The Arabidopsis dynamin-like GTPase RHD3 has membrane fusion activity needed to form a proper endoplasmic reticulum network, and this activity is regulated by phosphorylation of its C terminus.
Abstract
The endoplasmic reticulum (ER) consists of dynamically changing tubules and cisternae. In animals and yeast, homotypic ER membrane fusion is mediated by fusogens (atlastin and Sey1p, respectively) that are membrane-associated dynamin-like GTPases. In Arabidopsis (Arabidopsis thaliana), another dynamin-like GTPase, ROOT HAIR DEFECTIVE3 (RHD3), has been proposed as an ER membrane fusogen, but direct evidence is lacking. Here, we show that RHD3 has an ER membrane fusion activity that is enhanced by phosphorylation of its C terminus. The ER network was RHD3-dependently reconstituted from the cytosol and microsome fraction of tobacco (Nicotiana tabacum) cultured cells by exogenously adding GTP, ATP, and F-actin. We next established an in vitro assay system of ER tubule formation with Arabidopsis ER vesicles, in which addition of GTP caused ER sac formation from the ER vesicles. Subsequent application of a shearing force to this system triggered the formation of tubules from the ER sacs in an RHD-dependent manner. Unexpectedly, in the absence of a shearing force, Ser/Thr kinase treatment triggered RHD3-dependent tubule formation. Mass spectrometry showed that RHD3 was phosphorylated at multiple Ser and Thr residues in the C terminus. An antibody against the RHD3 C-terminal peptide abolished kinase-triggered tubule formation. When the Ser cluster was deleted or when the Ser residues were replaced with Ala residues, kinase treatment had no effect on tubule formation. Kinase treatment induced the oligomerization of RHD3. Neither phosphorylation-dependent modulation of membrane fusion nor oligomerization has been reported for atlastin or Sey1p. Taken together, we propose that phosphorylation-stimulated oligomerization of RHD3 enhances ER membrane fusion to form the ER network.
In eukaryotic cells, the endoplasmic reticulum (ER) is the organelle with the largest membrane area. The ER consists of an elaborate network of interconnected membrane tubules and cisternae that is continually moving and being remodeled (Friedman and Voeltz, 2011). In plant cells, ER movement and remodeling is primarily driven by the actin-myosin XI cytoskeleton (Sparkes et al., 2009; Ueda et al., 2010; Yokota et al., 2011; Griffing et al., 2014) and secondarily by the microtubule cytoskeleton (Hamada et al., 2014). Several factors involved in creating the ER architecture have been also identified (Anwar et al., 2012; Chen et al., 2012; Goyal and Blackstone, 2013; Sackmann, 2014; Stefano et al., 2014a; Westrate et al., 2015). Among them, ER membrane-bound GTPases, animal atlastins and yeast Sey1p (Synthetic Enhancement of Yop1), function as ER fusogens to form the interconnected tubular network (Hu et al., 2009; Orso et al., 2009; Anwar et al., 2012). Atlastin molecules on the two opposed membranes have been proposed to transiently dimerize to attract the two membranes to each other (Bian et al., 2011; Byrnes and Sondermann, 2011; Morin-Leisk et al., 2011; Moss et al., 2011; Lin et al., 2012; Byrnes et al., 2013). Closely attracted lipid bilayers are supposed to be destabilized by an amphipathic helical domain at the atlastin C terminus to facilitate membrane fusion (Bian et al., 2011; Liu et al., 2012; Faust et al., 2015). Knockdown of atlastins leads to fragmentation of the ER and unbranched ER tubules, while overexpression of atlastins enhances ER membrane fusion, which enlarges the ER profiles (Hu et al., 2009; Orso et al., 2009).
An Arabidopsis (Arabidopsis thaliana) protein, ROOT HAIR DEFECTIVE3 (RHD3), has been proposed as a fusogen because (1) when it is disrupted, the ER network is modified into large cable-like strands of poorly branched membranes (Zheng et al., 2004; Chen et al., 2011; Stefano et al., 2012), (2) it shares sequence similarity with the above-mentioned fusogen Sey1p (Hu et al., 2009), and (3) it has structural similarity to atlastin and Sey1p, with a functional GTPase domain at the N-terminal cytosolic domain (Stefano et al., 2012) followed by two transmembrane domains and a cytosolic tail. RHD3 has a longer cytosolic C-terminal tail than do atlastin and Sey1p (Stefano and Brandizzi, 2014). It contains not only an amphipathic region but also a Ser/Thr-rich C terminus.
Arabidopsis has two RHD3 isoforms called RHD3-Like 1 and RHD3-Like 2. Fluorescently tagged RHD3 and RHD3-Like 2 localize to the ER (Chen et al., 2011; Stefano et al., 2012; Lee et al., 2013). RHD3 and the two RHD3-Like proteins likely have redundant roles in ER membrane fusion (Zhang et al., 2013). Overexpression of either RHD3 or RHD3-Like 2 with a defective GTPase domain phenocopies the aberrant ER morphology in rhd3-deficient mutants (Chen et al., 2011; Lee et al., 2013).
In this study, we show that the Ser/Thr-rich C terminus enhances ER membrane fusion following phosphorylation of its C terminus. We propose a model in which phosphorylation and oligomerization of RHD3 is required for efficient ER membrane fusion. Our findings clarify the mechanisms that regulate RHD3 and consequently the homeostasis of membrane fusion in the ER.
RESULTS
A Deficiency of RHD3 Causes a Defect in Interconnection of the ER Membranes
A T-DNA insertion mutant (designated as rhd3-10) had no detectable RHD3 protein (Supplemental Figure S1, A and B). rhd3-10, like other rhd3 mutant alleles (Zheng et al., 2004; Chen et al., 2011; Stefano et al., 2012), had less branched ER and thick ER strands (Fig. 1A). In addition, both rhd3-1 and rhd3-10 had aberrant ER structures with diameters of several micrometers (Fig. 1A). We next performed fluorescence recovery after photobleaching (FRAP) analyses, in which the ER-luminal GFP fluorescence was recovered rapidly with the mobile fraction of 82.4 ± 12.3% (sd; n = 20; Fig. 1, B and E), as reported previously (Stefano et al., 2014b). The aberrant ER structures in rhd3-1 were separated into two populations (Fig. 1E): A large number of the structures showed little fluorescence recovery with the mobile fraction of only 3.0 ± 2.7% (sd; n = 22; Fig. 1C), and a small number of them showed the fluorescence recovery with the mobile fraction of 69.5 ± 12.4% (sd; n = 7; Fig. 1D). These results indicate that the most of aberrant ER structures in rhd3-1 are isolated from the ER network. In rhd3, tubules grew from the aberrant ER structures (Fig. 1F; Supplemental Movie S1), and tubules occasionally contracted into the aberrant ER structures (Fig. 1G; Supplemental Movie S1). On the other hand, the tubule elongation velocity in rhd3 was only approximately 40% of that in the wild type (Fig. 1H; Supplemental Fig. S2) and the number of successful fusion events of tubules in rhd3 was only approximately 40% of that in the wild type (Fig. 1I). These results indicate that a deficiency of RHD3 causes a defect in the interconnection of the membranes, resulting in the occurrence of many ER fragments in the rhd3 cells.
Figure 1.
In vivo requirement of RHD3 for continuous network of the ER. A, ER network patterns in petiole cells of 7-d-old cotyledons of wild-type and rhd3 mutant alleles expressing ER-luminal GFP. Arrowheads indicate aberrant ER structures. B to E, Analysis of the ER network continuity by FRAP in epidermal cells of 8-d-old wild-type and rhd3-1 cotyledons expressing ER-luminal GFP. Samples were treated with latrunculin B to inhibit ER movement. B to D, Examples of fluorescent images and curves of fluorescence recovery after photobleaching. Magenta circles indicate sites of spot photobleaching. E, Distributions of individual rate of fluorescent recovery (%), which is shown by average for nine time points during postbleach (from 67–72 s in FRAP curves). Horizontal bars represent the mean of 20 (wild type) and 29 (rhd3-1) independent bleaching experiments. Error bars represent ± sd. F and G, Time-lapse images of the rhd3-1 ER show a tubule emerging from the aberrant structure (arrowheads in F) and a tubule contracting into the aberrant structure (arrowheads in G). Images were obtained with a spinning-disk confocal microscope. Numbers indicate time in seconds. See Supplemental Movie S1. H, ER tubule elongation velocity in the wild type and rhd3-7. Error bars represent ± se. ***P < 0.0001 by Student’s t test (n = 30). I, Estimated number of fusion events in an area of 18.75 × 18.75 µm during the 39.4-s time-lapse sequence. Error bars represent ± se. ***P < 0.0001 by Student’s t test (n = 30).
In Vitro Requirement of RHD3 for ER Network Formation
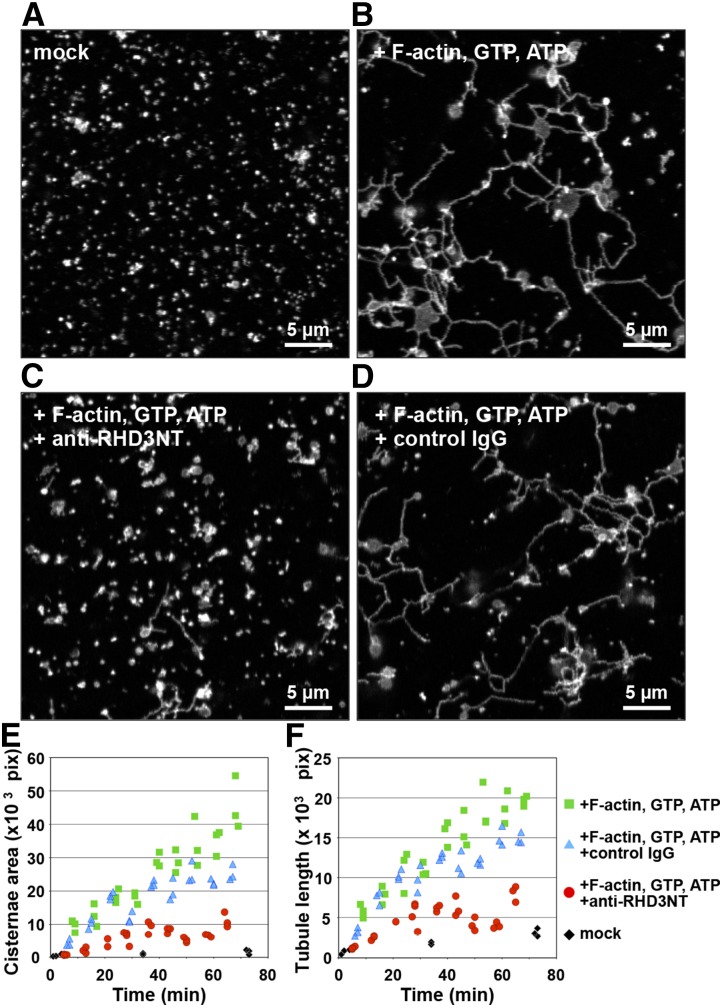
Fusogenic activity of RHD3 has been previously demonstrated in vitro with artificial liposomes and in yeast (Zhang et al., 2013). However, the extent of the involvement of RHD3 in the formation of the ER network structure is unknown. To address this question, we performed an in vitro reconstitution assay of the ER network by using the S12 fraction of tobacco (Nicotiana tabacum) cultured BY-2 cells expressing ER-luminal GFP (Supplemental Fig. S3; Yokota et al., 2011). The S12 fraction is a subcellular fraction that is rich in microsomes and cytosol. The cytosol includes a large amount of myosin XI, which is necessary for reconstitution of the ER network, in which it acts as a driving force with exogenously added F-actin and ATP to elongate ER tubules (Yokota et al., 2011). In our experimental setup, the S12 fraction contained small fluorescent ER microsomes, which formed hardly any ER tubules or ER cisternae during an approximately 70 min incubation in the absence of F-actin and the nucleotides GTP and ATP (Fig. 2A, mock). When the S12 fraction was incubated with F-actin and the nucleotides GTP and ATP for approximately 70 min, ER cisternae and ER tubules were formed and they interconnected to each other leading to the formation of a membranous network resembling that of the ER in cells (Fig. 2B). ER cisterna areas and the ER tubule lengths were measured with a plug-in package (MorphoER) that we developed for ImageJ software (Supplemental Fig. S4). Both the ER cisterna area (Fig. 2E) and the ER tubule length (Fig. 2F) increased during incubation with F-actin and nucleotides, while they hardly increased in their absence (Fig. 2, E and F, mock).
Figure 2.
RHD3 is required for ER network formation in vitro. A and B, An in vitro ER network formation assay. The cytosol-microsome fraction (S12 fraction) from cultured tobacco cells expressing ER-luminal GFP was incubated for approximately 70 min in the absence (mock in A) or in the presence of actin filaments (F-actin) and the nucleotides GTP and ATP (B) and then was inspected with a confocal laser scanning microscope. Note that the ER tubules and cisternae were efficiently generated from the ER microsomes by incubating with F-actin and the nucleotides, resulting in creating the ER network in vitro. C and D, An S12 fraction that was preincubated with either anti-RHD3NT (C) or control IgG (D) was subjected to the in vitro ER network formation assay. Note that anti-RHD3NT, but not control IgG, abolished the formation of ER tubules and cisternae, which prevented ER network formation. E and F, Quantitative analyzes of ER cisterna formation (E) and ER tubule formation (F) during incubation of ER microsomes with F-actin and nucleotides. The areas of ER cisternae and the lengths of ER tubules were calculated by MorphoER software at the indicated time points. The experiments were repeated at least three times.
To establish the contribution of RHD3 to ER network formation, a reconstitution assay was performed with the antiserum anti-RHD3NT, in which the antigen was a 19-amino acid peptide located adjacent to the conserved motif G3 in the N-terminal GTPase domain of Arabidopsis RHD3. Anti-RHD3NT recognized tobacco RHD3 in BY-2 cells as well as Arabidopsis RHD3 (Supplemental Figure S1, C–F). Preincubation of the S12 fraction with anti-RHD3NT significantly suppressed ER network formation (Fig. 2C). The increases of the ER cisterna area and ER tubule length occurred in the presence of F-actin and nucleotides were abolished by anti-RHD3NT (Fig. 2, E and F), but not by control IgG (Fig. 2, D–F). These results support the idea that RHD3 is required for ER network reconstitution from cell extracts through forming ER cisternae and tubules and interconnecting them.
RHD3 Is Necessary for GTP-Dependent ER Membrane Fusion to Form ER Sacs from ER Vesicles and Subsequent ER Tubule
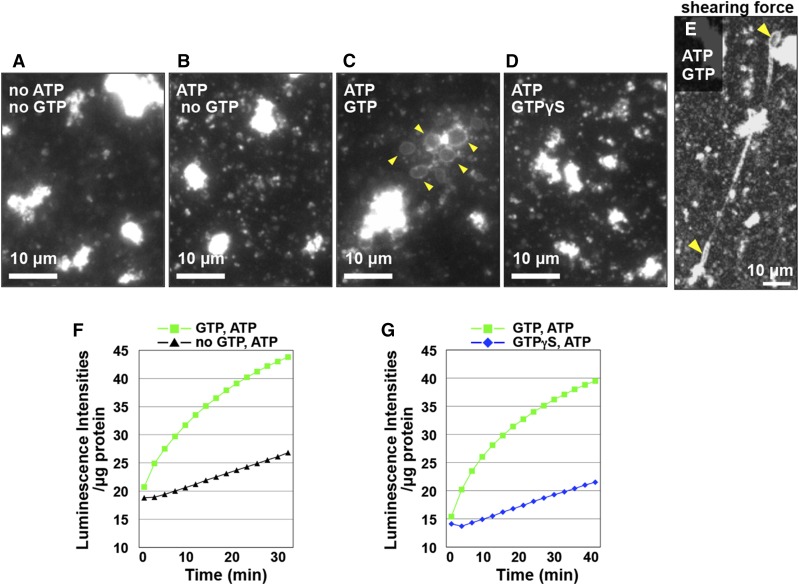
To determine the role of RHD3 in the formation of the ER network in greater detail, we first established an in vitro ER-tubule-elongation assay system with Arabidopsis-cultured MM2d cells (Supplemental Fig. S3A), as previously described (Yokota et al., 2011). The vesicle fraction that was prepared from the S12 fraction of the cultured cells enriched ER marker proteins and RHD3 (Supplemental Fig. S3B), indicating that the vesicles are derived from the ER. In the absence of ATP and GTP, no ER cisternae or tubules formed from ER vesicles (Fig. 3A). Similarly, none of them formed in the presence of ATP, which is required for myosin motor activity (Fig. 3B). However, adding GTP caused the formation of ER sacs through membrane fusion of the ER vesicles (Fig. 3C). A nonhydrolyzable analog of GTP (GTPγS) had no such effect (Fig. 3D). Hence, in this assay system, GTP-dependent membrane fusion caused the formation of ER sacs, but not ER tubules. The F-actin-induced increase of ER tubule length (Fig. 2) raised the possibility that some driving force was necessary for ER tubule elongation. To examine this possibility, we applied a shearing force by placing a cover slip on a drop of the mixture on a glass slide. An applied shearing force caused tubule emergence from ER sacs and elongation of tubules (Fig. 3E), indicating that the shearing force produces a driving force for ER tubule elongation and that the ER sacs serve as a membrane reservoir for the elongating tubules. These results show that we succeeded in establishing the in vitro assay system of ER tubule elongation from the Arabidopsis ER vesicles in the presence of the nucleotides and shearing force.
Figure 3.
Two in vitro assay systems for ER membrane fusion with ER vesicles. A to E, In vitro ER tubule elongation assay system. Fluorescent images of ER Tracker-stained ER vesicles from cultured Arabidopsis MM2d cells. The ER vesicle preparation was incubated with ER Tracker dye for 30 min in the absence (A) or presence of nucleotides as indicated in each panel (B–D). GTPγS indicates a nonhydrolyzable GTP analog. Note that many ER sacs were generated from the ER vesicles by adding GTP and ATP (arrowheads in C). E, In vitro ER tubule elongation under a shearing force. An ER vesicle preparation was incubated with a mixture of GTP, ATP, and ER Tracker dye for 10 min and then was exposed to a shearing force on a slide glass by placing a cover glass on top. Note that ER tubules emerged from ER sacs (arrowheads) in response to the applied shearing force. F and G, Ca2+-efflux-based assay system. The calcium ion efflux produced by ER vesicles was monitored by aequorin. Luminescent intensities per microgram of protein were plotted against time after addition of ATP without or with GTP or GTPγS. Note that the luminescent intensity was significantly increased by adding GTP and ATP (green), but not by adding ATP only (black in F) or by adding ATP and GTPγS (blue in G). See Supplemental Table S2 for statistical analyses of increasing luminescent intensity in each condition.
ER sac formation from ER vesicles has been associated with the ER structural changes through membrane fusion, resulting in Ca2+ release from the ER lumen (Voeltz et al., 2006). The Ca2+ release can be monitored by luminescence of the calcium sensor aequorin (Voeltz et al., 2006). We measured the luminescence intensities in the ER sac formation process from the ER vesicles in the absence of shearing force, in which no tubule formation occurred (Fig. 3C; Supplemental Fig. S5A). Adding GTP to ER vesicles increased the luminescence intensity (Fig. 3F). However, adding GTPγS, which caused no effects on the ER vesicles (Fig. 3D), did not significantly increase the luminescence intensity (Fig. 3G). These results indicate that membrane fusion involving ER sac formation from the ER vesicles can be evaluated by both the in vitro ER tubule elongation assay and the Ca2+-efflux-based assay.
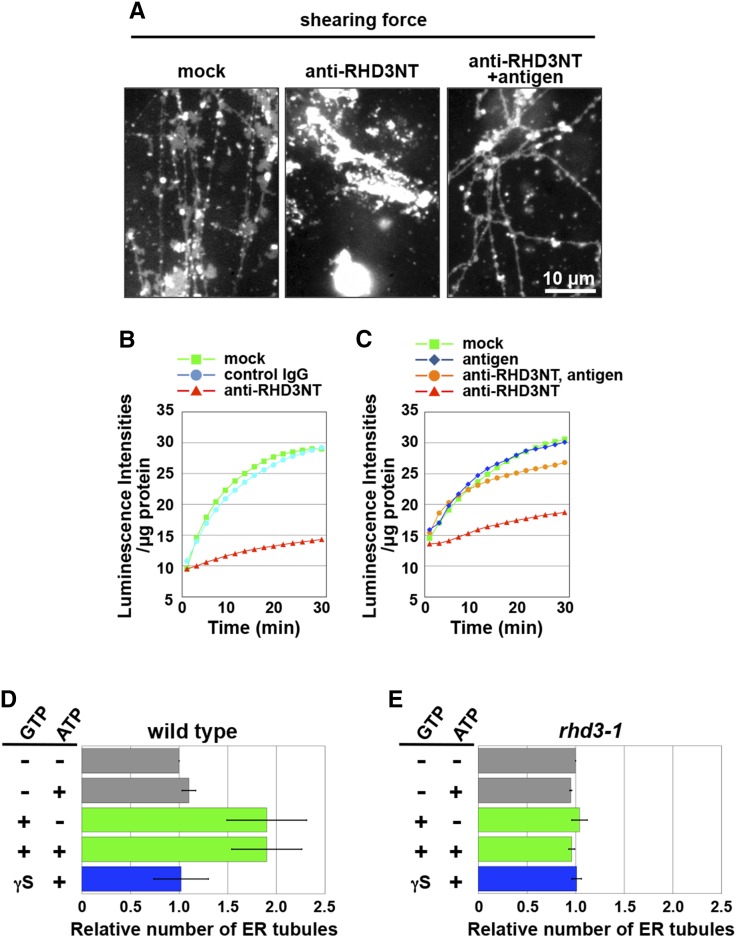
Next, anti-RHD3NT was applied to both assays. Preincubation of the ER vesicles with anti-RHD3NT noticeably inhibited both ER tubule formation (Fig. 4A, middle) and the increase of luminescence intensity (Fig. 4B). Premixing the antibody with its antigen peptide suppressed the inhibitory activity of the antibody on the ER tubule formation (Fig. 4A, right) and the increase of luminescence intensity (Fig. 4C). These results indicate that RHD3 is required for membrane fusion activity on the ER vesicles. To further test whether RHD3 is involved in ER tubule formation, we prepared ER vesicles from wild-type and rhd3-1 plants expressing ER-luminal GFP. The number of tubules produced by wild-type vesicles in the presence of an applied shearing force was greater in the presence of GTP than in the presence of GTPγS or in the absence of GTP (Fig. 4D). On the other hand, the number of tubules produced by the rhd3 vesicles was not affected by adding GTP (Fig. 4E). Together these results indicate that RHD3 is necessary for GTP-dependent ER membrane fusion to form ER sacs and subsequent ER tubule by an applied shearing force.
Figure 4.
In vitro requirement of RHD3 for membrane fusion and subsequent tubule formation. A, An ER vesicle preparation from cultured Arabidopsis MM2d cells was preincubated with each of buffer solution (mock) and anti-RHD3NT (no antigen) and preadsorbed anti-RHD3NT with the antigen peptide (2.5 µm, antigen) and was then subjected to the in vitro ER tubule elongation assay under a shearing force. Note that anti-RHD3NT abolished ER tubule formation. B and C, Ca2+-efflux-based assay. ER vesicles prepared from cultured Arabidopsis MM2d cells were preincubated with each of buffer solution (mock), control IgG, and anti-RHD3NT (B), and another preparation was preincubated with each of buffer solution (mock), the antigen peptide, anti-RHD3NT, and preadsorbed anti-RHD3NT with the antigen peptide (C). Luminescent intensities per microgram of protein were plotted against time after addition of ATP and GTP. Note that anti-RHD3NT abolished the GTP-dependent enhancement of luminescent intensity. See Supplemental Table S3 for statistical analyses of increasing intensity in each condition. D and E, Quantitative analysis of in vitro ER tubule formation from ER vesicles prepared from wild-type (D) and rhd3-1 (E) seedlings that expressed ER-luminal GFP. ER vesicle preparations that were treated with nucleotides were subjected to an in vitro ER tubule elongation assay under the shearing force. The x axis indicates the number of ER tubules formed relative to the number ER tubules formed in the absence of the nucleotides. The experiments were repeated at least three times. Error bars indicate sd. See Supplemental Table S4 for statistical analyses of the number of ER tubule in each condition.
The C Terminus of RHD3 Is Multiphosphorylated
A unique feature of the RHD3 polypeptide is a C-terminal stretch rich in Ser and Thr residues (Fig. 5A). Similar Ser/Thr-rich stretches were found in the C-terminal tails of RHD3 homologs of wide variety of plants from the green alga Chlamydomonas reinhardtii to land plants (Fig. 5A), although no Ser/Thr-rich stretches were found in yeast Sey1p or mammalian atlastins. Based on the structural homology between RHD3 and the fusogens atlastin and Sey1p, the Ser/Thr-rich stretch in RHD3 appears to be located in the cytosol. This raises the possibility that the Ser/Thr-rich stretch is a Ser/Thr-kinase phosphorylation site. To test whether the C terminus of RHD3 is phosphorylated, we prepared a solubilized fraction from ER vesicles from MM2d cells and applied it to a metal affinity column that retains phosphorylated proteins. RHD3 was detected in the fraction bound to the affinity column (Fig. 5B), suggesting that RHD3 is a phosphoprotein. Next, the solubilized fraction from the ER vesicles was subjected to two-dimensional electrophoresis. In an immunoblot of the gel with anti-RHD3NT, RHD3 appeared in eight acidic spots in the pI range 2.5 to 5.0 (Fig. 5C, untreated), corresponding to eight different charge states. Treatment with a Ser/Thr-kinase increased the signal intensities of the spots, especially spot 7 (Fig. 5C, kinase), whereas phosphatase treatment markedly decreased the intensities of spots 3 to 8 (Fig. 5C, phosphatase). These results indicate that RHD3 has multiple phosphorylation sites.
Figure 5.
The C terminus of RHD3 is multiphosphorylated. A, Sequence alignment of the C terminus of the RHD3/Sey1 family. Ser, Thr, and Tyr residues are indicated in magenta. An antigen peptide for raising anti-RHD3CT is underlined. RHD3 homologs are GSVIVP00035518001 in Vitis vinifera (a eudicot); Os01g0575000, Os11g0582300, and Os12g0604600 in Oryza sativa (a monocot); Sb03g025850, Sb04g019800, and Sb05g022650 in Sorghum bicolor (a monocot); Sm073603 and Sm439743 in Selaginella moellendorffii (a lycophyte); Pp206384, Pp226877, and Pp234372 in Physcomitrella patens (a moss); Ot010531 in Ostreococcus tauri (a green alga); and Cr281843 in C. reinhardtii (a green alga). B, The ER vesicle preparation from cultured MM2d cells was subjected to metal affinity chromatography. Total ER (input) and bound fractions were subjected to SDS-PAGE followed by Coomassie Brilliant Blue staining (CBB) or by immunoblot with anti-RHD3NT. C, Immunoblots with anti-RHD3NT of kinase-treated, alkaline phosphatase-treated, and untreated ER vesicles that were separated by two-dimensional electrophoresis. D to F, Electrophoretic gels and an immunoblot showing GFP-RHD3 protein immunoprecipitated with anti-GFP MicroBeads. CBB staining (D), silver staining (E), and immunoblot (F) with anti-GFP. Numbers in F indicate the relative amounts loaded. G, Frequency of detection (by mass spectrometry) of phosphorylation at each residue in the Ser cluster of the C terminus of RHD3 (see Table I).
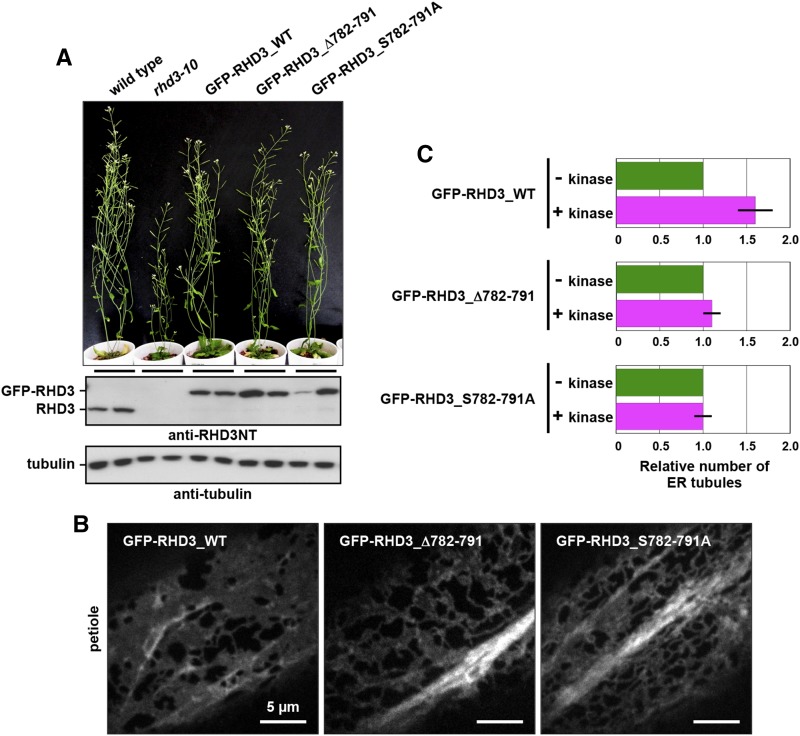
To identify the phosphorylation sites of RHD3, we used a functional GFP-RHD3 fusion protein (GFP-RHD3_WT), which restores defects of an rhd3 null allele (rhd3-10) for plant growth (Fig. 9A) and ER architecture (compare Fig. 1A with Fig. 9B). We purified the GFP-RHD3 protein from transgenic plants with anti-GFP beads (Fig. 5, D–F). The trypsin and Glu-C digests of GFP-RHD3 were then applied to reverse-phase liquid chromatography followed by tandem mass spectrometry by using two different injection methods and two different mass spectrometers (Orbitrap XL and TOF 5600). Repeated analyses isolated phosphorylated peptide fragments derived from the C terminus (Table I) and identified eight phosphorylated Ser residues and two phosphorylated Thr residues (Fig. 5G), indicating that the C terminus is a phosphorylation target of some Ser/Thr-kinase(s).
Figure 9.
The Ser cluster in the RHD3 C terminus is required for kinase-dependent enhancement of ER membrane fusion. A and B, Rescue of rhd3-10 dwarfism by expressing GFP-RHD3_WT, GFP-RHD3_Δ782-791, or GFP-RHD3_S782-791A. A, Seven-week-old plants of the wild type (Col-0), rhd3-10, and rhd3-10 expressing GFP-RHD3_WT, GFP-RHD3_Δ782-791, and GFP-RHD3_S782-791A. Immunoblot of total extracts (two samples for each genotype) prepared from 12-d-old seedlings with either anti-RHD3NT or anti-α-tubulin. B, Epidermal cells of cotyledon petioles from 7-d-old rhd3-10 plants that expressed GFP-RHD3_WT, GFP-RHD3_Δ782-791, or GFP-RHD3_S782-791A under control of the endogenous promoter. Fluorescent images were captured with a spinning-disk confocal microscope. C, Quantitative analysis of in vitro ER tubule formation under a shearing force using kinase-treated and untreated ER vesicles from seedlings of rhd3-10 expressing each of GFP fusion with RHD3 (GFP-RHD3_WT) and with modified RHD3 versions (GFP-RHD3_Δ782-791 and GFP-RHD3_S782-791A). The x axis indicates the number of ER tubules formed relative to the number ER tubules formed in the presence of the nucleotides without kinase. The experiments were repeated at least three times. Error bars indicate sd.
Table I. List of Identified Phosphopeptides in the C Terminus of GFP-RHD3.
| Peptide Number | Sequencea | Experiment Number | Machine | Injection Methodb | Search Engine |
|---|---|---|---|---|---|
| 1 | SSSSS(pS)SGS(pS)PAKNVPIDTSA | First | Orbitrap XL | TE | SEQUEST |
| 2 | SSSSSSSGS(pS)PAKNVPIDTSA | First | Orbitrap XL | TE | SEQUEST |
| 3 | SSSSS(pS)SGSSPAKNVPIDTSA | First | Orbitrap XL | TE | Mascot |
| 4 | VTT(daN)GESSSSSSSG(pS)(pS)PAK | First | Orbitrap XL | TE | Mascot |
| 5 | VTTNGESSSSSSSGS(pS)PAK | Second | Orbitrap XL | TE | SEQUEST |
| 6 | VTTNGESSSSSSSG(pS)SPAK | Second | Orbitrap XL | DI | SEQUEST |
| 7 | VTTNGESSSS(pS)(pS)SGSSPAK | Second | Orbitrap XL | DI | SEQUEST |
| 8 | VTT(daN)GESSSSSSSGS(pS)PAK | Second | Orbitrap XL | DI | Mascot |
| 9 | VTT(daN)GESS(pS)(pS)SSSGSSPAK | Second | Orbitrap XL | DI | Mascot |
| 10 | VTTNGESSSSSSSG(pS)SPAK | Second | Orbitrap XL | DI | Mascot |
| 11 | VTTNGESSSSSSSGS(pS)PAK | Third | TOF5600 | TE | ProteinPilot |
| 12 | V(pT)T(daN)GESSSSSSSGSSPAK | Third | TOF5600 | TE | ProteinPilot |
| 13 | VT(pT)(daN)GESSSSSSSGSSPAK | Third | TOF5600 | TE | ProteinPilot |
| 14 | V(pT)TNGESSSSSSSGSSPAK | Fourth | TOF5600 | DI | ProteinPilot |
| 15 | V(pT)T(daN)GESSSSSSSGSSPAK | Fourth | TOF5600 | DI | ProteinPilot |
| 16 | VT(pT)(daN)GESSSSSSSGSSPAK | Fourth | TOF5600 | DI | ProteinPilot |
| 17 | Q(pS)EVTT(oN)GESSSSSSSGSSPAK | Fourth | TOF5600 | DI | ProteinPilot |
| 18 | VT(pT)(daN)GESSSSSSSGSSPAK | Fourth | TOF5600 | DI | ProteinPilot |
| 19 | V(pT)T(daN)GESSSSSSSGSSPAK | Fourth | TOF5600 | TE | ProteinPilot |
| 20 | VT(pT)(daN)GESSSSSSSGSSPAK | Fourth | TOF5600 | TE | ProteinPilot |
| 21 | V(pT)T(daN)GESS(dhS)SSSSGSSPAK | Fourth | TOF5600 | TE | ProteinPilot |
| 22 | V(pT)TNGESSSSSSSGSSPAK | Fourth | TOF5600 | DI | Mascot |
Amino acids within brackets indicate the modified residues. p, phosphorylation; da, deamidation; dh, dehydration; and o, oxidation.
TE, Trap and elute; DI, direct injection.
Kinase-Dependent Enhancement of ER Membrane Fusion
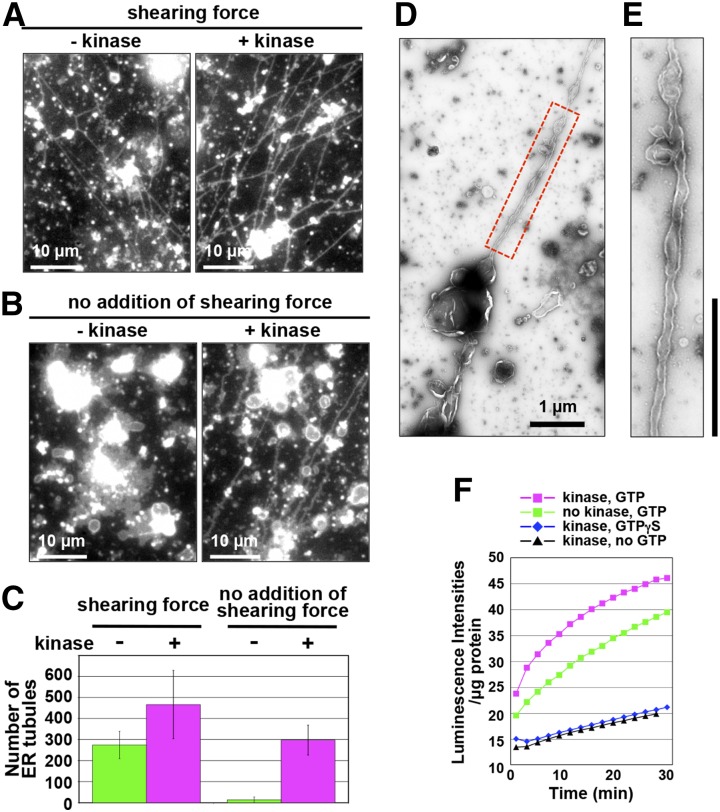
To gain insights into the role of phosphorylation in ER membrane remodeling, we applied Ser/Thr-kinase (a catalytic subunit of cAMP-dependent protein kinase) to the in vitro ER tubule elongation assay as described above. In the presence of an applied shearing force, Ser/Thr-kinase treatment enhanced ER tubule formation from the ER vesicles (Fig. 6A). Quantitative data are shown in Figure 6C (left). A negative-stain electron micrograph of the kinase-treated ER vesicles (Fig. 6, D and E) and the movie of the kinase-treated ER vesicles (Supplemental Movie S2) confirmed that the ER tubules emerged from ER sacs is a continuous structure, but not a long stretch of linear aggregates of small vesicles. In the absence of an applied shearing force, ER sacs were formed, but no ER tubules were formed from ER sacs (Figs. 3C and 6B, left). Surprisingly, however, after 30-min incubation of ER vesicles, kinase treatment triggered ER tubule formation from ER sacs, which appeared to be more abundant after 20-min incubation (Fig. 6, B and C, right; Supplemental Fig. S5).
Figure 6.
Kinase-dependent enhancement of ER membrane fusion and ER tubule formation activities. A to C, In vitro ER tubule formation from the kinase-treated and untreated ER vesicles with (A) or without (B) an applied shearing force. The assay is described in Figure 3E. In B, to minimize shearing forces during the assay, the specimen was prepared on ice. Note that kinase treatment enabled the ER vesicles to form ER tubules without an applied shearing force. C, Number of the ER tubules under the condition described in A and B was counted. D and E, Negative-staining electron micrographs of an ER tubule emerging from a kinase-treated ER vesicle. Boxed region in D was enlarged in E. Bars = 1 µm. F, Ca2+-efflux-based assay of an ER vesicle preparation from cultured Arabidopsis MM2d cells. An ER vesicle preparation was treated with kinase in the presence of GTP or a nonhydrolyzable GTP analog (GTPγS). The calcium ion efflux produced by the ER vesicles was monitored by aequorin. Luminescent intensities per microgram of protein were plotted against time after addition of ATP and GTP. Note that luminescent intensity was increased by adding GTP.
Ser/Thr-kinase was next applied to the Ca2+-efflux-based assay. In the presence of GTP, the luminescence intensities of the ER vesicles with Ser/Thr-kinase treatment were higher than those without the treatment (Fig. 6F). On the other hand, in the presence of GTPγS or in the absence of GTP, Ser/Thr-kinase treatment did not significantly increase the luminescence intensity (Fig. 6F). These results indicate that Ser/Thr phosphorylation promotes ER membrane fusion in a GTP-hydrolysis-dependent manner. Hence, phosphorylation of some protein(s) might facilitate membrane fusion and subsequent tubule formation from ER vesicles.
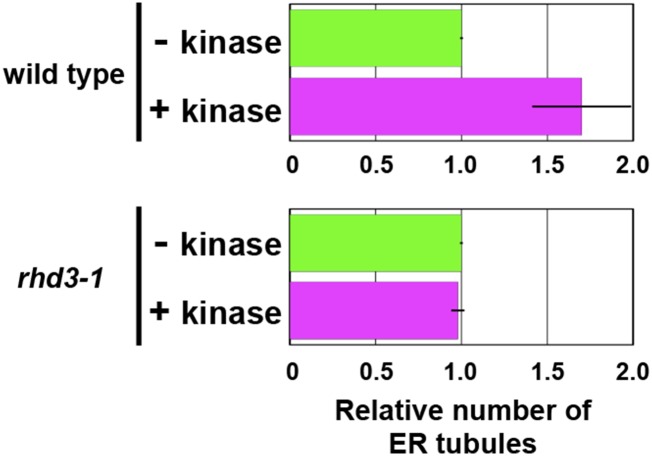
A Ser Cluster of the RHD3 C Terminus Is Required for Kinase-Dependent Enhancement of ER Membrane Fusion
To determine whether RHD3 is involved in the Ser/Thr-kinase-dependent enhancement of the membrane fusion, we performed an in vitro ER tubule elongation assay with ER vesicle preparations of whole wild-type and rhd3-1 seedlings. In the ER vesicles from wild-type seedlings, pretreatment of the vesicles with Ser/Thr-kinase increased the number of tubules (Fig. 7). On the other hand, in the ER vesicles from rhd3-1 seedlings, pretreatment with Ser/Thr-kinase did not affect the number of tubules (Fig. 7). These results indicate that RHD3 has a critical role in the Ser/Thr-kinase-enhanced ER tubule formation.
Figure 7.
RHD3 is required for kinase-dependent enhancement of ER tubule formation. Quantitative analysis of in vitro ER tubule formation under a shearing force using kinase-treated and untreated ER vesicles from seedlings of the wild type and rhd3-1. The x axis indicates the number of ER tubules formed relative to the number ER tubules formed in the presence of the nucleotides without kinase. The experiments were repeated at least three times. Error bars indicate sd. See Supplemental Table S5 for statistical analyses of the number of ER tubule in each condition.
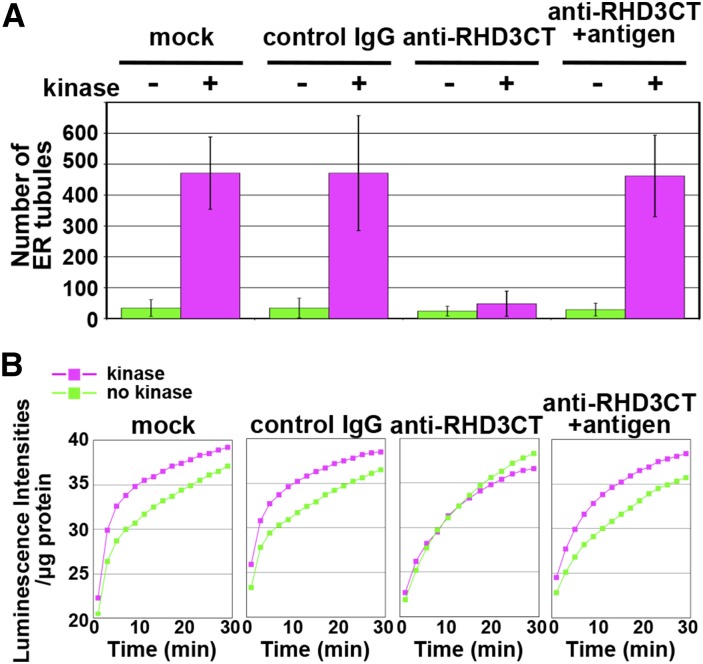
Next, to explore the function of the Ser/Thr-rich C terminus of RHD3, we generated a specific antibody against an unphosphorylated peptide corresponding to the C-terminal region of RHD3 (782SSSSSSSGSSPAKNVPIDTS801) and designated it as anti-RHD3CT (Fig. 5A; Supplemental Fig. S1, E and F). Anti-RHD3CT was applied to both the in vitro ER tubule elongation assay and the Ca2+-efflux-based assay. Preincubation of ER vesicles with anti-RHD3CT inhibited kinase-enhanced tubule formation (Fig. 8A) and kinase-dependent increase of the luminescence intensities (Fig. 8B). On the other hand, the antibody preadsorbed with the antigen peptide did not inhibit them (Fig. 8, A and B). It should be noted that anti-RHD3CT did not suppress the increase of the luminescence intensity as did control IgG (Fig. 8B), which is in contrast to the finding that anti-RHD3NT clearly suppresses it (Fig. 4, A–C), suggesting that anti-RHD3CT inhibits the kinase dependently enhanced membrane fusion, while anti-RHD3NT, which recognizes the N-terminal GTPase domain of RHD3, inhibits the membrane fusion itself. These results indicate that the Ser/Thr-kinase requires access to the C terminus of RHD3 to enhance membrane fusion from ER vesicles and subsequent tubule formation.
Figure 8.
The C terminus of RHD3 is required for kinase-dependent enhancement of ER membrane fusion. A, In vitro ER tubule formation without an applied shearing force using kinase-treated and untreated ER vesicles from cultured MM2d cells. The vesicles were preincubated with each of buffer (mock), control IgG, and anti-RHD3CT and preadsorbed anti-RHD3CT with the antigen peptide before the in vitro ER tubule elongation assay as described in Figure 3E. B, Ca2+-efflux-based assay of each ER vesicle preparation in A. The assay is described in Figure 3, F and G. Luminescent intensities per microgram of protein were plotted against time after addition of ATP and GTP. The experiments were repeated at least three times (3 ≤ n ≤ 8). Note that anti-RHD3CT abolished the kinase-dependent enhancement of luminescence intensity.
To establish whether the Ser cluster (782SSSSSSSGSS791) in the C terminus is involved in the Ser/Thr-kinase-enhanced membrane fusion, we expressed GFP fusions of the modified RHD3 proteins under the endogenous RHD3 promoter in a null mutant allele, rhd3-10. The modified RHD3 proteins included the wild-type RHD3 (GFP-RHD3_WT), RHD3 lacking the C-terminal Ser cluster 782SSSSSSSGSS791 (RHD3_Δ782-791), and RHD3, in which the Ser cluster was replaced with the Ala cluster 782AAAAAAAAGAA791 (GFP-RHD3_S782-791A). Expression of each of the GFP fusions rescued the dwarf phenotype of rhd3-10 (Fig. 9A) and the aberrant ER network architecture phenotype of rhd3-10 (compare Fig. 1A with Fig. 9B), indicating that these GFP-fusions are functional in the cells. Interestingly, however, in vitro assays demonstrated that ER vesicles from plants expressing GFP-RHD3_Δ782-791 or GFP-RHD3_S782-791A did not exhibit the kinase-enhanced tubule formation, while the ER vesicles from complemented plants expressing GFP-RHD3_WT clearly exhibited it (Fig. 9C). These results indicate that the C-terminal Ser cluster of RHD3 has a critical role in the kinase-enhanced membrane fusion and subsequent tubule formation.
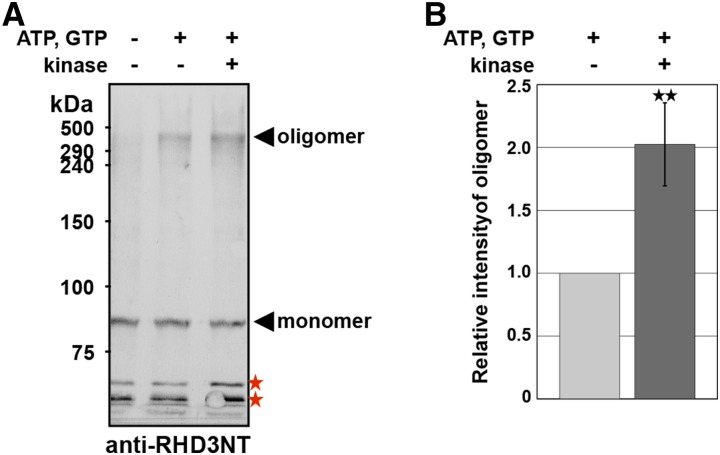
Kinase Treatment Enhances Oligomerization of RHD3
We next asked whether phosphorylation causes a change in the quaternary structure of RHD3. To answer this question, ER vesicles prepared from MM2d cells were treated with a cross-linking chemical, ethylene glycol bis(succinimidylsuccinate) (EGS). The vesicles provided a faint signal of an approximately 450-kD oligomer in addition to a signal of 90-kD monomer of RHD3 (Fig. 10A, left lane). The oligomer level was increased when the vesicles were treated with ATP and GTP (Fig. 10A, middle lane) and was further increased when the vesicles were treated with Ser/Thr-kinase, ATP, and GTP (Fig. 10A, right lane). Densitometry of the signal intensities revealed that the concentration of the oligomer in the kinase-treated vesicles was twice as high as that in the kinase-untreated vesicles (Fig. 10B). Because the phosphorylated RHD3 level in the kinase- and ATP/GTP-treated vesicles was much higher than that in the untreated vesicles (see Figure 5C), these results support the idea that phosphorylation of RHD3 promotes its oligomerization.
Figure 10.
Kinase treatment enhances oligomerization of RHD3. A, Effects of ATP, GTP, and kinase on the formation of RHD3 oligomers. The ER vesicle preparation from cultured MM2d cells was treated with kinase and/or the nucleotides and then was incubated with the cross-linking reagent (EGS). The cross-linked samples were subjected to SDS-PAGE followed by immunoblot with anti-RHD3NT. Asterisks indicate nonspecific bands. B, Relative intensities of oligomer bands in the immunoblots. Error bars represent ± sd. **P < 0.001 by Student’s t test (n = 4).
DISCUSSION
These results from three in vitro assay systems (ER tubule elongation assay, Ca2+-efflux-based assay, and ER-network reconstitution assay) provide a conceptual advance in the understanding how the ER network is established in plant cells. Establishing the ER network constitutes three processes: The first process is creation of membrane reservoir through membrane fusion, the second process is tubule formation from the membrane reservoir followed by tubule elongation, and the third process is interconnection between tubules and cisternae through membrane fusion. The first and third processes are mediated by the GTPase RHD3. On the other hand, the second process is driven by the actin-myosin XI cytoskeleton (Sparkes et al., 2009; Yokota et al., 2011; Griffing et al., 2014) and by a shearing force in vitro (this study; Yokota et al., 2011).
Our in vitro assay systems uncovered a yet undefined regulatory mechanism of ER membrane fusion, in which the fusion activity was enhanced by phosphorylation of RHD3. It should be noted that Ser/Thr-kinase treatment enables the ER vesicles to form tubules without a shearing force (Fig. 6). Kinase treatment may increase the sensitivity of ER vesicles to a shearing force so that kinase-treated ER vesicles may produce ER tubules through membrane fusion even when the shearing force is negligible. Our results support the ideas that RHD3 is a target of the exogenously added kinase and that the phosphorylation of its Ser/Thr stretch plays a critical role in regulating ER membrane fusion. The C-terminal cytosolic tail of atlastin contains an amphipathic helix, which promotes membrane fusion by interacting directly with and perturbing the lipid bilayer (Liu et al., 2012; Faust et al., 2015). Similarly, RHD3 has a putative amphipathic helix at the former half of the C-terminal tail (Stefano and Brandizzi, 2014). Anti-RHD3CT, which recognizes the Ser/Thr-rich stretch at the latter half of the tail, did not suppress ER membrane fusion and subsequent tubule formation, although it suppressed the kinase-dependent enhancement of membrane fusion and subsequent tubule formation (Fig. 8). These results provide a model that the C-terminal tail of RHD3 has two independent roles: a role of membrane fusion via the amphipathic helix and a regulatory role of enhancement of membrane fusion via Ser/Thr-rich stretch.
In vitro analyses appear to be well suited for studying the regulatory mechanism of ER morphogenesis because they enable us to observe biological events undetectable in vivo. The C-terminally modified RHD3 proteins clearly abolished the kinase-dependent enhancement of ER tubule formation in vitro. However, the modified RHD3 proteins showed no defects in vivo. The discrepancy might be come from the difference of membrane reservoir levels: The in vitro systems with ER vesicles have little membrane reservoir, while the differentiated petiole cells have a large amounts of the membrane reservoir. In the in vitro systems, the kinase-dependent enhancement of membrane fusion primarily might occur to create membrane reservoir. Thus, a defect in the enhancement might be easily detected in the in vitro systems than in the cells.
Although RHD3 has long been assumed to act as an ER fusogen, no conclusive evidence has been reported to date. This study demonstrates that RHD3 is indispensable for GTP-dependent membrane fusion in planta. We show that the frequency of ER membrane fusion is reduced in rhd3 mutants, in which a slowly elongated tubule reaches a cisterna but fails to fuse and a disconnected tubule contracts into the cisterna (Fig. 1). Consequently, the ER membranes of the mutants become fragmented. However, ER membrane fusion events did not completely disappear in rhd3 loss-of-function mutants. One possible explanation is that RHD3-Like proteins function as ER fusogens in RHD3-deficient cells. RHD3 and RHD3-Like proteins have redundant roles in plant development and viability (Zhang et al., 2013). We could not rule out the presence of other ER fusion machinery in plant cells because tubules formed in a GTP-independent manner in the ER vesicle preparation from rhd3 (Fig. 4). In yeast cells, ER SNAREs have been reported to be involved in ER membrane fusion independently of Sey1p (Anwar et al., 2012).
The chemical cross-linking assay showed that the RHD3 oligomer has a molecular mass of approximately 450 kD (Fig. 10). Atlastin also forms a high molecular mass oligomer (Liu et al., 2012). Atlastin self-assembles through its middle domain (Pendin et al., 2011) and/or its transmembrane domain (Moss et al., 2011; Liu et al., 2012). We found that the oligomerization of RHD3 was promoted by kinase treatment (Fig. 10). Based on these results, we propose a phosphorylation-enhanced ER membrane fusion model, in which phosphorylation-induced RHD3 assembly generates the RHD3-rich ER subdomains where ER membrane fusion may preferentially occur. In this model, ER membrane fusion is spatially modulated by phosphorylation of the cytosolic Ser/Thr-rich region of RHD3 through the oligomerization of RHD3. This might improve the efficiency of membrane fusion during the dynamic remodeling of the ER network. The lack of conservation of the C-terminal sequence of RHD3 in non-plant ER-shaping GTPases indicates that plants may have evolved unique mechanisms to control ER membrane fusion and shape.
MATERIALS AND METHODS
Plant Materials, Culture Cells, and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used as the wild-type plant. Transgenic Arabidopsis lines GFP-h (Matsushima et al., 2003a) and ER-YK (Nelson et al., 2007) were previously established. T-DNA-tagged mutant of RHD3 (rhd3-10) was established in this study (Supplemental Fig. S1). The rhd3-1 and rhd3-7 mutants were reported previously (Wang et al., 1997; Stefano et al., 2012). To visualize ER, rhd3-1 and rhd3-10 were crossed with GFP-h, and rhd3-7 was crossed with ER-YK. Arabidopsis seeds were surface-sterilized and then sown onto 0.5% Gellan Gum (Wako) containing 1% Suc and Murashige and Skoog medium. Plants were grown at 22°C. Suspension-cultured tobacco BY-2 cells (Nicotiana tabacum cv Bright Yellow 2) expressing ER-luminal GFP (Mitsuhashi et al., 2000) and suspension-cultured Arabidopsis MM2d cells (Menges and Murray, 2002) were cultured as described previously (Nagata et al., 1981).
Microscopy
We used a spinning-disk confocal microscope consisting of a fluorescence microscope (Axio Observer.Z1; Zeiss) equipped with a confocal laser-scanning unit (CSU X1; Yokogawa Electric) and a laser combiner system (Andor). The fluorescent images were captured using an EM-CCD camera (iXon3; ANDOR) with a 100× 1.45 numerical aperture oil-immersion objective. We also used a confocal laser-scanning microscope consisting of LSM780 or LSM510 META (Zeiss) with either a 100× 1.45 numerical aperture oil-immersion objective or a 63× 1.2 numerical aperture water-immersion objective. Images were processed using Adobe Photoshop (Adobe Systems) or ImageJ (National Institutes of Health).
FRAP Analysis
Continuity of the ER network was analyzed by FRAP as reported previously (Stefano et al., 2014b) using an LSM780 confocal laser-scanning microscope equipped with a 63× 1.2 numerical aperture water-immersion objective. We observed adaxial epidermal cells of 8-d-old cotyledons in wild-type or rhd3-1 expressing ER-luminal GFP. The cotyledons were incubated in 20 µm latrunculin B for 30 min prior to imaging to inhibit ER movement. Ten prebleach scans were captured using settings for GFP with the 488-nm laser transmission set to 1.5% transmission prior to bleaching of a 2.5 µm2 spot using 30 interactions of a 405/458/488/514-nm laser lines set to 100% transmission.
Analyses of ER Tubule Dynamics and Fusion Events
To analyze the velocity of ER tubule elongation, 30 time-lapse sequences with 200 frames (39.4 s) per sequence were recorded in the cortical region of 12-d-old cotyledon pavement cells. Time-lapse frames at 256 × 256 pixel resolution (18.75 × 18.75 µm) were captured at low laser power (i.e. 1–5% of an argon 514-nm laser line) to avoid photobleaching and 3× digital zoom using an EC Plan-Neofluar 40× 1.30 objective. Imaging settings for single fluorochromes were described previously (Brandizzi et al., 2002). Velocities of ER tubule elongation in ER-YK (control) and rhd3-7/ER-YK mutant were analyzed using the Manual Tracking plug-in available through Image J. The number of ER tubules analyzed in Col-0 and rhd3-7/ER-YK was 382 and 113, respectively. ER tubule velocity values used to build the graph represent the average velocity of each ER tubule tracked in the 200 (256 × 256) frames of each time-lapse sequence for each sample. For ER tubule fusion analysis in vivo, ER in the cell cortex area (18.75 × 18.75 μm) was imaged during 39.4 s. Tubules that anchored to a homotypic membrane and remained attached during the time of the observation were considered fused, whereas tubules that did not remain attached to other tubules were not considered fused. Statistical analysis was based on Student’s t test.
Preparation of Antibodies
Anti-RHD3NT was raised against 98TDGRERGEDDTAFEKQSALF117 peptide, as reported previously (Matsushima et al., 2003b), and purified by the immunoadsorption (Yokota et al., 1995) on an Immobilon-PSQ membrane (Millipore) on which the antigen peptide was immobilized. Anti-RHD3CT was raised against 782SSSSSSSGSSPAKNVPIDTS801 peptide, as reported previously (Yokota et al., 2011).
Isoelectric Focusing, SDS-PAGE, and Immunoblotting
Two-dimensional PAGE was carried out according to protocols reported previously (O’Farrell, 1975); isoelectric focusing was performed in gels containing Pharmalyte 2.5-5 (GE Healthcare). SDS-PAGE and immunoblotting were performed as described previously (Shimada et al., 2003). Antibodies used in this study were anti-RHD3NT (affinity-purified, diluted 100-fold), anti-RHD3CT (diluted 500-fold), anti-α-tubulin (diluted 2,000-fold; b-5-1-2; Sigma-Aldrich), anti-GFP (diluted 2,500-fold; JL-8; Clontech), anti-BiP (diluted 10,000-fold; Hatano et al., 1997), anti-RGP1 (diluted 10,000-fold; Dhugga et al., 1997), and anti-AtPIP2b (diluted 1,000-fold; Rose Biotechnology).
In Vitro ER Network Reconstitution Assay
The S12 fraction containing cytosol and microsomes was prepared from BY-2 cells expressing ER-luminal GFP, and the ER network reconstitution assay was performed as reported previously (Yokota et al., 2011). For the inhibition assay, IgG was purified from anti-RHD3NT antiserum using a HiTrap Protein A column (GE Healthcare). Normal rabbit IgG (Sigma) and purified anti-RHD3NT were dialyzed against a solution containing 40 mm PIPES-NaOH (pH 7.0), 8 mm EGTA, 1 mm MgCl2, and 0.3 m Suc. The equilibrated solution outside of the dialyzing bag was used as a control (labeled as “mock” in Figure 2A). The S12 fraction was premixed with the equilibrated solution, normal IgG, or anti-RHD3NT (final concentrations 3 mg/mL) on ice. After 1 h, ATP (final concentration 4 mm), GTP (final concentration 2 mm), and F-actin (final concentration 16 µg/mL) were added to the mixtures. Fluorescent images were randomly captured at each time point by a confocal laser-scanning microscope (LSM510 META) with a 100× 1.45 numerical aperture oil-immersion objective.
Quantitative Analysis of ER Network Formation in Vitro Using Image Processing
To analyze the ER network morphology in vitro, we developed an image analysis program named MorphoER, which extracted ER regions from confocal images and measured their shapes including area and length (see Supplemental Methods). We classified the analytical targets as enlarged ER vesicles and ER tubules. MorphoER automatically extracted the targets by their shape and fluorescent intensity (Supplemental Fig. S4). As an alternative algorithmic approach, morphometric image processing to separate tubules from cisternae was reported (Sparkes et al., 2009; Griffing et al., 2014).
Ca2+-Efflux-Based Assay
Aequorin luminescence assays were performed as reported (Voeltz et al., 2006), with some modifications. ER vesicles were prepared from MM2d cells as reported previously (Yokota et al., 2011). For the inhibition assay, anti-RHD3 and normal IgG were dialyzed against solution A (0.25 m Suc, 2.5 mm MgCl2, 0.2 m KCl, and 50 mm HEPES-KOH, pH 7.5). ER vesicles in the solution A were premixed with the equilibrated solution (mock in Fig. 4, B and C), normal IgG (final concentration 250 µg/mL), or anti-RHD3NT (final concentrations 325 µg/mL) for 20 min at 25°C. After addition of aequorin (final concentration 12.5 µg/mL), ATP (final concentration 1 mm), and GTP (final concentration 1 mm) at 0 min, chemiluminescence intensities were measured every 2 min using a luminometer Gene Light GL-200 (Microtec). To examine the effect of antigen peptide, anti-RHD3NT was premixed with antigen peptide (final concentration 25 µm) at 4°C overnight. For kinase treatment, ER vesicles were premixed with Ser/Thr-kinase (a catalytic subunit of cAMP-dependent protein kinase; final concentration 4 µg/mL; Promega) and ATP (final concentration 1 mm) at 25°C for 10 min, and then GTP (final concentration 1 mm) and aequorin were added at 0 min. The anti-RHD3CT antiserum dialyzed against solution A was preadsorbed without or with the same volume of 80 µm antigen peptide on ice overnight and then mixed with ER vesicles. After 15 min, the mixture was treated with ATP and kinase for 10 min, and then GTP and aequorin were added at 0 min. The antiserum with the antigen peptide was diluted 80-fold in the final volume.
In Vitro ER Tubule Elongation Assay
This assay was performed as reported previously (Yokota et al., 2011) using ER vesicles prepared from MM2d cells or 2- to 3-week-old seedlings of Arabidopsis. ER vesicle preparation was preincubated with antibodies, Ser/Thr-kinase, or both similarly as for the aequorin luminescence assay, except for the addition of ER-Tracker (Invitrogen) instead of aequorin. After incubation for 20 to 40 min, the mixtures were mounted on a glass slide and exposed to shearing force by placing a cover slip. To avoid shearing force, specimens were prepared on ice and incubated at 25°C for 20 to 40 min.
For negative staining electron microscopy, the mixture was mounted on a carbon-coated copper grid and stained with 2% (w/v) uranyl acetate. The specimen was observed with an electron microscope (JEM-1200 EX II; JEOL) operated at 80 kV.
Immobilized Metal-Affinity Chromatography
ER vesicles isolated from MM2d cells were solubilized with 1% Triton X-100 and subjected to immobilized metal affinity chromatography according to the manufacturer’s instructions (PhosphoCruz Protein Purification System; Santa Cruz Biotechnology). Eluted fractions were subjected to SDS-PAGE followed by immunoblotting with anti-RHD3NT.
Plasmid Construction and Transformation
Translational fusion between synthetic GFP(S65T) cDNA and RHD3 genomic fragment was generated by fluorescent tagging of full-length proteins (Tian et al., 2004). The DNA fragment of ProRHD3:GFP-RHD3 was amplified using primers RHD3-IF-F, RHD3-5U-G-R, RHD3-5U-G-F, G-RHD3-R, G-RHD3-F, and RHD3-IF-R (Supplemental Table S1) and was subcloned into pENTR by an In-Fusion Advantage PCR cloning kit (Takara). GFP cDNA was inserted upstream of the RHD3 start codon. The amino acid linker between GFP and RHD3 was Gly-Gly-Gly. Gateway system (Invitrogen) LR reaction was used to introduce the construct into the destination vector pHGW (Plant System Biology) for plant expression. The vector was transformed into the rhd3-10 mutant mediated by Agrobacterium tumefaciens (strain GV3101) using the floral dip method (Clough and Bent, 1998).
Identification of RHD3 Phosphorylation Site
Transgenic plants expressing GFP-RHD3 were homogenized in 50 mm Tris-HCl (pH 8.0) containing 150 mm NaCl, 10 mm EGTA, 10 mm NaF, protease inhibitor cocktail (Roche Diagnostics), and 1% Triton X-100. The homogenate was centrifuged at 10,000g for 20 min at 4°C to remove cellular debris. The supernatant was incubated with anti-GFP MicroBeads (Miltenyi Biotech) for 30 min on ice and then applied to a µColumn (Miltenyi Biotech). After extensive washing, immunoprecipitate was eluted with SDS-PAGE sample buffer. GFP-RHD3 bands detected by silver staining were excised and digested with trypsin, and peptides were extracted from the bands as reported previously (Tamura et al., 2010). Trypsin-digested peptides were treated with 50 µg/mL Glu-C (Promega) in 50 mm ammonium bicarbonate at 37°C for 16 h. After evaporation with a vacuum concentrator, phosphorylated peptides were isolated with TiO2 beads (Phos-trap; Perkin-Elmer) and subjected to mass spectrometry analysis.
Mass Spectroscopy Methods, Instruments, and Database Searches
Samples were analyzed by nano-flow reverse-phase liquid chromatography followed by tandem mass spectrometry using an LTQ Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific) and Triple TOF 5600+ (AB SCIEX; see Supplemental Methods). Mass spectra of phosphopeptides derived from C-terminal region of GFP-RHD3 are shown in Supplemental Figure S6.
Cross-Linking with EGS
ER vesicles prepared from MM2d cells were incubated at 25°C in the presence or absence of 1 mm ATP, 1 mm GTP, or 16 µg/mL catalytic subunit of A-kinase. After 40 min, the mixture was further treated with 0.1 mm EGS (Pierce) for 30 min at 25°C and then subjected to SDS-PAGE and immunoblotting. Blots were digitized on a flat-bed scanner, and band intensity was quantified by densitometry using ImageJ software. Values were normalized to the background.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number At3g13870 (RHD3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. rhd3 mutants and anti-RHD3 antibodies used in this study.
Supplemental Figure S2. Defects in ER dynamics in the Arabidopsis rhd3 mutant.
Supplemental Figure S3. ER network reconstitution assay and ER tubule elongation assay.
Supplemental Figure S4. Development of MorphoER software.
Supplemental Figure S5. The large vesicle and tubule formation from the ER vesicles without applied shearing force in the absence or presence of kinase.
Supplemental Figure S6. Tandem mass spectra of phosphopeptides derived from C-terminal region of GFP-RHD3.
Supplemental Table S1. Primer sequences used in this study.
Supplemental Table S2. Statistical analysis of relative initial velocity of increasing luminescence intensity estimated from Figures 3, F and G, and 6F.
Supplemental Table S3. Statistical analysis of relative initial velocity of increasing luminescence intensity estimated from Figure 4, B and C.
Supplemental Table S4. Statistical analysis of relative number of ER tubules shown in Figure 4, D and E.
Supplemental Table S5. Statistical analysis of relative number of ER tubules shown in Figure 7.
Supplemental Movie S1. ER fragments of rhd3-1 show a tubule emerging from the ER fragment and a tubule contracting into the ER fragment
Supplemental Movie S2. In vitro ER tubule formation from the kinase-treated ER vesicles
Supplementary Material
Acknowledgments
We thank Peter M. Ray and Kanwarpal S. Dhugga (Stanford University) for providing anti-pea RGP1 and Tomoko Mori (National Institute for Basic Biology) for help with peptide synthesis. We are grateful to the Arabidopsis Biological Resource Center for providing the Arabidopsis mutant lines rhd3-1 and rhd3-10.
Glossary
- FRAP
fluorescence recovery after photobleaching
- EGS
ethylene glycol bis(succinimidylsuccinate)
Footnotes
Articles can be viewed without a subscription.
This work was supported by Specially Promoted Research of Grant-in-Aid for Scientific Research to I.H.-N. (no. 22000014), by Grants-in-Aid for Scientific Research to H.U. (nos. 25440132 and 15KT0151), E.Y. (no. 24576057), Te.S. (no. 23247009), and I.H.-N. (no. 15H05776) from the Japan Society for the Promotion of Science (JSPS), and by the National Science Foundation (MCB 1243792 to F.B.).
References
- Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport TA, Prinz WA (2012) The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol 197: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J (2011) Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci USA 108: 3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes LJ, Singh A, Szeto K, Benvin NM, O’Donnell JP, Zipfel WR, Sondermann H (2013) Structural basis for conformational switching and GTP loading of the large G protein atlastin. EMBO J 32: 369–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes LJ, Sondermann H (2011) Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci USA 108: 2216–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Doyle C, Qi X, Zheng H (2012) The endoplasmic reticulum: a social network in plant cells. J Integr Plant Biol 54: 840–850 [DOI] [PubMed] [Google Scholar]
- Chen J, Stefano G, Brandizzi F, Zheng H (2011) Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J Cell Sci 124: 2241–2252 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM (1997) A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA 94: 7679–7684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust JE, Desai T, Verma A, Ulengin I, Sun TL, Moss TJ, Betancourt-Solis MA, Huang HW, Lee T, McNew JA (2015) The Atlastin C-terminal tail is an amphipathic helix that perturbs the bilayer structure during endoplasmic reticulum homotypic fusion. J Biol Chem 290: 4772–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Voeltz GK (2011) The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol 21: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal U, Blackstone C (2013) Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta 1833: 2492–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing LR, Gao HT, Sparkes I (2014) ER network dynamics are differentially controlled by myosins XI-K, XI-C, XI-E, XI-I, XI-1, and XI-2. Front Plant Sci 5: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Ueda H, Kawase T, Hara-Nishimura I (2014) Microtubules Contribute to Tubule Elongation and Anchoring of Endoplasmic Reticulum, Resulting in High Network Complexity in Arabidopsis. Plant Physiol 166: 1869–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano K, Shimada T, Hiraiwa N, Nishimura M, Hara-Nishimura I (1997) A rapid increase in the level of binding protein (BiP) is accompanied by synthesis and degradation of storage proteins in pumpkin cotyledons. Plant Cell Physiol 38: 344–351 [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C (2009) A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138: 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sparkes I, Gattolin S, Dzimitrowicz N, Roberts LM, Hawes C, Frigerio L (2013) An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol 197: 481–489 [DOI] [PubMed] [Google Scholar]
- Lin S, Sun S, Hu J (2012) Molecular basis for sculpting the endoplasmic reticulum membrane. Int J Biochem Cell Biol 44: 1436–1443 [DOI] [PubMed] [Google Scholar]
- Liu TY, Bian X, Sun S, Hu X, Klemm RW, Prinz WA, Rapoport TA, Hu J (2012) Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc Natl Acad Sci USA 109: E2146–E2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Hayashi Y, Yamada K, Shimada T, Nishimura M, Hara-Nishimura I (2003a) The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol 44: 661–666 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I (2003b) A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J 33: 493–502 [DOI] [PubMed] [Google Scholar]
- Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N, Shimada T, Mano S, Nishimura M, Hara-Nishimura I (2000) Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol 41: 993–1001 [DOI] [PubMed] [Google Scholar]
- Morin-Leisk J, Saini SG, Meng X, Makhov AM, Zhang P, Lee TH (2011) An intramolecular salt bridge drives the soluble domain of GTP-bound atlastin into the postfusion conformation. J Cell Biol 195: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss TJ, Andreazza C, Verma A, Daga A, McNew JA (2011) Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc Natl Acad Sci USA 108: 11133–11138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Okada K, Takebe I, Matsui C (1981) Delivery of tobacco mosaic virus RNA into plant protoplasts mediated by reverse-phase evaporation vesicle (liposomes). Mol Gen Genet 184: 161–165 [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250: 4007–4021 [PMC free article] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, Daga A (2009) Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460: 978–983 [DOI] [PubMed] [Google Scholar]
- Pendin D, Tosetto J, Moss TJ, Andreazza C, Moro S, McNew JA, Daga A (2011) GTP-dependent packing of a three-helix bundle is required for atlastin-mediated fusion. Proc Natl Acad Sci USA 108: 16283–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann E. (2014) Endoplasmatic reticulum shaping by generic mechanisms and protein-induced spontaneous curvature. Adv Colloid Interface Sci 208: 153–160 [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, et al. (2003) Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J Biol Chem 278: 32292–32299 [DOI] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Hawes C, Griffing L (2009) Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 21: 3937–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G, Brandizzi F (2014) Unique and conserved features of the plant ER-shaping GTPase RHD3. Cell Logist 4: e28217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G, Hawes C, Brandizzi F (2014a) ER - the key to the highway. Curr Opin Plant Biol 22: 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G, Renna L, Brandizzi F (2014b) The endoplasmic reticulum exerts control over organelle streaming during cell expansion. J Cell Sci 127: 947–953 [DOI] [PubMed] [Google Scholar]
- Stefano G, Renna L, Moss T, McNew JA, Brandizzi F (2012) In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J 69: 957–966 [DOI] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I (2010) Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al. (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I (2010) Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci USA 107: 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11: 799–811 [DOI] [PubMed] [Google Scholar]
- Westrate LM, Lee JE, Prinz WA, Voeltz GK (2015) Form follows function: the importance of endoplasmic reticulum shape. Annu Rev Biochem 84: 791–811 [DOI] [PubMed] [Google Scholar]
- Yokota E, Mcdonald AR, Liu B, Shimmen T, Palevitz BA (1995) Localization of a 170 kDa myosin heavy chain in plant cells. Protoplasma 185: 178–187 [Google Scholar]
- Yokota E, Ueda H, Hashimoto K, Orii H, Shimada T, Hara-Nishimura I, Shimmen T (2011) Myosin XI-dependent formation of tubular structures from endoplasmic reticulum isolated from tobacco cultured BY-2 cells. Plant Physiol 156: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu F, Shi J, Zhu Y, Zhu Z, Gong Q, Hu J (2013) ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol 163: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I (2004) A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J 37: 398–414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.