Abstract
IMPORTANCE
In schizophrenia, working memory deficits appear to reflect abnormalities in the generation of gamma oscillations in the dorsolateral prefrontal cortex. The generation of gamma oscillations requires the phasic excitation of inhibitory parvalbumin-containing interneurons. Thus, gamma oscillations depend, in part, on the number of synaptic glutamate receptors on parvalbumin interneurons. However, little is known about the molecular factors that regulate glutamate receptor–mediated excitation of parvalbumin interneurons in schizophrenia.
OBJECTIVE
To quantify in individuals with schizophrenia the expression of immediate early genes (NARP, ARC, and SGK1) regulating glutamate synaptic neurotransmission.
DESIGN, SETTING, AND PARTICIPANTS
Postmortem brain specimens (n = 206) were obtained from individuals with schizophrenia, bipolar disorder, or major depressive disorder and from well-matched healthy persons (controls). For a study of brain tissue, quantitative polymerase chain reaction, in situ hybridization, or microarray analyses were used to measure transcript levels in the dorsolateral prefrontal cortex at gray matter, laminar, and cellular levels of resolutions. This study was conducted between January 1, 2013, and November 30, 2014.
MAIN OUTCOMES AND MEASURES
Expression levels for NARP, ARC, and SGK1 messenger RNA (mRNA) were compared between specimens from individuals with schizophrenia and controls. Diagnostic specificity was assessed by quantifying NARP mRNA levels in specimens from individuals with mood disorders.
RESULTS
By quantitative polymerase chain reaction, levels of NARP mRNA were significantly lower by 25.6%in specimens from individuals with schizophrenia compared with the controls (mean [SD], 0.036 [0.018] vs 0.049 [0.015]; F1,114 = 21.0; P < .001). Levels of ARC (F1,112 = 0.93; P = .34) and SGK1 (F1,110 = 2.52; P = .12) were not significant. These findings were supported by in situ hybridization (NARP; individuals with schizophrenia vs controls: 40.1% lower [P = .003]) and microarray analyses (NARP; individuals with schizophrenia vs controls: 12.2%lower in layer 3 [P = .11] and 14.6%lower in layer 5 pyramidal cells [P = .001]). In schizophrenia specimens, NARP mRNA levels were positively correlated with GAD67 mRNA (r = 0.55; P < .001); the expression of GAD67 mRNA in parvalbumin interneurons is activity dependent. The NARP mRNA levels were also lower than healthy controls in bipolar disorder (−18.2%; F1,60 = 11.39; P = .001) and major depressive disorder (−21.7%; F1,30 = 5.36; P = .03) specimens, especially those from individuals with psychosis. In all 3 diagnostic groups, NARP mRNA levels were positively correlated (all r ≥ 0.53; all P ≤ .02) with somatostatin mRNA, the expression of which is activity dependent.
CONCLUSIONS AND RELEVANCE
Given the role of NARP in the formation of excitatory inputs to parvalbumin (and perhaps somatostatin) interneurons, our findings suggest that lower NARP mRNA expression contributes to lower excitatory drive onto parvalbumin interneurons in schizophrenia. This reduced excitatory drive may lead to lower synthesis of γ-aminobutyric acid in these interneurons, contributing to a reduced capacity to generate the gamma oscillations required for working memory.
The neural substrate for cognitive functions, including working memory, involves synchronization of cortical neuronal activity at gamma frequency oscillations.1–4 Consequently, gamma oscillation abnormalities in the dorsolateral prefrontal cortex (DLPFC) are thought to contribute to working memory deficits in schizophrenia.5–8 Gamma oscillations require the synchronized inhibition of neighboring populations of pyramidal neurons by the parvalbumin-containing basket cell subclass of γ-aminobutyric acid interneurons.9–11 Specifically, excitatory input from pyramidal neurons activates parvalbumin basket neurons, which furnish feedback inhibition to pyramidal neurons.12 Because the axons of parvalbumin basket neurons are highly divergent,13 this feedback inhibition simultaneously hyperpolarizes multiple neighboring pyramidal neurons. The fast and synchronous decay of this inhibition enables the simultaneous firing of these pyramidal neurons at gamma band frequency.14 Thus, given their importance in gamma oscillations, excitatory inputs onto DLPFC parvalbumin interneurons might be a key component in the neural circuitry basis of working memory.
The strong coupling of the phasic excitation of parvalbumin interneurons with gamma oscillation frequency15 suggests that the composition of synaptic glutamate receptors on parvalbumin interneurons is critical for gamma oscillations.16 The α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid/N-methyl-D-aspartate receptor (AMPAR/NMDAR) ratio in synapses onto parvalbumin interneurons is approximately 3 times greater than onto cortical pyramidal neurons and other γ-aminobutyric acid interneurons.17–19 Selective knockout of the AMPAR subunits GluR1 or GluR4 from parvalbumin interneurons reduces both their phasic excitatory drive and the power of gamma oscillations.20 Furthermore, fast AMPAR-mediated excitation of parvalbumin interneurons is sufficient to support gamma oscillations.19,21 Collectively, these data suggest that alterations of the AMPAR-mediated excitatory inputs onto parvalbumin interneurons could contribute to impaired gamma oscillations in schizophrenia.
One potential regulator of AMPAR-mediated excitatory input onto parvalbumin interneurons is the immediate early gene, neuronal activity−regulated pentraxin (NARP; GenBank NM_002523), which is prominently expressed in pyramidal neurons in response to neuronal activation22 and is secreted at presynaptic axon terminals in glutamate synapses onto parvalbumin interneurons.23 At these synapses, NARP binding helps to cluster GluR4-containing AMPARs (Figure 1A) and enhances excitatory input to parvalbumin interneurons.23,24 Consistent with these findings, mice homozygous for a NARP gene knockout exhibit reduced excitatory inputs onto parvalbumin interneurons.24 These findings suggest that deficient NARP messenger RNA (mRNA) expression contributes to lower AMPAR-mediated excitation of parvalbumin interneurons in schizophrenia.
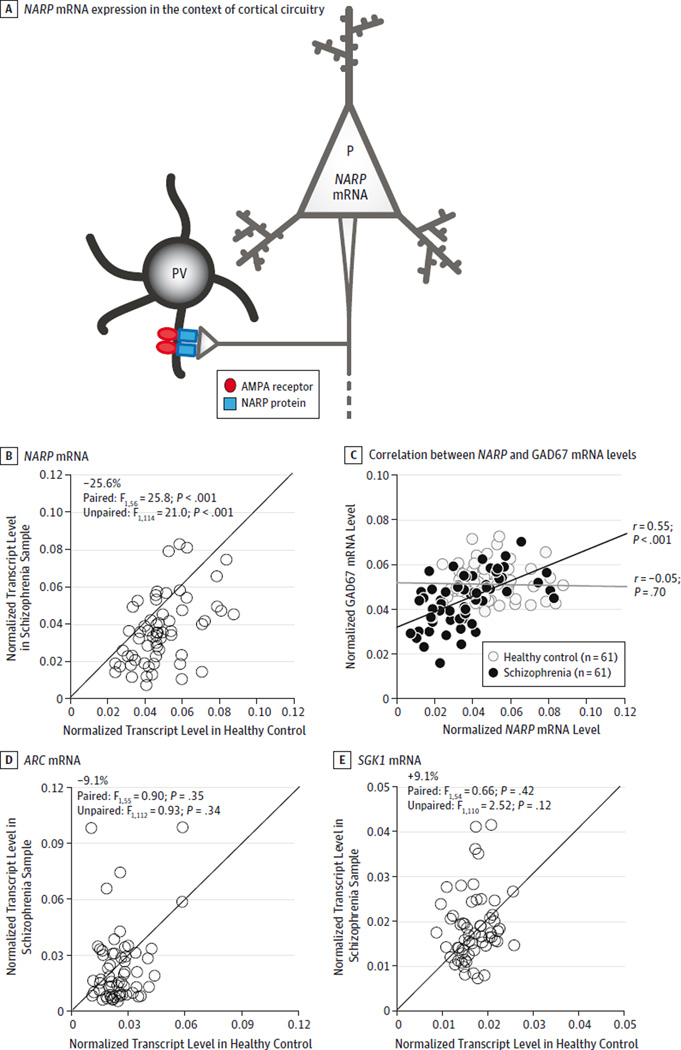
Figure 1. Polymerase Chain Reaction Determination of Relative Messenger RNA (mRNA) Levels for NARP, ARC, and SGK1 in Schizophrenia and Healthy Control Samples.
NARP mRNA is expressed in pyramidal cells (P), and NARP protein is secreted at excitatory synapses on the dendrites of parvalbumin interneurons (PV) (A). At these synapses, NARP contributes to the clustering of AMPA receptors. The levels of NARP (B), ARC (D), and SGK1 (E) mRNA for each control and schizophrenia sample in a pair. Data points below the diagonal unity line indicate lower mRNA levels in the schizophrenia sample relative to its matched control and vice versa. B, Mean NARP mRNA level was significantly lower in schizophrenia samples relative to matched controls. C, NARP mRNA levels were positively correlated with GAD67 mRNA levels in schizophrenia samples (black line) but not in controls (gray line). D and E, Mean levels of ARC and SGK1 mRNA did not differ significantly between groups.
To test this hypothesis, we used quantitative polymerase chain reaction (qPCR), in situ hybridization, and microarray analyses to quantify the expression of NARP mRNA at the gray matter, laminar, and cellular levels in the DLPFC from a large cohort of brain tissue samples from individuals with schizophrenia as well as healthy comparison individuals (controls). To determine the molecular specificity of altered NARP expression, we examined them RNA levels of 2 other immediate early genes that regulate AMPAR-mediated excitatory synaptic transmission through different mechanisms: activity-regulated, cytoskeleton-associated protein (ARC; GenBank NM_015193) and serum/glucocorticoid-regulated kinase 1 (SGK1; GenBank NM_001143677). Unlike NARP, ARC regulates homeostatic scaling of AMPARs specifically on pyramidal neurons,25 whereas SGK1 affects glutamate transmission without cell-type specificity.26 To test the disease process specificity of altered NARP expression, we examined NARP mRNA levels in the DLPFC in brain tissue samples from individuals with bipolar disorder or major depressive disorder and in samples from monkeys with long-term exposure to antipsychotic medications. Finally, because the expression of glutamic acid decarboxylase 67 kD (GAD67), the major enzyme of γ-aminobutyric acid synthesis in the cortex, is regulated by neuronal activity27,28 and is lower in parvalbumin interneurons in the DLPFC of people with schizophrenia,29,30 we also determined whether NARP expression predicted GAD67 mRNA levels in the DLPFC.
Methods
Human Tissue Samples
Brain specimens (n = 206) were obtained at the Allegheny County Office of the Medical Examiner (Pittsburgh, Pennsylvania) after consent was obtained from next of kin. Diagnoses were made using DSM-IV-R for each patient,31 and tissue was collected from DLPFC area 9, as described previously30,32 (eMethods in the Supplement). All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research and Clinical Trials Involving the Dead and Institutional Review Board for Biomedical Research. The study was conducted between January 1, 2013, and November 30, 2014.
To control for experimental variance and reduce biological variance between groups, each sample from a person with schizophrenia or schizoaffective disorder (n = 62), bipolar disorder (n = 35) (21 met the criteria for bipolar 1 disorder), or major depressive disorder (n = 19) was matched with a sample from 1 control (n = 90) for the donor’s sex and as closely as possible for age (Table; eTable 1 in the Supplement provides details on each person); some control samples were matched to more than 1 sample from an individual with a psychiatric illness. Consequently, to compare the transcript levels between groups, individual data are presented in scatterplots that show the values for both members in a sample pair. For each set of disease and comparison samples, group means did not differ significantly for age, postmortem interval, RNA integrity number (Agilent Bioanalyzer), brain pH, or tissue storage time at −80°C with one exception: brain pH significantly differed (t61 = 2.68; P = .01) between groups in the schizophrenia cohort, but the mean difference was small (0.1 pH unit) and of uncertain biological relevance.
Table.
Summary of Demographic and Postmortem Characteristics of Individuals Contributing Tissue Samples
| Characteristic | Controls (n = 62) |
Schizophrenia (n = 62) |
Controls (n = 35) |
Bipolar Disorder (n = 35) |
Controls (n = 19) |
Major Depressive Disorder (n = 19) |
|---|---|---|---|---|---|---|
| Sex, No. (%) | ||||||
| Male | 47 (76) | 47 (76) | 20 (57) | 20 (57) | 10 (53) | 10 (53) |
| Female | 15 (24) | 15 (24) | 15 (43) | 15 (43) | 9 (47) | 9 (47) |
| Race, No. (%) | ||||||
| White | 52 (84) | 46 (74) | 32 (91) | 34 (97) | 18 (95) | 18 (95) |
| Black | 10 (16) | 16 (26) | 3 (9) | 1 (3) | 1 (5) | 1 (5) |
| Age, mean (SD), y | 48.7 (13.8) | 47.7 (12.7) | 46.6 (12.7) | 45.5 (12.2) | 47.8 (10.4) | 46.3 (9.5) |
| PMI, mean (SD), h | 18.8 (5.5) | 19.2 (8.5) | 19.1 (5.1) | 20.5 (7.0) | 19.3 (5.3) | 21.3 (6.6) |
| Brain pH, mean (SD) | 6.7 (0.2) | 6.6 (0.3) | 6.7 (0.3) | 6.6 (0.3) | 6.6 (0.2) | 6.6 (0.2) |
| RIN, mean (SD) | 8.2 (0.6) | 8.1 (0.6) | 8.1 (0.6) | 8.0 (0.5) | 8.0 (0.6) | 8.0 (0.4) |
| Storage time, mean (SD), moa | 125.8 (54.5) | 122.0 (58.8) | 106.8 (49.8) | 113.7 (45.1) | 124.7 (43.4) | 140.5 (28.8) |
Abbreviations: PMI, postmortem interval; RIN, RNA integrity number.
Storage at −80°C.
Quantitative PCR
Quantitative PCR was performed to determine relative expression levels of each target transcript (eTable 2A in the Supplement), as described previously33 (eMethods in the Supplement). Three internal reference transcripts (β-actin, cyclophilin A, and glyceraldehyde-3-phosphate dehydrogenase), selected based on their stable expression across the subjects in this cohort regardless of diagnoses,34,35 were used to normalize the target transcripts (eMethods in the Supplement).
In Situ Hybridization
In situ hybridization procedures for NARP mRNA (eTable 2B in the Supplement) were performed with 3 tissue sections from each donor33 (eMethods in the Supplement). The NARP mRNA levels in area 9 were measured at total gray matter, laminar, and cellular levels of resolution (eMethods and eFigure 1 in the Supplement).
Microarray Analyses
For each sample, 200 pyramidal cells in deep layer 3 and in layer 5 of area 9 were individually dissected and then pooled per layer per subject (LMD 6500; Leica Microsystems), as described previously.36 The synthesized complementary DNA was then loaded onto an array plate (Affymetric GeneChip HT HG-U133+ PM; Affymetrix).36
Antipsychotic-Exposed Monkeys
The effect of long-term exposure to antipsychotic medication on altered NARP mRNA levels in schizophrenia was examined using macaque monkeys exposed for a long duration to haloperidol, olanzapine, or placebo (6 monkeys per group)32,37 (eMethods and eTable 2A in the Supplement). All procedures followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the University of Pittsburgh’s Institutional Animal Care and Use Committee.
Statistical Analysis
qPCR and In Situ Hybridization
Paired and unpaired analyses of covariance models, including covariates of sex, age, postmortem interval, brain pH, RNA integrity number, and storage time, were used to test the effects of diagnostic group on gene expression levels determined by qPCR and for total, laminar, and cellular NARP mRNA levels determined by in situ hybridization (eMethods in the Supplement). The results from both models are illustrated in this report, and only the results from the unpaired model are reported in the text except when the P values differed regarding significance. Within each group, the samples with transcript levels that were greater than 3 SDs from the group mean were considered as outliers for that measure and removed. Consequently, reported degrees of freedom differ slightly across analyses.
The influence of comorbid factors on NARP mRNA expression within each diagnostic group was assessed with the unpaired analysis of covariance model (eMethods in the Supplement). The relationship between each immediate early gene and GAD67 mRNA levels was assessed by Pearson correlation. For the antipsychotic-exposed monkeys, analyses of covariance models were used with treatment group as a main effect and triad as a blocking factor. All P values were 2-tailed, and the significance level was set at .05.
Microarray Analysis
Probe sets from the array were filtered, and paired t tests were performed using the random intercept model, as described previously.36 Discovery of differentially expressed genes was conducted using meta-analysis, and an adaptively weighted Fisher method was applied for each transcript. Meta-analyzed P values from the adaptively weighted testing were then adjusted by the Benjamini-Hochberg procedure for multiple comparisons to control false discovery rate.36
Results
qPCR for NARP, ARC, and SGK1 mRNA in Schizophrenia
The mean level of NARP mRNA (Figure 1B) was significantly lower by 25.6% in the schizophrenia samples compared with their matched controls (F1,114 = 21.0; P < .001). The NARP mRNA levels were lower in the schizophrenia samples for 51 of the 62 pairs (Figure 1B). However, mean mRNA levels of ARC and SGK1 (Figure 1D and E) did not differ significantly between groups (ARC: F1,112 = 0.93; P = .34; and SGK1: F1,110 = 2.52; P = .12).
In the unpaired analysis of covariance model, age was a significant determinant of NARP mRNA expression in the gray matter (F1,114 = 41.2; P < .001), and NARP mRNA levels were similarly negatively correlated with age in the schizophrenia (r = −0.49; P < .001) and control (r = −0.55; P < .001) groups (eFigure 2A in the Supplement). Levels of NARP mRNA in schizophrenia samples did not differ significantly (all F <2.91; all P > .09) as a function of sex; diagnosis of schizoaffective disorder; history of substance dependence or abuse; nicotine use at the time of death; use of antipsychotics, antidepressants or benzodiazepines, and/or sodium valproate at the time of death; or death by suicide (eFigure 2B in the Supplement). Finally, levels of NARP mRNA did not differ significantly (F2,10 = 0.42; P = .67) among monkeys with long-term exposure to haloperidol, olanzapine, or placebo (eFigure 3 in the Supplement).
We examined the relationship between changes in NARP mRNA expression and GAD67 mRNA in the DLPFC using GAD67 mRNA levels previously reported in the same 62 subject pairs.33 Levels of NARP and GAD67 mRNA as determined by qPCR were positively correlated in schizophrenia samples (r = 0.55; P < .001) but not in the control samples (r = −0.05; P = .70) (Figure 1C). In contrast, neither ARC nor SGK1 mRNA levels were significantly correlated with GAD67 mRNA levels in either the schizophrenia or control samples (all r <0.18; all P > .17).
In Situ Hybridization for NARP mRNA in Schizophrenia
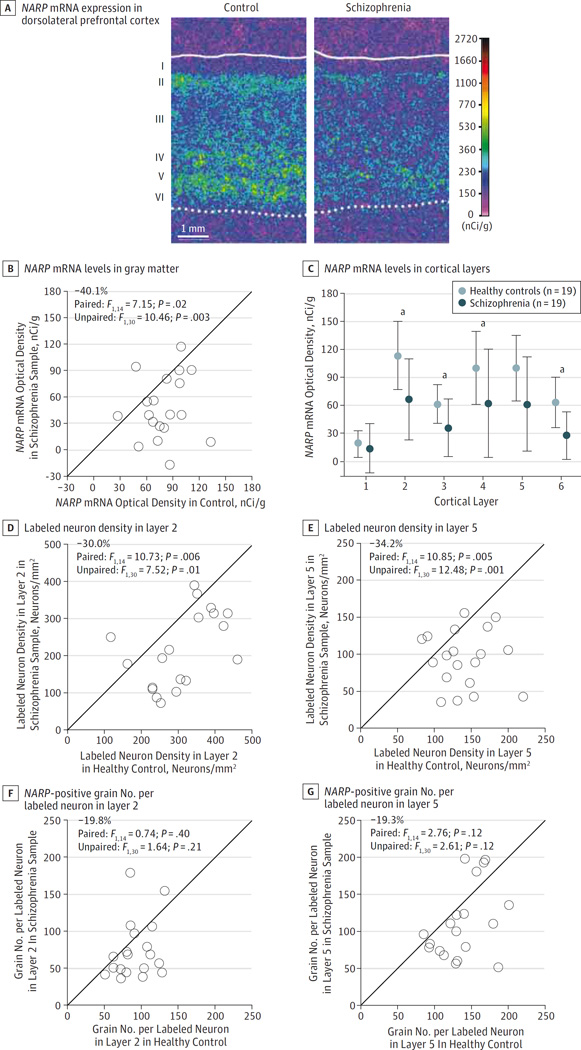
In situ hybridization (with a probe directed against a different portion of NARP mRNA than in the primer set used in the qPCR study) was performed to quantify mRNA levels in the DLPFC from 20 sample pairs with a sufficient number of available tissue sections (eTable 1 in the Supplement). Mean total gray matter levels of NARP mRNA were significantly (F1,30 = 10.46; P = .003) lower by 40.1% in schizophrenia samples compared with matched controls (mean [SD], 47.6 [35.8] vs 79.4 [24.7] nCi/g) (Figure 2A and B), a difference similar to that found by qPCR (−34.1%) in the same samples. Furthermore, levels of NARP mRNA quantified by qPCR or in situ hybridization were highly correlated (r = 0.77; P < .001) across all samples (n = 38). Analysis by cortical layers revealed that NARP mRNA expression was significantly lower in layers 2 (−41.7%;F1,30 = 11.5;P = .002), 3 (−42.0%;F1,30 = 9.80;P = .004), 4 (−38.2%; F1,30 = 6.91; P = .013), and 6 (−56.2%; F1,30 = 15.9; P = .001) and nearly so in layer 5 (−38.8%; paired: F1,14 = 3.78; P = .07; unpaired: F1,30 = 6.31; P = .02) (Figure 2C).
Figure 2. In Situ Hybridization Analyses for NARP Messenger RNA (mRNA) Between Schizophrenia and Healthy Control Samples.
A through C, In situ hybridization film analysis. A, Representative pseudocolored film autoradiographs of dorsolateral prefrontal cortex sections illustrating NARP mRNA expression levels in 1 pair of schizophrenia and control tissue samples. The solid and dotted lines indicate the pial surface and the gray-white matter border, respectively. B, Mean NARP mRNA levels across the gray matter for each control and schizophrenia sample in a pair. Data points below the diagonal unity line indicate lower NARP mRNA levels in the schizophrenia sample relative to the matched control and vice versa. Mean NARP mRNA levels in schizophrenia samples were significantly lower relative to those of the matched controls. C, Mean NARP mRNA levels in each cortical layer in schizophrenia and healthy comparison subjects. Mean (SD) NARP mRNA levels were significantly lower in layers 2, 3, 4, and 6 and nearly so in layer 5 in schizophrenia samples relative to matched controls (see the In Situ Hybridization for NARP mRNA in Schizophrenia subsection of the Results section for statistical results). D through G, Cellular grain counting analysis. D and F, NARP-positive neurons per millimeters squared. E and G, Grains per positive neuron in layers 2 and 5 for each control and schizophrenia sample in a pair. Data points below the diagonal unity lines indicate lower NARP mRNA levels in the schizophrenia sample relative to its matched control and vice versa. The mean number of NARP-positive neurons per millimeter squared in schizophrenia samples was lower in both layers 2 and 5 compared with matched controls. In contrast, the difference in mean grain density per NARP-positive neuron in layers 2 and 5 did not differ significantly between groups.
a P < .01.
At the cellular level, the mean number of NARP-positive neurons per millimeters squared was significantly lower (Figure 2D and E) in both layer 2 (−30.0%; F1,30 = 7.52; P = .01) and layer 5 (−34.2%; F1,30 = 12.48; P = .001) in schizophrenia samples relative to the matched control samples. In contrast, the mean grain density per NARP-positive neuron did not significantly differ in either layer 2 or 5 (all F1,30 <2.62; all P > .11) between groups (Figure 2F and G).
Microarray Analyses for NARP mRNA in Schizophrenia
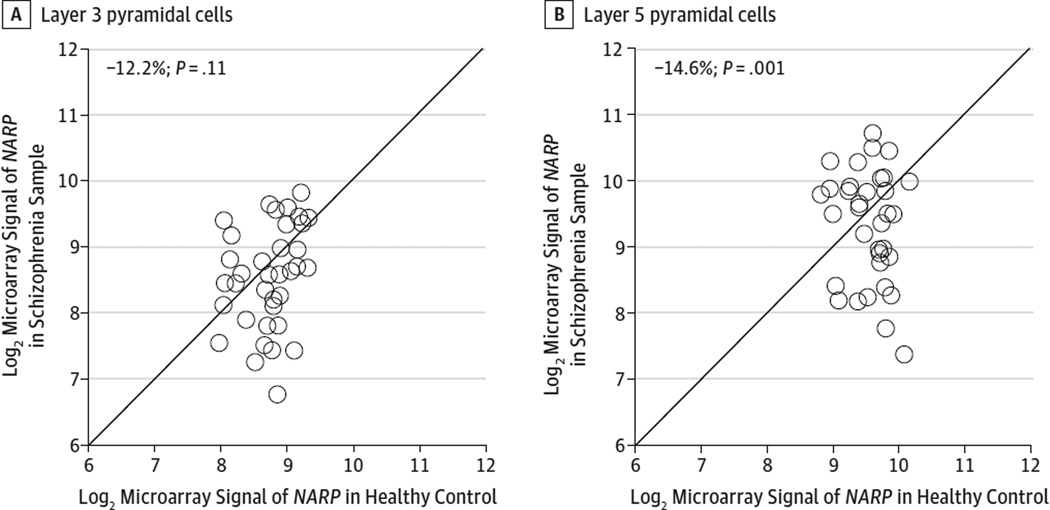
Recently, Arion et al36 performed microarray analyses from pools of individually dissected pyramidal neurons (eTable 1 in the Supplement) and neurons labeled with Vicia villosa agglutinin,38 which is present in the perineuronal nets that surround parvalbumin interneurons. In controls, NARP mRNA expression as determined by microarray was approximately 16 times higher in pyramidal than parvalbumin cells in area 9 from the same samples. In pyramidal neurons, mean levels of NARP mRNA were significantly lower (Figure 3A and B) in layer 5 (−14.6%; P = .001) and nearly so in layer 3 (−12.2%; P = .11) in the schizophrenia samples compared with the matched controls; these findings show the same direction of change to the 21% reduction in mean NARP mRNA levels in total gray matter as determined by qPCR for the same 36 pairs of subjects. Finally, consistent with the gray matter qPCR findings, mean levels of ARC and SGK1 mRNA in layer 3 or 5 pyramidal cells did not differ significantly between schizophrenia and control samples (all P > .58).
Figure 3. Microarray Analyses of NARP Messenger RNA (mRNA) Levels in Nissl-Stained Pyramidal Neurons in Matched Pairs of Schizophrenia and Control Samples.
Log2-transformed microarray signals of NARP mRNA in layer 3 (A) and layer 5 (B) for schizophrenia samples relative to matched controls are plotted for each pair. The data points below the diagonal unity line indicate lower mRNA signals in the schizophrenia sample relative to its matched control and vice versa.
qPCR for NARP mRNA in Mood Disorders
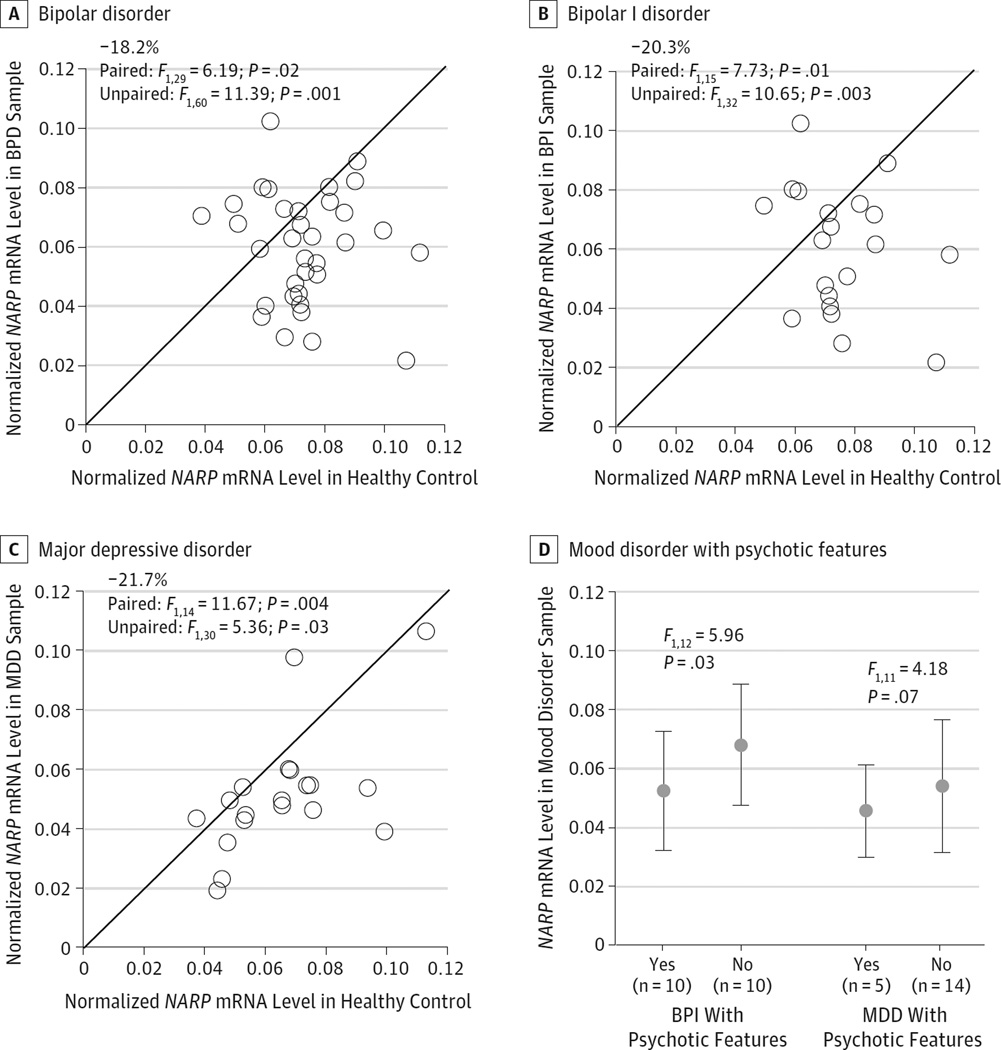
Mean NARP mRNA levels were significantly lower in samples from individuals with bipolar disorder (−18.2%; F1,60 = 11.39; P = .001), bipolar 1 disorder only (−20.3%; F1,32 = 10.65; P = .003), and major depressive disorder (−21.7%; F1,30 = 5.36; P = .03) relative to their matched controls (Figure 4A–C). To explore the relationship between NARP and GAD67 mRNA levels, we used GAD67 mRNA levels previously reported35 in matched 18 triads of bipolar disorder, major depressive disorder, and control samples included in the present study. As determined by qPCR, NARP and GAD67 mRNA levels were positively correlated in the bipolar disorder and major depressive disorder samples (all r >0.47; all P < .048) but not in their matched controls (all r < 0.25; all P > .35). In addition, 10 bipolar disorder samples (all from individuals who had bipolar I disorder) and 5 major depressive disorder samples were from individuals with a history of psychotic features (eTable 1 in the Supplement). Compared with those without a history of psychotic features, samples from those with a history of psychotic features had lower NARP mRNA levels (Figure 4D) for both the bipolar 1 disorder (−23.1%;F1.12 = 5.96;P = .03) and major depressive disorder (−15.9%; F1.11 = 4.18; P = .07) groups.
Figure 4. Quantitative Polymerase Chain Reaction Determination of Relative NARP Messenger RNA (mRNA) Levels in Mood Disorders and Effect of Psychotic Features on NARP mRNA Expression Levels in Bipolar I (BPI) Disorder and Major Depressive Disorder (MDD).
Scatterplots indicate the levels of NARP mRNA for each healthy control sample and bipolar disorder (BPD) (A), BPI disorder (B), or MDD (C) subject in a pair. Data points below the diagonal unity line indicate lower mRNA levels in the mood disorder subject relative to the matched healthy control and vice versa. Mean NARP mRNA level was significantly lower in subjects with BPD, BPI disorder, and MDD with matched healthy controls. Mean NARP mRNA levels for BPI disorder and individuals with MDD are grouped by psychotic features (D).
Discussion
Using total gray matter, layer, and cellular levels of resolution and different techniques (qPCR, in situ hybridization, and microarray), we found convergent evidence of lower NARP mRNA expression in the DLPFC of brain specimens from individuals with schizophrenia that was most prominent in a subset of pyramidal neurons. Our finding of a lower density of NARP mRNA-positive neurons is unlikely to represent a deficit in the number of NARP-expressing neurons in schizophrenia based on several findings. First, a stereologic study39 reported no significant difference in the total number of neurons in the prefrontal cortex samples from individuals with schizophrenia compared with controls. Second, the density of pyramidal neurons was reported to be slightly increased across cortical layers40 or to be unchanged in layer 3 of the DLPFC.41 In concert, these findings suggest that lower NARP is attributable to a downregulation of NARP mRNA in existing pyramidal neurons and not to a deficit in the number of these neurons.
Given the role of secreted NARP in regulating AMPAR clustering on parvalbumin interneurons23 (Figure 1A), lower NARP mRNA expression might contribute to lower AMPAR-mediated excitatory synaptic inputs onto parvalbumin interneurons in schizophrenia. Because GAD67 expression is activity dependent,27,28 disease-related alterations in expression of the activity-dependent gene NARP could contribute to altered levels of GAD67 mRNA in individuals with schizophrenia. This hypothesis was supported by the significant positive correlation between GAD67 and NARP mRNA levels in the schizophrenia samples, indicating that the larger range of both GAD67 and NARP mRNA levels in these samples revealed the predicted positive correlation between 2 transcripts. Furthermore, although other populations of γ-aminobutyric acid interneurons may be affected, only parvalbumin interneurons have been directly shown to have lower levels of GAD67 mRNA29 and protein42 in schizophrenia. Together, these findings support a coupling effect of lower NARP expression in pyramidal cells, with a downstream, activity-dependent deficit in expression of GAD67 in parvalbumin interneurons. However, studies characterizing NARP protein levels specifically at pyramidal cell inputs to parvalbumin interneurons in individuals with schizophrenia are needed to validate this hypothesis.
The regulation of AMPAR-mediated excitatory synaptic transmission differs among NARP, ARC, and SGK1, with all expression being activity dependent.23,25,26 In contrast to our NARP findings, mRNA levels of ARC and SGK1 did not differ significantly between groups and did not correlate with levels of GAD67 mRNA in the schizophrenia or control samples. Thus, lower expression of NARP mRNA in pyramidal neurons might be a specific aspect of the disease process of schizophrenia and not a general consequence of a hypoactive cortical circuit that would be expected to be associated with lower expression levels of multiple immediate early genes.
Lower levels of NARP mRNA in people with schizophrenia appear to be due to the disease process rather than to factors frequently associated with the illness. First, among the 62 samples from individuals with schizophrenia studied by qPCR, none of the examined comorbid factors accounted for lower NARP mRNA expression. Second, NARP mRNA expression was unaltered in the DLPFC of monkeys with long-term exposure to either haloperidol or olanzapine. Third, age and NARP mRNA expression were similarly negatively correlated in both the control and schizophrenia samples, with the regression line for schizophrenia samples parallel to and shifted downward from that for the controls. In a previous study43 using the same tissue samples as in the present study, neither illness duration nor age accounted for lower levels of GAD67 expression in schizophrenia. Thus, the lower NARP and GAD67 mRNA levels in schizophrenia samples seen across specimens from individuals aged 17 to 83 years in the present study are unlikely to be a consequence of illness progression and may reflect blunting or incomplete developmental trajectories of these transcripts. However, we cannot rule out a decline in NARP mRNA expression during the early stages of clinical illness because we do not have data on the people immediately before and after the onset of psychosis.
Mean levels of NARP mRNA were also significantly lower in specimens from individuals with bipolar disorder or major depressive disorder, especially those with psychotic features, suggesting that lower NARP mRNA levels might contribute to the greater cognitive impairments observed in bipolar disorder or major depressive disorder with psychotic features.44,45 However, GAD67 mRNA levels were unchanged in the specimens from these subjects independent of the presence or absence of psychotic features.35 Thus, although lower NARP mRNA levels could be a conserved feature across major psychiatric illnesses, achieving deficits in cortical GAD67 levels might require factors in addition to fewer NARP mediated glutamatergic inputs from pyramidal neurons. For example, allelic variants in the GAD1 gene associated with increased risk for schizophrenia,46 altered chromatin structures at the GAD1 promoter region,47,48 or lower levels of the GAD67 transcription factor Zif26833 could also be required for deficits in GAD67 expression to emerge in schizophrenia.
Alternatively, lower expression of NARP in schizophrenia, bipolar disorder, and major depressive disorder could be upstream of other alterations that are common to all 3 diagnoses. For example, NARP protein was recently localized to somatostatin interneurons where it is thought to regulate excitatory synapse development.49 Because somatostatin gene expression is activity dependent,50,51 lower NARP expression could contribute to the deficits in somatostatin mRNA levels observed in schizophrenia,32,52–55 bipolar disorder,35,53 and major depressive disorder.35 Consistent with this interpretation, NARP mRNA levels were similarly positively correlated with previously determined somatostatin levels in the specimens from individuals with schizophrenia (r = 0.74; P < .01 [n = 61]),56 bipolar disorder, (r = 0.76; P < .01 [n = 17]),35 and major depressive disorder (r = 0.53; P = .02 [n = 18])35 examined in the present study.
Conclusions
The present findings suggest that lower NARP mRNA expression in DLPFC pyramidal neurons may lead to reduced clustering of AMPAR at excitatory synapses on parvalbumin interneurons, thus contributing to lower excitatory drive to these neurons. In individuals with schizophrenia, this reduced excitatory drive could result in lower GAD67 expression and presumably lower γ-aminobutyric acid synthesis in parvalbumin interneurons. Given the role of parvalbumin interneurons in the generation of gamma oscillations, altered NARP expression could contribute to the molecular basis for altered gamma oscillations and impaired cognition in schizophrenia. Across individuals with psychosis, lower NARP mRNA levels might also lead to less clustering of AMPAR at excitatory synapses on somatostatin interneurons, perhaps contributing to the dysfunction of these neurons seen across diagnoses.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by National Institutes of Health grants MH043784 and MH084053 (Dr Lewis) from the National Institute of Mental Health and Nara Medical University (Dr Kimoto).
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Kimoto and Lewis had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Kimoto, Lewis.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Kimoto, Zaki, Lewis.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Kimoto, Zaki.
Obtained funding: Kimoto, Lewis.
Administrative, technical, or material support: Bazmi, Lewis.
Study supervision: Bazmi, Lewis.
Conflict of Interest Disclosures: Dr Lewis reports receiving investigator-initiated research support from Bristol-Myers Squibb and Pfizer and in 2012–2014 served as a consultant in the areas of target identification and validation and new compound development to Autifony, Bristol-Myers Squibb, and Concert Pharmaceuticals. No other disclosures are reported.
Additional Contributions: Mary Brady, BS (University of Pittsburgh), assisted with editing the graphics, and Dominique Arion, PhD (University of Pittsburgh), assisted in providing the results of microarray analyses. There was no financial compensation.
REFERENCES
- 1.Herrmann CS, Munk MH, Engel AK. Cognitive functions of gamma-band activity: memory match and utilization. Trends Cogn Sci. 2004;8(8):347–355. doi: 10.1016/j.tics.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Howard MW, Rizzuto DS, Caplan JB, et al. Gamma oscillations correlate with working memory load in humans. Cereb Cortex. 2003;13(12):1369–1374. doi: 10.1093/cercor/bhg084. [DOI] [PubMed] [Google Scholar]
- 3.Roux F, Wibral M, Mohr HM, Singer W, Uhlhaas PJ. Gamma-band activity in human prefrontal cortex codes for the number of relevant items maintained in working memory. J Neurosci. 2012;32(36):12411–12420. doi: 10.1523/JNEUROSCI.0421-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto J, Suh J, Takeuchi D, Tonegawa S. Successful execution of working memory linked to synchronized high-frequency gamma oscillations. Cell. 2014;157(4):845–857. doi: 10.1016/j.cell.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Cho RY, Konecky RO, Carter CS. Impairments in frontal cortical γ synchrony and cognitive control in schizophrenia. Proc Natl Acad Sci U S A. 2006;103(52):19878–19883. doi: 10.1073/pnas.0609440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lesh TA, Niendam TA, Minzenberg MJ, Carter CS. Cognitive control deficits in schizophrenia: mechanisms and meaning. Neuropsychopharmacology. 2011;36(1):316–338. doi: 10.1038/npp.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35(1):57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2010;11(2):100–113. doi: 10.1038/nrn2774. [DOI] [PubMed] [Google Scholar]
- 9.Bartos M, Vida I, Jonas P. Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci. 2007;8(1):45–56. doi: 10.1038/nrn2044. [DOI] [PubMed] [Google Scholar]
- 10.Cardin JA, Carlén M, Meletis K, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459(7247):663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459(7247):698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis DA, Gonzalez-Burgos G. Neuroplasticity of neocortical circuits in schizophrenia. Neuropsychopharmacology. 2008;33(1):141–165. doi: 10.1038/sj.npp.1301563. [DOI] [PubMed] [Google Scholar]
- 13.Packer AM, Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J Neurosci. 2011;31(37):13260–13271. doi: 10.1523/JNEUROSCI.3131-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Burgos G, Lewis DA. GABA neurons and the mechanisms of network oscillations: implications for understanding cortical dysfunction in schizophrenia. Schizophr Bull. 2008;34(5):944–961. doi: 10.1093/schbul/sbn070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hájos N, Paulsen O. Network mechanisms of gamma oscillations in the CA3 region of the hippocampus. Neural Netw. 2009;22(8):1113–1119. doi: 10.1016/j.neunet.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Burgos G, Lewis DA. NMDA receptor hypofunction, parvalbumin-positive neurons, and cortical gamma oscillations in schizophrenia. Schizophr Bull. 2012;38(5):950–957. doi: 10.1093/schbul/sbs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hull C, Isaacson JS, Scanziani M. Postsynaptic mechanisms govern the differential excitation of cortical neurons by thalamic inputs. J Neurosci. 2009;29(28):9127–9136. doi: 10.1523/JNEUROSCI.5971-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korotkova T, Fuchs EC, Ponomarenko A, von Engelhardt J, Monyer H. NMDA receptor ablation on parvalbumin-positive interneurons impairs hippocampal synchrony, spatial representations, and working memory. Neuron. 2010;68(3):557–569. doi: 10.1016/j.neuron.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Rotaru DC, Yoshino H, Lewis DA, Ermentrout GB, Gonzalez-Burgos G. Glutamate receptor subtypes mediating synaptic activation of prefrontal cortex neurons: relevance for schizophrenia. J Neurosci. 2011;31(1):142–156. doi: 10.1523/JNEUROSCI.1970-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs EC, Zivkovic AR, Cunningham MO, et al. Recruitment of parvalbumin-positive interneurons determines hippocampal function and associated behavior. Neuron. 2007;53(4):591–604. doi: 10.1016/j.neuron.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 21.Caputi A, Fuchs EC, Allen K, Le Magueresse C, Monyer H. Selective reduction of AMPA currents onto hippocampal interneurons impairs network oscillatory activity. PLoS One. 2012;7(6):e37318. doi: 10.1371/journal.pone.0037318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsui CC, Copeland NG, Gilbert DJ, Jenkins NA, Barnes C, Worley PF. NARP, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J Neurosci. 1996;16(8):2463–2478. doi: 10.1523/JNEUROSCI.16-08-02463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang MC, Park JM, Pelkey KA, et al. NARP regulates homeostatic scaling of excitatory synapses on parvalbumin-expressing interneurons. Nat Neurosci. 2010;13(9):1090–1097. doi: 10.1038/nn.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Y, Huang S, Chang MC, Worley P, Kirkwood A, Quinlan EM. Obligatory role for the immediate early gene NARP in critical period plasticity. Neuron. 2013;79(2):335–346. doi: 10.1016/j.neuron.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shepherd JD, Rumbaugh G, Wu J, et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52(3):475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang F, Strutz-Seebohm N, Seebohm G, Lang UE. Significance of SGK1 in the regulation of neuronal function. J Physiol. 2010;588(pt 18):3349–3354. doi: 10.1113/jphysiol.2010.190926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson DL, Huntsman MM, Jones EG. Activity-dependent changes in GAD and preprotachykinin mRNAs in visual cortex of adult monkeys. Cereb Cortex. 1994;4(1):40–51. doi: 10.1093/cercor/4.1.40. [DOI] [PubMed] [Google Scholar]
- 28.Lau CG, Murthy VN. Activity-dependent regulation of inhibition via GAD67. J Neurosci. 2012;32(25):8521–8531. doi: 10.1523/JNEUROSCI.1245-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto T, Volk DW, Eggan SM, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23(15):6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical γ-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57(3):237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 31.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th. Washington, DC: American Psychiatric Association; 2000. text revision. [Google Scholar]
- 32.Volk DW, Matsubara T, Li S, et al. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry. 2012;169(10):1082–1091. doi: 10.1176/appi.ajp.2012.12030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimoto S, Bazmi HH, Lewis DA. Lower expression of glutamic acid decarboxylase 67 in the prefrontal cortex in schizophrenia: contribution of altered regulation by Zif268. Am J Psychiatry. 2014;171(9):969–978. doi: 10.1176/appi.ajp.2014.14010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165(4):479–489. doi: 10.1176/appi.ajp.2007.07081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sibille E, Morris HM, Kota RS, Lewis DA. GABA-related transcripts in the dorsolateral prefrontal cortex in mood disorders. Int J Neuropsychopharmacol. 2011;14(6):721–734. doi: 10.1017/S1461145710001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arion D, Corradi JP, Tang S, et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder [published online January 6, 2015] Mol Psychiatry. doi: 10.1038/mp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorph-Petersen KA, Pierri JN, Perel JM, Sun Z, Sampson AR, Lewis DA. The influence of chronic exposure to antipsychotic medications on brain size before and after tissue fixation: a comparison of haloperidol and olanzapine in macaque monkeys. Neuropsychopharmacology. 2005;30(9):1649–1661. doi: 10.1038/sj.npp.1300710. [DOI] [PubMed] [Google Scholar]
- 38.Georgiev D, Arion D, Enwright JF, et al. Lower gene expression for KCNS3 potassium channel subunit in parvalbumin-containing neurons in the prefrontal cortex in schizophrenia. Am J Psychiatry. 2014;171(1):62–71. doi: 10.1176/appi.ajp.2013.13040468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thune JJ, Uylings HB, Pakkenberg B. No deficit in total number of neurons in the prefrontal cortex in schizophrenics. J Psychiatr Res. 2001;35(1):15–21. doi: 10.1016/s0022-3956(00)00043-1. [DOI] [PubMed] [Google Scholar]
- 40.Selemon LD, Rajkowska G, Goldman-Rakic PS. Abnormally high neuronal density in the schizophrenic cortex: a morphometric analysis of prefrontal area 9 and occipital area 17. Arch Gen Psychiatry. 1995;52(10):805–818. doi: 10.1001/archpsyc.1995.03950220015005. [DOI] [PubMed] [Google Scholar]
- 41.Pierri JN, Volk CL, Auh S, Sampson A, Lewis DA. Somal size of prefrontal cortical pyramidal neurons in schizophrenia: differential effects across neuronal subpopulations. Biol Psychiatry. 2003;54(2):111–120. doi: 10.1016/s0006-3223(03)00294-4. [DOI] [PubMed] [Google Scholar]
- 42.Curley AA, Arion D, Volk DW, et al. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am J Psychiatry. 2011;168(9):921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoftman GD, Volk DW, Bazmi HH, Li S, Sampson AR, Lewis DA. Altered cortical expression of GABA-related genes in schizophrenia: illness progression vs developmental disturbance. Schizophr Bull. 2015;41(1):180–191. doi: 10.1093/schbul/sbt178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bora E, Yücel M, Pantelis C. Cognitive impairment in affective psychoses: a meta-analysis. Schizophr Bull. 2010;36(1):112–125. doi: 10.1093/schbul/sbp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill SK, Reilly JL, Keefe RS, et al. Neuropsychological impairments in schizophrenia and psychotic bipolar disorder: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study. Am J Psychiatry. 2013;170(11):1275–1284. doi: 10.1176/appi.ajp.2013.12101298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straub RE, Lipska BK, Egan MF, et al. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007;12(9):854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- 47.Huang HS, Matevossian A, Whittle C, et al. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1–regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27(42):11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bharadwaj R, Jiang Y, Mao W, et al. Conserved chromosome 2q31 conformations are associated with transcriptional regulation of GAD1 GABA synthesis enzyme and altered in prefrontal cortex of subjects with schizophrenia. J Neurosci. 2013;33(29):11839–11851. doi: 10.1523/JNEUROSCI.1252-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spiegel I, Mardinly AR, Gabel HW, et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type–specific gene programs. Cell. 2014;157(5):1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez-Muñoz I, Sánchez-Franco F, Vallejo M, et al. Activity-dependent somatostatin gene expression is regulated by cAMP-dependent protein kinase and Ca2+-calmodulin kinase pathways. J Neurosci Res. 2010;88(4):825–836. doi: 10.1002/jnr.22264. [DOI] [PubMed] [Google Scholar]
- 51.Tolón RM, Sánchez Franco F, de los Frailes MT, Lorenzo MJ, Cacicedo L. Effect of potassium-induced depolarization on somatostatin gene expression in cultured fetal rat cerebrocortical cells. J Neurosci. 1994;14(3, pt 1):1053–1059. doi: 10.1523/JNEUROSCI.14-03-01053.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 2010;167(12):1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- 53.Fung SJ, Fillman SG, Webster MJ, Shannon Weickert C. Schizophrenia and bipolar disorder show both common and distinct changes in cortical interneuron markers. Schizophr Res. 2014;155(1–3):26–30. doi: 10.1016/j.schres.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 54.Hashimoto T, Arion D, Unger T, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13(2):147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris HM, Hashimoto T, Lewis DA. Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder. Cereb Cortex. 2008;18(7):1575–1587. doi: 10.1093/cercor/bhm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Volk DW, Chitrapu A, Edelson JR, Lewis DA. Chemokine receptors and cortical interneuron dysfunction in schizophrenia. Schizophr Res. 2014 doi: 10.1016/j.schres.2014.10.031. S0920-9964(14)00600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.