This infection is an increasing threat to aging populations.
Keywords: Streptococcus dysgalactiae subsp. equisimilis, emm type, multilocus sequence typing, bacteria, Japan, streptococci
Abstract
We collected β-hemolytic streptococci (1,611 isolates) from patients with invasive streptococcal infections in Japan during April 2010–March 2013. Streptococcus dysgalactiae subsp. equisimilis (SDSE) was most common (n = 693); 99% of patients with SDSE infections were elderly (mean age 75 years, SD ±15 years). We aimed to clarify molecular and epidemiologic characteristics of SDSE isolates and features of patient infections. Bacteremia with no identified focus of origin and cellulitis were the most prevalent manifestations; otherwise, clinical manifestations resembled those of S. pyogenes infections. Clinical manifestations also differed by patient’s age. SDSE isolates were classified into 34 emm types; stG6792 was most prevalent (27.1%), followed by stG485 and stG245. Mortality rates did not differ according to emm types. Multilocus sequence typing identified 46 sequence types and 12 novel types. Types possessing macrolide- and quinolone-resistance genes were 18.4% and 2.6%, respectively; none showed β-lactam resistance. Among aging populations, invasive SDSE infections are an increasing risk.
Streptococcus dysgalactiae subspecies equisimilis (SDSE) belongs to the pyogenic group of streptococci first designated by Vandamme et al. in 1996 as a new subspecies within the species S. dysgalactiae (1). Previously isolated from humans, as commensal microorganisms, these streptococci have been designated β-hemolytic groups C and G because they are agglutinated by serum against Lancefield group C or G antigens. On blood agar plates, SDSE typically appears as large glossy colonies surrounded by a broad zone of strong β-hemolysis (2). For SDSE to be distinguished according to current taxonomy (3), specific biochemical properties need to be ascertained.
Although SDSE has long been considered much less virulent than S. pyogenes, many clinical and epidemiologic studies have determined that SDSE can cause a variety of severe invasive infections resembling those caused by S. pyogenes (4–12). These include not only cellulitis and deep abscesses but also streptococcal toxic shock syndrome (STSS) (13), necrotizing fasciitis, meningitis, endocarditis, and others. In addition, severity of invasive SDSE (iSDSE) infection approximates that seen with invasive S. pyogenes infection (6,9).
SDSE and S. pyogenes are considered to be closely related phylogenetically and may have originated from a common precursor (14). Moreover, recent genomic research has demonstrated that many pathogenically notable virulence factors in SDSE, including M protein, streptokinase, and streptolysin, were all encoded by genes highly homologous with those identified in S. pyogenes (15–17). However, SDSE lack several virulence factors, such as a cysteine protease (designated erythrogenic toxin B); a hyaluronic acid capsule (hasA and hasB); and an inhibitor of complement activation (sic) (17), in addition to many superantigens (18,19).
Despite this absence of some virulence factors, clinical (4,13,20) and epidemiologic reports (5,6,8–10,21) indicate that SDSE is pathogenic for humans, particularly, elderly persons with coexisting conditions. Surveillance that we conducted in 2006 implicated SDSE as a major causative pathogen in invasive β-streptococcal infections affecting the elderly in Japan (22). In industrialized countries, SDSE infections are frequent among elderly persons, especially among those with underlying medical conditions (23,24).
In Japan, we have organized large-scale epidemiologic surveillance for β-streptococci that are causing invasive infections and have identified SDSE as the most prevalent β-streptococcal pathogen since 2003 (22,25). However, information is limited regarding molecular characteristics of isolates and early indicators of prognosis for patients with these infections.
On the basis of emm genes that show polymorphisms similar to S. pyogenes (26), gene sequence analysis has been applied to emm typing for epidemiologic study of SDSE. According to the Centers for Disease Control and Prevention (CDC; http://www2a.cdc.gov/ncidod/biotech/strepblast.asp), >90 emm types have been recognized among SDSE. We previously reported that in Japan, stG485 and stG6792 were more prevalent in isolates from iSDSE infections, whereas stG10 and stG6 were more prevalent in noninvasive strains (22). Predominance of emm types also has been found to vary by geographic region.
In this study, we aimed to clarify molecular and epidemiologic characteristics of isolates from patients with iSDSE infections and the clinical features of these infections. The analysis included assessing clinical manifestations according to specific patient age group, conducting emm typing and multilocus sequence typing (MLST), and determining antimicrobial agent susceptibility and mechanisms of resistance to antimicrobial agents.
Materials and Methods
Study Design and Case Definition
We conducted nationwide surveillance of iSDSE infections during April 2010–March 2013, supported by a grant from the Japanese Ministry of Health, Labour and Welfare. After we obtained written permission from the laboratory director or hospital director, 341 general hospitals with a clinical microbiology laboratory participated in this surveillance project. Participating hospitals were located throughout Japan. Surveillance for iSDSE was carried out in parallel with 3 other investigations concerning invasive pneumococcal diseases (27), invasive S. pyogenes diseases, and invasive S. agalactiae diseases (28).
Infections with iSDSE were defined as cases in which SDSE was isolated from normally sterile clinical samples such as blood, cerebrospinal fluid, joint fluid, or pus obtained from within a closed space. Strains were sent by the various participating institutions when SDSE was re-identified by β-hemolysis on sheep blood agar (Becton Dickinson, Tokyo, Japan) and met the following criteria: agglutination results indicated Lancefield group A, C, or G; resistance to bacitracin; lack of L-pyrrolidonyl arylamidase, according to the Manual of Clinical Microbiology (2); and, for some isolates, 16S rRNA sequencing results consistent with SDSE. Isolates were stored at −80°C in 10% skim milk until use (Becton Dickinson, Sparks, MD, USA).
Requested Information
We asked attending physicians to complete and anonymously submit questionnaires along with iSDSE isolates. Requested data included patient age at onset, patient sex, origin of sample, clinical manifestation or diagnosis, underlying diseases, prior administration of antimicrobial agents, antimicrobial agent used for the infection, clinical laboratory data obtained at hospitalization, and outcome at discharge. Clinical manifestations and diagnoses were verified by a pulmonologist, according to the diagnostic criteria for sepsis based on the guidelines of the American College of Chest Physicians and the Society of Critical Care Medicine (29,30), as well as input from attending physicians, in the context of the definition of STSS established by CDC (31).
emm Typing and MLST
Typing of the emm gene was performed as described (22,25), by amplification by PCR, after which resulting PCR fragments were sequenced. Each emm type was identified by using the CDC emm sequence database (http://www2a.cdc.gov/ncidod/biotech/strepblast.asp).
MLST was performed according to the method of Ahmad et al. (32). First, 7 housekeeping genes, gki (glucose kinase), gtr (glutamine transport protein), murI (glutamate racemase), mutS (DNA mismatch repair protein), recP (transketolase), xpt (xanthine phosphoribosyl transferase), and atoB (acetoacetyl-coathioloase) were amplified, and all amplified DNA fragments were sequenced. Sequencing results for the 7 housekeeping genes in every strain each were assigned a sequence type (ST) by using the MLST website (http://sdse.mlst.net/). Relationships of each ST were analyzed by eBURST version 3.1 (http://eburst.mlst.net/v3/).
Antimicrobial Agent Susceptibility
Susceptibilities to 8 oral and 7 parenteral antimicrobial agents for SDSE strains were determined by agar-dilution methods by using Mueller-Hinton agar supplemented with 5% defibrinated sheep blood. Antimicrobial agents were obtained from their respective manufacturers. We used the following breakpoints recommended by the Clinical Laboratory Standards Institute (33): penicillin G (susceptible, ≤0.12 μg/mL); cefotaxime (susceptible, ≤0.25 μg/mL); meropenem (susceptible, ≤0.5 μg/mL); vancomycin (susceptible, ≤1 μg/mL); clarithromycin (susceptible, ≤0.25 μg/mL; intermediate, 0.5 μg/mL; resistant, ≥1 μg/mL); clindamycin (susceptible, ≤0.25 μg/mL; intermediate, 0.5 μg/mL; resistant, ≥1 μg/mL); and levofloxacin (susceptible, ≤2 μg/mL; intermediate, 4 μg/mL; resistant, ≥8 μg/mL).
Identification of Antimicrobial Resistance Determinants
Three macrolide-resistant genes, erm(A), erm(B), and mef(A), were identified in iSDSE strains by PCR methods as described (25,34). To determine fluoroquinolone resistance, quinolone-resistant determining regions of gyrA, gyrB, parC, and parE, we sequenced genes and deduced amino acid substitutions (34,35).
Statistical Analysis
We assessed statistical significance of differences for age group and specific infectious disease, macrolide or quinolone resistance, and emm type. We performed χ2 tests or the Fisher exact test using Ekuseru-Toukei 2012 software for statistics (Social Survey Research Information, Tokyo, Japan).
Results
Age Distributions of Patients
Age distributions of patients with invasive β-streptococcal infection caused by iSDSE, S. pyogenes, and S. agalactiae are shown in Figure 1. Infections caused by iSDSE were most prevalent (n = 693). During 3 successive periods, iSDSE infections accounted for the following numbers of cases: 231 during April 2010–March 2011, 216 during April 2011–March 2012, and 246 during April 2012–March 2013. Of all patients infected with iSDSE, 687 were adults >18 years of age (99.1%); only 6 were children. The mean age of adult patients with iSDSE infection was 75 years (SD ± 15 years), significantly older than those infected with S. pyogenes (61 years, SD ± 17 years) and S. agalactiae (70 years, SD ± 15 years) (p<0.001 for each).
Figure 1.
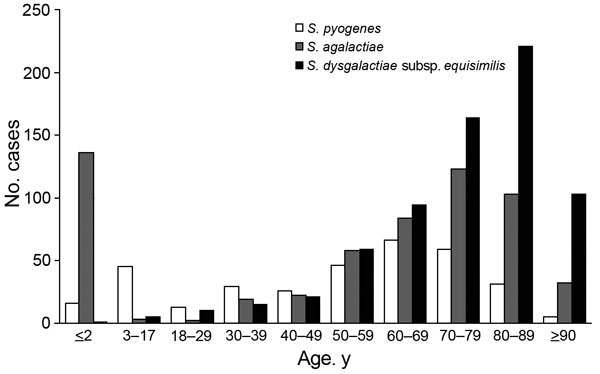
Age distribution of patients with invasive β-streptococcal infections, Japan, April 2010–March 2013. Streptococcus pyogenes, n = 336; Streptococcus agalactiae, n = 582; Streptococcus dysgalactiae subsp. equisimilis, n = 693. Means and SDs of ages in patients ≥18 years of age for each pathogen were the following: S. pyogenes (mean 61 years, SD ± 17), S. agalactiae (mean 70 years, SD ± 15), and S. dysgalactiae subsp. equisimilis (mean, 75 years, SD ± 15).
Relationships between Age Group and Clinical Manifestations
Relationships between age group and clinical manifestations in patients with iSDSE are shown in Table 1. Comorbid conditions, including a variety of underlying diseases, were found in 76.8% of patients: diabetes (22.7%), malignancies (16.7%), cardiac diseases (21.4%), and liver or renal dysfunction (16.3%). The male-to-female ratio was 1.2:1.
Table 1. Clinical manifestations and age of patients with invasive Streptococcus dysgalactiae subsp. equisimilis infection, Japan, April 2010–March 2013*.
| Clinical manifestation | No. (%) cases by age, y |
Total no. (%) cases | p value† | |||||
|---|---|---|---|---|---|---|---|---|
| <18 | 18–59 | ≥60 | ≥70 | ≥80 | ≥90 | |||
| Cellulitis | 1 (0.4) | 36 (15.4) | 26 (11.1) | 59 (25.2) | 78 (33.3) | 34 (14.5) | 234 (33.8) | 0.702 |
| Pneumonia | 2 (4.9) | 4 (9.8) | 5 (12.2) | 18 (43.9) | 12 (29.3) | 41 (5.9) | 0.006 | |
| Arthritis | 1 (2.1) | 11 (23.4) | 9 (19.1) | 13 (27.7) | 8 (17.0) | 5 (10.6) | 47 (6.8) | 0.076 |
| Abscess, noncutaneous | 1 (3.2) | 13 (41.9) | 5 (16.1) | 5 (16.1) | 5 (16.1) | 2 (6.5) | 31 (4.5) | <0.001 |
| Endocarditis | 3 (27.3) | 5 (45.5) | 2 (18.2) | 1 (9.1) | 11 (1.6) | – | ||
| Meningitis | 1 | 1 | 3 | 1 | 6 (0.9) | – | ||
| STSS | 1 | 1 | 1 | 3 (0.4) | – | |||
| Necrotizing fasciitis | 1 (6.3) | 3 (18.8) | 3 (18.8) | 3 (18.8) | 4 (25.0) | 2 (12.5) | 16 (2.3) | 0.803 |
| Cholangitis/peritonitis | 2 (14.3) | 1 (7.1) | 5 (35.7) | 5 (35.7) | 1 (7.1) | 14 (2.0) | 0.740 | |
| Osteomyelitis/spondylitis | 2 (14.3) | 5 (35.7) | 4 (28.6) | 3 (21.4) | 14 (2.0) | – | ||
| Bacteremia without primary focus | 1 (0.4) | 30 (11.1) | 39 (14.4) | 58 (21.5) | 97 (35.9) | 45 (16.7) | 270 (39.0) | 0.058 |
| Others‡ |
|
1 |
1 |
3 |
1 |
|
6 (0.9) |
– |
| Total | 6 (0.9) | 105 (15.2) | 94 (13.6) | 164 (23.7) | 221 (31.9) | 103 (14.9) | 693 (100) | |
*STSS, streptococcal toxic shock syndrome; –, not determined because of small number of strains. Blank cells indicate 0. †p values were calculated for differences between the 5 age groups, except the <18 y group. ‡Lymphangitis (n = 5) and keratitis (n = 1).
SDSE caused a variety of invasive infections. Most common was bacteremia without an identified primary focus (39.0%), followed by cellulitis (33.8%) and septic arthritis (6.8%); pneumonia with a positive blood culture accounted for 5.9%. STSS (0.4%) and necrotizing fasciitis (2.3%) occurred infrequently, as did endocarditis (1.6%), cholangitis/peritonitis (2.0%), and osteomyelitis/spondylitis (2.0%). Pneumonia occurred in patients >80 years of age (p = 0.006); in contrast, septic arthritis and noncutaneous abscesses tended to occur in patients <59 years of age (p = 0.076 and p<0.001, respectively).
emm Type, Clonal Complex, and ST
Correlations between emm type and clonal complex (CC) in iSDSE strains are shown in Table 2. The emm types were classified into 34 groups. The most prevalent was stG6792, which accounted for 27.1% of isolates, followed by stG485 (13.3%), stG245 (10.7%), stG652 (6.8%), stG10 (6.2%), and stG6 (5.5%). These 6 emm types accounted for 69.6% of types in all strains. Among strains typed as stG485 or stG245, 6 had Lancefield group A antigen rather than C or G.
Table 2. Correlation with emm type and clonal complex among Streptococcus dysgalactiae subsp. equisimilis isolates from invasive infections, Japan, April 2010–March 2013*.
| emm type | Clonal complex, no. (%) |
Total no. (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC17 | CC25 | CC29 | CC128 | CC15 | CC129 | ST138/153 | ST78/130 | Singleton | Novel ST† | ||
| stG6792 | 183 | 1 | 1 | 3 | 188 (27.1) | ||||||
| stG485 | 3 | 50 | 37 | 2 | 92 (13.3) | ||||||
| stG245 | 1 | 68 | 5 | 74 (10.7) | |||||||
| stG652 | 6 | 19 | 7 | 1 | 3 | 2 | 9 | 47 (6.8) | |||
| stG10 | 2 | 41 | 43 (6.2) | ||||||||
| stG6 | 2 | 26 | 10 | 38 (5.5) | |||||||
| stG653 | 25 | 1 | 1 | 27 (3.9) | |||||||
| stG2078 | 26 | 26 (3.8) | |||||||||
| stC36 | 9 | 1 | 14 | 1 | 25 (3.6) | ||||||
| stC74a | 18 | 18 (2.6) | |||||||||
| stG166b | 1 | 12 | 4 | 17 (2.5) | |||||||
| stG480 | 7 | 6 | 3 | 16 (2.3) | |||||||
| stG5420 | 1 | 14 | 15 (2.2) | ||||||||
| stC6979 | 13 | 13 (1.9) | |||||||||
| stG4974 | 10 | 10 (1.4) | |||||||||
| stG4222 | 4 | 6 | 10 (1.4) | ||||||||
| Others‡ |
7 |
4 |
1 |
2 |
|
1 |
|
2 |
16 |
1 |
34 (4.9) |
| Total | 287 (41.4) | 149 (21.5) | 77 (11.1) | 45 (6.5) | 49 (7.1) | 15 (2.2) | 3 (0.4) | 2 (0.3) | 45 (6.5) | 21 (3.0) | 693 (100) |
*ST, sequence type. Blank cells indicate 0. †Allele numbers for novel STs are being requested. ‡Others include stG11, stG211, stG2691, stG495, stG5345, stG97, stG643, stG62647, stC1400, stC46, stC5345, stC9431, stC10, stC839, emmG2, emmG3, stGM220, and stL1929.
MLST performed for all iSDSE strains yielded 46 STs and 12 novel STs. Allele numbers are being requested for the novel STs. Novel STs accounted for 3.0% of strains.
Results of eBURST analysis are shown in Figure 2. These STs were classified into 8 CCs and 10 singletons. Among them, CC17 was most prevalent (41.4%, n = 287), followed by CC25 (21.5%, n = 149) and CC29 (11.1%, n = 77).
Figure 2.
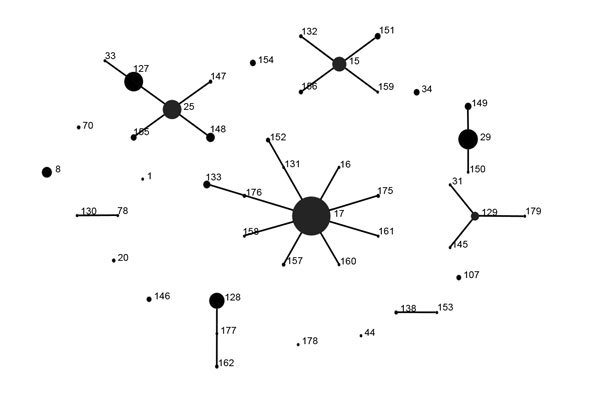
eBURST analysis (http://eburst.mlst.net/v3/) of Streptococcus dysgalactiae subsp. equisimilis from invasive infections, Japan, April 2010–March 2013. Eight clonal complexes (CC) were identified: CC15, CC17, CC25, CC29, CC128, CC129, ST78/ST130, and ST138/ST153.
To clarify relationships between emm type and CC, we identified a dominant CC for each emm type. Except for stG652 strains, CCs in almost all strains in stG6792 and stG653, and all of those in stG2078 and stG4974, were identified as CC17. Similarly, strains in stG245, stG6, stG166b, and stG5420 were assigned to CC25, stC74a to CC29, stG10 to CC15, and stC6979 to CC129. Several emm types, stG485, stG652, stC36, stG480, and stG4222, belonged to >2 different CCs. No significant correlation was found between emm type and fatality rate for infected patients (Technical Appendix Figure 1).
Identification of Novel emm Types
We identified 2 novel emm types, emmG2.0 and emmG3.0, among iSDSE strains (Technical Appendix Figure 2). G2.0 was a new emm type in which 21-bp deletions occurred in stG245.0, and emmG3.0 had a chimeric structure derived from stG3251.0 and stG485.0. In these novel emm type strains, the STs were ST33 and ST128, which belonged to CC25 and CC128, respectively. These findings suggest that strains in emmG2.0 and emmG3.0 were derived from those with stG245-CC25 and stG485-CC128.
Antimicrobial Agent Resistance and emm Type
Relationships between macrolide resistance or quinolone resistance and emm type are shown in Table 3. A total of 18.5% (n = 128) of the strains showed macrolide resistance mediated by 3 genes. Resistance conferred by the erm(A) gene, representing inducible resistance to macrolides, lincosamide, and streptogramin B, was found in 7.9% of isolates; that arising from the erm(B) gene conferring constitutive macrolide resistance was found in 8.9%; and that mediated by the mef(A) gene conferring intermediate resistance to macrolides was found in 1.6%. Resistant strains were distributed among >10 emm types. In particular, 51.4% of the stG245 strains and 44.2% of stG10 strains showed macrolide resistance mediated by the erm(A) or erm(B) gene. These types were related significantly to macrolide resistance (p<0.001).
Table 3. Correlation of emm type and macrolide or quinolone resistance genes among Streptococcus dysgalactiae subsp. equisimilis isolates, Japan, April 2010–March 2013*.
| emm type | Total no. strains | No. (%) macrolide resistance |
Total no. (%) resistance | p value | No. (%) quinolone resistance† |
Total no. (%) resistance | p value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| erm(A) | erm(B) | mef(A) | gyrA + parC | parC | ||||||
| stG6792 | 188 | 10 (5.3) | 7 (3.7) | 7 (3.7) | 24 (12.8) | 0.018 | 2 (1.1) | 3 (1.6) | 5 (2.7) | 0.950 |
| stG485 | 92 | 12 (13.0) | 1 (1.1) | 2 (2.2) | 15 (16.3) | 0.565 | 4 (4.3) | 4 (4.3) | 0.257 | |
| stG245 | 74 | 1 (1.4) | 37 (50.0) | 38 (51.4) | <0.001 | 1 (1.4) | 1 (1.4) | 0.476 | ||
| stG652 | 47 | 6 (12.8) | 1 (2.1) | 7 (14.9) | 0.513 | 3 (6.4) | 3 (6.4) | 0.091 | ||
| seG10 | 43 | 10 (23.3) | 8 (18.6) | 1(2.3) | 19 (44.2) | <0.001 | 1 (2.3) | 1 (2.3) | 0.908 | |
| stG6 | 38 | 2 (5.3) | 1 (2.6) | 1 (2.6) | 4 (10.5) | 0.194 | 0 | – | ||
| stG653 | 27 | 0 | – | 1 (3.7) | 1 (3.7) | 0.712 | ||||
| stG2078 | 26 | 6 (23.1) | 1 (3.8) | 7 (26.9) | 0.656 | 1 (3.8) | 1 (3.8) | 0.725 | ||
| stC36 | 25 | 1 (4.0) | 1 (4.0) | 2 (8.0) | 0.169 | 0 | – | |||
| stC74a | 18 | 2 (11.1) | 2 (11.1) | 0.415 | 0 | – | ||||
| stG166b | 17 | 0 | – | 0 | – | |||||
| stG480 | 16 | 2 (12.5) | 2 (12.5) | 0.534 | 0 | – | ||||
| stG5420 | 15 | 0 | – | 1 (6.7) | 1 (6.7) | 0.316 | ||||
| stC6979 | 13 | 1 (7.7) | 1 (7.7) | 0.312 | 0 | – | ||||
| stG4222 | 10 | 0 | – | 0 | – | |||||
| stG4974 | 10 | 2 (20.0) | 2 (20.0) | 0.900 | 1 (10.0) | 1 (10.0) | 0.138 | |||
| Others | 34 | 4 (11.8) | 1 (2.9) | 5 (14.7) | 0.562 | 0 | – | |||
| Total | 693 | 55 (7.9) | 62 (8.9) | 11 (1.6) | 128 (18.5) | 11 (1.6) | 7 (1.0) | 18 (2.6) | ||
*Dashes indicate p value not determined because of the small number of strains. Blank cells indicate 0. †Mutations in gyrA gene: Ser81Phe (n = 8) and Ser81Tyr (n = 3). Mutation in parC genes: Ser79Phe (n = 16), Ser79Tyr (n = 1), Asp83Gly (n = 1).
Fluoroquinolone-nonsusceptible strains accounted for 2.6% of isolates (n = 18). These possessed amino acid substitutions in quinolone resistance-determining regions of GyrA and ParC, encoded by gyrA and parC genes, respectively. Strains (n = 11) for which levofloxacin MICs were at least 16 μg/mL had both substitutions of Ser81Phe or Ser81Tyr in GyrA and Ser79Phe in ParC, whereas remaining strains for which levofloxacin MICs were 4–8 μg/mL had Ser79Phe (n = 5), Ser79Tyr (n = 1), or Asp83Gly (n = 1) in ParC. These fluoroquinolone-resistant strains were distributed in 9 emm types, including stG6792 (n = 5) and stG485 (n = 4).
Susceptibilities to 8 oral and 7 parenteral antimicrobial agents, among 693 iSDSE strains, are shown in Technical Appendix Table 1. Except for cefazolin and cefotiam, which were preferred by physicians in Japan, the antibacterial activity of β-lactam agents was superior; MIC for 90% of strains tested ranged from 0.004 to 0.125 μg/mL. MICs of β-lactam agents or vancomycin were not excessive for any strain.
Discussion
We analyzed molecular characteristics of SDSE strains from invasive infections, including emm typing, MLST, and antimicrobial resistance determinants, together with clinical features. Molecular epidemiologic surveillance showed that the most prevalent emm type was stG6792, which has been true for iSDSE infection since 2003 (22,25). Surprisingly, this type has not been prevalent in other countries (6–11). The reason for variation in dominant emm type between countries remains to be determined.
MLST analysis indicated that CC17, particularly consisting of ST17, was the most prevalent CC, which was identified in a variety of emm types. S. pyogenes strains belonging to a single emm type usually have shown the same CC with only single- and double-locus variants (28). In contrast, SDSE strains included a variety of CCs (STs), a fundamental difference from S. pyogenes strains. Data reported by McMillan et al. indicated that SDSE strains belonging to stG2078 were classified as ST17 (CC17), whereas those belonging to stG6792 were assigned to ST4 (CC4) (36). These findings may indicate that CC17 in SDSE conveyed high virulence and that emm gene findings have recently become more apparent.
Genomic analysis suggests that SDSE obtained several virulence genes from S. pyogenes by horizontal transfer. Our results also show the possibility of novel emm types arising from recombination events among emm genes in SDSE, indicating that SDSE still is undergoing change. Cross-species transmission between SDSE and other streptococci suggests diversification of SDSE and evolution of highly pathogenic SDSE.
SDSE strains in this study were uniformly susceptible to β-lactam agents, and MICs of these agents were excellent, except for those of some cephalosporin agents. In contrast, macrolide resistance was found in 18.5% of strains, an increase from our previous findings (25). We also previously reported that macrolide resistance increased among S. pyogenes strains, exceeding 50% in invasive infections (28) and 60% in noninvasive infections (34). Similarly, macrolide-resistant strains may increase among SDSE strains. Although quinolone resistance was uncommon, we predict that its prevalence will increase with increasing quinolone administration.
SDSE isolates were collected at the same time as strains of S. pyogenes, S. agalactiae, and S. pneumoniae (27) for 3 years throughout Japan. The mean age of patients with iSDSE infections was greater than of those with S. pyogenes and S. agalactiae infection. As expected, the fatality rate was significantly higher for elderly patients, especially those with pneumonia, severe sepsis, septic shock, or disseminated intravascular coagulation (data not shown). Our results identify iSDSE as a common cause of community-acquired infections in an aging society. Immunologic senescence associated with aging as well as underlying diseases are suspected to contribute to risk. Differences in virulence factors between S. pyogenes and SDSE, including superantigens and cysteine proteases, may be key causative factors. Further clarification of the contribution of virulence factors present at onset is needed because severe SDSE infections will probably become more frequent.
In Japan, community-acquired iSDSE infections first drew attention in 2003 (12,13), when persons >65 years of age accounted for nearly 20% of the total population. According to 2013 Japanese population statistics, the segment of the population >65 years of age had exceeded 25%, with Japan becoming the highest-ranking country in terms of average life expectancy (Statistics Bureau Japan, Ministry of Internal Affairs and Communications; http://www.stat.go.jp/english/index.htm). Given the relationship between age and iSDSE, we believe that our population dynamics particularly predispose the country to increases in iSDSE infection that may not yet be present in other countries.
In conclusion, SDSE may become a global concern as a causative pathogen with the potential for high mortality rates among elderly persons with community-acquired infections, especially in industrialized countries. Global surveillance of invasive SDSE infection is needed.
Susceptibilities to 15 antimicrobial agents among 693 invasive Streptococcus dysgalactiae subsp. equisimilis (SDSE) strains and relationships between outcomes of patients with invasive SDSE infection and emm type of the isolate and type-specific regions in 2 novel emm types of emmG2.0 and emmG3.0 identified in SDSE strains, Japan, April 2010–March 2013.
Acknowledgments
We thank everyone who participated actively in this large surveillance activity covering 3 periods. We also thank Madoka Naito and Shinji Masuyoshi for laboratory assistance.
This study was funded in part by a grant under the category, “Research on Emerging and Re-emerging Infectious Diseases” (H22-013).
Biography
Dr. Wajima is an assistant professor at Tokyo University of Pharmacy and Life Sciences. His research interests are molecular epidemiology and pathogenicity of β-hemolytic streptococci. Dr. Morozumi is an assistant professor at Keio University School of Medicine. Her research interests include molecular epidemiology as well as severe infections caused by S. agalactiae and Mycoplasma pneumoniae.
Footnotes
Suggested citation for this article: Wajima T, Morozumi M, Hanada S, Sunaoshi K, Chiba N, Iwata S, et al. Molecular characterization of invasive Streptococcus dysgalactiae subsp. equisimilis, Japan. Emerg Infect Dis. 2016 Feb [date cited]. http://dx.doi.org/10.3201/eid2202.141732
These authors contributed equally to this article.
References
- 1.Vandamme P, Pot B, Falsen E, Kersters K, Devriese LA. Taxonomic study of Lancefield streptococcal groups C, G, and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int J Syst Bacteriol. 1996;46:774–81. 10.1099/00207713-46-3-774 [DOI] [PubMed] [Google Scholar]
- 2.Spellerberg B, Brandt C. Streptococcus. In: Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW, editors. Manual of clinical microbiology, 10th ed. Washington (DC): ASM Press; 2011. p. 331–49. [Google Scholar]
- 3.Vieira VV, Teixeira LM, Zahner V, Momen H, Facklam RR, Steigerwalt AG, et al. Genetic relationships among the different phenotypes of Streptococcus dysgalactiae strains. Int J Syst Bacteriol. 1998;48:1231–43. 10.1099/00207713-48-4-1231 [DOI] [PubMed] [Google Scholar]
- 4.Humar D, Datta V, Bast DJ, Beall B, De Azavedo JC, Nizet V. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet. 2002;359:124–9. 10.1016/S0140-6736(02)07371-3 [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Poradosu R, Jaffe J, Lavi D, Grisariu-Greenzaid S, Nir-Paz R, Valinsky L, et al. Group G Streptococcal bacteremia in Jerusalem. Emerg Infect Dis. 2004;10:1455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekelund K, Skinhøj P, Madsen J, Konradsen HB. Invasive group A, B, C and G streptococcal infections in Denmark 1999–2002: epidemiological and clinical aspects. Clin Microbiol Infect. 2005;11:569–76. 10.1111/j.1469-0691.2005.01169.x [DOI] [PubMed] [Google Scholar]
- 7.Liao CH, Liu LC, Huang YT, Teng LJ, Hsueh PR. Bacteremia caused by group G streptococci, Taiwan. Emerg Infect Dis. 2008;14:837–40. 10.3201/eid1405.070130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broyles LN, Van Beneden C, Beall B, Facklam R, Shewmaker PL, Malpiedi P, et al. Population-based study of invasive disease due to β-hemolytic streptococci of groups other than A and B. Clin Infect Dis. 2009;48:706–12. 10.1086/597035 [DOI] [PubMed] [Google Scholar]
- 9.Rantala S, Vuopio-Varkila J, Vuento R, Huhtala H, Syrjänen J. Clinical presentations and epidemiology of β-haemolytic streptococcal bacteraemia: a population-based study. Clin Microbiol Infect. 2009;15:286–8. 10.1111/j.1469-0691.2008.02672.x [DOI] [PubMed] [Google Scholar]
- 10.Kittang BR, Bruun T, Langeland N, Mylvaganam H, Glambek M, Skrede S. Invasive group A, C and G streptococcal disease in western Norway: virulence gene profiles, clinical features and outcomes. Clin Microbiol Infect. 2011;17:358–64. 10.1111/j.1469-0691.2010.03253.x [DOI] [PubMed] [Google Scholar]
- 11.Loubinoux J, Plainvert C, Collobert G, Touak G, Bouvet A, Poyart C, et al. Adult invasive and noninvasive infections due to Streptococcus dysgalactiae subsp. equisimilis in France from 2006 to 2010. J Clin Microbiol. 2013;51:2724–7. 10.1128/JCM.01262-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikebe T, Murayama S, Saitoh K, Yamai S, Suzuki R, Isobe J, et al. Surveillance of severe invasive group-G streptococcal infections and molecular typing of the isolates in Japan. Epidemiol Infect. 2004;132:145–9. 10.1017/S0950268803001262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashikawa S, Iinuma Y, Furushita M, Ohkura T, Nada T, Torii K, et al. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J Clin Microbiol. 2004;42:186–92. 10.1128/JCM.42.1.186-192.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen A, Kilian M. Delineation of Streptococcus dysgalactiae, its subspecies, and its clinical and phylogenetic relationship to Streptococcus pyogenes. J Clin Microbiol. 2012;50:113–26. 10.1128/JCM.05900-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimomura Y, Okumura K, Murayama SY, Yagi J, Ubukata K, Kirikae T, et al. Complete genome sequencing and analysis of a Lancefield group G Streptococcus dysgalactiae subsp. equisimilis strain causing streptococcal toxic shock syndrome (STSS). BMC Genomics. 2011;12:17. 10.1186/1471-2164-12-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki H, Lefébure T, Hubisz MJ, Pavinski Bitar P, Lang P, Siepel A, et al. Comparative genomic analysis of the Streptococcus dysgalactiae species group: gene content, molecular adaptation, and promoter evolution. Genome Biol Evol. 2011;3:168–85. 10.1093/gbe/evr006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe S, Kirikae T, Miyoshi-Akiyama T. Complete genome sequence of Streptococcus dysgalactiae subsp. equisimilis 167 carrying Lancefield group C antigen and comparative genomics of S. dysgalactiae subsp. equisimilis strains. Genome Biol Evol. 2013;5:1644–51. 10.1093/gbe/evt117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okumura K, Shimomura Y, Murayama SY, Yagi J, Ubukata K, Kirikae T, et al. Evolutionary paths of streptococcal and staphylococcal superantigens. BMC Genomics. 2012;13:404. 10.1186/1471-2164-13-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai CT, Chi CY, Ho CM, Lin PC, Chou CH, Wang JH, et al. Correlation of virulence genes to clinical manifestations and outcome in patients with Streptococcus dysgalactiae subspecies equisimilis bacteremia. J Microbiol Immunol Infect. 2014;47:462–8. 10.1016/j.jmii.2013.08.019 [DOI] [PubMed] [Google Scholar]
- 20.Rantala S, Tuohinen S. Two cases of cardiac device-related endocarditis due to Streptococcus dysgalactiae subsp. equisimilis (group C or G streptococci). BMC Infect Dis. 2014;14:174. 10.1186/1471-2334-14-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambertsen LM, Ingels H, Schønheyder HC, Hoffmann S. Danish Streptococcal Surveillance Collaboration Group 2011. Nationwide laboratory-based surveillance of invasive β-haemolytic streptococci in Denmark from 2005 to 2011. Clin Microbiol Infect. 2014;20:O216–23. 10.1111/1469-0691.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi T, Sunaoshi K, Sunakawa K, Fujisima S, Watanabe H, Ubukata K, et al. Clinical aspects of invasive infections with Streptococcus dysgalactiae ssp. equisimilis in Japan: differences with respect to Streptococcus pyogenes and Streptococcus agalactiae infections. Clin Microbiol Infect. 2010;16:1097–103. 10.1111/j.1469-0691.2009.03047.x [DOI] [PubMed] [Google Scholar]
- 23.Brandt CM, Spellerberg B. Human infections due to Streptococcus dysgalactiae subspecies equisimilis. Clin Infect Dis. 2009;49:766–72. 10.1086/605085 [DOI] [PubMed] [Google Scholar]
- 24.Rantala S. Streptococcus dysgalactiae subsp. equisimilis bacteremia: an emerging infection. Eur J Clin Microbiol Infect Dis. 2014;33:1303–10. 10.1007/s10096-014-2092-0 [DOI] [PubMed] [Google Scholar]
- 25.Sunaoshi K, Murayama SY, Adachi K, Yagoshi M, Okuzumi K, Chiba N, et al. Molecular emm genotyping and antibiotic susceptibility of Streptococcus dysgalactiae subsp. equisimilis isolated from invasive and non-invasive infections. J Med Microbiol. 2010;59:82–8. 10.1099/jmm.0.013201-0 [DOI] [PubMed] [Google Scholar]
- 26.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiba N, Morozumi M, Shouji M, Wajima T, Iwata S, Ubukata K, et al. Changes in capsule and drug resistance of pneumococci after introduction of PCV7, Japan, 2010–2013. Emerg Infect Dis. 2014;20:1132–9. 10.3201/eid2007.131485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wajima T, Morozumi M, Chiba N, Shouji M, Iwata S, Sakata H, et al. Associations of macrolide and fluoroquinolone resistance with molecular typing in Streptococcus pyogenes from invasive infections, 2010–2012. Int J Antimicrob Agents. 2013;42:447–9. 10.1016/j.ijantimicag.2013.06.022 [DOI] [PubMed] [Google Scholar]
- 29.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–55. 10.1378/chest.101.6.1644 [DOI] [PubMed] [Google Scholar]
- 30.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250–6. 10.1097/01.CCM.0000050454.01978.3B [DOI] [PubMed] [Google Scholar]
- 31.Stevens DL. Streptococcal toxic-shock syndrome: spectrum of disease, pathogenesis, and new concepts in treatment. Emerg Infect Dis. 1995;1:69–78. 10.3201/eid0103.950301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad Y, Gertz RE Jr, Li Z, Sakota V, Broyles LN, Van Beneden C, et al. Genetic relationships deduced from emm and multilocus sequence typing of invasive Streptococcus dysgalactiae subsp. equisimilis and S. canis recovered from isolates collected in the United States. J Clin Microbiol. 2009;47:2046–54. 10.1128/JCM.00246-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, M100–S23. Wayne (PA): The Institute; 2013. [Google Scholar]
- 34.Wajima T, Chiba N, Morozumi M, Shouji M, Sunaoshi K, Sugita K, et al. Prevalence of macrolide resistance among group A streptococci isolated from pharyngotonsillitis. Microb Drug Resist. 2014;20:431–5. 10.1089/mdr.2013.0213 [DOI] [PubMed] [Google Scholar]
- 35.Wajima T, Murayama SY, Sunaoshi K, Nakayama E, Sunakawa K, Ubukata K. Distribution of emm type and antibiotic susceptibility of group A streptococci causing invasive and noninvasive disease. J Med Microbiol. 2008;57:1383–8. 10.1099/jmm.0.2008/002642-0 [DOI] [PubMed] [Google Scholar]
- 36.McMillan DJ, Bessen DE, Pinho M, Ford C, Hall GS, Melo-Cristino J, et al. Population genetics of Streptococcus dysgalactiae subspecies equisimilis reveals widely dispersed clones and extensive recombination. PLoS ONE. 2010;5:e11741. 10.1371/journal.pone.0011741 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Susceptibilities to 15 antimicrobial agents among 693 invasive Streptococcus dysgalactiae subsp. equisimilis (SDSE) strains and relationships between outcomes of patients with invasive SDSE infection and emm type of the isolate and type-specific regions in 2 novel emm types of emmG2.0 and emmG3.0 identified in SDSE strains, Japan, April 2010–March 2013.


