Phloem-specific methionine recycling is sufficient to overcome sulfur-deficient growth conditions that would otherwise result in polyamine shortage and impair flower and seed development.
Abstract
The Yang or Met Cycle is a series of reactions catalyzing the recycling of the sulfur (S) compound 5′-methylthioadenosine (MTA) to Met. MTA is produced as a by-product in ethylene, nicotianamine, and polyamine biosynthesis. Whether the Met Cycle preferentially fuels one of these pathways in a S-dependent manner remained unclear so far. We analyzed Arabidopsis (Arabidopsis thaliana) mutants with defects in the Met Cycle enzymes 5-METHYLTHIORIBOSE-1-PHOSPHATE-ISOMERASE1 (MTI1) and DEHYDRATASE-ENOLASE-PHOSPHATASE-COMPLEX1 (DEP1) under different S conditions and assayed the contribution of the Met Cycle to the regeneration of S for these pathways. Neither mti1 nor dep1 mutants could recycle MTA but showed S-dependent reproductive failure, which was accompanied by reduced levels of the polyamines putrescine, spermidine, and spermine in mutant inflorescences. Complementation experiments with external application of these three polyamines showed that only the triamine spermine could specifically rescue the S-dependent reproductive defects of the mutant plants. Furthermore, expressing gene-reporter fusions in Arabidopsis showed that MTI1 and DEP1 were mainly expressed in the vasculature of all plant parts. Phloem-specific reconstitution of Met Cycle activity in mti1 and dep1 mutant plants was sufficient to rescue their S-dependent mutant phenotypes. We conclude from these analyses that phloem-specific S recycling during periods of S starvation is essential for the biosynthesis of polyamines required for flowering and seed development.
Sulfur (S) deficiency greatly impacts flower development and seed yield of different plant species (Hell, 2008; Marschner and Marschner, 2012; D’Hooghe et al., 2013). Shoots and flowers of S-deprived plants appear pale yellow and seeds show reduced germination efficiency (Higgins et al., 1986; Nikiforova et al., 2003). In particular, Brassica species have high S demands, presumably because of the large amounts of Cys-rich storage proteins in their cotyledons (Shewry and Casey, 1999) and the production of glucosinolates, which mostly derive from Met (Windsor et al., 2005). Both sulfate transport and assimilation pathways are highly regulated by S availability, and the expression and activity levels of the corresponding proteins are efficiently adjusted under low S availability (Saito, 2004; Koprivova and Kopriva, 2014). S deficiency promotes the synthesis of transport proteins of the SULFATE TRANSPORTER (SULTR) family to increase root sulfate uptake (Shinmachi et al., 2010; Maruyama-Nakashita et al., 2015) or sulfate efflux from storage vacuoles (Kataoka et al., 2004), which supports the remobilization of sulfate from source to sink tissues. Moreover, plants increase the efficiency of S utilization by inducing S recycling pathways. The Met Cycle, also known as Yang cycle or 5′-methylthioadenosine (MTA) cycle, is the major S recycling pathway in plants and consists of a series of reactions that convert MTA back to Met (Sauter et al., 2013). MTA is generated as a by-product during ethylene, polyamine, and nicotianamine synthesis. However, the quantitative contribution of these three pathways to MTA formation and their relative importance for Met regeneration via the Met Cycle are still unclear.
The existence of a recycling pathway for Met was first postulated by Baur and Yang (1972), and the first enzymatic activities of plant Met Cycle enzymes, 5′-methylthioribose kinase (MTK) and 5′-methylthioadenosine nucleosidase (MTN), were found 5 years later in extracts from lupin seeds (Guranowski, 1983). The first genes encoding plant Met Cycle enzymes (MTK from Arabidopsis [Arabidopsis thaliana] and MTK1 and MTK2 from rice [Oryza sativa]) were cloned 20 years later (Sauter et al., 2004). In the following years, genes for acidoreductone dioxygenases (ARD1-4) as well as for MTA nucleosidases (MTN1 and MTN2) were identified (Sauter et al., 2005; Rzewuski et al., 2007). The number of the remaining enzymatic steps and genes of the Met Cycle in Arabidopsis remained unclear, as the conversion of 5-methyl-thioribulose-1-P (MTRu-1-P) to 1,2-dihydroxy-3-keto-5-methylthiopentene (DHKMP) is catalyzed by two enzymes in certain organisms and by three enzymes in others (Sekowska et al., 2004). Recently, two novel higher plant Met Cycle genes, 5-METHYLTHIORIBOSE-1-PHOSPHATE ISOMERASE1 (MTI1) and DEHYDRATASE-ENOLASE-PHOSPHATASE-COMPLEX1 (DEP1), have been identified (Pommerrenig et al., 2011). DEP1 is a trifunctional enzyme that catalyzes the conversion of MTRu-1-P to DHKMP. MTI1 has a surprising and high similarity to eukaryotic translation initiation factor eIF-2B family proteins. Phylogenetic analyses and complementation of a yeast strain with a defect in the corresponding MRI1 gene confirmed the enzymatic conversion of MTR-1P to MTRu-1-P by the MTI1 protein (Pommerrenig et al., 2011).
An essential role of the Met Cycle for plant S nutrition has been proposed for plants and plant organs exhibiting high ethylene synthesis, for example, during germination and seedling growth, but also during periods of hypoxia or fruit ripening. During these phases, elevated Met Cycle activity helps to replenish the Met pool (Baur and Yang, 1972; Bürstenbinder et al., 2007; Rzewuski et al., 2007). Utilizing a mtk mutant, the ethylene overproducing mutant eto3, and the mtk/eto3 double mutant, Bürstenbinder et al. (2007) could show that the Met Cycle is important during periods of high ethylene synthesis in seedlings. In contrast, in adult plants, the overall ethylene synthesis is low; thus, an elevated S requirement for ethylene may be restricted to plants that naturally produce or need to produce large quantities of the hormone for a prolonged period of time (Rzewuski et al., 2007; Sauter et al., 2013).
However, Met Cycle activities are not restricted to seedlings and fruits, since the levels of both mRNA of Met Cycle genes and Met Cycle-related metabolites were found to accumulate in the vasculature of adult rosette leaves of Arabidopsis and Plantago major (Pommerrenig et al., 2011). The specific expression of Met Cycle genes in the vasculature is in line with the second essential function of the Met Cycle, which is the degradation of MTA, the by-product of ethylene, nicotianamine, and polyamine biosynthesis. Mutants lacking MTA nucleosidase activity (Bürstenbinder et al., 2010; Waduwara-Jayabahu et al., 2012) also showed hyperproliferation of xylem elements in their vasculature and impaired flower development. These effects have been attributed to elevated MTA levels and inhibited polyamine and nicotianamine (NA) biosynthesis.
Polyamines are positively charged polycations, which occur in all living organisms and fulfill important functions in cellular metabolism. In Arabidopsis, the main polyamines are putrescine, spermidine, spermine, and thermospermine. All polyamines have the ability to bind DNA but also contribute to plant tolerance to biotic and abiotic stresses (Jimènez-Bremont et al., 2014; Minocha et al., 2014). Spermine synthase (SPMS) has been shown to protect plants during salt stress. Additionally, thermospermine, which is synthesized by thermospermine synthase (ACL5), functions in vascular development by repressing xylem differentiation (Vera-Sirera et al., 2010; Takano et al., 2012), and spermidine has been proven important for plant reproduction (Imai et al., 2004; Deeb et al., 2010). The acl5/spms double mutant was shown to be hypersensitive to salt stress but could be rescued by the exogenous application of spermine (Yamaguchi et al., 2006). Overexpression of spermidine and spermine biosynthesis or exogenous supply of spermine have been reported to increase the tolerance to drought (Capell et al., 2004) or heat stress (Sagor et al., 2013).
Whether the Met Cycle has a role apart from the degradation of MTA, most notably in sustaining S supply for efficient polyamine biosynthesis during reproductive growth, and to what extent the Met Cycle contributes to S nutrition remained unclear so far. For clarification of this issue, we isolated Arabidopsis mutants defective in the expression of MTI1 and DEP1 and studied their growth and metabolism under S-limiting conditions. To this end, we were able to characterize MTA-independent effects of altered Met Cycle activities. During our analyses we discovered strong S-dependent reproductive defects in mti1 and dep1 mutants that had not been reported for other genes of the Met Cycle. Our physiological analyses of mti1 and dep1 mutants further illustrate the importance of sustained polyamine synthesis for plant reproduction during S deficiency.
RESULTS
MTI1 and DEP1 Are Met Cycle Unigenes in Arabidopsis
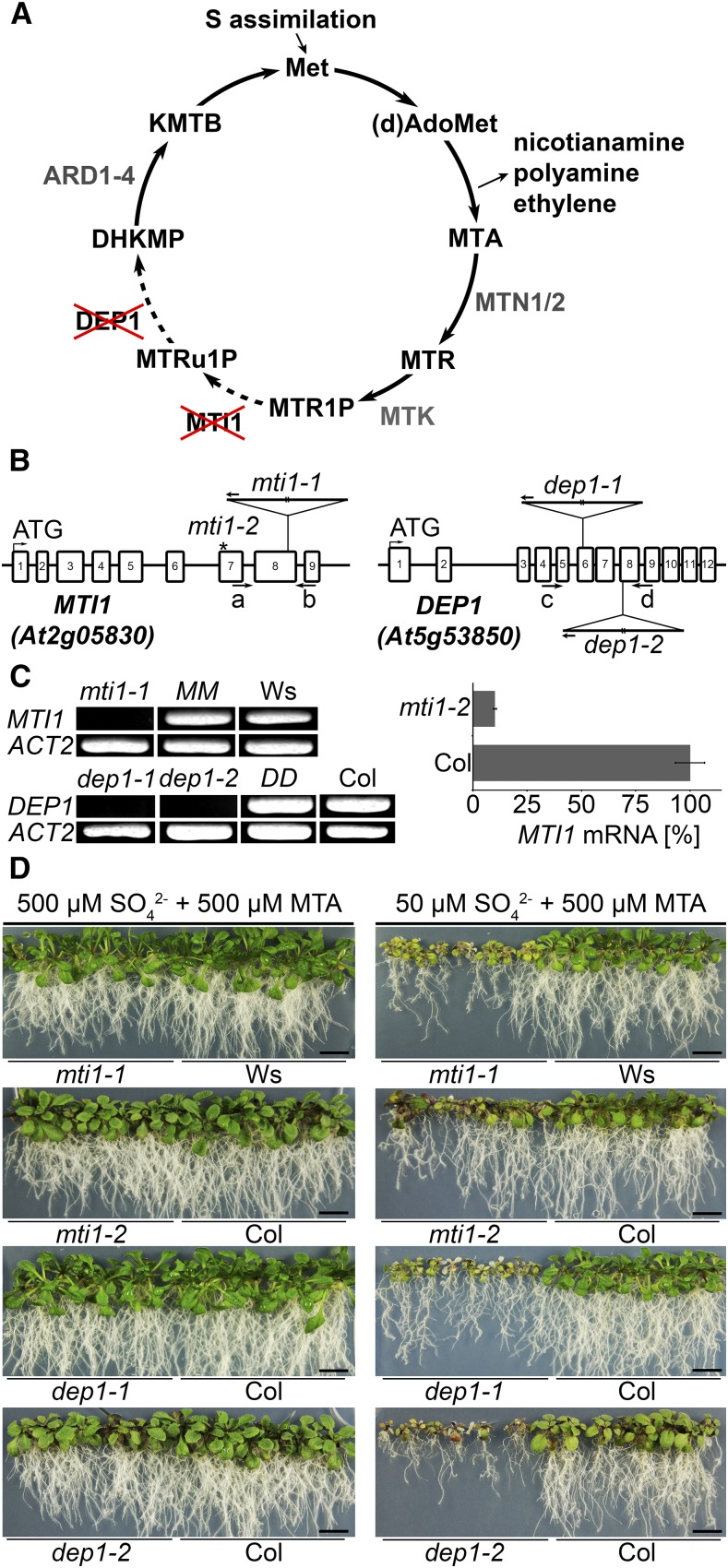
In the Met Cycle, MTA is recycled to Met. Therefore, mutants with a block in one or more steps of the cycle are unable to use MTA as S source (Sauter et al., 2004; Bürstenbinder et al., 2007; Bürstenbinder et al., 2010). We had previously shown that MTI1 and DEP1 complemented the growth reduction of yeast mutants defective in the conversion of MTR1-P to MTRu1-P or of MTRu-1-P to DHKMP (Fig. 1A) on medium with MTA as sole S source (Pommerrenig et al., 2011). To analyze the function of both enzymes also in planta, we isolated mti1 and dep1 mutants and used them for growth assays on agar medium containing MTA or on hydroponic culture with different levels of S supply.
Figure 1.
Characterization of mti1 and dep1 plants as Met Cycle mutants. A, Schematic depiction of the Met Cycle. (d)AdoMet, (deoxy)S-adenosyl-Met; MTR, methylthioribose; MTR1P, MTR-1-phosphate; MTRu1P, methylthioribulose-1-phosphate; KMTB, 2-keto-4-methylthiobutyrate; MTN, MTA-NUCLEOSIDASE; MTK, MTR-KINASE; MTI1, MTR1P-ISOMERASE1; DEP1, DEHYDRATASE-ENOLASE-PHOSPHATASE-COMPLEX1; ARD, ACIDOREDUCTONE DIOXYGENASE. B, Scheme of the MTI1 and DEP1 genes and mutant alleles. The T-DNA insertions are represented as triangles. Arrows a to d represent primers used for specific amplification of MTI1 or DEP1 transcripts (see Supplemental Table S1). The asterisk marks the amiRNA binding site on the MTI1 mRNA in the analyzed mti1-2 lines. C, RT-PCR analysis of MTI1 and DEP1 mRNAs in mutants, wild-type, and complemented lines and qRT-PCR analysis of the amiRNA line mti1-2. D, Growth of 3-week-old mti1 and dep1 mutant plants together with corresponding wild-type plants on half-strength Murashige and Skoog medium with either 50 µm sulfate and 500 µm MTA or with 500 µm sulfate and 500 µm MTA as sulfur sources. Bars = 1 cm.
The mti1-1 mutant allele (FLAG_145B05) carries a T-DNA insertion in the eighth exon of the MTI1 gene (Fig. 1B). Full-length mRNA of MTI1 was absent in the mti1-1 mutant but clearly detectable in Wassilewskija (Ws) wild-type and complemented mutant lines (Fig. 1C). The complemented mti1-1 mutant lines carried the wild-type MTI1 allele under transcriptional control of a 1925-bp fragment upstream of the start codon of the MTI1 open reading frame (ORF; line MM). We generated a second mti1 mutant line by introducing an artificial microRNA (amiRNA) targeting the MTI1 gene into the Columbia (Col) background. The MTI1-amiRNA was under transcriptional control of the CaMV35S promoter. The resulting T1 lines exhibited a residual expression level of the MTI1 gene between 10% and 40%. We selected the line with the strongest reduction in MTI1 expression (mti1-2) for further analysis of the T2 generation (Fig. 1C). The two dep1 mutant alleles carry T-DNA insertions either in the sixth exon (dep1-1, SALK_091578C) or at the start of the eighth exon of the DEP1 gene (dep1-2, WiscDsLox489-492G6). Full-length transcripts of DEP1 were absent in both dep1 mutants and could only be amplified from Col wild type or from complemented mutant lines (Fig. 1C). The complemented dep1-1 mutant lines carried the wild-type DEP1 allele under transcriptional control of a fragment containing a 1649-bp upstream sequence of the DEP1 ORF (line DD). To analyze the dependence of these mutants on the Met Cycle, their growth was tested on medium containing either 500 µm sulfate and 500 µm MTA or 50 µm sulfate and 500 µm MTA (Fig. 1D). When 500 µm sulfate and 500 µm MTA were supplied, only a weak growth reduction of mutant plants could be observed. On sulfate-deficient medium with MTA as main S source, both mutant and wild-type lines exhibited reduced root and shoot growth compared with growth on S-sufficient medium. However, mti1-1, mti1-2, dep1-1, and dep1-2 mutant plants displayed stronger S deficiency symptoms as leaves suffered more severely from chlorosis and root and shoot growth was more reduced than in the corresponding wild type (Fig. 1D). Mutant lines complemented with the corresponding wild-type alleles showed wild-type-like growth when grown with 500 µm MTA as sole S source (Supplemental Fig. S1). These results show that mti1 and dep1 mutant plants cannot use MTA as S source. Therefore, MTI1 and DEP1 are components of the Met Cycle and contribute to the recycling of MTA in Arabidopsis.
MTI1 and DEP1 Are Cytosolic Enzymes Localized in Vascular and Developing Tissues
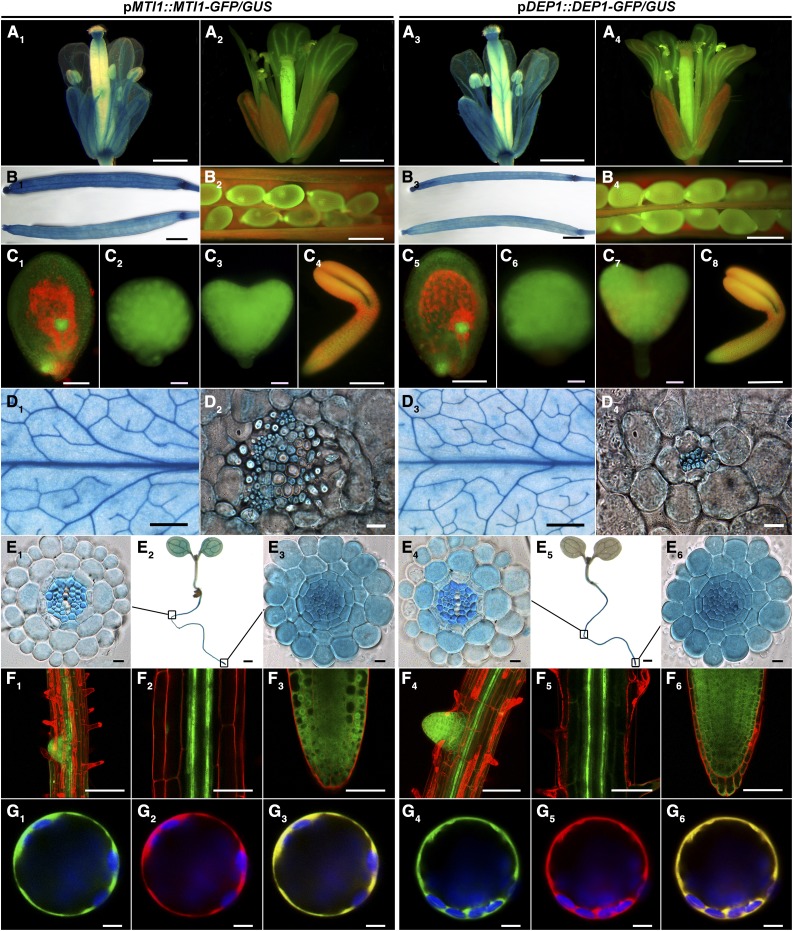
On the basis of an expression analysis of MTI1 and DEP1 in rosette leaves (Pommerrenig et al., 2011), we used reporter lines carrying promoter-gene-GFP and promoter-gene-GUS fusions of MTI1 and DEP1 for a more comprehensive expression analysis. These analyses revealed tissue-specific and overlapping localization patterns of MTI1 and DEP1. We observed GUS staining and GFP fluorescence in reproductive organs, specifically in the vasculature of sepals, petals, and anthers (Fig. 2, A1–A4), in siliques (Fig. 2, B1–B4), in seeds (Fig. 2, B2 and B4), and throughout embryo development (Fig. 2, C1–C8) in all reporter lines. Again, both genes showed the previously reported expression in the vasculature of rosette leaves (Fig. 2, D1–D4). In roots of 7-d-old plants, GUS staining and GFP fluorescence was observed in the vasculature of the differentiation zone (Fig. 2, E1, E4, F1, F2, F4, and F5) and in all cells of primary and lateral root tips (Fig. 2, E3, E6, F1, F3, F4, and F6). For subcellular localization of the MTI1 and DEP1 enzymes, Arabidopsis mesophyll protoplast were cotransformed with C-terminal GFP fusion constructs under the control of the CaMV35S promoter together with a control construct coding for soluble mCherry. GFP-dependent fluorescence derived from fusions with either MTI1 or DEP1 filled a broad zone at the border of the protoplasts that included chloroplasts and could be assigned to the cytosol (Fig. 2, G1, G2, G4, and G5). Fluorescence overlay (Fig. 2, G3 and G6) showed complete colocalization with mCherry. These observations showed that MTI1 and DEP1 are cytosolic enzymes and indicated that the Met Cycle is predominantly active in the vasculature of all plant organs.
Figure 2.
Tissue- and cell compartment-specific expression of MTI1 and DEP1. A, B, D, and E, GUS staining of flowers (A1 and A3), unopened siliques (B1 and B3), rosette leaves (D1 and D3), 7-d-old seedlings (E2 and E5), cross sections of leave vascular bundles (D2 and D4), cross sections of root differentiation zones (E1 and E4), and cross sections of root tips (E3 and E6). A, B, C, F, and G, GFP fluorescence of flowers (A2 and A4), seeds (B2 and B4), embryos (C1 and C5, early globular stage and funiculus; C2 and C6, globular stage; C3 and C7, heart stage; C4 and C8, torpedo stage), roots (F1, F2, F4, and F5, root differentiation zones; F3 and F6, root tips) and Arabidopsis mesophyll protoplasts expressing C-terminal GFP fusions (G1–3, MTI1; G4–6, DEP1). All constructs were cotransformed with a cytosolic mCherry expression construct. G1 and G4, GFP fluorescence is shown in green. G2 and G5, mCherry fluorescence is shown in red. G3 and G6, Merged fluorescence is shown in yellow. Chloroplasts are shown in blue. Bars = 5 mm for D1 and D3, 2 mm for B1 and B3, 1 mm for A1 to A4, 200 µm for E2 and E5, 50 µm for B2, B4, and F1–6, 10 µm for C1, C4, C5, and C8, 5 µm for G1–6, and 1 µm for C2, C3, C6, C7, D2, D4, E1, E3, E4, and E6.
Sulfur Deficiency Causes Reproductive Defects in mti1 and dep1 Mutants
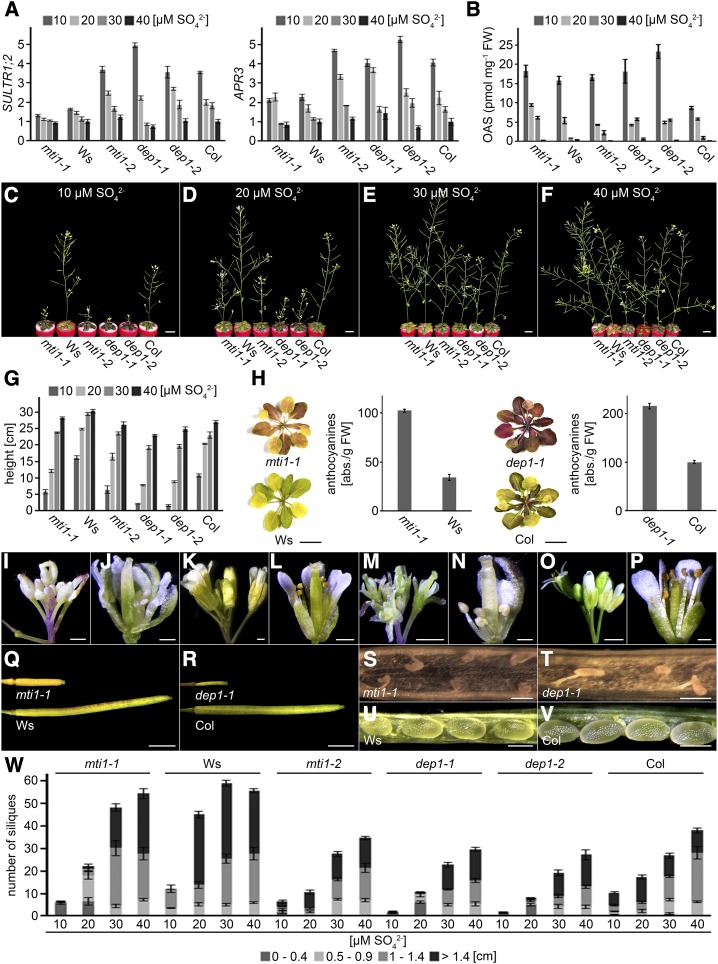
To dissect which organs or tissues depend most on Met Cycle activity, the impact of limiting S supply on growth and fertility of Met Cycle mutants and wild-type plants was analyzed. For these analyses, a hydroponic growth system was chosen as it allows precise adjustment of the sulfate concentration. In contrast to plant growth on plates, 50 µm sulfate did not result in S deficiency and clear S deficiency responses could only be observed below 40 µm sulfate (Supplemental Fig. S2). Therefore, hydroponic growth analyses were carried out with 10 to 40 µm sulfate. Monitoring mRNA levels of SULTR1;2 and APR3 to assess the S nutritional status of the tissue (Takahashi et al., 2011) showed that expression of both genes increased gradually with decreasing sulfate supply to the culture medium (Fig. 3A). The expression of SULTR1;2 and APR3 in roots was 4 to 5 times higher at 10 µm sulfate than at 40 µm sulfate in mti1-2, dep1-1, and dep1-2 and their corresponding Col wild type. In the mutant mti1-1 and its Ws wild type, the sulfate-dependent expression of SULTR1;2 and APR3 was less pronounced so that SULTR1;2 and APR3 transcript levels remained largely similar in mutants and wild-type plants. These results confirmed that all lines responded to different sulfate supplies with changes in the transcriptional regulation of the sulfate uptake/assimilation pathway and that a defect in the Met Cycle did not feed-back on the regulation of the sulfate uptake/assimilation pathway (Fig. 3A). In addition, measurements of inflorescence O-acetylserine (OAS) concentrations revealed OAS accumulation in plants grown in Hoagland medium containing 10 µm sulfate. When the medium was supplemented with higher sulfate concentrations, OAS levels strongly declined and were hardly detectable in plants grown in 40 µm sulfate (Fig. 1B). These results demonstrated that plants grown in 10 to 30 µm sulfate experienced concentration dependent S deficiency, while plants grown in 40 µm sulfate did not. However, phenotypic analysis of the mutants and the wild type revealed different growth phase and S-dependent responses. During the vegetative growth phase, no phenotypic differences were observed between mutant and wild-type plants. Mutants had the same number of rosette leaves and their root biomass was comparable to that of the wild type (Supplemental Fig. S3). Phenotypic differences between mutant and wild-type plants first appeared during emergence of the inflorescence and became stronger over time (Supplemental Fig. S4). In all plants, the development of flowers suffered from decreasing sulfate supply. In comparison to wild-type plants, however, growth of mutant inflorescences was reduced even further (Fig. 3, C–G). Moreover, rosette leaves of mutant plants grown on 10 or 20 µm sulfate became violet and had a more than 2-fold higher anthocyanin content than leaves from wild-type plants grown under the same conditions (Fig. 3H). In contrast to the wild type, mutant plants grown on 10 or 20 µm sulfate displayed severe floral defects such as abnormally shaped sepals and petals, deformed ovaries, and, most notably, short anthers without pollen (Fig. 3, I–P). Mutant plants also had fewer and shorter siliques than the wild type, and these shorter siliques only contained highly deformed seeds. Wild-type siliques however, contained fully developed seeds (Fig. 3, Q–W). These data showed that mti1 and dep1 mutants were affected to a greater extent by low S concentrations than the wild type and that in the mutants limiting S availability resulted in developmental defects of the inflorescence and in male sterility.
Figure 3.
Growth of mutant and wild-type lines during S deficiency. A, Relative expression of SULTR1;2 and APR3 in plants grown in Hoagland medium containing 10 to 40 µm sulfate. Bars represent mean values and standard errors calculated with data from three independent measurements. B, OAS measurements of mutant and wild-type plants grown in Hoagland medium containing 10 to 40 µm sulfate. Bars represent mean values and standard errors calculated with data from three independent measurements. C to F, Growth of 5-week-old mutant and wild-type plants in Hoagland medium containing 10 (C), 20 (D), 30 (E), or 40 µm (F) sulfate. G, Inflorescence height of mutant and wild-type plants grown in Hoagland medium containing 10 to 40 µm sulfate. Bars represent mean values and standard errors calculated with data from eight individual plants. H, Rosette leaves and their relative anthocyanin concentration of mti1-1 and Ws or dep1-1 and Col plants grown in Hoagland medium containing 10 µm sulfate. Bars represent mean values and standard errors calculated with data from three individual measurements. I to P, Flowers of mti1-1 (I and J), Ws (K and L), dep1-1 (M and N), and Col (O and P) plants grown in Hoagland medium containing 10 µm sulfate. Q and R, Closed siliques of mti1-1 and Ws plants (Q) or dep1-1 and Col plants (R) grown in Hoagland medium containing 10 µm sulfate. S to V, Opened siliques of mti1-1 (S) and Ws (U) or dep1-1 (T) and Col (V) plants grown in Hoagland medium containing 10 µm sulfate. W, Silique number and length of mutant and wild-type plants grown in Hoagland medium containing 10 to 40 µm sulfate. Siliques were grouped into four categories (0–0.4 cm, 0.5–0.9 cm, 1–1.4 cm, and >1.4 cm). Bars represent mean values and standard errors calculated with data from eight plants per genotype. Bars = 1 cm in C to F, G, and H, 1 mm in Q and R, 500 µm in I to L, U, and V, 200 µm in M to P, and 100 µm for S and T.
Levels of Met and Met-Related Metabolites Are Reduced in mti1 and dep1 Plants
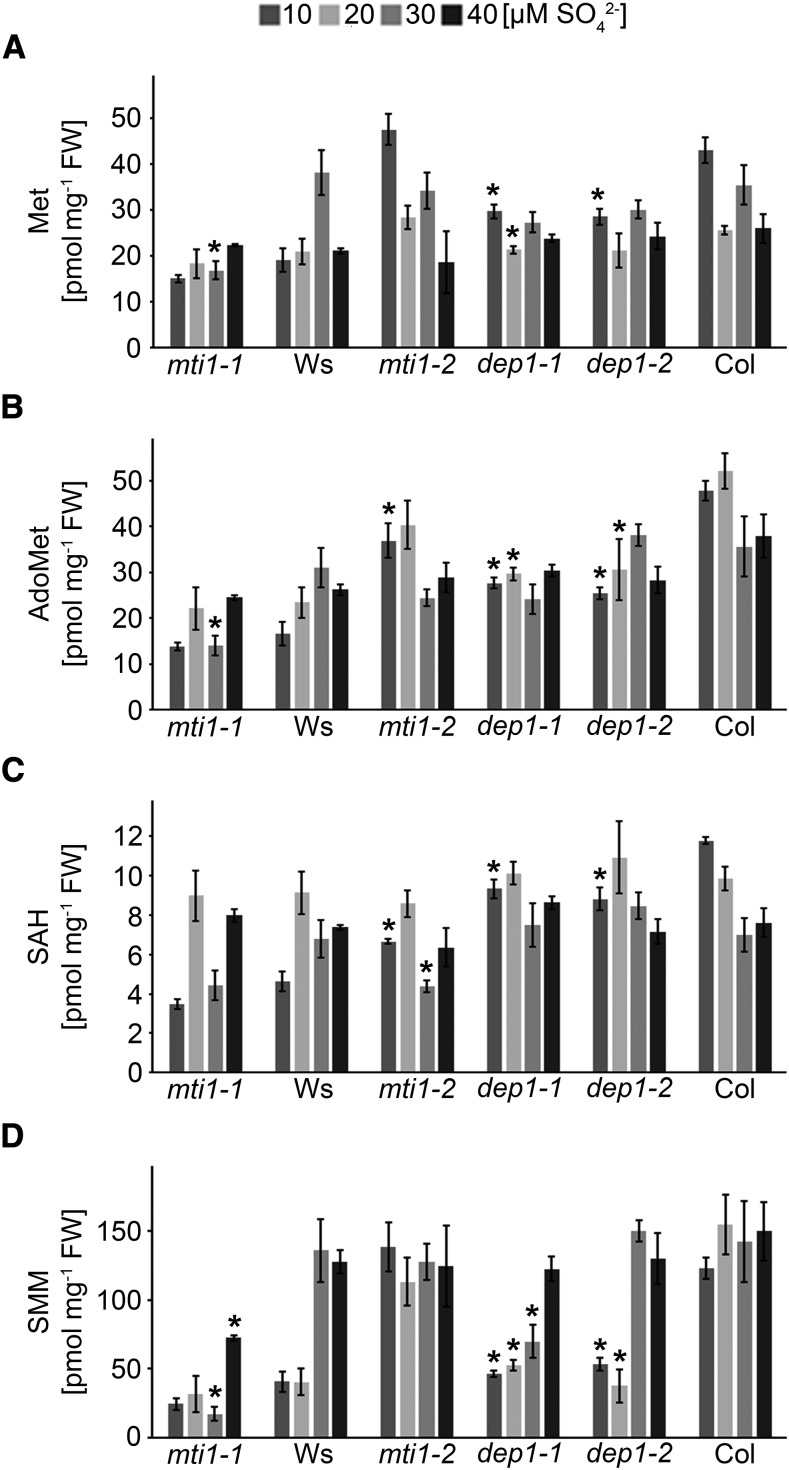
In cells with a high activity of MTA-producing biosynthetic pathways, the freely available Met pool is constantly replenished through the activity of the Met Cycle. This function becomes increasingly important during periods of S starvation when primary sulfate assimilation is limited. To analyze the impact of missing Met Cycle activity on the levels of Met and Met-related metabolites during S deficiency, Met, S-adenosyl-Met (AdoMet), S-adenosylhomo-Cys (SAH), and S-methyl-Met (SMM) were measured in mutant and wild-type inflorescences (Fig. 4). Met and SAH concentrations ranged between 2 and 4 ng mg−1 fresh weight (FW) in Ws and Col ecotypes. Overall Met and SAH concentrations were similar between the different S conditions, with dep1 mutant plants showing significant reductions in comparison to Col plants. AdoMet levels ranged between 5 and 10 ng mg−1 FW and 10 to 20 ng mg−1 FW in Ws and Col ecotypes, respectively. In comparison to the Col wild type, the Col-related mutant plants mti1-2, dep1-1, and dep1-2 all showed significant reductions by approximately 25% or 50% of their AdoMet levels when grown in 10 or 20 µm sulfate. Strong differences were also found in the phloem-related metabolite SMM, which serves as storage and transport form of sulfate (Bourgis et al., 1999). In comparison to wild-type plants, SMM levels were reduced by approximately 60% in dep1 mutant plants when grown in 10 or 20 µm sulfate. In contrast, a reduction of SMM (and also of Met) in mti1-2 mutant plants could not be detected under these conditions, most likely because the smaller amount of MTI1 mRNA in these amiRNA lines was sufficient to maintain wild-type-like levels of these two metabolites. However, the significant reduction of AdoMet and SAH in mti1-2 plants showed that missing or reduced Met Cycle activity leads to reductions in Met or Met-related metabolites.
Figure 4.
Met, AdoMet, SAH, and SMM concentrations in mutant and wild-type lines. A to D, Met (A), AdoMet (B), SAH (C), and SMM (D) concentrations in inflorescences of mti1-1, Ws, mti1-2, dep1-1, dep1-2, and Col plants grown in Hoagland medium containing 10 to 40 µm sulfate. Bars represent mean values and standard errors calculated with data from three independent measurements. Significances were tested using t test with mutant and corresponding wild-type values within the same S treatment. Asterisks indicate P-values < 0.05.
Phloem-Specific Expression of MTI1 and DEP1 Alleviates Plants from S Deficiency
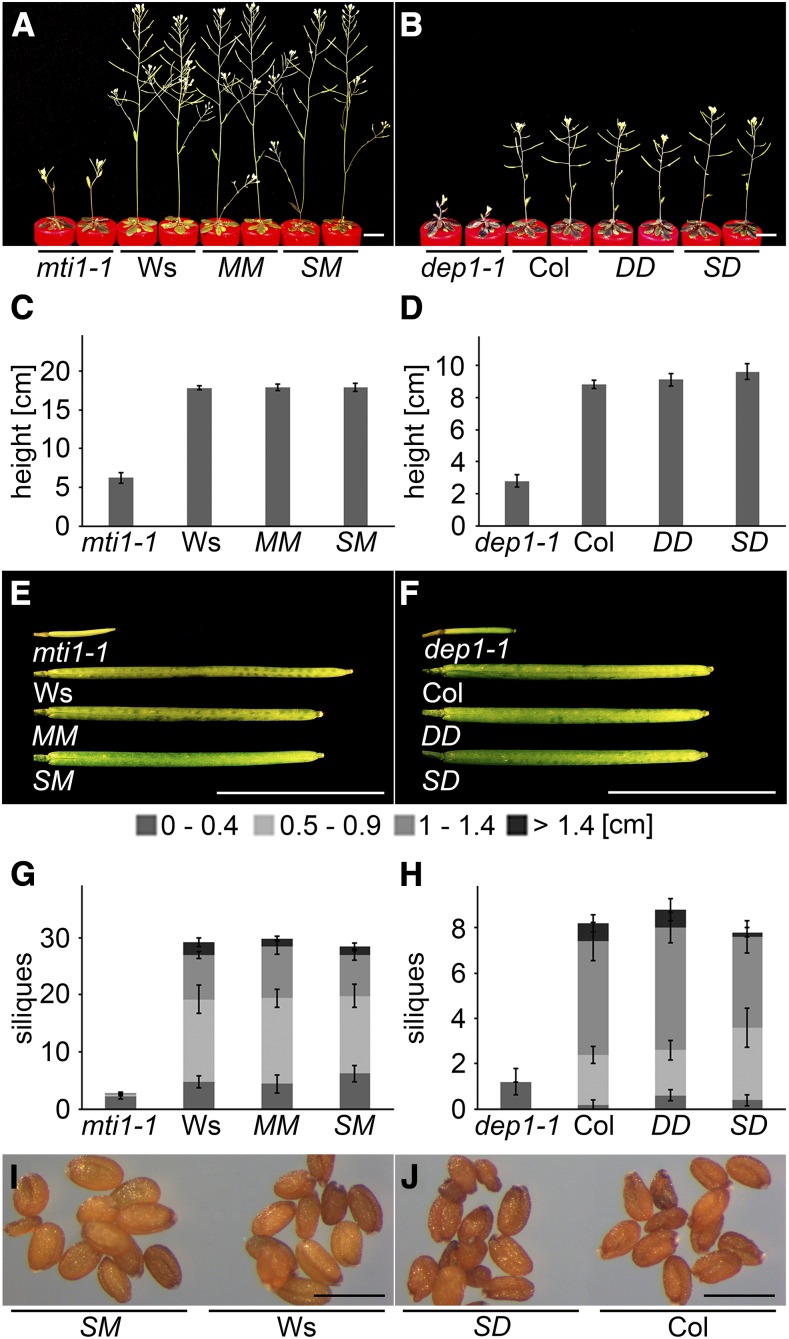
Since S deficiency caused reproductive defects in mti1 and dep1 mutant plants and since the corresponding genes were primarily expressed in the vasculature of vegetative and reproductive organs, we investigated whether the S-dependent mutant phenotypes could be rescued by vascular-specific expression of these genes. We generated constructs that expressed the ORFs of MTI1 and DEP1 under transcriptional control of the companion cell-specific SUC2 promoter (Truernit and Sauer, 1995; Stadler and Sauer, 1996; Imlau et al., 1999) and introduced them into the mti1-1 or dep1-1 mutant background (SM = pSUC2::MTI1; SD = pSUC2::DEP1). The complemented lines MM and DD, carrying the respective wild-type alleles under control of their own promoter, were used as controls. When grown in hydroponic culture with 10 µm sulfate, both mutants displayed S deficiency symptoms, i.e. reduced inflorescence height (Fig. 5, A–D) and short siliques without seeds (Fig. 5, E–H). By contrast, the two complemented lines, including the SM and SD lines, had the same phenotype as the wild type. These lines had a similar height (Fig. 5, C and D) and the same silique number and length as wild-type plants (Fig. 5, E–H). Furthermore, SM and SD lines produced mature and fertile seeds (Fig. 5, I and J). These results clearly showed that a localized expression of these two Met Cycle genes in companion cells is required and sufficient to restore wild-type-like growth in the mti1 and dep1 mutants under the conditions applied here and substantiated the importance of the Met Cycle in the phloem to overcome S deficiency.
Figure 5.
Phloem-specific complementation of the S deficiency mutant phenotype. A to J, Mutant, wild-type, and complementation lines were grown in Hoagland medium containing 10 µm sulfate. Mutant plants were complemented by expressing the wild-type alleles under their own (MM and DD) or the companion cell-specific SUC2 promoter (SM and SD). A and B, Habitus of mti1-1, Ws, MM, and SM plants (A) or dep1-1, Col, DD, and SD plants (B). C and D, Inflorescence height of mti1-1, Ws, MM, and SM plants (C) or dep1-1, Col, DD, and SD plants (D). E and F, Siliques of mti1-1, Ws, MM, and SM plants (E) or dep1-1, Col, DD, and SD plants (F). G and H, Silique number and length of mti1-1, Ws, MM, and SM plants (G) or dep1-1, Col, DD, and SD plants (H). Siliques were grouped into four categories (0–0.4 cm, 0.5–0.9 cm, 1–1.4 cm, and >1.4 cm). Bars represent mean values and standard errors calculated with data from five plants per genotype. I, Mature seeds of SM and Ws plants. J, Mature seeds of SD and Col plants. Bars = 1 cm in A, B, E, and F and 500 µm in I and J.
S-Deficient mti1 and dep1 Mutants Display Low NA, But Not Low Metal Levels
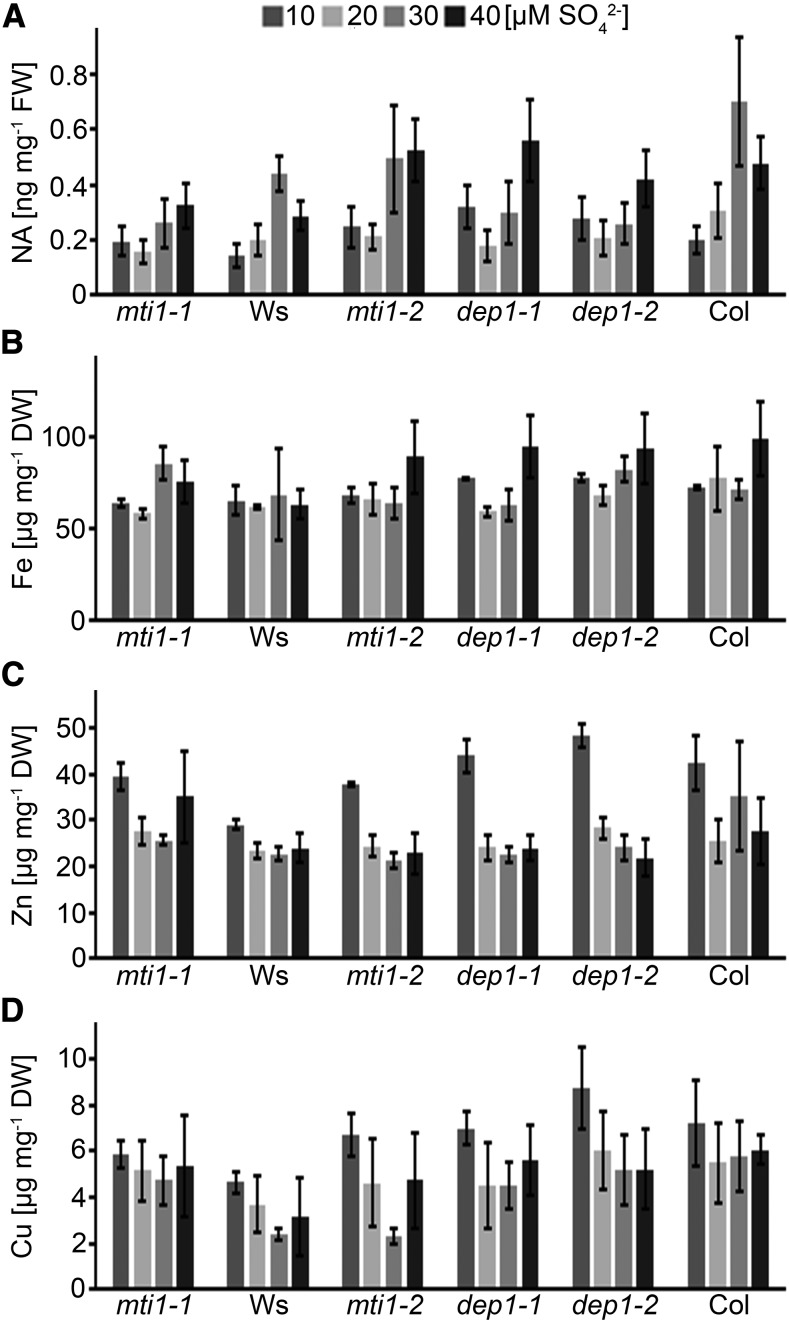
Since the Met Cycle recycles MTA produced during the synthesis of NA, polyamines, and ethylene, the observed growth suppression of the mutants under S deficiency was likely to be connected to one or more of these metabolic pathways. To assess the impact of the Met Cycle on the production of NA during S deficiency, we measured NA levels of the inflorescences of mutant and wild-type plants grown at different sulfate concentrations. Accumulation of NA depended on sufficient S supply in all genotypes. NA levels were reduced by up to 50%, when plants were grown at 10 or 20 µm sulfate in comparison to 30 or 40 µm sulfate. No significant changes could be detected (t test, P < 0.05) between wild-type and mutant plants for a particular S condition (Fig. 6A). To test if the reduced NA concentrations resulted in altered metal homeostasis, concentrations of Fe, Zn, and Cu ions in inflorescences were measured. The concentrations of iron ranged from 60 to 80 µg g−1 dry weight and were not significantly altered by the S status in any of the tested genotypes or conditions (Fig. 6B). Zinc levels were also similar in all genotypes and nutritional conditions with the exception of plants grown in 10 µm sulfate, which accumulated more Zn (Fig. 5C). Copper levels ranged from 2 to 8 µg g−1 dry weight and were also comparable among all tested genotypes and conditions (Fig. 6D). In conclusion, NA accumulation in wild-type and mutant plants depended on sulfate supply in a similar manner. However, lower NA levels did not result in reduced metal delivery to the floral organs of the mutants or the wild type. Therefore, the reproductive defects observed only in mutant plants under low S were likely not caused by a suppression of NA biosynthesis.
Figure 6.
NA, Fe, Zn, and Cu ion concentrations in mutant and wild-type lines. A to D, NA (A), iron (B), zinc (C), and copper (D) ion concentrations in inflorescences of mti1-1, Ws, mti1-2, dep1-1, dep1-2, and Col plants grown in Hoagland medium containing 10 to 40 µm sulfate. Bars represent mean values and standard errors calculated with data from three independent measurements.
S-Deficient dep1 Mutant Plants Display Reduced Putrescine and Spermidine Levels
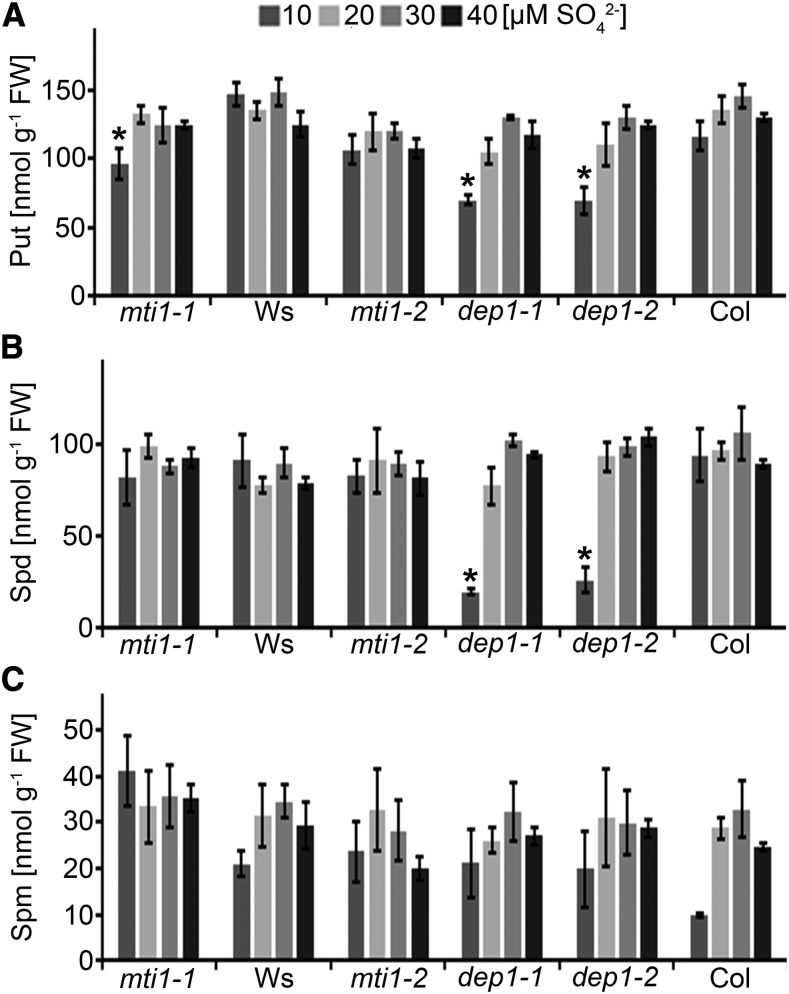
To analyze the S dependency of polyamine biosynthesis in mutant and wild-type plants, we measured the concentration of the polyamines (PAs) putrescine, spermidine, and spermine in the same inflorescences that had been used for NA and metal analyses. The PA levels we measured in the inflorescences of wild-type and mutant plants were in the same order of magnitude as reported previously (Naka et al., 2010; Waduwara-Jayabahu et al., 2012). Putrescine levels were similar in all genotypes and conditions and at approximately 120 nmol g−1 FW with the exception of mti1-1, dep1-1, and dep1-2 grown in 10 µm sulfate. In mti1-1, putrescine levels were reduced by 35% (t test, P < 0.049) and by 40% in dep1-1 (t test, P < 0.015) and dep1-2 (t test, P < 0.030; Fig. 7A). Spermidine levels were similar in all genotypes and conditions and at approximately 80 nmol g−1 FW with the exception of dep1-1 and dep1-2 grown in 10 µm sulfate. In dep1-1, spermidine levels were reduced by 80% (t test, P < 0.006) and by 72% in dep1-2 (t test, P < 0.012; Fig. 7B). Spermine levels were similar in all genotypes when grown in 20, 30, or 40 µm sulfate. Interestingly, spermine levels of wild-type plants grown in 10 µm sulfate were reduced compared to wild-type plants grown in 20 to 40 µm sulfate. Spermine levels were reduced by 30% in Ws wild type and by 65% in Col wild type. Spermine levels of mti1-1, mti1-2, dep1-1, and dep1-2 were approximately twice as high as in their respective wild types, although the differences were not significant due to high standard errors (Fig. 7C). Nevertheless, the reduction in putrescine and spermidine and the decrease of spermine in wild-type plants grown in 10 µm sulfate indicated insufficient polyamine metabolism in plants grown in 10 µm sulfate.
Figure 7.
Polyamine concentrations in mutant and wild-type lines. A to C, Putrescine (A), spermidine (B), and spermine (C) concentrations in inflorescences of mti1-1, Ws, mti1-2, dep1-1, dep1-2, and Col plants grown in Hoagland medium containing 10 to 40 µm sulfate. Bars represent mean values and standard errors calculated with data from three independent measurements. Significances were tested using t test with mutant and corresponding wild-type values within the same S treatment. Asterisks indicate P-values < 0.05.
Spermine But Not Putrescine or Spermidine Can Partially Complement the Reproductive Defects in S-Deficient mti1 and dep1 Mutants
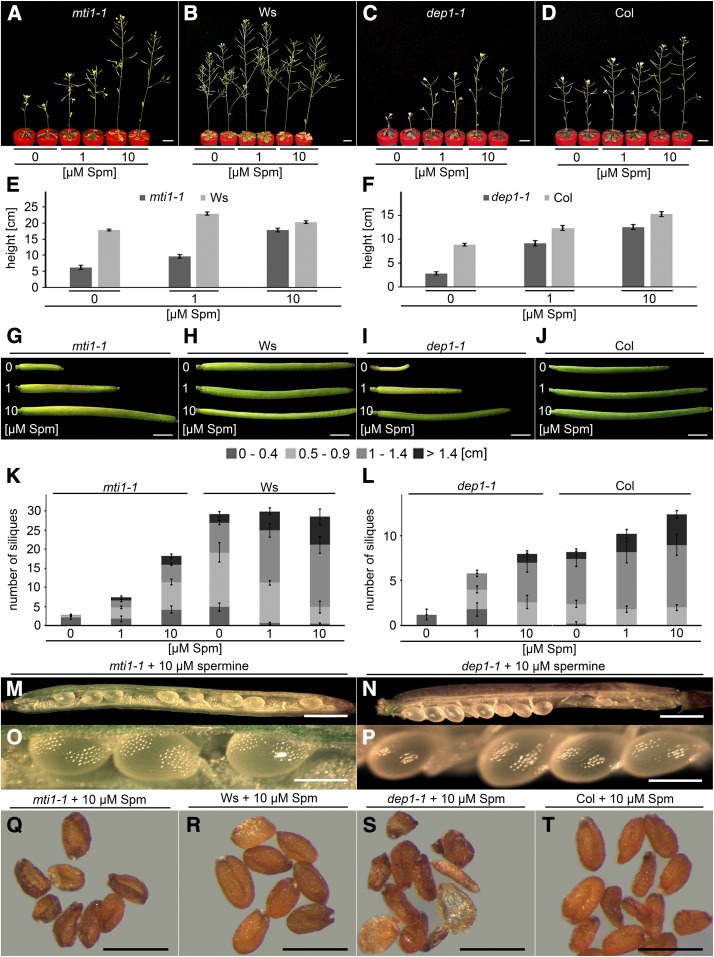
Since mti1 and dep1 displayed altered polyamine levels in inflorescences, we tried to complement their S deficiency-induced phenotypes by external application of different concentrations and forms of polyamines. For this purpose, the hydroponic medium containing 10 µm sulfate was supplemented with 0, 1, 10, or 100 µm putrescine, spermidine, or spermine. As the supplementation with 100 µm of either of these polyamines turned out to be detrimental to plants, rescue analyses were conducted with 0, 1, or 10 µm supply. Neither 1 nor 10 µm putrescine or spermidine had a positive effect on the fertility of the mutants at 10 µm sulfate (Supplemental Figs. S5 and S6). However, when spermine was added to the culture medium, plant growth improved in a concentration-dependent manner. Already 1 µm spermine substantially improved inflorescence height and silique production in the mutants. When 10 µm spermine were added, mutant inflorescences almost reached wild-type height (Fig. 8, A–F) and silique number and length was increased even further (Fig. 8, K and L). In contrast to the aborted mutant seeds, found in plants grown without spermine supplementation (Fig. 3, S and T), the addition of spermine enabled mutant plants to develop seeds (Fig. 8, M–P). At maturity, however, mutant seeds were deformed (Fig. 8, Q–S) and their germination rate was only 7% (mti1-1 + 10 µm spermine) or 12% (dep1-1 + 10 µm spermine) of wild-type seeds. Interestingly, supplementation with 1 or 10 µm spermine proved beneficial even to wild-type plants as they showed slightly better growth (Fig. 8, B and D–F) and longer siliques (Fig. 8, K and L) than control plants without additional spermine supply. These results showed that external application of spermine can partially rescue the S deficiency-induced reproductive phenotype of the Met Cycle mutants, indicating that the mutants suffered from spermine deficiency when grown under low sulfate conditions.
Figure 8.
Complementation of mutant phenotypes with externally applied spermine. A to T, Plants were grown in Hoagland medium containing 10 µm sulfate and supplemented with either 0, 1, or 10 µm spermine. A to D, Habitus of mti1-1 (A), Ws (B), dep1-1 (C), or Col (D) plants. E and F, Inflorescence height of mti1-1 and Ws plants (E) or dep1-1 and Col plants (F). G to J, Siliques of mti1-1 (G), Ws (H), dep1-1 (I), or Col (J) plants. K and L, Silique number and length of mti1-1 and Ws plants (K) or dep1-1 and Col plants (L). Siliques were grouped into four categories (0–0.4 cm, 0.5–0.9 cm, 1–1.4 cm, and >1.4 cm). Bars represent mean values and standard errors calculated with data from five plants per genotype and condition. M to P, Opened siliques of mti1-1 (M and O) or dep1-1 (N and P) plants supplemented with 10 µm spermine. Q to T, Mature seeds of mti1-1 (Q) and Ws (R) plants or dep1-1 (S) and Col (T) plants supplemented with 10 µm spermine. Bars = 1 cm for A to D, 1 mm for G to J, M, and N, 500 µm for Q to T, and 200 µm for O and P.
DISCUSSION
Previous studies showed that the Met Cycle plays an essential role for the biosynthesis of ethylene, NA, and PAs (Bürstenbinder et al., 2007; Rzewuski et al., 2007; Waduwara-Jayabahu et al., 2012) and that in Arabidopsis the enzymes MTI1 and DEP1 were able to complement yeast mutants defective in the Met Cycle (Pommerrenig et al., 2011). So far, a characterization of the functional importance of the two enzymes in planta and their involvement in any of the Met Cycle-dependent pathways was lacking. Here, we show that MTI1 and DEP1 take over an essential function in polyamine biosynthesis under S-deficient growth conditions in planta. Under growth conditions in which MTA served as the sole S source, we observed a strong growth reduction of mti1-1, mti1-2, dep1-1, and dep1-2 mutant plants, demonstrating their inability to utilize MTA as S source. Supplying MTA plus sulfate resulted only in a slight reduction of root growth of the mutant plants when compared to the wild type. This reduction was likely caused by the direct exposure of roots to elevated MTA concentrations, which have been shown to inhibit root growth (Waduwara-Jayabahu et al., 2012). Of course, growth on MTA as sole S source can only serve for the functional characterization of Met Cycle enzymes, whereas it does not necessarily describe the role of the Met Cycle for S metabolism and for the plant’s response to environmental changes affecting the S status of a cell or tissue. To address these issues, we performed growth experiments with mti1 and dep1 and wild-type plants at different S concentrations. During the vegetative growth stage, no S-dependent differences between mutant and wild-type plants were visible, and lacking Met Cycle activity during this phase, even at lowest S supply, did not hamper growth to a considerable extent (Supplemental Fig. S3), maybe because the S requirement for PA and NA biosynthesis was much lower during vegetative growth (Naka et al., 2010). With the onset of flowering, however, phenotypic differences between mutants and wild type became progressively stronger in an S-dependent manner. At this developmental stage, anthocyanin accumulation in mutant plants under low sulfate conditions (Fig. 3, D and E) indicated a disturbance in primary metabolism, typically resulting in enhanced anthocyanin biosynthesis as a result of hampered amino acid biosynthesis and C overflow (Nikiforova et al., 2003, 2005). S deficiency-induced stress symptoms increased in the same way as inflorescence length, and silique number decreased with S availability in the culture medium. However, wild-type plants were still able to produce fertile seeds at the lowest level of sulfate supply (10 µm).
The developmental defects observed here in seeds were similar to those reported for mtn1/mtn2 double knockout mutants grown under nonlimiting S conditions. In these plants, the missing MTN activity resulted in an accumulation of MTA, which in turn severely reduced NA and PA levels through feedback inhibition of NA and PA synthesis enzymes (Waduwara-Jayabahu et al., 2012). In both dep1 and mti1 mutants, however, the block in the Met Cycle occurs after degradation of MTA by MTN and further conversion to MTR-1P by MTK (Fig. 1A). Mutant plants defective in MTK, catalyzing the step preceding MTI1, were unable to grow on MTA as a sole S source but did not otherwise display defects or changes in fresh weight when compared to the wild type (Sauter et al., 2004; Bürstenbinder et al., 2007). There, the growth of mtk plants under S deficiency was only analyzed during vegetative growth, when mtk mutants did not display higher MTA levels than wild-type plants (Bürstenbinder et al., 2007). Therefore, an inhibition of NA and PA synthesis in dep1 and mti1 mutants caused by MTA accumulation through feedback inhibition of Met Cycle intermediates is unlikely. Instead, the observed reduction of PA in the mutant plants rather results from reduced free Met pools than from high MTA concentrations.
Plants without a functional Met Cycle have lower Met, AdoMet, SAH, and SMM levels than wild-type plants (Fig. 4). Especially, concentrations of AdoMet and SMM were reduced at S-limiting conditions. AdoMet is synthesized from Met by AdoMet synthetase (SAMS). There are four SAMS isoforms in Arabidopsis (SAMS1 to SAMS4). SAMS3 and SAMS4 are expressed in trichomes (Wienkoop et al., 2004), while SAMS1 has been shown to be expressed in the vascular tissue (Peleman et al., 1989). Vascular-specific proteome analyses revealed abundance of SAMS1 and SAMS2 in the phloem sap of Arabidopsis and Brassica napus (Schad et al., 2005; Giavalisco et al., 2006). AdoMet itself is a major component of Arabidopsis phloem sap (Bourgis et al., 1999). SMM is the phloem-mobile transport form of Met and has a major role for S supply of sink tissues (Bourgis et al., 1999; Tan et al., 2010). These reports are in line with the phloem-specific expression of the Met Cycle genes and indicate that the reduction of AdoMet and SMM in the dep1 and mti1 mutants most likely represents a reduction of reduced S compounds in the phloem.
To further elucidate the connection between the S deficiency-induced symptoms in the mti1 and dep1 mutants and the observed vascular specificity of the Met Cycle (Fig. 2), we introduced a pSUC2::MTI1 or pSUC2::DEP1 construct into the mti1-1 or dep1-1 mutant, respectively. The SUC2 promoter was shown to be exclusively active in companion cells throughout the plant phloem, including sink tissues like roots and flowers (Truernit and Sauer, 1995; Stadler and Sauer, 1996; Imlau et al., 1999). In both mutants, the companion cell-specific expression of the Met Cycle genes could fully revert the S deficiency-induced symptoms, indicating that the observed morphological and developmental defects were caused by a phloem-located factor. Most likely, this factor was connected to one of the pathways producing MTA, which are either NA, PA, or ethylene biosynthesis.
Previously, ethylene biosynthesis has been shown to be several times higher in seedlings and in young plants than in mature plants (Rodrigues-Pousada et al., 1993; Bürstenbinder et al., 2007). Consistent with this, expression of ACS1 was shown to be high in young tissues and switched off in mature tissues (Rodrigues-Pousada et al., 1993). In this study, 1-Aminocyclopropanecarboxylate (ACC) could not be detected in inflorescences of both mutant and wild-type plants with neither ultraperformance liquid chromatography (UPLC) nor ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry (UPLC-ESI-MS/MS) measurements. Relative to the net demand of the other pathways, we concluded that ethylene biosynthesis did not require considerable amounts of Met for the development of flowers and seeds.
NA is a ubiquitously occurring metal chelator in plants with important functions in metal scavenging, mobilization, long-distance transport, and metal supply to floral organs. Regarding the latter, NA is most important for the phloem mobility of Fe, Zn, and Cu (Klatte et al., 2009; Deinlein et al., 2012; Haydon et al., 2012; Schuler et al., 2012; Zheng et al., 2012). Furthermore, several phloem localized NA transporters have been identified (Koike et al., 2004; Chu et al., 2010; Zheng et al., 2012), some of which appeared to mediate source-to-sink translocation of Fe and other metals during plant senescence (DiDonato et al., 2004). To our knowledge, cell type-specific expression of NA synthases has not been shown directly in Arabidopsis. To assess if the S deficiency-induced reproductive failure of mti1 and dep1 could be caused by a lack of NA, NA and metal concentrations of inflorescences were measured. NA levels were in the same order of magnitude as reported elsewhere (Klatte et al., 2009; Waduwara-Jayabahu et al., 2012) and apparently dependent on the supplied sulfate concentration. However, also under the least S concentration supplied, plants were still able to synthesize a minimum of NA. Compatible with this, we found that the inflorescence levels of Fe, Zn, and Cu were largely unaffected by the S amount supplied to the plants. This is supported by analyses of nas4x quadruple knockouts (Schuler et al., 2012). One of the quadruple mutants, nas4x-1, had a leaky nas2-1 allele with residual NAS activity in contrast to the complete loss of NA in the nas4x-2 mutants. Whereas nas4x-2 plants were chlorotic and sterile, the residual NA in nas4x-1 was sufficient so sustain plant viability and reproduction, illustrating that only small amounts of NA were needed to complete the life cycle. In line with these results, mti1 and dep1 mutants did not display interveinal chlorosis, which is typical for NA deficiency and was first reported in the NA-less tomato (Solanum lycopersicum) mutant chloronerva (Scholz et al., 1992). We therefore concluded that the S deficiency-induced reproductive defects of mti1 and dep1 were not caused by insufficient metal supply through a lack of NA.
In contrast to NA, many enzymes of the PA biosynthesis pathway have been reported to be active in the vasculature, including ACL5 (Clay and Nelson, 2005), BUD2 (Ge et al., 2006), SPDS2 (Hewezi et al., 2010), and SAMDC2 (Pommerrenig et al., 2011). Additionally, cell type-specific expression analysis (eFP browser; http://efp.ucr.edu/) predicts phloem-specific expression of basically all genes of the PA biosynthesis pathway (ADC1, AIH, NLP1, SPDS1, SPDS2, SPMS, and SAMDC1-4). The phloem-specific detection of the expression of the Met Cycle genes and the phloem-specific complementation of the S deficiency-induced mutant phenotype are in line with these reports and expression data. Obviously, Met Cycle activity in the phloem of adult plants was sufficient for the replenishment of the Met pool required for the synthesis of PA in this tissue. Naka et al. (2010) reported that the levels of putrescine, spermidine, and spermine were about twice as high in flowers as in stems. Since we measured whole inflorescences, and since mti1 and dep1 mutant plants, when grown in 10 µm sulfate, formed very short inflorescences, it is possible that the PA levels increased due to a concentration effect. Nevertheless, we found that in the Col background putrescine and spermidine levels were significantly reduced when plants were grown in 10 µm sulfate. In contrast, there were no significant differences in PA levels in the Ws background due to S supply (Fig. 7). We assume that due to the higher tolerance of the Ws ecotype to S deficiency, the level of free Met was sufficient to maintain PA synthesis. Interestingly, wild-type plants grown in 10 µm sulfate displayed a reduction in spermine concentrations, indicating that even in presence of a functional Met Cycle, spermine synthesis is restricted under such low S supplies. Compared to the wild type, spermine levels were higher in mutant plants when grown in 10 µm sulfate (Fig. 7). However, this increase was not significant and could very well be a result of afore-mentioned concentration effects.
Since PA levels were strongly compromised in mti1 and dep1 mutants under S deficiency, we performed growth experiments by supplementing different polyamines externally. Root application of PA has been previously shown to complement PA-associated growth defects (Waduwara-Jayabahu et al., 2012), and recently PA transporters have been identified from rice and Arabidopsis (Fujita et al., 2012; Mulangi et al., 2012a, 2012b), suggesting uptake of polyamines by the root system. We added varying amounts of the non-S-containing PA putrescine, spermidine, or spermine directly to the 10 µm sulfate-containing medium and observed that neither putrescine nor spermidine improved the growth of the mti1 and dep1 mutant plants (Supplemental Figs. S5 and S6). This is in contrast to the results of Waduwara-Jayabahu et al. (2012), in which the addition of spermidine had the strongest positive effect. While the mtn1/mtn2 double knockout plants accumulated MTA, which caused feedback inhibition of PA synthases and ultimately lead to thermospermine deficiency with thermospermine-dependent vascular defects, the mti1 and dep1 mutants rather suffered from a lack of spermine and displayed stress-related phenotypes. This is supported by the fact that the vascular organization of mti1 and dep1 mutant plants, grown in 10 µm sulfate, was not different compared to the wild type (Supplemental Fig. S7). We conclude from these results that the S deficiency-induced defects of mti1 and dep1 are not caused by the lack of thermospermine but rather by the lack of spermine.
Different reports have shown correlations between PAs and abiotic stress protection, and although the precise mechanism of the individual polyamines is still unknown, they have been implicated in several biological processes. Among many other roles, PAs can function as compatible solutes, protective substances for nucleotides and membranes, as scavengers for reactive oxygen species, and as promoters for the production of antioxidant enzymes and metabolites (Minocha et al., 2014). Several reports also demonstrated the importance of spermine, particularly during long-term stress treatments. In this study, spermine was able to protect plants from the negative effects caused by S deficiency. A metabolic canalization from putrescine toward spermine was reported under drought stress in rice, Arabidopsis, and the resurrection plant Craterostigma plantagineum (Alcázar et al., 2011; Do et al., 2013). However, despite the metabolic canalization, free spermine levels did not increase in these studies. A similar situation might have occurred in the S-deficient mutant plants observed in this study. Our results show that S deficiency during the reproductive phase ultimately leads to a shortage of polyamines, most importantly spermine and that this spermine shortage is responsible for S deficiency-induced infertility in the Met Cycle mutants mti1 and dep1. We assume that during S deficiency, large amounts of spermine are used for binding and protection of nucleic acids and proteins. For the biosynthesis of spermine, two molecules of AdoMet are required, whereas only one is required for the biosynthesis of spermidine. Functional Met recycling can compensate for the S deficiency and replenish low Met and consecutively low AdoMet pools required for the conversion of spermidine to spermine. How exactly spermine interacts with the regulatory network of S metabolism and whether additional regulatory mechanisms exist beyond the assimilation and recycling of S remain to be uncovered in future experiments.
MATERIALS AND METHODS
Plant Growth Conditions
Growth experiments with MTA containing media were performed as described by Sauter et al. (2004). For hydroponic growth experiments, seeds were surface-sterilized for 5 min in a household bleach/Tween 20 mixture and washed three times with water. After stratification, single seeds were sown onto PCR tubes containing Hoagland medium with 0.5% agar. Seeds were germinated and seedlings were grown under short-day conditions (12 h light, 22°C; 12 h dark, 18°C). After 14 d plants were transferred to Falcon tubes containing Hoagland solutions with different sulfate concentrations and grown under long-day conditions (16 h light, 22°C; 8 h dark, 18°C). Hoagland medium contained 88 µm (NH4)2HPO4, 400 µm Ca(NO3)2, 600 µm KNO3, 200 µm MgSO4, 4.62 µm H3BO3, 0.032 µm CuSO4, 0.92 µm MnCl2, 0.076 µm ZnSO4, 0.011 µm MoO3, and 5 µm Fe-[N,N'-di(2-hydroxybenzyl)ethylenediamine-N,N'-diacetic acid monohydrochloride]. For Hoagland medium with defined sulfate concentrations, sulfate salts were replaced with chloride salts and MgSO4 was added to the desired sulfate concentration. The pH value was adjusted to pH 5.7 with nitric acid. Hoagland medium was exchanged weekly. For all measurements, three independent sets of plants were grown and whole inflorescences were harvested and used for the analyses. Inflorescences were harvested at 10 to 12 d after flowering, corresponding to growth stage 6.5 when approximately 50% of flower buds were open (Boyes et al., 2001). The inflorescences of at least 10 hydroponically grown plants per set and genotype were harvested, immediately frozen in liquid nitrogen, and powdered prior to analysis.
Plant Materials and Mutant Characterization
Arabidopsis (Arabidopsis thaliana) T-DNA insertion line FLAG_145B05 (Samson et al., 2002) and the Ws wild type were obtained from the INRA Arabidopsis thaliana Resource Centre. Salk_091578C (Alonso et al., 2003) and WiscDsLox489-492G6 (Woody et al., 2007) and Col wild type were obtained from the Nottingham Arabidopsis Stock Centre. Homozygous null mutants were identified by PCR/RT-PCR using the primers listed in Supplemental Table S1.
Generation of an AmiRNA Targeting MTI1
AmiRNA targeting MTI1 was designed and cloned according to the WMD3 Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). The resulting amiRNA construct was inserted into pENTR/D-TOPO (Life Technologies) and recombined with pEarlygate100 (Earley et al., 2006). Col-0 plants were transformed with the resulting p35S::MTI1 amiRNA construct using floral dip (Clough and Bent, 1998).
Construction of Reporter Gene Lines
pMTI1::MTI1 (1925-bp promoter) and pDEP1::DEP1 (1649-bp promoter) were amplified from genomic Arabidopsis DNA using the primers MTI1-1925f and MTI1+2018r or DEP1-1649f and DEP1+3660r, respectively. The 5′ end of the MTI1 promoter sequence is in 11.918 bp distance to the neighboring gene At2g05820 (uncharacterized transposon). The promoter of DEP1 has an 1114-bp overlap into the ORF of At5g53860 (uncharacterized gene, involved in embryogenesis). PCR products were inserted into pENTR/D-TOPO, which was recombined with pMDC107 (Curtis and Grossniklaus, 2003), yielding pMTI1::MTI1-GFP and pDEP1::DEP1-GFP, respectively. Cloning of pMTI1::MTI1-GUS and pDEP1::DEP1-GUS was described previously (Pommerrenig et al., 2011).
Construction of Complementation Lines
pMTI1::MTI1-GFP or pDEP1::DEP1-GFP was introduced into mti1-1 or dep1-1 plants by floral dip, respectively. Transformed plants were selected with hygromycin B. Resistant plants were validated visually and with PCR and named MM or DD, respectively. To create the pSUC2::MTI1 and pSUC2::DEP1 constructs, MTI1 (1125 bp) and DEP1 (1524 bp) coding sequences (CDS) were amplified and inserted into pENTR/D-TOPO, which was recombined with a destination vector containing a 2125-bp-long fragment of the SUC2 promoter (pSUC2-dest; Thompson and Wolniak, 2008). The pSUC2::MTI1 and pSUC2::DEP1 constructs were introduced into mti1-1 and dep1-1 plants by floral dip, respectively, yielding the lines SM and SD.
Chemical Complementation of Mutants
The mti1-1, Ws wild type, MM, SM, dep1-1, Col wild type, DD, and SD plants were grown in Hoagland medium containing 10 µm sulfate. At the onset of flowering 0, 1, or 10 µm putrescine, spermidine, or spermine were added to the medium. Putrescine dihydrochloride (P7505), spermidine trihydrochloride (85578), and spermine tetrahydrochloride (85605) were obtained from Sigma-Aldrich.
Microscopy
Confocal images were taken on a TCS SP5 (Leica Microsystems) using 488-nm (GFP) and 561-nm (mCherry) laser light for excitation. Detection windows were 495 to 545 nm (GFP), 575 to 600 nm (mCherry), or 675 to 700 nm (chlorophyll). Light microscopy and nonconfocal images were taken on a Zeiss Axioskop HBO 50 or a Leica MZFLIII fluorescence stereomicroscope.
GUS Staining, Embedding, and Cross-Sectioning
GUS staining was performed as described by Weingartner et al. (2011), and Technovit embedding was performed as described by Ühlken et al. (2014). The 25-µm cross sections of rosette leaves or roots were cut using a Leica RM 2135 BioCut rotary microtome.
Subcellular Localization of MTI1 and DEP1 Proteins
MTI1 and DEP1 coding sequences were amplified from Arabidopsis cDNA using the primers MTI1_CDS+1f and MTI1_CDS+1122r or DEP1_CDS+1f and DEP1_CDS+1521r. PCR products were inserted into pENTR/D-TOPO, which was recombined with pMDC83 (Curtis and Grossniklaus, 2003), yielding pCaMV35S::MTI1-GFP or pCaMV35S::DEP1-GFP. pCaMV35S::CaM2-mCherry (Fischer et al., 2013) was used as cytosolic control. Arabidopsis mesophyll protoplasts were generated and transformed as previously described (Abel and Theologis, 1994; Drechsel et al., 2011).
Extraction and Measurement of Soluble Amino Acids
Soluble amino acids were extracted from 100 mg of plant powder with 1 mL of 80% ethanol for 1 h at 80°C. After centrifugation, 0.8 mL of supernatant was concentrated and the resulting pellet was resuspended with water. Standards and samples were derivatized with AQC reagent (Waters) according to the manufacturer’s instructions. Separation of soluble amino acids was performed using UPLC (AcQuity H-Class; Waters). Separation was carried out on a C18 reversed phase column (ACCQ Tag Ultra C18, 1.7 µm, 2.1 × 100 mm) with a flow rate of 0.7 mL per min and duration of 10.2 min. The column was heated at 50°C during the whole run. The detection wavelengths were 266 nm for excitation and 473 nm for emission. The gradient was accomplished with four solutions prepared from two different buffers purchased from Waters (eluent A concentrate and eluent B for amino acid analysis). Eluent A was pure concentrate, eluent B was a mixture of 90% LCMS water (Th. Geyer) and 10% eluent B concentrate, eluent C was pure concentrate (eluent B for amino acid analysis), and eluent D was LCMS water (Th. Geyer). The column was equilibrated with eluent A (10%) and eluent C (90%) for at least 30 min. The gradient was produced as follows: 0 min 10% A and 90% C/0.29 min 9.9% A and 90.1% C/5.49 min 9% A, 80% B, and 11% C/7.1 min 8% A, 15.6% B, 57.9% C, and 18.5% D/7.3 min 8% A, 15.6% B, 57.9% C, and 18.5% D/7.69% 7.8% A, 70.9% C, and 21.3% D/7.99 min 4% A, 36.3% C, and 59.7% D/8.68 min 10% A, 90% C/10.2 min 10% A, and 90% C. For accurate identification of the OAS peak, different samples were spiked with purest OAS and measured under the same conditions.
Detection of Sulfur Metabolism Metabolites by UPLC-ESI-MS/MS
Reagents and Reference Materials
Acetonitrile (ACN) used for extraction and liquid chromatography was purchased from Th. Geyer and formic acid (FA) from Biosolve Chimie (France). Deionized distilled water was from the Milli-Q Reference System, Merck. The chemical standards were purchased from Sigma-Aldrich (Supplemental Table S2).
Sample Extraction
Approximately 100 mg fresh tissue was crushed to fine powder under liquid nitrogen. Samples were transferred immediately in a 2-mL Eppendorf tube, mixed, and centrifuged in 300 µL solution containing 45/45% ACN/water and 10% FA (v/v) and stored at −80°C until extraction. With the extraction, two steel balls and 700 µL 50/50 ACN/water (v/v) were added to each sample. All further steps were conducted on ice or at 4°C. The mixture was sonicated for 5 min, extracted with an overhead shaker for 15 min, and centrifuged for 10 min at 14000 rpm and the supernatant transferred to a new Eppendorf tube. For reextraction, 0.5 mL 99/1% water/FA (v/v) was added and the mixture was sonicated for 5 min, extracted with an overhead shaker for 15 min, and 0.5 mL 99/1% ACN/FA (v/v) was added. The mixture was extracted again at 5 min by overhead shaking. After centrifugation for 10 min at 14000 rpm, the supernatants were combined and filtered with a 0.45-µm polyvinylidene difluoride syringe filter (Merck, Millex-HV). For UPLC-ESI-MS/MS, an aliquot of 100 µL sample was mixed with 20 µL internal standard containing 500 ng mL−1 13C-MET and 80 µL 94.5/94.5% ACN/water and 1% FA (v/v) and transferred in a vial for analysis.
Liquid Chromatography-Tandem Mass Spectrometry Analysis
Ten microliters of sample was injected into an Acquity ultraperformance liquid chromatography system (Waters) coupled with a Xevo TQ MS mass spectrometer (Waters). The samples were separated on an Acquity UPLC BEH Amide 2.1 × 100-mm, 1.7 µm (Waters) coupled with a VanGuard precolumn BEH Amide 1.7µm , 2.1 mm × 5 mm (Waters). The column temperature was set to 40°C and the autosampler was set to 4°C. The mobile phases were water containing 0.1% FA (A) and ACN containing 0.1% FA. The mobile phase flow was 0.4 mL min−1. The gradient conditions were as follows: A, 15% in 0 to 1.0 min; 15 to 50% in 1.0 to 7.0 min; 50 to 99% in 7.0 to 7.5 min; 99 to 99% in 7.5 to 8.5 min; 99 to 15% in 8.5 to 9.0 min; 15% in 9.0 to 11.0 min. The Xevo TQ MS operated in ESI+ ion mode. The electrospray capillary voltage was 2.7 kV with a cone voltage of 20 V. The cone and desolvation gas flows were set to 20 and 1000 L h−1. The cone and desolvation gas flows were set to 50 and 1000 L h−1. The source and desolvation temperature were 150°C and 650°C. The collision energy of the MS/MS was between 5 and 50 eV. For the quantification and qualification of the analytes, we used three fragment ions (Supplemental Table S2). Mass spectrometry data processing was done using TargetLynx V4.1 SCN 904. The peak area of the diagnostic product ion was used for quantification.
Measurements of Cu, Fe, and Zn
Elemental analyses were performed with 40 mg plant material as described by Gruber et al. (2013).
Extraction and Measurement of NA
NA was extracted from 100 mg plant powder with 1 mL 80% ethanol for 1 h at 80°C. After centrifugation, 0.8 mL supernatant was concentrated. The resulting pellet was resuspended with 0.6 mL HPLC-H2O. After centrifugation for 5 min at 14.000 rpm and filtration with Multiscreen 10KD plates (Bio-Rad) for 90 min at 4°C and 5000 rpm, samples were used for liquid chromatography-tandem mass spectrometry measurements. For separation and detection of NA, an Agilent UPLC 1290 system connected to an Agilent triple Quad MS 6490 mass spectrometer equipped with an easy Jet Spray was employed. Ten microliters of sample was separated on an Acquity UPLC HSS T3 column (1.8 µm, 2.1 × 150 mm; Waters) with a flow rate of 0.4 mL min−1. Solvents were water (A) and acetonitrile (B), both acidified with 0.1% (v/v) formic acid. The binary gradient used was as follows: 0 to 0.5 min at 99.9:0.1% (A:B), 0.5 to 3.1 min at 95:5%, 3.1 to 4.5 min at 30:70%, 4.5 to 4.6 at 99.9:0.1%, and 4.6 to 5 min at 99.9:0.1%. The mass spectrometer was operated in the ESI V+ mode and multiple reaction monitoring mode. The following instrument parameters were used: nitrogen gas flow = 12 l min−1, ion spray voltage = 3000 V, and auxiliary gas temperature = 300°C. Dwell time for each transition was 20 ms. For accurate identification of the NA peak and to exclude sample matrix effects, different samples were spiked with purest NA and measured under the same conditions. Quantification was performed on the respective reconstructed ion traces of the protonated molecular ions using the Mass Hunter (version B.04.00).
Extraction and Measurement of Polyamines
PAs were extracted and dansylated as described elsewhere (Minocha et al., 1990, 1994) and measured with UPLC. Separation was carried out on a reversed phase column (BEH C18 column, 1.7 µm, 2.1 × 50 mm; Waters) with a flow rate of 0.4 mL min−1 for 4.5 min. The column temperature was set to 40°C. As solvents, HPLC-grade water (A) and LCMS-grade acetonitrile (B) were used. The binary gradient used was as follows: 0 to 0.06 min at 10:90% (A:B), 0.06 to 1.7 min at 42:58%, 1.7 to 3.17 min at 100:0%, 3.17 to 3.29 min at 42:58%, and 3.29 to 4.50 min at 10:90%. Detection wavelengths were 254 and 360 nm as single wavelengths and a spectrum between 200 and 300 nm with a bandwidth of 4.8 and a sampling rate of 20 points per second.
Extraction and Measurement of Anthocyanin
Whole shoots of three representative plants grown in Hoagland solution containing 10 µm sulfate were harvested, frozen in liquid nitrogen, and ground. One hundred milligrams of plant material was extracted using a buffer containing 45% methanol and 5% acetic acid. After extraction for 5 min, cell debris was removed by centrifugation and relative anthocyanin concentrations were determined using an absorption-based method previously described by Matsui et al. (2004).
RNA Isolation, cDNA Synthesis, and qRT-PCR Analyses
RNA was isolated from 20 mg of plant tissue using Trizol reagent (Life Technologies). cDNAs were synthesized from 1 µg of RNA using the QuantiTect reverse transcription kit (Qiagen). Primers used for qRT-PCR analyses were designed using QuantPrime (Arvidsson et al., 2008) and are listed in Supplemental Table S1. Primer specificity was validated by visualizing amplified PCR products on agarose gels. AtUBQ10 was used as reference gene, since its expression was shown to be stable throughout development (Czechowski et al., 2005). Measurements were performed on a Rotor-Gene Q cycler (Qiagen) using Brilliant III Ultra Fast SYBR Green QPCR Master Mix (Agilent). cDNAs from all biological samples were run in triplicates. Transcripts were quantified by using the ∆∆Cq method.
Accession Numbers
Arabidopsis Genome Initiative gene codes for the Arabidopsis genes described in this study are as follows: MTI1, At2g05830; and DEP1, At5g53850.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. MTI1 and DEP1 wild-type alleles complement mutant growth phenotypes on MTA-containing media.
Supplemental Figure S2. S deficiency response in wild-type plants.
Supplemental Figure S3. Number of rosette leaves and dry weight.
Supplemental Figure S4. Time course of mutant growth during S deficiency.
Supplemental Figure S5. Chemical complementation of mutant phenotype with putrescine.
Supplemental Figure S6. Chemical complementation of mutant phenotype with spermidine.
Supplemental Figure S7. Stem cross sections of plants grown in 10 µm sulfate.
Supplemental Table S1. Oligonucleotides.
Supplemental Table S2. Standards used during UPLC-ESI-MS/MS measurements.
Supplementary Material
Acknowledgments
We thank Susanne Reiner (IPK Gatersleben) for inductively coupled plasma optical emission spectroscopy analysis and Cornelia Fischer (Molecular Plant Physiology Erlangen) for providing pCaMV35S::CaM2-mCherry.
Glossary
- MTA
5′-methylthioadenosine
- MTRu-1-P
5-methyl-thioribulose-1-P
- DHKMP
1,2-dihydroxy-3-keto-5-methylthiopentene
- NA
nicotianamine
- Ws
Wassilewskija
- ORF
open reading frame
- amiRNA
artificial microRNA
- Col
Columbia
- AdoMet
S-adenosyl-Met
- SAH
S-adenosylhomo-Cys
- SMM
S-methyl-Met
- FW
fresh weight
- PA
polyamine
- UPLC
ultraperformance liquid chromatography
- UPLC-ESI-MS/MS
ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry
- ACN
acetonitrile
- FA
formic acid
Footnotes
Articles can be viewed without a subscription.
References
- Abel S, Theologis A (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 5: 421–427 [DOI] [PubMed] [Google Scholar]
- Alcázar R, Bitrián M, Bartels D, Koncz C, Altabella T, Tiburcio AF (2011) Polyamine metabolic canalization in response to drought stress in Arabidopsis and the resurrection plant Craterostigma plantagineum. Plant Signal Behav 6: 243–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur AH, Yang SF (1972) Methionine metabolism in apple tissue in relation to ethylene biosynthesis. Phytochemistry 11: 3207–3214 [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL, Gage DA, Hanson AD (1999) S-methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11: 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürstenbinder K, Rzewuski G, Wirtz M, Hell R, Sauter M (2007) The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J 49: 238–249 [DOI] [PubMed] [Google Scholar]
- Bürstenbinder K, Waduwara I, Schoor S, Moffatt BA, Wirtz M, Minocha SC, Oppermann Y, Bouchereau A, Hell R, Sauter M (2010) Inhibition of 5′-methylthioadenosine metabolism in the Yang cycle alters polyamine levels, and impairs seedling growth and reproduction in Arabidopsis. Plant J 62: 977–988 [DOI] [PubMed] [Google Scholar]
- Capell T, Bassie L, Christou P (2004) Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc Natl Acad Sci USA 101: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HH, Chiecko J, Punshon T, Lanzirotti A, Lahner B, Salt DE, Walker EL (2010) Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Nelson T (2005) Arabidopsis thickvein mutation affects vein thickness and organ vascularization, and resides in a provascular cell-specific spermine synthase involved in vein definition and in polar auxin transport. Plant Physiol 138: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb F, van der Weele CM, Wolniak SM (2010) Spermidine is a morphogenetic determinant for cell fate specification in the male gametophyte of the water fern Marsilea vestita. Plant Cell 22: 3678–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U, Weber M, Schmidt H, Rensch S, Trampczynska A, Hansen TH, Husted S, Schjoerring JK, Talke IN, Krämer U, Clemens S (2012) Elevated nicotianamine levels in Arabidopsis halleri roots play a key role in zinc hyperaccumulation. Plant Cell 24: 708–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hooghe P, Escamez S, Trouverie J, Avice JC (2013) Sulphur limitation provokes physiological and leaf proteome changes in oilseed rape that lead to perturbation of sulphur, carbon and oxidative metabolisms. BMC Plant Biol 13: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato RJ, Jr., Roberts LA, Sanderson T, Eisley RB, Walker EL (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J 39: 403–414 [DOI] [PubMed] [Google Scholar]
- Do PT, Degenkolbe T, Erban A, Heyer AG, Kopka J, Köhl KI, Hincha DK, Zuther E (2013) Dissecting rice polyamine metabolism under controlled long-term drought stress. PLoS One 8: e60325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G, Bergler J, Wippel K, Sauer N, Vogelmann K, Hoth S (2011) C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. J Exp Bot 62: 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fischer C, Kugler A, Hoth S, Dietrich P (2013) An IQ domain mediates the interaction with calmodulin in a plant cyclic nucleotide-gated channel. Plant Cell Physiol 54: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Iuchi S, Yamada K, Kobayashi Y, Urano K, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2012) Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc Natl Acad Sci USA 109: 6343–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Cui X, Wang Y, Hu Y, Fu Z, Zhang D, Cheng Z, Li J (2006) BUD2, encoding an S-adenosylmethionine decarboxylase, is required for Arabidopsis growth and development. Cell Res 16: 446–456 [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Kapitza K, Kolasa A, Buhtz A, Kehr J (2006) Towards the proteome of Brassica napus phloem sap. Proteomics 6: 896–909 [DOI] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guranowski A. (1983) Plant 5-methylthioribose kinase: properties of the partially purified enzyme from yellow lupin (Lupinus luteus L.) seeds. Plant Physiol 71: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Kawachi M, Wirtz M, Hillmer S, Hell R, Krämer U (2012) Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis. Plant Cell 24: 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R. (2008) Sulfur Metabolism in Phototrophic Organisms. Springer, Dordrecht, the Netherlands [Google Scholar]
- Hewezi T, Howe PJ, Maier TR, Hussey RS, Mitchum MG, Davis EL, Baum TJ (2010) Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii. Plant Physiol 152: 968–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins TJ, Chandler PM, Randall PJ, Spencer D, Beach LR, Blagrove RJ, Kortt AA, Inglis AS (1986) Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds. J Biol Chem 261: 11124–11130 [PubMed] [Google Scholar]
- Imai A, Matsuyama T, Hanzawa Y, Akiyama T, Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D, et al. (2004) Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol 135: 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Bremont JF, Marina M, Guerrero-González MdeL, Rossi FR, Sánchez-Rangel D, Rodríguez-Kessler M, Ruiz OA, Gárriz A (2014) Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front Plant Sci 5: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H (2004) Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P (2009) The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol 150: 257–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39: 415–424 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S (2014) Molecular mechanisms of regulation of sulfate assimilation: first steps on a long road. Front Plant Sci 5: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H, Marschner P (2012) Marschner’s Mineral Nutrition of Higher Plants. Academic Press, London [Google Scholar]
- Maruyama-Nakashita A, Watanabe-Takahashi A, Inoue E, Yamaya T, Saito K, Takahashi H (2015) Sulfur-responsive elements in the 3′-nontranscribed intergenic region are essential for the induction of SULFATE TRANSPORTER 2;1 gene expression in Arabidopsis roots under sulfur deficiency. Plant Cell 27: 1279–1296 [DOI] [PMC free article] [PubMed]
- Matsui K, Tanaka H, Ohme-Takagi M (2004) Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol J 2: 487–493 [DOI] [PubMed] [Google Scholar]
- Minocha R, Majumdar R, Minocha SC (2014) Polyamines and abiotic stress in plants: a complex relationship. Front Plant Sci 5: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocha SC, Minocha R, Robie CA (1990) High-performance liquid chromatographic method for the determination of dansyl-polyamines. J Plant Growth Regul 13: 187–193 [Google Scholar]
- Minocha R, Shortle W, Long S, Minocha S (1994) A rapid and reliable procedure for extraction of cellular polyamines and inorganic ions from plant tissues. J Plant Growth Regul 13: 187–193 [Google Scholar]
- Mulangi V, Chibucos MC, Phuntumart V, Morris PF (2012a) Kinetic and phylogenetic analysis of plant polyamine uptake transporters. Planta 236: 1261–1273 [DOI] [PubMed] [Google Scholar]
- Mulangi V, Phuntumart V, Aouida M, Ramotar D, Morris P (2012b) Functional analysis of OsPUT1, a rice polyamine uptake transporter. Planta 235: 1–11 [DOI] [PubMed] [Google Scholar]
- Naka Y, Watanabe K, Sagor GH, Niitsu M, Pillai MA, Kusano T, Takahashi Y (2010) Quantitative analysis of plant polyamines including thermospermine during growth and salinity stress. Plant Physiol Biochem 48: 527–533 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van Montagu M, Inzé D (1989) Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell 1: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerrenig B, Feussner K, Zierer W, Rabinovych V, Klebl F, Feussner I, Sauer N (2011) Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis. Plant Cell 23: 1904–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pousada RA, De Rycke R, Dedonder A, Van Caeneghem W, Engler G, Van Montagu M, Van Der Straeten D (1993) The Arabidopsis 1-Aminocyclopropane-1-Carboxylate Synthase Gene 1 is expressed during early development. Plant Cell 5: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzewuski G, Cornell KA, Rooney L, Bürstenbinder K, Wirtz M, Hell R, Sauter M (2007) OsMTN encodes a 5′-methylthioadenosine nucleosidase that is up-regulated during submergence-induced ethylene synthesis in rice (Oryza sativa L.). J Exp Bot 58: 1505–1514 [DOI] [PubMed] [Google Scholar]
- Sagor GH, Berberich T, Takahashi Y, Niitsu M, Kusano T (2013) The polyamine spermine protects Arabidopsis from heat stress-induced damage by increasing expression of heat shock-related genes. Transgenic Res 22: 595–605 [DOI] [PubMed] [Google Scholar]
- Saito K. (2004) Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol 136: 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M, Cornell KA, Beszteri S, Rzewuski G (2004) Functional analysis of methylthioribose kinase genes in plants. Plant Physiol 136: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M, Lorbiecke R, Ouyang B, Pochapsky TC, Rzewuski G (2005) The immediate-early ethylene response gene OsARD1 encodes an acireductone dioxygenase involved in recycling of the ethylene precursor S-adenosylmethionine. Plant J 44: 718–729 [DOI] [PubMed] [Google Scholar]
- Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M (2013) Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 451: 145–154 [DOI] [PubMed] [Google Scholar]
- Schad M, Lipton MS, Giavalisco P, Smith RD, Kehr J (2005) Evaluation of two-dimensional electrophoresis and liquid chromatography--tandem mass spectrometry for tissue-specific protein profiling of laser-microdissected plant samples. Electrophoresis 26: 2729–2738 [DOI] [PubMed] [Google Scholar]
- Scholz G, Becker R, Pich A, Stephan UW (1992) Nicotianamine ‐ a common constituent of strategies I and II of iron acquisition by plants: A review. J Plant Nutr 15: 1647–1665 [Google Scholar]
- Schuler M, Rellán-Álvarez R, Fink-Straube C, Abadía J, Bauer P (2012) Nicotianamine functions in the Phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24: 2380–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska A, Dénervaud V, Ashida H, Michoud K, Haas D, Yokota A, Danchin A (2004) Bacterial variations on the methionine salvage pathway. BMC Microbiol 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Casey R (1999) Seed Proteins. Springer, Dordrecht, The Netherlands [Google Scholar]
- Shinmachi F, Buchner P, Stroud JL, Parmar S, Zhao FJ, McGrath SP, Hawkesford MJ (2010) Influence of sulfur deficiency on the expression of specific sulfate transporters and the distribution of sulfur, selenium, and molybdenum in wheat. Plant Physiol 153: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Sauer N (1996) The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot Acta 109: 299–306 [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Takano A, Kakehi J, Takahashi T (2012) Thermospermine is not a minor polyamine in the plant kingdom. Plant Cell Physiol 53: 606–616 [DOI] [PubMed] [Google Scholar]
- Tan Q, Zhang L, Grant J, Cooper P, Tegeder M (2010) Increased phloem transport of S-methylmethionine positively affects sulfur and nitrogen metabolism and seed development in pea plants. Plant Physiol 154: 1886–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MV, Wolniak SM (2008) A plasma membrane-anchored fluorescent protein fusion illuminates sieve element plasma membranes in Arabidopsis and tobacco. Plant Physiol 146: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Sauer N (1995) The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Ühlken C, Horvath B, Stadler R, Sauer N, Weingartner M (2014) MAIN-LIKE1 is a crucial factor for correct cell division and differentiation in Arabidopsis thaliana. Plant J 78: 107–120 [DOI] [PubMed] [Google Scholar]
- Vera-Sirera F, Minguet EG, Singh SK, Ljung K, Tuominen H, Blázquez MA, Carbonell J (2010) Role of polyamines in plant vascular development. Plant Physiol Biochem 48: 534–539 [DOI] [PubMed] [Google Scholar]
- Waduwara-Jayabahu I, Oppermann Y, Wirtz M, Hull ZT, Schoor S, Plotnikov AN, Hell R, Sauter M, Moffatt BA (2012) Recycling of methylthioadenosine is essential for normal vascular development and reproduction in Arabidopsis. Plant Physiol 158: 1728–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M, Subert C, Sauer N (2011) LATE, a C(2)H(2) zinc-finger protein that acts as floral repressor. Plant J 68: 681–692 [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Zoeller D, Ebert B, Simon-Rosin U, Fisahn J, Glinski M, Weckwerth W (2004) Cell-specific protein profiling in Arabidopsis thaliana trichomes: identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry 65: 1641–1649 [DOI] [PubMed] [Google Scholar]
- Windsor AJ, Reichelt M, Figuth A, Svatoš A, Kroymann J, Kliebenstein DJ, Gershenzon J, Mitchell-Olds T (2005) Geographic and evolutionary diversification of glucosinolates among near relatives of Arabidopsis thaliana (Brassicaceae). Phytochemistry 66: 1321–1333 [DOI] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ (2007) The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Takahashi Y, Berberich T, Imai A, Miyazaki A, Takahashi T, Michael A, Kusano T (2006) The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett 580: 6783–6788 [DOI] [PubMed] [Google Scholar]
- Zheng L, Yamaji N, Yokosho K, Ma JF (2012) YSL16 is a phloem-localized transporter of the copper-nicotianamine complex that is responsible for copper distribution in rice. Plant Cell 24: 3767–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.