Abstract
Currently, the gold standard to establish benign vs. malignant breast tissue diagnosis requires an invasive biopsy followed by tissue fixation for subsequent histopathological examination. This process takes at least 24 h resulting in tissues that are less suitable for molecular, functional, or metabolic analysis. We have recently conducted redox scanning (cryogenic NADH/flavoprotein fluorescence imaging) on snap-frozen breast tissue biopsy samples obtained from human breast cancer patients at the time of their breast cancer surgery. The redox state was readily determined by the redox scanner at liquid nitrogen temperature with extraordinary sensitivity, giving oxidized flavoproteins (Fp) an up to tenfold discrimination of cancer to non-cancer of breast in our preliminary data. Our finding suggests that the identified metabolic parameters could discriminate between cancer and non-cancer breast tissues without subjecting tissues to fixatives. The remainder of the frozen tissue is available for additional analysis such as molecular analysis and conventional histopathology. We propose that this novel redox scanning procedure may assist in tissue diagnosis in ex vivo tissues.
Keywords: Mitochondrial redox ratio, Fluorescence imaging, Metabolism, Tumor aggressiveness, Oxygenation
1 Introduction
Biochemical analysis of cancer core biopsies has little diagnostic value in the clinic so far. A novel approach to utilize the biochemistry of biopsies is afforded by our redox scanner [1, 2]. This diagnostic tool requires only a small tissue sample (1 mm in 2 dimensions) to obtain tissue mitochondrial redox states at a high spatial resolution down to 50 μm. In this study, we have discovered that redox scanning may provide a novel biochemical approach to identify breast cancer tissue in clinical samples. The differential redox state in breast cancer vs. normal tissues is presumably due to tissue reoxygenation which reactivates the electron transport activity in the mitochondria. In our previous studies [3–6], we discovered that the redox state of tumor tissue was accurately determined by the redox ratio, Fp/(Fp + NADH), a sensitive indicator of the tumor metabolic activity which correlates with the degree of invasiveness. In another study, we also showed that the premalignant pancreatic tissue is more heterogeneous in mitochondrial redox state than the normal one [7].
Currently, the gold standard to establish benign vs. malignant breast tissue diagnosis requires an invasive biopsy followed by tissue fixation for subsequent histological examination. The required tissue fixation step renders the tissue less suitable for additional molecular, functional, and metabolic analyses. So far the histological examination and redox scanning cannot be correlated as redox scanning requires viable tissue with a preserved metabolic state while histology evaluation often requires tissue fixation resulting in nonviable tissues. These two seemingly incompatible requirements are now met by our simple observation that indicates that tissue obtained from normal adjacent tissue in the periphery of the surgical specimen and from the centre of the tumor specimen obtained by core biopsy needle are well oxygenated and the mitochondrial electron transport metabolism is reactivated.
In this paper, we report the preliminary data on imaging the redox state of breast biopsies to discriminate the cancer to non-cancer tissues. Upon further confirmation with more patient samples and standardization of tissue sample collection procedures, this novel redox scanning procedure may assist in real-time tissue diagnosis in ex vivo tissues at the time of surgery.
2 Methods
Tissue collection was performed according to a protocol approved by the Internal Review Board of our institution. Our procedure involves excising a thin section from normal adjacent breast tissue at the periphery of the surgical specimen at the time of breast cancer surgery. Tissue from the central portion of the surgical specimen containing tumor tissue was collected using a 20-gauge core biopsy needle (~1 × 1 × 5 mm3). The redox scan requires mounting the snap-frozen tissue slice or core biopsies in the frozen mounting buffer (H2O:ethanol: glycerol = 10:30:60, freezing point –30°C), followed by redox scanning in two or three dimensions for the redox states of NADH and Fp. Unused portion of both normal and tumor tissues were preserved in the frozen mounting buffer and stored in liquid nitrogen tank for histopathology evaluation.
Breast tissues (normal adjacent tissue collected as a small tissue block and tumor tissue collected by core needle) collected from patients 1 and 2 were placed on saline-moistened paper at room temperature in the operating room. Both tissues were then brought outside the surgery room where they were immediately wrapped with aluminum foil and dipped into liquid N2. The estimated time interval between removing tissue from the body and snap-freezing was 5–10 min. The core needle samples of cancerous tissue from various locations in the surgical specimen from patient 1 were dipped into liquid nitrogen temporally in the following order: a, b, and d with a time interval of 30~60 s. For samples from patient 3, both cancerous and normal tissues were given as a thin tissue block of ~0.3 × 2 × 2 cm3. The estimated time interval between tissue removal from the patient and snap-freeze was ~5 min. All collected tissues, both normal and cancerous tissues, were from the affected cancer-bearing breasts.
The detailed procedures for embedding tissue samples for redox scanning have been reported elsewhere [5, 7–9]. Small portions of the frozen core biopsies were embedded. For tissue blocks, thin pieces were first sliced off from the snap-frozen tissue blocks using a handsaw on dry-ice-chilled metal surface. The thin piece was embedded in the mounting buffer in such a way that the starting layer for redox scanning was the surface exposed to the air before snap-freeze. NADH and Fp standards were first embedded with chilled glycerol/ethanol/water mounting buffer. The tissue was then placed adjacently using a pair of chilled forceps. Cold mounting buffer slush was added on top of the tissues to secure.
The embedded samples were carefully milled flat under liquid N2 by shaving off the top surfaces, ~50 μm from tumor tissue (core biopsy) and ~100–300 μm from normal tissue. The samples were then scanned under liquid N2 using the redox scanner. Two to four sections with spacing between 40 and 120 μm (depth range 0–240 μm) were scanned for each core biopsy sample. Two sections with spacing ~200–300 μm were scanned for each tissue block sample.
The acquired NADH and Fp signals were analyzed using Matlab® software which constructed all images. Both the NADH and Fp images were displayed as concentration maps, where concentration was in the unit of μM calibrated to the fluorescence from the corresponding standard. The redox ratio images were displayed as the concentration ratio of Fp/(Fp + NADH) in the range of 0 ~ 1.
For multiple tissue samples from patient 1, the redox scanning indices (Fp, NADH, and redox ratio) were averaged across tissue sections and then across tissue samples from various locations. Single factor ANOVA analysis was performed to compare the differences among the mean values of three core biopsies from different locations.
3 Results and Discussion
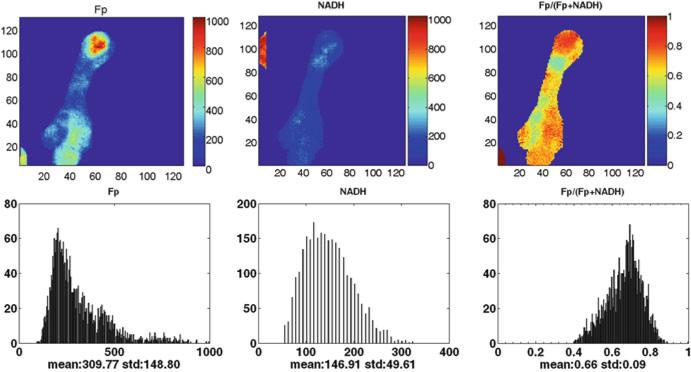
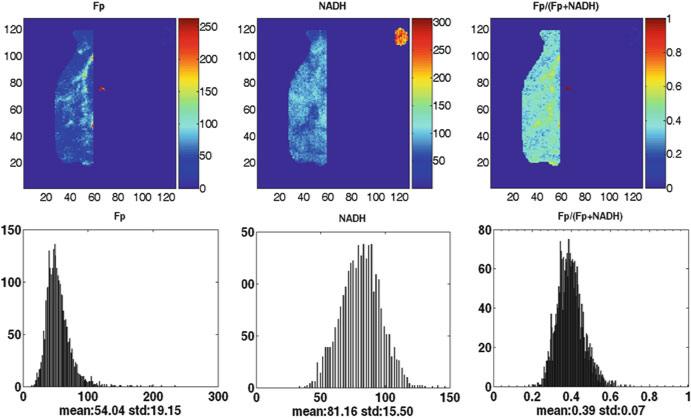
Figures 48.1 and 48.2 illustrate the typical findings of the redox images of breast tumor tissue and normal breast tissue. The results of redox scans of the human biopsy samples are shown in Table 48.1. The first and most notable finding is the high values of oxidized flavoprotein Fp, clearly indicating that the electron transfer chain to oxygen was operating efficiently prior to snap-freezing. Fp nominal concentration in cancerous tissue (Fpt = 652 ± 93 μM) is significantly higher than that in normal tissue (Fpn = 118 ± 89 μM) with p = 0.027. Their ratio, Fpt/Fpn, averaged over three patients is 7.7 ± 4.4.
Fig. 48.1.
Typical redox images of breast tumor tissue (patient 1) (image matrix 128 × 128, resolution 40 μm). The redox ratio ranges between 0 and 1; the Fp or NADH images are in the unit of μM. The x axes of the corresponding histograms represent the redox ratio or concentration. The y axes represent the number of pixels in the tumor section having a specific value of redox ratio or concentration
Fig. 48.2.
Typical redox images of normal breast tissue (patient 1) (image matrix 128 × 128, resolution 100 μm)
Table 48.1.
Redox indices of the cancerous and normal breast tissues from three patients
| Cancerous |
Normal |
||||||
|---|---|---|---|---|---|---|---|
| Patient | Redox ratiot | NADHt (μM) | Fpt | Redox ration | NADHn (μM) | Fpn (μM) | Fpt/Fpn |
| 1a | 0.71 | 153 | 569 | 0.42 | 86 | 65 | 9.2 |
| 2b | 0.52 | 699 | 759 | 0.31 | 153 | 69 | 11.1 |
| 3c | 0.59 | 295 | 601 | 0.48 | 174 | 221 | 2.7 |
| Mean | 0.61 | 384 | 652 | 0.40 | 138 | 118 | 7.7d |
| SD | 0.10 | 281 | 93 | 0.09 | 46 | 89 | 4.4 |
| p valuee | 0.062 | 0.248 | 0.027 | ||||
2.1 cm, invasive ductal carcinoma (IDC), triple negative (TN) receptors, negative nodes, poorly differentiated, 3 core biopsies, and 1 normal tissue block
3 cm, BRCA1 mutation, TN, negative nodes, poorly differentiated, 1 core biopsy, and 1 normal tissue block
10 cm, ER+, positive node, Arimidex treated for 6 months, 1 cancer and 1 normal tissue block
Average of Fpt/Fpn over the three patients
Comparison of the redox indices between cancerous and normal tissue of the three patients
In comparing the cancerous and normal tissues, we find a still greater difference, the redox ratio is different for all three normal (0.40 ± 0.09) and three cancerous tissues (0.61 ± 0.10), despite the borderline statistical difference which might be partially due to the very different disease histories. Patient 1 and 2 received no neo-adjuvant therapy, while patient 3 had been treated with Arimidex for 6 months before she had the surgery. In spite of the disparities in patient disease history and sample collection procedures, the cancerous tissue has strikingly higher Fp (almost tenfold higher). This cannot be fully accounted for by slightly higher O2 diffusion coefficient in tumor tissue than that in normal one [10–14]. The oxygen diffusion distance is more than 500 μm for a time window of 5 min assuming a diffusion constant of 10–5 cm2/s. The reported redox scanning data were from the top layers of both the tissue blocks (depth ~0–300 μm) and the core biopsy samples (depth ~0–240 μm). Thus, it is reasonable to assume that the reoxygenation due to oxygen diffusion should be similar for both normal and cancerous tissue samples.
For patient 1, several cancerous tissue core biopsies from different locations were collected and redox-scanned. Table 48.2 summaries the highly significant heterogeneity in the redox ratio, NADH, and Fp in her tumor tissue obtained by core needle in the following temporal order a→b→d. The highly significant difference in the redox indices is readily recognized. The temporal order of the sample snap-freezing a→b→d coincides with the increase of both NADH and Fp concentrations. Delayed snap-freeze led to longer air-exposure and likely more reoxygenation, which should result in increasing Fp and decreasing NADH.
Table 48.2.
Redox indices of the cancerous core biopsies from different locations of patient 1
| Tissue location | Redox ratio | NADHt (μM) | Fpt (μM) |
|---|---|---|---|
| a (2 sections) | 0.54 ± 0.01 | 45 ± 13 | 57 ± 20 |
| b (4 sections) | 0.69 ± 0.03 | 142 ± 30 | 351 ± 37 |
| d (5 sections) | 0.81 ± 0.03 | 218 ± 34 | 1,044 ± 337 |
| p valuea | 0.00001 | 0.0004 | 0.0017 |
Single factor ANOVA analysis was used to show the statistical difference between locations
It warrants further investigation whether the marked difference in redox indices between the normal and cancerous tissues of the three patients and/or the difference among the samples of patient 1 were mainly caused by the air-exposure difference or the intrinsic biological difference in these tissues. Sample collection procedures need to be standardized to minimize its possible contribution to the redox indices in the future. Analysis of more tissue samples is planned to confirm the preliminary results reported here.
4 Conclusions
In summary the mitochondrial redox states of human breast cancer tissue samples can be measured fluorometrically and show marked differences between cancer and non-cancer tissues. The redox scanning results clearly indicate aerobiosis of the samples. We report here, for the first time, the feasibility of redox cryo-imaging of breast biopsies which may open up new avenues to explore the correlation of histopathology and redox detection of cancer and perhaps to monitor efficacy of therapeutics. We propose that this novel redox scanning procedure may be used as a real-time tissue diagnosis tool in ex vivo tissues.
Acknowledgments
This work was supported by the Susan G. Komen Foundation Grant KG081069 (L.Z. Li), National Institutes of Health (NIH) grant R01CA155348 (L.Z.L.), the Center of Magnetic Resonance and Optical Imaging (CMROI)—an NIH supported research resource P41EB015893 (R. Reddy), the Small Animal Imaging Program (SAIR) 2U24-CA083105 (J. Glickson & L. Chodosh), and the Abramson Cancer Center Pilot Grant funded by the NCI Cancer Center Support Grant (J. Tchou)
Footnotes
This article is dedicated to the memory of Dr. Britton Chance, who devoted himself to the research process with sleepless nights and his profound insights into science as well as his great attention to detail until the last moment of his life.
Contributor Information
H. N. Xu, Department of Radiology, University of Pennsylvania, Philadelphia, PA, USA
J. Tchou, Department of Surgery, University of Pennsylvania, Philadelphia, PA, USA Rena Rowan Breast Center, Abramson Cancer Center, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
B. Chance, Department of Biochemistry and Molecular Biophysics, Johnson Research Foundation, University of Pennsylvania, Philadelphia, PA, USA
L. Z. Li, Department of Radiology, University of Pennsylvania, Philadelphia, PA, USA
References
- 1.Chance B, Schoener B, Oshino R, et al. Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem. 1979;254:4764–4771. [PubMed] [Google Scholar]
- 2.Quistorff B, Haselgrove JC, Chance B. High spatial resolution readout of 3-D metabolic organ structure: an automated, low-temperature redox ratio-scanning instrument. Anal Biochem. 1985;148:389–400. doi: 10.1016/0003-2697(85)90244-1. [DOI] [PubMed] [Google Scholar]
- 3.Li LZ, Zhou R, Zhong T, et al. Predicting melanoma metastatic potential by optical and magnetic resonance imaging. Adv Exp Med Biol. 2007;599:67–78. doi: 10.1007/978-0-387-71764-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li LZ, Zhou R, Xu HN, et al. Quantitative magnetic resonance and optical imaging biomarkers of melanoma metastatic potential. Proc Natl Acad Sci U S A. 2009;106:6608–6613. doi: 10.1073/pnas.0901807106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu HN, Nioka S, Glickson JD, et al. Quantitative mitochondrial redox imaging of breast cancer metastatic potential. J Biomed Opt. 2010;15:036010. doi: 10.1117/1.3431714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li LZ, Xu HN, Ranji M, et al. Mitochondrial redox imaging for cancer diagnostic and therapeutic studies. J Innov Opt Health Sci. 2009;2:325–341. doi: 10.1142/S1793545809000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu HN, Nioka S, Chance B, et al. Heterogeneity of mitochondrial redox state in premalignant pancreas in a PTEN null transgenic mouse model. Adv Exp Med Biol. 2011;201:207–213. doi: 10.1007/978-1-4419-7756-4_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu HN, Wu B, Nioka S, et al. Quantitative redox scanning of tissue samples using a calibration procedure. J Innov Opt Health Sci. 2009;2:375–385. doi: 10.1142/S1793545809000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu HN, Wu B, Nioka S, et al. Proceedings of Biomedical Optics in San Jose, CA, Jan. 24, Ed. Vol. 7174. SPIE; 2009. Calibration of redox scanning for tissue samples. p. 71742F. [Google Scholar]
- 10.Evans NTS, Naylor PFD, Quinton TH. The diffusion coef fi cient of oxygen in respiring kidney and tumour tissue. Respir Physiol. 1981;43:179. doi: 10.1016/0034-5687(81)90100-6. [DOI] [PubMed] [Google Scholar]
- 11.Groebe K, Vaupel P. Evaluation of oxygen diffusion distances in human breast cancer xenografts using tumor-specific in vivo data: role of various mechanisms in the development of tumor hypoxia. Int J Radiat Oncol Biol Phys. 1988;15:691. doi: 10.1016/0360-3016(88)90313-6. [DOI] [PubMed] [Google Scholar]
- 12.Krogh A. The rate of diffusion of gases through animal tissues, with some remarks on the coefficient of invasion. J Physiol. 1919;52:391–408. doi: 10.1113/jphysiol.1919.sp001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macdougall JDB, McCabe M. Diffusion coefficient of oxygen through tissues. Nature. 1967;215:1173. doi: 10.1038/2151173a0. [DOI] [PubMed] [Google Scholar]
- 14.Vaupel P, Mayer A, Briest S, et al. Hypoxia in breast cancer: role of blood flow, oxygen diffusion distances, and anemia in the development of oxygen depletion. Adv Exp Med Biol. 2005;566:333–342. doi: 10.1007/0-387-26206-7_44. [DOI] [PubMed] [Google Scholar]