Abstract
Purpose of review
To overview advances in the genetics of puberty based on studies in the general population, describe evidence for sex-specific genetic effects on pubertal timing, and briefly review possible mechanisms mediating sexually dimorphic genetic effects.
Recent findings
Pubertal timing is highly polygenic, and many loci are conserved among ethnicities. A number of identified loci underlie both pubertal timing and related traits such as height and body mass index (BMI). It is increasingly apparent that understanding the factors modulating the onset of puberty is important because the timing of this developmental stage is associated with a wider range of adult health outcomes than previously appreciated. While most of the genetic effects underlying the timing of puberty are common between boys and girls, some effects show sex-specificity and many are epigenetically modulated. Several potential mechanisms, including hormone-independent ones, may be responsible for observed sex differences.
Summary
Studies of pubertal timing in the general population have provided new knowledge about the genetic architecture of this complex trait. Increasing attention paid to sex-specific effects may provide key insights into the sexual dimorphism in pubertal timing and even into the associations between puberty and adult health risks by identifying common underlying biological pathways.
Keywords: pubertal timing, sex specificity, genome-wide association, epigenetics
Introduction
The onset of puberty is sexually dimorphic, occurring earlier in girls than in boys and exhibiting partly sex-specific impact on health. Recent large studies show strong evidence that pubertal timing is linked to a diverse range of adult health outcomes, some of which are shared while others are specific to either men or women (1). Investigation of the genetic underpinnings of pubertal timing within the general population may improve our understanding of male vs. female differences in onset and associations with health outcomes. Significant discoveries have been made in recent years by incorporating ever-larger sample sizes, samples with diverse ethnic backgrounds, and traits which also allow the inclusion of males. In particular, studies in both sexes allow investigation not only of the common genetic underpinnings, but also of the differences. In this review, we provide a brief update of the key developments in our understanding of the genetics of normal pubertal timing, explore some evidence for sex-specific genetic effects in pubertal traits, and speculate on potential mechanisms that may mediate these effects.
Genetic studies of age at menarche (AAM): larger sample sizes and diverse ethnicities
Recent studies have shed light on the genetic architecture underlying pubertal timing in the general population. Large-scale genome-wide association studies (GWAS) show that pubertal onset is highly polygenic, like other quantitative traits (2,3). However, known genome-wide significant variants for AAM (currently 123 SNPs at 106 loci, n > 180,000 women) still explain only 2.7% of the trait variance (2). Expanding the variants studied to include potentially functional low-frequency variants (with a minor allele frequency of 1–5%) and common variants on the X chromosome found an additional 5 and 7 associated loci, respectively, but these variants only explained 0.5% more of the variance (4). The small amount of explained variance suggests that other types of genetic variation, such as copy number changes and gene-gene and epigenetically mediated gene-environment interactions (5), should be investigated to help explain the missing heritability. It is also possible that variation in pubertal timing may be the result of hundreds or thousands of genetic variants with very small effect sizes.
Until recently, little overlap was seen between genes harboring rare, functionally damaging variants and genes near common variants known to influence puberty. However, with the publication of 106 AAM loci, a significant enrichment for signals in/near genes causing rare Mendelian puberty disorders was observed. Examples include MKRN3, underlying central precocious puberty (CPP), and genes associated with hypogonadism, such as LEPR-LEPROT, encoding the leptin receptor, and TACR3, encoding the neurokinin B receptor. In addition, a variant 10 kb away from GNRH1 was suggestively associated with AAM (2).
Other recent studies have examined puberty in non-European populations, notably in African American (AA) and Asian women (Table 1). These studies mainly seek to replicate European loci, although not necessarily at the same SNPs. Candidate studies in AA women have had some success (6,7); in one study, 25 of the 42 loci known at the time contained variants that were significantly associated in AA women. However, de novo GWAS in these samples failed to generate statistically significant novel loci, partly due to small sample sizes. Furthermore, the linkage disequilibrium (LD) structure differs between populations, with Europeans having longer stretches of LD than African populations (8), so widely used SNP-chips aimed at tagging common European haplotypes may miss important variation in other ethnicities. In other populations, including Koreans (9), Asians, Hispanics, and Native Hawaiians (10), and Filipinos (11), effect sizes for variants at the leading European loci (at LIN28B and 9q31/TMEM38B) do appear robust across populations, although effect sizes vary by ethnicity (Table 1). Taken together, these studies show that some known loci are important for pubertal onset regardless of ethnicity, and that larger sample sets are needed.
Table 1. (Original) Reported effect sizes for the leading European menarche-associated locus at LIN28B in diverse ethnicities.
Until 2013, only European-descent populations were assessed for age at menarche in GWAS studies. The leading locus in these studies at LIN28B has since been assessed in various ethnicities. In this table, the effect at the LIN28B locus is summarized for various ethnicities, including the effect size and P-value for the most recent European GWAS for reference (2). These studies show that while variants at LIN28B are the most robust in Europeans, heterogeneity exists in other populations. For example, a GWAS in Korean women (6) did not report variants at this locus as significantly associated, and many other ethnicities show varied effects.
| First author | Year | Study Type | Population | Sample size | LIN28B SNP Studied | Reported allele | Effect size (years) | P | If GWAS, was LIN28B the leading locus? | Was rs7759938 leading SNP at LIN28B locus? |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Perry | 2014 | GWAS | European | 182,416 | rs7759938 | C | 0.12 | 7.8 × 10−110 | Yes | |
|
| ||||||||||
| Pyun | 2013 | GWAS | Korean | 3,437 | Not reported | NA | NA | NA | No | Not reported |
|
| ||||||||||
| Croteau-Chonka | 2013 | Candidate | Filipino | 827 | rs7759938 | T | −0.0118 | 0.019 | NA | NA |
|
| ||||||||||
| Tanikawa | 2013 | GWAS + candidate | Japanese | 15,495 | rs364663 rs7759938 |
A T |
−0.089 −0.087 |
5.49 × 10−7 1.12 × 10−6 |
Yes | No |
|
| ||||||||||
| Demerath | 2013 | GWAS | African American | 18,089 | rs9386427 | T | 0.079 | 0.0025 | No | No |
|
| ||||||||||
| Carty | 2013 | Candidate | European | 24,819 | rs314280 | T | 0.08 | 2 × 10−8 | ||
|
|
||||||||||
| African American | 7,999 | 0.05 | 0.16 | |||||||
|
|
||||||||||
| Asians | 3,934 | 0.13 | 0.0012 | |||||||
|
|
||||||||||
| Hispanics | 3751 | 0.001 | 0.98 | |||||||
|
|
||||||||||
| American Indians | 231 | 0.25 | 0.14 | NA | NA | |||||
|
|
||||||||||
| Hawaiians | 728 | 0.22 | 0.02 | |||||||
|
| ||||||||||
| Spencer | 2013 | GWAS + Candidate | African American | 4,159 | rs7759938 | A | −0.02 | 0.61 | No | No |
|
| ||||||||||
| Delahanty | 2013 | Candidate | Chinese | 6,929 | rs7759938 | C | 0.12 | 3.8 × 10−6 | NA | NA |
Epigenetics: imprinting and parent-of-origin effects
The role of epigenetics is a recent key advance in our understanding of the mechanisms regulating pubertal onset (5). In the large-scale GWAS of AAM, six loci fell in imprinted genomic regions and four of these had parent-of-origin effects, i.e. only the paternal or maternal allele was associated with AAM (2). Of these, a paternally-inherited variant at MKRN3, an imprinted gene in the Prader-Willi syndrome critical region (chr 15q11-q13), was associated at a similar magnitude as variants at LIN28B (> 0.1 yrs per allele). Concurrently, a whole-exome sequencing study of 40 individuals from 15 families with CPP found 4 heterozygous mutations in MKRN3 (12). All individuals with CPP inherited the mutation from their fathers; individuals who inherited mutations from their mothers had normal puberty. Further studies revealed additional MKRN3 mutations in Brazilian (13), Greek (14), German (15), Ashkenazi-Sephardic Jewish (16), and Korean CPP cases (17).
It is unknown whether MKRN3 or another gene, MAGEL2, is responsible for the GWAS signal (2), as truncating mutations in MAGEL2 affecting paternal alleles have been reported in Prader-Willi cases with hypogonadism or delayed puberty (18). In mice, however, levels of Mkrn3 decreased in the arcuate nucleus of the hypothalamus immediately before puberty (12), and circulating MKRN3 also declines preceding pubertal onset in girls (19). Collectively, these data indicate that MKRN3 is part of the inhibitory brake that restrains puberty. It is interesting in this regard that when prepubertal rodent models were treated with chemicals modifying genomic epigenetic marks, each of three approaches showed results that were consistent with a mechanism in which epigenetically-mediated suppression is lifted at the onset of puberty (5,20).
The closest gene at 19 out of 32 AAM loci published in 2010 is a regulator of the epigenome (3,5), and genes in the Jmj-domain-containing lysine-specific demethylase family were highly enriched for association with AAM (enrichment P = 0.006) in the most recent GWAS (2). Many genes in this family encode proteins that epigenetically modify specific methylation marks. In female mice, DNA methylation of Polycomb group silencing complex genes leads to the enrichment of activating H3 lysine modifications, triggering pubertal onset (20). AAM has been inversely associated with global DNA methylation (21), providing further evidence that an epigenetic switch plays an important role in the onset of puberty (22,23).
Sex differences and genetic effects on puberty
Several recent GWAS of pubertal traits investigated the genetic overlap in the regulation of pubertal phenotypes between boys and girls. These studies mainly focused on the commonalities between the sexes. However, differences also exist, and may prove important (Fig. 1). For example, in a GWAS of the timing of the pubertal growth spurt, several variants were associated in only one sex in gender-specific analyses (24), including rs960273 (GNA12) in males (P for sexual heterogeneity (Psex-het) = 5.2 × 10−4) and rs7628864 (VGLL3) in females (Psex-het = 6.8 × 10−6). Of these, rs7628864 was also associated with pubertal timing, while rs960273 showed sex-specific associations with total postnatal linear growth but not pubertal timing. Additionally, another study found an association between an AAM-associated locus, rs480014 (CABLES1), and taller childhood height in boys (Psex-het = 0.04) but not girls, as well as other race- and sex-specific associations with weight and BMI changes in adolescence (29).
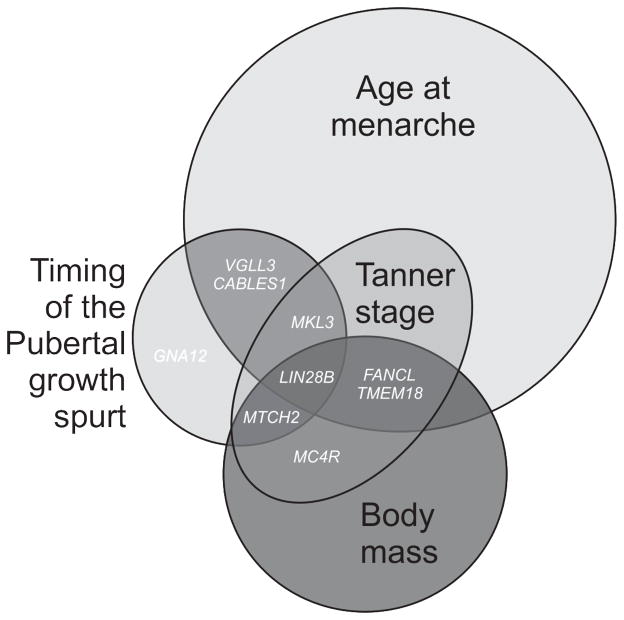
Fig. 1. (Original) Puberty-associated loci with sex-specific effects.
Genome-wide association studies have resulted in over 100 genome-wide significant loci for the pubertal traits of age at menarche, the timing of the pubertal growth spurt, and Tanner puberty staging. Body mass is a related trait that shares a genetic component with puberty. For AAM, 106 autosomal loci (2), 2 X-chromosome loci, and 5 low-frequency loci (4) are currently identified. There are 5 known loci which are significantly associated with the pubertal growth spurt and pubertal timing (24), and an additional 2 significant loci for Tanner staging (26). Gene names are shown for loci at which sex-specific effects have been seen. For example, variants near LIN28B have been associated with age at menarche (2,3), the timing of the pubertal growth spurt and postnatal growth (24,25), and Tanner stage (26), with stronger associations seen in girls. Additionally, these LIN28B variants were also associated with body mass traits in adult women but not adult men (27,28). Other pubertal loci at which sex-specific effects have been seen include GNA12 and VGLL3 (24), CABLES1 (29), and MKL3, MTCH2, MC4R, FANCL, and TMEM18 (26).
Two additional studies explored the overlap between menarche-associated variants and male genital and female breast development assessed by Tanner puberty scales (2,26). These studies concluded that much of the genetic architecture underlying pubertal timing is shared between the sexes, as the majority of alleles have concordant effect directions in males and females. However, further investigation of AAM-associated loci in male and female Tanner data revealed that not all menarche-associated loci behave the same way (Table 2; Supplemental figure 1). Some variants have the same effect direction but significantly different effect sizes, such as rs7759938 and rs2153127 at LIN28B and rs10453225 at TMEM38B; some loci show a strong association in one sex only, such as rs17233066 at SAT2B; and others have opposite effects, like rs1324913 at KLF12. It should be noted that the Tanner dataset was quite small, which could have resulted imprecise estimates and random effects. Further in-depth studies of loci with sex-specific associations are necessary to understand sexual dimorphism in pubertal timing.
Table 2. (Original) Sex-specific puberty effects.
123 menarche-associated variants were extracted from GWAS data on Tanner male genital stage and Tanner female breast stage (25). The effect sizes are shown for the menarche-delaying alleles. In the Tanner data, beta values are derived directly from the Tanner scale, where 1 is prepubertal and 5 is adult phenotype. Thus, a negative beta value corresponds to later-than-average pubertal timing, since it reflects a lower Tanner score. Only 4 variants had significant P-values for a difference between the sexes (Psex-dif) at a Bonferoni-corrected significance threshold of 0.0004 (corrected for assessing 123 menarche-associated variants). A) Variants with a stronger effect in one sex versus the other. In some cases the variant appears to be associated with Tanner stage in both sexes but to a stronger degree in one versus the other, and in other cases the effect size is close to zero for one sex but higher in the other. B) Variants showing potentially opposite directions of effect. It should be noted that the dataset was relatively small (n males = 3769; n females =6147), meaning that random effects may exist.
| SNP | Nearby genea | Chr | Positionb | Allele c | Male beta | Female beta | Psex-difd | Psex-hete |
|---|---|---|---|---|---|---|---|---|
| A. Stronger effect in one sex | ||||||||
| rs2153127 | LIN28B | 6 | 105455237 | T | −0.049 | −0.067 | 2.6 × 10−5 | 0.474 |
| rs7759938 | LIN28B | 6 | 105485647 | C | −0.063 | −0.088 | 1.5 × 10−7 | 0.344 |
| rs10453225 | TMEM38B | 9 | 107960041 | G | −0.074 | −0.053 | 9.7 × 10−5 | 0.411 |
| rs246185 | MKL2 | 16 | 14302933 | C | −0.131 | −0.019 | 1.9 × 10−6 | 4.2 × 10−4 |
| rs2274465 | KDM4A | 1 | 43894144 | C | −0.071 | 0.006 | 0.014 | 0.010 |
| rs2947411 | TMEM18 | 2 | 604168 | A | 0.014 | −0.060 | 0.017 | 0.046 |
| rs1400974 | SATB2 | 2 | 199346935 | A | 0.005 | −0.046 | 0.027 | 0.081 |
| rs4895808 | CENPW | 6 | 126823127 | C | 0.001 | −0.041 | 0.044 | 0.141 |
| rs6964833 | GTF2I | 7 | 73739845 | T | −0.021 | −0.057 | 0.007 | 0.262 |
| rs12915845 | DET1 | 15 | 86843471 | C | −0.073 | −0.022 | 0.003 | 0.072 |
| rs12446632 | GPRC5B | 16 | 19842890 | A | −0.085 | −0.045 | 0.006 | 0.303 |
| rs8050136 | FTO | 16 | 52373776 | C | 0.001 | −0.042 | 0.041 | 0.139 |
| rs10423674 | CRTC1 | 19 | 18678903 | A | −0.002 | −0.045 | 0.033 | 0.145 |
| C. Opposite effects | ||||||||
| rs268067 | BCL11A | 2 | 59734549 | A | 0.034 | −0.039 | 0.101 | 0.048 |
| rs2687729 | EEFSEC | 3 | 129377916 | G | −0.037 | 0.031 | 0.082 | 0.031 |
| rs4840086 | SIM1 | 6 | 100315159 | A | 0.031 | −0.033 | 0.060 | 0.027 |
| rs7821178 | PEX2 | 8 | 78256392 | C | −0.049 | 0.011 | 0.108 | 0.044 |
| rs1324913 | KLF12 | 13 | 73533589 | G | 0.050 | −0.038 | 0.011 | 0.003 |
| rs9635759 | CA10 | 17 | 46968784 | A | 0.039 | −0.046 | 0.014 | 0.007 |
Best nearby candidate gene according to Perry, 2014.
Position shown for genome build 37.
Menarche-delaying allele.
P-value for sex difference is defined as the combined P-value of both sexes assuming different effect sizes between them (2 degrees of freedom), according to http://www.well.ox.ac.uk/gwama/tutorial.shtml and Mägi, Lindgren, and Morris, 2010.
P-value for sex heterogeneity is defined as the amount of heterogeneity in allelic effects between the sexes (1 degree of freedom), according to http://www.well.ox.ac.uk/gwama/tutorial.shtml and Mägi, Lindgren, and Morris, 2010 [31].
Sex-specific effects at LIN28B have been investigated in humans and mouse models. In humans, LIN28B variants have sex-specific associations with postnatal growth (25), and are associated with adiposity traits in adult women, but not men (27, 28). LIN28A and LIN28B participate in a negative feedback loop with the let-7 family of microRNAs. In mouse models of this pathway, Lin28b loss of function (LOF) and reciprocal let-7 gain of function (GOF) mice both displayed sex-specific effects on pubertal timing and growth (Corre, et al., submitted). Such data suggest that the LIN28-let-7 system is likely to have complex, partly sex-specific influences on growth and pubertal timing and that further study of this pathway in humans is warranted.
Explaining sex differences
Sex-specific genetic effects are common in model organisms (30,31), and recent GWAS show that common genetic variation can influence traits or diseases in a sex-specific manner in humans (see (32) for a recent review). Puberty is a dimorphic trait that displays sex-specific genetic effects. The mechanisms underlying sex-specificity remain largely speculative. While sex hormones (testosterone and estrogen) are the primary drivers of differential gene expression, resulting in different trait manifestations and disease risks (33), these hormones do not explain all sex differences. For example, in blastocysts, which are pre-gonadal and lack influence from sex hormones, almost a third of detected gene transcripts showed sexually dimorphic gene expression (34), pointing toward an alternative mechanism prior gonadal steroid synthesis.
The most fundamental difference between male and female cells is the sex chromosome complement (Fig. 2). The X and Y chromosomes are structurally heteromorphic (37) but share a pseudoautosomal region containing 29 genes (38); the human X and Y chromosomes have about 1400 (39) and 27 (40) unique genes, respectively. In humans, Turner (45-XO) and Kleinfelter (47-XXY) syndromes illustrate the implications of having the correct complement of sex chromosomes (41). In mice, the Four Core Genotypes (FCG) model has been used to test the contribution of the sex chromosomes versus the gonads (42). In this model, a normal XX female is mated with a XY−Sry male (with the Sry region translocated from the Y chromosome to an autosome), creating offspring with four potential genotypes: XX females, XY− females with normal autosomes and no Sry, XXSry males, and XY−Sry males with Sry on an autosome. In studies utilizing this model, X vs Y sex chromosome effects have been seen for many traits regardless of the hormonal milieu, including adiposity (43), metabolism (44), HDL cholesterol (45), food intake (46,47), hypothalamic neuronal development (48), brain structure and function (49), and juvenile behavior (50).
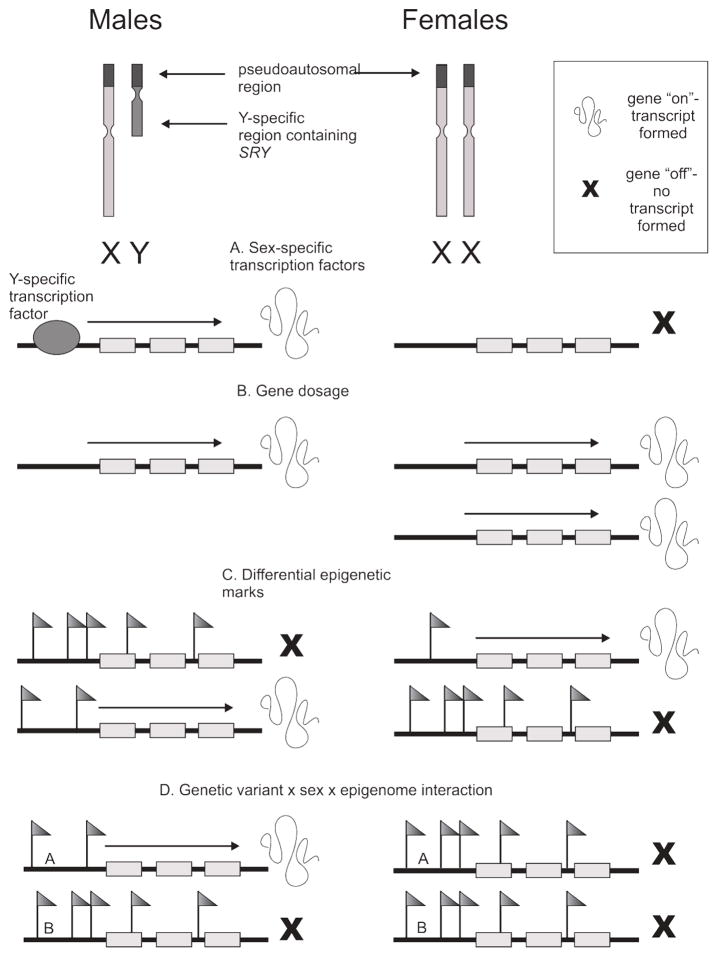
Fig. 2. (Original) Schematic representation of several mechanisms of sex-specific gene expression.
The most fundamental difference between male and female cells is the sex chromosome complement; males possess an X and a Y chromosome, while females have two X chromosomes. Some examples of how male and female cells may experience differential gene expression include A) sex-specific transcription factors, such as the male-specific SRY gene on the Y chromosome; B) gene dosage due to incomplete X-inactivation in females, resulting in higher transcript levels in females for some genes; and C) sex-differentiated epigenetic marks, which may cause differential regulatory element usage, expression of different isoforms in each sex, or even differential gene expression entirely. In D) a real example of a genetic variant × sex × epigenome interaction is schematically represented. In (35,36), GWAS-associated genetic variants at 7q12-q21 were associated with childhood-onset asthma in boys only, via sex-specific DNA methylation in response to smoking exposure. In the figure, A and B represent two DNA sequence variants in the associated region. Only boys with variant A have active gene expression in the presence of sex-specific DNA methylation.
One way the sex chromosomes influence autosomal gene expression is through sex-linked genes acting as transcription factors, such as SRY (51,52) (Fig. 2a). Additionally, the sex chromosomes can reportedly affect epigenetic regulation of gene expression on the autosomes (53). In Drosophila, the large heterochromatic Y chromosome directly impacts autosomal gene expression through effects on the epigenetic status of other chromosomes (54–57). The heterochromatic inactive X may also bias autosomal expression (52,53).
Gene dosage is another contributor to sex-specific gene expression. Most genes on the second female X chromosome are randomly switched off, but around 15% escape X-inactivation and are expressed at twice the level found in males (58). Mammalian embryos may have sex-specific gene expression before random X-inactivation (33), which may have later-life effects on gene expression. In addition, 6–10% of autosomal genes are monoallelically expressed (59), and in these cases, parent-of-origin effects such as at MKRN3 can occur where a variant is expressed when it comes from one parent and not the other.
Epigenetic marks can be sex-specific and have lasting impact on sex-differentiated gene expression. For example, an X-linked gene was found that had significantly lower expression in males and was reduced further by in utero maternal stress (60). This gene had long-term effects on metabolic and neurodevelopmental programming (61). Several studies showed long-lasting sex-specific epigenetic changes with transcriptional implications for nearby genes in the adult brain and liver, some of which are hormone-dependent (62,63). However, a small but significant increase in global autosomal methylation is associated with being male (64), apparently not driven by sex hormones (65). Epigenetic mechanisms are likely to play a large role in how certain genetic polymorphisms regulate gene expression differently in males and females. In lymphoblast cell lines grown without sex hormones, 12–15% of autosomal expression quantitative trait loci (eQTLs), genomic variants associated with variation in mRNA expression levels, were expressed in a sex-specific manner (66) due to differential usage of regulatory elements or isoforms (67,68).
In GWAS, mechanisms underlying sex-specific genetic associations remain mostly unknown. In two cases, a sex-specific eQTL overlapped a previously reported disease SNP, one for eosinophilic esophagitis and one for Crohn’s disease, two conditions more common in males (66). In other studies, epigenetic mechanisms, such as sex-specific DNA methylation in response to environmental cues, may be sequence-dependent and have been highlighted (63,64).
Conclusions
Genetic investigation of pubertal timing is an active field of research with implications for understanding the biological basis for how pubertal maturation is triggered. In future research, more emphasis should be placed on sex-specific effects, as these may provide keys to understanding sexual dimorphism in pubertal onset and perhaps the associations between timing and later life health outcomes, some of which are also sex-specific.
Supplementary Material
Quadrants a and d represent loci that show opposite directions of effect in males vs. females, while quadrants b and c are loci that have effect sizes in the same direction in both sexes. While the majority of menarche-delaying SNPs also associate with later sexual development in both sexes as assessed by the Tanner scale, there are notable exceptions for specific loci (see Table 2).
Key Points.
Significant discoveries into the genetic background of pubertal timing have been made in recent years by incorporating ever-larger sample sizes, samples with diverse ethnic backgrounds, and traits which also enable the study of puberty in males.
Studies in both sexes show that many genetic effects are similar between boys and girls, but differences also exist that may be important for understanding sexual dimorphism in pubertal timing and the associations between the timing of puberty and adult health.
Many mechanisms, including hormone-independent ones, may be responsible for sex-specific effects.
Acknowledgments
Financial support
Dr. Palmert acknowledges past support from the National Institutes of Health and current funding from the Hospital for Sick Children and the Canadian Institutes of Health Research. Dr. Widen acknowledges the Academy of Finland and Finska Läkaresällskapet for their financial support.
Footnotes
Conflicts of interest
There are no conflicts of interest to declare.
References
- 1**.Day FR, Elks CE, Murray A, Ong KK, Perry JRB. Puberty timing associated with diabetes, cardiovascular disease and also diverse health outcomes in men and women: the UK Biobank study. Sci Rep. 2015;5:11208. doi: 10.1038/srep11208. The largest study of its kind to date, this study investigated the correlation between pubertal timing and adverse adult health outcomes in 250,000 women and nearly 200,000 men and noted both previously observed and novel associations with 48 health outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Perry JRB, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014 Oct 2;514(7520):92–7. doi: 10.1038/nature13545. This paper is a large genome-wide association study of age at menarche in 180,000 women of European origin which described 106 loci associated with the trait. Of note is the enrichment for loci that show parent-of-origin imprinting effects, where only the maternally or paternally inherited variant is associated with menarche. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elks CE, Perry JRB, Sulem P, Chasman DI, Franceschini N, He C, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010 Dec;42(12):1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4**.Lunetta KL, Day FR, Sulem P, Ruth KS, Tung JY, Hinds DA, et al. Rare coding variants and X-linked loci associated with age at menarche. Nat Commun. 2015;6:7756. doi: 10.1038/ncomms8756. Investigating two previously overlooked areas, this study published novel significantly associated loci for age at menarche in low-frequency protein coding variants and on the X chromosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5*.Rzeczkowska PA, Hou H, Wilson MD, Palmert MR. Epigenetics: a new player in the regulation of mammalian puberty. Neuroendocrinology. 2014;99(3–4):139–55. doi: 10.1159/000362559. This paper overviews recent developments in our understanding of the importance of epigenetics in the timing of puberty. [DOI] [PubMed] [Google Scholar]
- 6.Spencer KL, Malinowski J, Carty CL, Franceschini N, Fernández-Rhodes L, Young A, et al. Genetic variation and reproductive timing: African American women from the Population Architecture using Genomics and Epidemiology (PAGE) Study. PloS One. 2013;8(2):e55258. doi: 10.1371/journal.pone.0055258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demerath EW, Liu C-T, Franceschini N, Chen G, Palmer JR, Smith EN, et al. Genome-wide association study of age at menarche in African-American women. Hum Mol Genet. 2013 May 1; doi: 10.1093/hmg/ddt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert CA, Tishkoff SA. Genetic structure in African populations: implications for human demographic history. Cold Spring Harb Symp Quant Biol. 2009;74:395–402. doi: 10.1101/sqb.2009.74.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyun J-A, Kim S, Cho NH, Koh I, Lee J-Y, Shin C, et al. Genome-wide association studies and epistasis analyses of candidate genes related to age at menarche and age at natural menopause in a Korean population. Menopause N Y N. 2014 May;21(5):522–9. doi: 10.1097/GME.0b013e3182a433f7. [DOI] [PubMed] [Google Scholar]
- 10.Carty CL, Spencer KL, Setiawan VW, Fernandez-Rhodes L, Malinowski J, Buyske S, et al. Replication of genetic loci for ages at menarche and menopause in the multi-ethnic Population Architecture using Genomics and Epidemiology (PAGE) study. Hum Reprod Oxf Engl. 2013 Jun;28(6):1695–706. doi: 10.1093/humrep/det071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croteau-Chonka DC, Lange LA, Lee NR, Adair LS, Mohlke KL. Replication of LIN28B SNP association with age of menarche in young Filipino women. Pediatr Obes. 2013 Oct;8(5):e50–e53. doi: 10.1111/j.2047-6310.2013.00178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, Gill JC, et al. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013 Jun 27;368(26):2467–75. doi: 10.1056/NEJMoa1302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macedo DB, Abreu AP, Reis ACS, Montenegro LR, Dauber A, Beneduzzi D, et al. Central precocious puberty that appears to be sporadic caused by paternally inherited mutations in the imprinted gene makorin ring finger 3. J Clin Endocrinol Metab. 2014 Jun;99(6):E1097–1103. doi: 10.1210/jc.2013-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Settas N, Dacou-Voutetakis C, Karantza M, Kanaka-Gantenbein C, Chrousos GP, Voutetakis A. Central Precocious Puberty in a Girl and Early Puberty in Her Brother Caused by a Novel Mutation in the MKRN3 Gene. J Clin Endocrinol Metab. 2014 Apr;99(4):E647–651. doi: 10.1210/jc.2013-4084. [DOI] [PubMed] [Google Scholar]
- 15.Schreiner F, Gohlke B, Hamm M, Korsch E, Woelfle J. MKRN3 mutations in familial central precocious puberty. Horm Res Pædiatrics. 2014;82(2):122–6. doi: 10.1159/000362815. [DOI] [PubMed] [Google Scholar]
- 16.De Vries L, Gat-Yablonski G, Dror N, Singer A, Phillip M. A novel MKRN3 missense mutation causing familial precocious puberty. Hum Reprod Oxf Engl. 2014 Dec;29(12):2838–43. doi: 10.1093/humrep/deu256. [DOI] [PubMed] [Google Scholar]
- 17.Lee HS, Jin H-S, Shim YS, Jeong HR, Kwon E, Choi V, et al. Low Frequency of MKRN3 Mutations in Central Precocious Puberty Among Korean Girls. Horm Metab Res Horm Stoffwechselforschung Horm Metab. 2015 May 4; doi: 10.1055/s-0035-1548938. [DOI] [PubMed] [Google Scholar]
- 18.Schaaf CP, Gonzalez-Garay ML, Xia F, Potocki L, Gripp KW, Zhang B, et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet. 2013 Nov;45(11):1405–8. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Hagen CP, Sørensen K, Mieritz MG, Johannsen TH, Almstrup K, Juul A. Circulating MKRN3 levels decline prior to pubertal onset and through puberty: a longitudinal study of healthy girls. J Clin Endocrinol Metab. 2015 May;100(5):1920–6. doi: 10.1210/jc.2014-4462. This study substantiated previous evidence from rodents that MKRN3 level declines prior to puberty, supporting the notion that MKRN3 acts as a brake that must be lifted for puberty to proceed. [DOI] [PubMed] [Google Scholar]
- 20.Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosch M, Kaidar G, et al. Epigenetic control of female puberty. Nat Neurosci. 2013 Mar;16(3):281–9. doi: 10.1038/nn.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demetriou CA, Chen J, Polidoro S, van Veldhoven K, Cuenin C, Campanella G, et al. Methylome analysis and epigenetic changes associated with menarcheal age. PloS One. 2013;8(11):e79391. doi: 10.1371/journal.pone.0079391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann A, Zimmermann CA, Spengler D. Molecular epigenetic switches in neurodevelopment in health and disease. Front Behav Neurosci [Internet] 2015 May 13; doi: 10.3389/fnbeh.2015.00120. [cited 2015 Aug 3];9. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4429584/ [DOI] [PMC free article] [PubMed]
- 23.Ojeda SR, Lomniczi A. Puberty in 2013: Unravelling the mystery of puberty. Nat Rev Endocrinol. 2014 Feb;10(2):67–9. doi: 10.1038/nrendo.2013.233. [DOI] [PubMed] [Google Scholar]
- 24.Cousminer DL, Berry DJ, Timpson NJ, Ang W, Thiering E, Byrne EM, et al. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum Mol Genet. 2013 Mar 21; doi: 10.1093/hmg/ddt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Widén E, Ripatti S, Cousminer DL, Surakka I, Lappalainen T, Järvelin M-R, et al. Distinct variants at LIN28B influence growth in height from birth to adulthood. Am J Hum Genet. 2010 May 14;86(5):773–82. doi: 10.1016/j.ajhg.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cousminer DL, Stergiakouli E, Berry DJ, Ang W, Groen-Blokhuis MM, Körner A, et al. Genome-wide association study of sexual maturation in males and females highlights a role for body mass and menarche loci in male puberty. Hum Mol Genet. 2014 Apr 28; doi: 10.1093/hmg/ddu150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leinonen JT, Surakka I, Havulinna AS, Kettunen J, Luoto R, Salomaa V, et al. Association of LIN28B with adult adiposity-related traits in females. PloS One. 2012;7(11):e48785. doi: 10.1371/journal.pone.0048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong KK, Elks CE, Wills AK, Wong A, Wareham NJ, Loos RJF, et al. Associations between the pubertal timing-related variant in LIN28B and BMI vary across the life course. J Clin Endocrinol Metab. 2011 Jan;96(1):E125–129. doi: 10.1210/jc.2010-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu W, Wagner EK, Eckert GJ, Yu Z, Hannon T, Pratt JH, et al. Associations between menarche-related genetic variants and pubertal growth in male and female adolescents. J Adolesc Health Off Publ Soc Adolesc Med. 2015 Jan;56(1):66–72. doi: 10.1016/j.jadohealth.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008 Dec;9(12):911–22. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magi R, Lindgren CM, Morris AP. Meta-analysis of sex-specific genome-wide association studies. Genet Epidemiol. 2010;34(8):846–53. doi: 10.1002/gepi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Gilks WP, Abbott JK, Morrow EH. Sex differences in disease genetics: evidence, evolution, and detection. Trends Genet TIG. 2014 Oct;30(10):453–63. doi: 10.1016/j.tig.2014.08.006. This paper provides a review of recent studies reporting sex-specific genetic associations, as well as a discussion about the evolutionary implications. [DOI] [PubMed] [Google Scholar]
- 33.Arnold AP, Chen X, Itoh Y. What a difference an X or Y makes: sex chromosomes, gene dose, and epigenetics in sexual differentiation. Handb Exp Pharmacol. 2012;(214):67–88. doi: 10.1007/978-3-642-30726-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci U S A. 2010 Feb 23;107(8):3394–9. doi: 10.1073/pnas.0913843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naumova AK, Al Tuwaijri A, Morin A, Vaillancourt VT, Vaillancout VT, Madore A-M, et al. Sex- and age-dependent DNA methylation at the 17q12-q21 locus associated with childhood asthma. Hum Genet. 2013 Jul;132(7):811–22. doi: 10.1007/s00439-013-1298-z. [DOI] [PubMed] [Google Scholar]
- 36.Xie P, Kranzler HR, Zhang H, Oslin D, Anton RF, Farrer LA, et al. Childhood adversity increases risk for nicotine dependence and interacts with α5 nicotinic acetylcholine receptor genotype specifically in males. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2012 Feb;37(3):669–76. doi: 10.1038/npp.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Cox KH, Bonthuis PJ, Rissman EF. Mouse model systems to study sex chromosome genes and behavior: relevance to humans. Front Neuroendocrinol. 2014 Oct;35(4):405–19. doi: 10.1016/j.yfrne.2013.12.004. This article reviews the mouse model systems used to distinguish between sex chromosome and hormonal effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, et al. The DNA sequence of the human X chromosome. Nature. 2005 Mar 17;434(7031):325–37. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Disteche CM. Sex differences in brain expression of X- and Y-linked genes. Brain Res. 2006 Dec 18;1126(1):50–5. doi: 10.1016/j.brainres.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 40.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003 Jun 19;423(6942):825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 41.Hong DS, Hoeft F, Marzelli MJ, Lepage J-F, Roeltgen D, Ross J, et al. Influence of the X-chromosome on neuroanatomy: evidence from Turner and Klinefelter syndromes. J Neurosci Off J Soc Neurosci. 2014 Mar 5;34(10):3509–16. doi: 10.1523/JNEUROSCI.2790-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold AP, Chen X. What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol. 2009 Jan;30(1):1–9. doi: 10.1016/j.yfrne.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, McClusky R, Chen J, Beaven SW, Tontonoz P, Arnold AP, et al. The number of x chromosomes causes sex differences in adiposity in mice. PLoS Genet. 2012;8(5):e1002709. doi: 10.1371/journal.pgen.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Link JC, Chen X, Arnold AP, Reue K. Metabolic impact of sex chromosomes. Adipocyte. 2013 Apr 1;2(2):74–9. doi: 10.4161/adip.23320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Link JC, Chen X, Prien C, Borja MS, Hammerson B, Oda MN, et al. Increased High-Density Lipoprotein Cholesterol Levels in Mice With XX Versus XY Sex Chromosomes. Arterioscler Thromb Vasc Biol. 2015 Aug;35(8):1778–86. doi: 10.1161/ATVBAHA.115.305460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Wang L, Loh DH, Colwell CS, Taché Y, Reue K, et al. Sex differences in diurnal rhythms of food intake in mice caused by gonadal hormones and complement of sex chromosomes. Horm Behav. 2015 Jul 29;75:55–63. doi: 10.1016/j.yhbeh.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seu E, Groman SM, Arnold AP, Jentsch JD. Sex chromosome complement influences operant responding for a palatable food in mice. Genes Brain Behav. 2014 Jul;13(6):527–34. doi: 10.1111/gbb.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scerbo MJ, Freire-Regatillo A, Cisternas CD, Brunotto M, Arevalo MA, Garcia-Segura LM, et al. Neurogenin 3 mediates sex chromosome effects on the generation of sex differences in hypothalamic neuronal development. Front Cell Neurosci. 2014;8:188. doi: 10.3389/fncel.2014.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corre C, Friedel M, Vousden DA, Metcalf A, Spring S, Qiu LR, et al. Separate effects of sex hormones and sex chromosomes on brain structure and function revealed by high-resolution magnetic resonance imaging and spatial navigation assessment of the Four Core Genotype mouse model. Brain Struct Funct. 2014 Dec 2; doi: 10.1007/s00429-014-0952-0. [DOI] [PubMed] [Google Scholar]
- 50.Cox KH, Rissman EF. Sex differences in juvenile mouse social behavior are influenced by sex chromosomes and social context. Genes Brain Behav. 2011 Jun;10(4):465–72. doi: 10.1111/j.1601-183X.2011.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubin RA, Ostrer H. Sry is a transcriptional activator. Mol Endocrinol Baltim Md. 1994 Sep;8(9):1182–92. doi: 10.1210/mend.8.9.7838151. [DOI] [PubMed] [Google Scholar]
- 52.Wijchers PJ, Yandim C, Panousopoulou E, Ahmad M, Harker N, Saveliev A, et al. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev Cell. 2010 Sep 14;19(3):477–84. doi: 10.1016/j.devcel.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011 Jan 4;27(4):132–40. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 54.Jiang P-P, Hartl DL, Lemos B. Y not a dead end: epistatic interactions between Y-linked regulatory polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics. 2010 Sep;186(1):109–18. doi: 10.1534/genetics.110.118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci U S A. 2010 Sep 7;107(36):15826–31. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemos B, Branco AT, Jiang P-P, Hartl DL, Meiklejohn CD. Genome-wide gene expression effects of sex chromosome imprinting in Drosophila. G3 Bethesda Md. 2014 Jan;4(1):1–10. doi: 10.1534/g3.113.008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Francisco FO, Lemos B. How Do Y-Chromosomes Modulate Genome-Wide Epigenetic States: Genome Folding, Chromatin Sinks, and Gene Expression. J Genomics. 2014 May 1;2:94–103. doi: 10.7150/jgen.8043. This review looks at mechanisms by which repetitive elements on the Drosophila Y chromosome may modulate autosomal gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005 Mar 17;434(7031):400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 59*.Massah S, Beischlag TV, Prefontaine GG. Epigenetic events regulating monoallelic gene expression. Crit Rev Biochem Mol Biol. 2015 Jul;9:1–22. doi: 10.3109/10409238.2015.1064350. This paper reviews the current knowledge about monoallelic expression via imprinting and stochastic mechanisms. [DOI] [PubMed] [Google Scholar]
- 60.Howerton CL, Morgan CP, Fischer DB, Bale TL. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):5169–74. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howerton CL, Bale TL. Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc Natl Acad Sci U S A. 2014 Jul 1;111(26):9639–44. doi: 10.1073/pnas.1401203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62*.Tyler CR, Hafez AK, Solomon ER, Allan AM. Developmental exposure to 50 parts-per-billion arsenic influences histone modifications and associated epigenetic machinery in a region- and sex-specific manner in the adult mouse brain. Toxicol Appl Pharmacol. 2015 Jul 17;288(1):40–51. doi: 10.1016/j.taap.2015.07.013. This study is an example of an environmental exposure that leads to long-lasting sex-specific epigenetic alterations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito S, Hirabayashi K, Moriishi K, Matsui Y, Moriya K, Koike K, et al. Novel sex-dependent differentially methylated regions are demethylated in adult male mouse livers. Biochem Biophys Res Commun. 2015 Jul 10;462(4):332–8. doi: 10.1016/j.bbrc.2015.04.137. [DOI] [PubMed] [Google Scholar]
- 64.McCarthy NS, Melton PE, Cadby G, Yazar S, Franchina M, Moses EK, et al. Meta-analysis of human methylation data for evidence of sex-specific autosomal patterns. BMC Genomics. 2014;15:981. doi: 10.1186/1471-2164-15-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iwasaki M, Ono H, Kuchiba A, Kasuga Y, Yokoyama S, Onuma H, et al. Association of postmenopausal endogenous sex hormones with global methylation level of leukocyte DNA among Japanese women. BMC Cancer. 2012;12:323. doi: 10.1186/1471-2407-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dimas A, Nica A, Montgomery S, Stranger B, Raj T, Buil A, et al. Sex-biased genetic effects on gene regulation in humans. Genome Res. 2012 Sep 7; doi: 10.1101/gr.134981.111. gr.134981.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010 Feb;20(2):180–9. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su W-L, Modrek B, GuhaThakurta D, Edwards S, Shah JK, Kulkarni AV, et al. Exon and junction microarrays detect widespread mouse strain- and sex-bias expression differences. BMC Genomics. 2008;9:273. doi: 10.1186/1471-2164-9-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quadrants a and d represent loci that show opposite directions of effect in males vs. females, while quadrants b and c are loci that have effect sizes in the same direction in both sexes. While the majority of menarche-delaying SNPs also associate with later sexual development in both sexes as assessed by the Tanner scale, there are notable exceptions for specific loci (see Table 2).