA-containing MS medium to investigate the response of seedling growth to salt or ABA for 10 days.
Statistical analysis was performed in all the experiments. At least 300 seeds of each line were used for analyzing seed germination rate and cotyledon greening rate, and 30–60 seedlings in vertical cultivation were used for measuring root length. The experiments were repeated at least three times with three technical replications.
Subcellular localization of GhDi19-1 and GhDi19-1 protein
The coding sequences of GhDi19-1, GhDi19-1(S116D), GhDi19-1(S116A), GhDi19-1(T114A), GhDi19-1(S/T-A/A), GhDi19-2, GhDi19-2(S114D), GhDi19-2(S114A), GhDi19-2(T112A) and GhDi19-2(S/T-A/A) were cloned into pBI121-eGFP vector to generate GhDi19-1:eGFP, GhDi19-1(S116D):eGFP, GhDi19-1(S116A):eGFP, GhDi19-1(T114A):eGFP, GhDi19-1(S/T-A/A):eGFP, GhDi19-2:eGFP, GhDi19-2(S114D):eGFP, GhDi19-2(S114A):eGFP, GhDi19-2(T112A):eGFP and GhDi19-2(S/T-A/A):eGFP construct, respectively, and then introduced into Arabidopsis by the floral dip method. The harvested seeds were germinated on selective MS medium for selecting transgenic plants. Subsequently, GFP fluorescence in root cells of the transgenic seedlings was observed under a SP5 Meta confocal laser microscope (Leica, Germany) with a filter set (488 nm for excitation and 506 ∼ 538 nm for emission). SP5 software (Leica, Germany) was employed to record and process the digital images taken. Primers used in vector construction are as follows: GhDi19-1 5′-CTTGGATCCATGGATGCTGATTCATGGAG-3′ and 5′-GGGTCTAGATTATAAAATTTCATCAGGC-3′; GhDi19-2 5′-CTTGGATCCATGGATGCTGATCCATGGAC-3′ and 5′-GGGTCTAGATCATAAAACATCATCAAGAA-3′; GhDi19-1(S116D) 5′-CTCATTCCGACCTTGACCTTCTGAGGA-3′ and 5′-TCCTCAGAAGGTCAAGGTCGGAATGAG-3′; GhDi19-2(S114D) 5′-CATTCAGACCTTGACCTTCTGAGGAAAG-3′ and 5′-CTTTCCTCAGAAGGTCAAGGTCTGAATG-3′; GhDi19-1(S116A) 5′-TCCTCAGAAGAGCAAGCGTGGAATGAG-3′ and 5′-CTCATTCCACGCTTGCTCTTCTGAGGA-3′; GhDi19-2(S114A) 5′-GAAGAGAAAGCGCTGAATGAGATCCAC-3′ and 5′-GTGGATCTCATTCAGCGCTTTCTCTTC-3′; GhDi19-1(T114A) 5′-CAGAAGAGAAAGCGCGGAATGAGATCC-3′ and 5′-GGATCTCATTCCGCGCTTTCTCTTCTG-3′; GhDi19-2(T112A) 5′-CTTTCCTCAGAAGAGCAAGCGTTGAATGAG-3′ and 5′-CTCATTCAACGCTTGCTCTTCTGAGGAAAG-3′; GhDi19-1(S/T-A/A) 5′-CCTCAGAAGAGCAAGCGCGGAATGA-3′ and 5′-TCATTCCGCGCTTGCTCTTCTGAGG-3′; GhDi19-2(S/T-A/A) 5′-CCTCAGAAGAGCAAGCGCTGAATGAGATC-3′ and 5′-GATCTCATTCAGCGCTTGCTCTTCTGAGG-3′.
In vitro phosphorylation assay
The coding sequences of AtCPK11, GhDi19-1, GhDi19-1(S116A), GhDi19-2 and GhDi19-2(S114A) were inserted downstream the malE gene, which encodes maltose-binding protein (MBP), in pMAL-c2X vector for expressing MBP-AtCPK11, MBP-GhDi19-1, MBP-GhDi19-1(S116A), MBP-GhDi19-2 and MBP-GhDi19-2(S114A) fusion proteins, respectively. In vitro phosphorylation assay was performed for the fusion proteins and MBP protein (control) purified from Escherichia coli strain BL21 by MBP′s affinity for maltose (NEW ENGLAND), according to the method described previously48,49. Primers used in vector construction are as follows: AtCPK11 5′-CTTGGATCCATGGAGACGAAGCCAAACCCTAG-3′ and 5′-GGGGTCGACTCAGTCATCAGATTTTTCACCA-3′; GhDi19-1 5′-CTTGGATCCATGGATGCTGATTCATGGAG-3′ and 5′-GGGTCTAGATTATAAAATTTCATCAGGC-3′; GhDi19-2 5′-CTTGGATCCATGGATGCTGATCCATGGAC-3′ and 5′-GGGTCTAGATCATAAAACATCATCAAGAA-3′; GhDi19-1(S116A) 5′-CTCATTCCACGCTTGCTCTTCTGAGGA-3′ and 5′-TCCTCAGAAGAGCAAGCGTGGAATGAG-3′; GhDi19-2(S114A) 5′-CTCATTCAACGCTTGCTCTTCTGAGGAAAG-3′ and 5′-CTCATTCAACGCTTGCTCTTCTGAGGAAAG-3′.
Additional Information
How to cite this article: Qin, L.-X. et al. Phosphorylation of serine residue modulates cotton Di19-1 and Di19-2 activities for responding to high salinity stress and abscisic acid signaling. Sci. Rep. 6, 20371; doi: 10.1038/srep20371 (2016).
Acknowledgments
This work was supported by the Project of Transgenic Research from the Ministry of Agriculture of China (Grant No. 2014ZX08009-27B) and National Natural Science Foundation of China (Grant No. 31471542).
Footnotes
Author Contributions X.B.L. and L.X.Q. conceived and designed the research. L.X.Q., X.Y.N., R.H., G.L. and W.L.X. performed the experiments. L.X.Q., W.L.X. and X.B.L. analyzed the data and wrote the article.
References
- Xiong L. M., Schumaker K. S. & Zhu J. K. Cell signaling during cold, drought, and salt stress. Plant Cell 14, 165–183 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab G. et al. Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 139, 267–274 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. & Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417 (2003). [DOI] [PubMed] [Google Scholar]
- Huang J. et al. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol. Biol. 80, 337–350 (2012). [DOI] [PubMed] [Google Scholar]
- Belin C., Megies C., Hauserová E. & Lopez-Molina L. Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21, 2253–2268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M. & Zhang J. Cross-talk between calcium and reactive oxygen species originated from NADPH oxidase in abscisic acidinduced antioxidant defense in leaves of maize seedlings. Plant Cell Environ. 26, 929–939 (2003). [DOI] [PubMed] [Google Scholar]
- Zhang A., Jiang M., Zhang J., Tan M. & Hu X. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 141, 475–487 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Jia W. & Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54, 440–451 (2008). [DOI] [PubMed] [Google Scholar]
- Shi B. et al. OsDMI3-mediated activation of OsMPK1 regulates the activities of antioxidant enzymes in abscisic acid signaling in rice. Plant Cell Environ. 37, 341–352 (2014). [DOI] [PubMed] [Google Scholar]
- Ding Y. F. et al. ZmCPK11 is involved in abscisic acid-induced antioxidant defense and functions upstream of ZmMPK5 in the abscisic acid signaling in maize. J. Exp. Bot. 64, 871–884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S. Y. et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19, 3019–3036 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K. & Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803 (2006). [DOI] [PubMed] [Google Scholar]
- Nakashima K., Ito Y. & Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaira K. S. et al. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 157, 742–756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemose S., O’Shea C., Jensen M. K. & Skriver K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14, 5842–5878 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Z., Creelman R. A. & Zhu J. K. From laboratory to field: using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 135, 615–621 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K. & Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227 (2007). [DOI] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger proteins, the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078 (1999). [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Peisach E. & Grant R. A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 70, 313–340 (2001). [DOI] [PubMed] [Google Scholar]
- Wolfe S. A., Nekludova L. & Pabo C. O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29, 183–212 (2000). [DOI] [PubMed] [Google Scholar]
- Agarwal P. et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485 (2007). [DOI] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S. & Mittler R. The zinc finger network of plants. Cell Mol. Life Sci. 65, 1150–1160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H. et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold and high-salinity stress conditions. Plant Physiol. 136, 2734–2746 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. et al. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580, 6537–6542 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Wang X. & Chen J. Zinc finger protein 1 (ThZF1) from salt cress (Thellungiella halophila) is a Cys-2/His-2-type transcription factor involved in drought and salt stress. Plant Cell Rep. 26, 497–506 (2007). [DOI] [PubMed] [Google Scholar]
- Xu D. Q. et al. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Lett 582, 1037–1043 (2008). [DOI] [PubMed] [Google Scholar]
- Sun S. J. et al. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 61, 2807–2818 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Y., Chao D. Y., Gao J. P., Zhu M. Z., Shi M. & Lin H. X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23, 1805–1817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F., Bertauche N., Vartanian N. & Giraudat J. Abscisic acid-dependent and-independent regulation of gene expression by progressive drought in Arabidopsis thaliana. Mol. Gen. Genet. 246, 10–18 (1995). [DOI] [PubMed] [Google Scholar]
- Milla M. A. R., Townsend J., Chang I. F. & Cushman J. C. The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways. Plant Mol. Biol. 61, 13–30 (2006a). [DOI] [PubMed] [Google Scholar]
- Liu W. X., Zhang F. C., Zhang W. Z., Song L. F., Wu W. H. & Chen Y. F. Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 6, 1487–1502 (2013). [DOI] [PubMed] [Google Scholar]
- Qin L. X., Li Y., Li D. D., Xu W. L., Zheng Y. & Li X. B. Arabidopsis drought-induced protein Di19-3 participates in plant response to drought and high salinity stresses. Plant Mol. Biol. 86, 609–625 (2014). [DOI] [PubMed] [Google Scholar]
- Milla M. A. R. et al. A novel yeast two-hybrid approach to identify CDPK substrates: characterization of the interaction between CPK11 and AtDi19, a nuclear zinc finger protein. FEBS Lett. 580, 904–911 (2006b). [DOI] [PubMed] [Google Scholar]
- Li G. et al. Two cotton Cys2/His2-type zinc-finger proteins, GhDi19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling. Plant Mol. Biol. 74, 437–452 (2010). [DOI] [PubMed] [Google Scholar]
- Seki M. et al. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A. et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149 (2008). [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K. & Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K. & Yamaguchi-Shinozaki K. Molecular response to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223 (2000). [PubMed] [Google Scholar]
- Zhu J. K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J. & Kim S. Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730 (2000). [DOI] [PubMed] [Google Scholar]
- Johnson R. R., Wagner R. L., Verhey S. D. & Walker-Simmons M. K. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 130, 837–846 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C. P. et al. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T. et al. Abscisic aciddependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103, 1988–1993 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R., Umezawa T., Muzogushi T., Takahashi S., Takahashi F. & Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281, 5310–5318 (2006). [DOI] [PubMed] [Google Scholar]
- Ma S. Y. & Wu W. H. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 65, 511–518 (2007). [DOI] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Li X. B., Fan X. P., Wang X. L., Cai L. & Yang W. C. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17, 859–875 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G. H., Meng X. Z., Liu Y. D., Zheng Z. Y., Chen Z. X. & Zhang S. Q. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ren F., Zhou L., Wang Q. Q., Zhong H. & Li X. B. The Brassica napus Calcineurin B-Like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signaling. J. Exp. Bot. 63, 6211–6222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]