A tissue-specific inducible system allows a combination of inducible promoters, genes, reporters, and plant selection markers in a single cloning step.
Abstract
A powerful method to study gene function is expression or overexpression in an inducible, cell type-specific system followed by observation of consequent phenotypic changes and visualization of linked reporters in the target tissue. Multiple inducible gene overexpression systems have been developed for plants, but very few of these combine plant selection markers, control of expression domains, access to multiple promoters and protein fusion reporters, chemical induction, and high-throughput cloning capabilities. Here, we introduce a MultiSite Gateway-compatible inducible system for Arabidopsis (Arabidopsis thaliana) plants that provides the capability to generate such constructs in a single cloning step. The system is based on the tightly controlled, estrogen-inducible XVE system. We demonstrate that the transformants generated with this system exhibit the expected cell type-specific expression, similar to what is observed with constitutively expressed native promoters. With this new system, cloning of inducible constructs is no longer limited to a few special cases but can be used as a standard approach when gene function is studied. In addition, we present a set of entry clones consisting of histochemical and fluorescent reporter variants designed for gene and promoter expression studies.
In the postgenomic era in plant research, the challenge is to discover functions of every gene in the plant genome and to understand how the encoded gene products interact to regulate plant development and physiology. One of the most useful molecular genetic techniques is the expression of a gene of interest under the control of its native promoter in a mutant background. Often, the gene product is fused to an enzymatic or fluorescent protein to provide for visualization of the expression domain. However, mutant phenotypes reflect long-term developmental impacts of loss of function, and expression levels are difficult to control. Temporal regulation provided by inducible gene overexpression or silencing provides a more targeted means of identifying the primary role of a regulator. Additionally, inducible systems permit gene functional studies when constitutive gene overexpression, or loss-of-function of a gene, leads to lethality. A cell type-specific system provides an additional benefit, as deleterious effects of ectopic gene expression can be avoided. During the past two decades, several different inducible vector systems have been developed (for review, see Moore et al., 2006; Borghi, 2010). Typically, these systems consist of genetic components from heterologous systems to avoid interference with plant signaling networks that are induced by chemical interaction with a transcriptional activator (Moore et al., 2006). Perhaps the most widely used system is the estrogen-inducible XVE system (Zuo et al., 2000).
The XVE vector system exhibits very low levels of constitutive or baseline activation, and there are only minimal physiological or developmental abnormalities caused by the use of estrogen or the system (Zuo et al., 2000). The XVE elements have been transferred into a single-site Gateway-compatible destination vector to allow high-throughput cloning of genes under the control of the inducible, ubiquitous promoter pG10-90 (Ishige et al., 1999; Curtis and Grossniklaus, 2003). Although useful, this vector does not allow easy cloning of other promoters to drive inducible expression. To avoid that problem, the XVE elements were divided into two different Gateway destination vectors to allow for high-throughput cloning of both genes and promoters (Brand et al., 2006). However, the use of two independent vectors necessarily requires crossing of lines containing the two parts of the system to achieve inducible gene expression. Additionally, this vector system is limited to a single plant selection marker and does not allow easy addition of promoter or protein fusions into transformants. We now describe a system that utilizes MultiSite Gateway technology to generate a versatile cell type-specific, inducible vector system that enables selective cloning of inducible promoters, genes, plant selection markers, and, where desired, reporter fusions in a single cloning step.
RESULTS
Construction of the MultiSite Gateway-Compatible Inducible System
MultiSite Gateway technology allows the assembly of up to four DNA fragments into one construct in a single cloning step. For the purposes of the system described herein, this typically combines promoter (first box), gene of interest (second box), and terminator (third box) Gateway entry clones with binary destination vectors in a MultiSite LR Clonase reaction (Fig. 1B). This construction principle was used to insert a modified version of the XVE inducible system (Zuo et al., 2000) into the first box entry clone to replace constitutive promoters (Fig. 1). The inducible promoter in the first box entry clone consists of a native cell type-specific promoter, the chimeric transcriptional activator XVE, two terminators (e9T and nosT), and multiple copies of XVE-binding sites (LexA operator sequence) fused with a minimal cauliflower mosaic virus 35S promoter (together called pLexA in Fig. 1). The promoter of interest drives XVE expression constitutively, and after estrogen application, XVE binds to pLexA to promote the transcription of a gene cloned in the second box only in the tissues where the promoter of interest is active (Fig. 1). The genes in the second box are provided from the entry clones containing attL1 and attL2 recombination sites. These entry clones are identical to the widely used single-site Gateway entry clones; therefore, the wide selection of existing entry clones, such as the genome-scale collection of Arabidopsis (Arabidopsis thaliana) transcription factors (Pruneda-Paz et al., 2014), can be utilized when constructing inducible lines with our system. New inducible promoters can be created by cloning promoter sequences into the promoterless version of the inducible system first box entry clone, p1R4-ML:XVE (Fig. 1A). This entry construct has unique cloning sites for several restriction endonucleases to allow promoter cloning with traditional digestion-ligation techniques (Fig. 1). After cloning of the promoters into p1R4-ML:XVE, new cell type-specific inducible constructs can be assembled in a single cloning step (Fig. 1). This entry clone and its derivatives have been used to create a number of cell type-specific and ubiquitously inducible promoters (Table I). The original XVE inducible system utilizes a hygromycin selection marker (hygromycin phosphotransferase II [HPT] coding sequence), and it is difficult to exchange this for another selection marker (Zuo et al., 2000). In the system described herein, selection marker genes have been preinserted into a set of binary destination vectors containing attR4 and attR3 recombination sites (such as the pBm43GW, pHm43GW, and pKm43GW vectors described by Karimi et al. [2005] and pCAM-hyg-R4R3 and pCAM-kan-R4R3 here), making it easy to choose the selection marker of need. To achieve this versatility, the original XVE-pLexA cassette (Zuo et al., 2000) was modified by removing the nopaline synthase promoter (pNOS) and HPT originally positioned between e9T and nosT (Fig. 1).
Figure 1.
Schematic representation of the inducible expression system. A, Promoters of interest are cloned with digestion-ligation techniques into one or two of the unique cloning sites located in the promoterless version of the first box entry clone, p1R4-ML:XVE. In the example, the unique XhoI site was utilized to clone a promoter. B, Generation of an inducible binary vector in a single cloning step. Entry clones for the XVE inducible promoter in the first box (L4-R1), gene of interest (GOI) in the second box (L1-L2), and terminator- or reporter-encoding sequences in the third box (R2-L3) are recombined with a destination vector containing attR4 and attR3 sites in a single MultiSite Gateway reaction. L4, Gateway recombination site attL4; R1, attR1; L1, attL1; L2, attL2; R2, attR2; L3, attL3; B4, attB4; B1, attB1; B2, attB2; B3, attB3; XVE, chimeric transcription factor containing the DNA-binding domain of LexA, the transcriptional activation domain of VP16, and the regulatory domain of the human estrogen receptor; e9T, rbcS E9 poly(A) addition sequence; nosT, nopaline synthase poly(A) addition sequence; pLexA, eight copies of the LexA operator sequence together with a minimal cauliflower mosaic virus 35S promoter.
Table I. Cell type-specific XVE inducible entry clones.
All clones are ampicillin resistant in bacteria.
| Entry Clone Names | Tissue Specificity of the Corresponding Published Constitutive Promoter/Name of the Corresponding Gene/Arabidopsis Genome Initiative Code (Reference) | Shows Correct Expression Pattern after Induction? |
|---|---|---|
| p1R4-ML:XVE | Promoterless version for cloning new promoters (Fig. 1A) | – (contains no promoter) |
| p1R4-p35S:XVE | Strong ubiquitous expression/cauliflower mosaic virus 35S promoter (35S; Odell et al., 1985) | Yes (data not shown) |
| p1R4-p6xUAS:XVE | Upstream activation sequence containing several GAL4-binding elements and minimal 35S (Brand and Perrimon, 1993; Johnson et al., 2005) | Yes (Vatén et al., 2011) |
| p1R4-pAHP6:XVE | Protoxylem + protoxylem-pericycle, organ primordia in shoot apical meristem/ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN6 (AHP6)/AT1G8010 (Mähönen et al., 2006; Bartrina et al., 2011; Besnard et al., 2014) | Yes (Mähönen et al., 2014) |
| p1R4-pANT:XVE | Developing ovules, primordia of all lateral shoot organs/AINTEGUMENTA/AT4G37750 (Elliott et al., 1996) | Yes (data not shown) |
| p1R4-pAPL:XVE | Phloem (developing sieve elements and companion cells)/ALTERED PHLOEM DEVELOPMENT (APL)/AT1G79430 (Bonke et al., 2003) | Yes (Bishopp et al., 2011b; Vatén et al., 2011) |
| p1R4-pAtSUC2:XVE | Phloem companion cells/ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER2/AT1G22710 (Truernit and Sauer, 1995) | Yes (data not shown) |
| p1R4-pCALS3:XVE | Vascular bundle, root stem cell niche/CALLOSE SYNTHASE3 (CALS3)/AT5G13000 (Vatén et al., 2011) | Yes (Vatén et al., 2011) |
| p1R4-pCCS52A1:XVE | Root elongation zone, lateral roots/CELL CYCLE SWITCH PROTEIN52 A1 (CCS52A1)/AT4G22910 (Vanstraelen et al., 2009) | Yes (data not shown) |
| p1R4-pCO2:XVE | Cortex/CORTEX2 (CO2)/AT1G62500 (Heidstra et al., 2004) | Yes (Dhonukshe et al., 2010; this article) |
| p1R4-pCYP79B3:XVE | Meristem quiescent center and the surrounding stem cells/CYTOCHROME P450, FAMILY 79, SUBFAMILY B3 (CYP79B3)/AT2G22330 (Ljung et al., 2005) | Yes (data not shown) |
| p1R4-pG1090:XVE | Strong ubiquitous expression/G10-90 (artificial promoter containing repeats of G-box; Ishige et al., 1999) | Yes (Dhonukshe et al., 2010; Mähönen et al., 2014, this article) |
| p1R4-pGC1:XVE | Guard cell/GC1/AT1G22690 (Yang et al., 2008) | Not tested |
| p1R4-pIRX8:XVE | Interfascicular fibers and developing xylem cells/IRREGULAR XYLEM8 (IRX8)/AT5G54690 (Peña et al., 2007; Persson et al., 2007) | Yes (data not shown) |
| p1R4-pLFY:XVE | Floral buds, lateral primordia/LEAFY (LFY)/AT5G61850 (Weigel et al., 1992) | Not tested |
| p1R4-pLexA | Entry clone containing only the LexA-binding sites and the minimal 35S (i.e. lacking PROM:XVE:T elements); can be used if several genes need to be induced simultaneously (by crossing pLexA:gene-of-interest with any XVE inducible lines) | Yes (data not shown) |
| p1R4-pRCH1:XVE | Root meristem/ROOT CLAVATA1 HOMOLOG (RCH1)/AT5G48940 (Casamitjana-Martínez et al., 2003) | Not tested |
| p1R4-pS17:XVE | Phloem pole pericycle cells/AT2G22850 (Lee et al., 2006) | Yes (data not shown) |
| p1R4-pSCR:XVE | Endodermis/SCARECROW (SCR)/AT3G54220 (Di Laurenzio et al., 1996) | Yes (this article) |
| p1R4-pTHE:XVE | Root elongation zone, vasculature, cotyledons, shoot apical meristem/THESEUS1/AT5G54380 (Hématy et al., 2007) | Yes (data not shown) |
| p1R4-pTMO5:XVE | Hypophysis-adjacent embryo cells, embryo vasculature and cotyledons, xylem precursor cells/TARGET OF MP5 (TMO5)/AT3G25710 (Schlereth et al., 2010; De Rybel et al., 2013) | Not tested |
| p1R4-pUBQ10:XVE | Moderate level, ubiquitous expression/UBIQUITIN10 (UBQ10)/AT4G05320 (Geldner et al., 2009) | Not tested |
| p1R4-pWER:XVE | Nonhair cells in epidermis/WEREWOLF (WER)/AT5G14750 (Lee and Schiefelbein, 1999) | Yes (Dhonukshe et al., 2010; Mähönen et al., 2014; this article) |
| p1R4-pWOL:XVE | Root meristem procambium, vascular bundle in general, shoot apical meristem/WOODEN LEG/CRE1/AHK4 (WOL)/AT2G01830 (Mähönen et al., 2000, 2006; Gordon et al., 2009) | Yes (Bishopp et al., 2011a; Vatén et al., 2011; Ursache et al., 2014; this article) |
| p1R4-pWOX5:XVE | Root meristem quiescent center/WUSCHEL-RELATED HOMEOBOX5 (WOX5)/AT3G11260 (Haecker et al., 2004) | Yes (data not shown) |
| p1R4-pZCP4:XVE | Immature tracheary elements/ZINNIA CYSTEINE PROTEASE4 (ZCP4)/AB091070 (Pyo et al., 2004) | Yes (data not shown) |
Stringency of the Modified Inducible System in Transgenic Plants
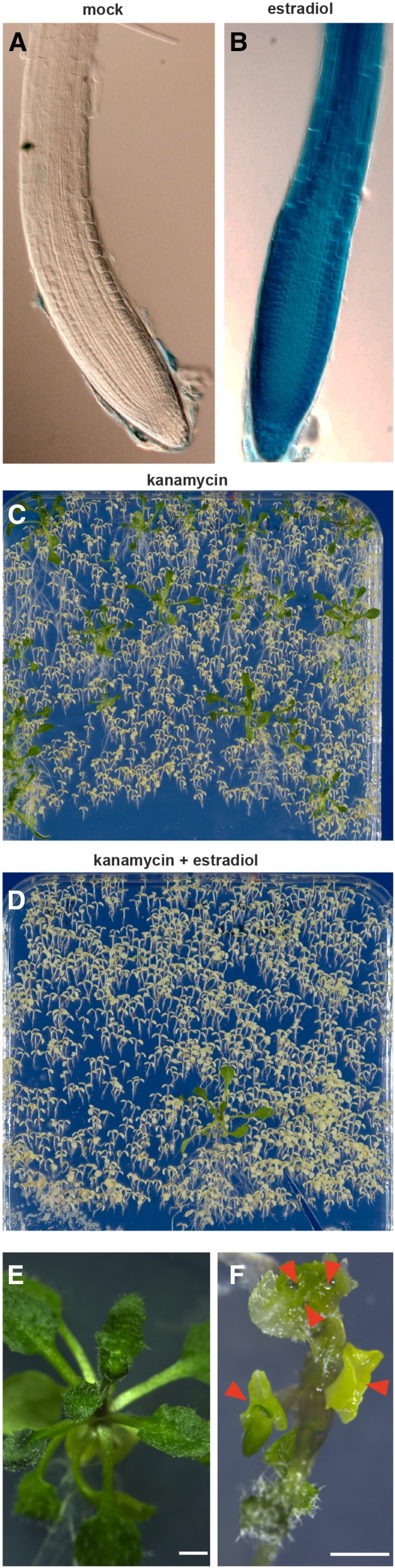
In order to determine if any loss of stringency would result from the removal of the pNOS::HPT sequences from the original XVE system (Zuo et al., 2000), induction by 17-β-estradiol, a synthetic derivative of estrogen, was tested in the revised system. Experiments with a pG10-90:XVE>>GUS transformant line, in which the ubiquitous G10-90 promoter (Ishige et al., 1999) drives inducible GUS expression, showed GUS expression that is highly dependent on 17-β-estradiol (Fig. 2, A and B). In a further stringency test, the gene encoding the diphtheria toxin A-chain (DTA; Harrison et al., 1991) was placed under the control of pG10-90:XVE. Diphtheria toxin kills cells by inhibiting protein synthesis (Collier, 1967) and has been utilized successfully to ablate specific tissues in plants (Day et al., 1995; Weijers et al., 2003). Wild-type plants transformed with pG10-90:XVE>>DTA were screened in the presence or absence of 17-β-estradiol. In the absence of the inducer, 23 independent transformants, comparable to the number of transformants obtained when the same binary vector is used to deliver the expression of nontoxic genes, were obtained (Fig. 2C; data not shown). However, when an equal number of seeds from pG10-90:XVE>>DTA transformants were plated on selection medium containing 17-β-estradiol, only a single transformant survived (Fig. 2D). We next asked whether the conditionality in the inducible system can be used to gain insights into the functions of key regulators by misexpressing them. Toward this end, we placed PLT5/AIL5, which encode double AP2 domain-containing transcription factors under the control of pG10-90:XVE, and examined the phenotypes in transformed seedlings upon estradiol induction. While induction of PLT5/AIL5 expression resulted in ectopic somatic embryo-like structures on the surface of seedlings, as demonstrated before (Tsuwamoto et al., 2010), the untreated control seedlings displayed normal growth (Fig. 2, E and F). These data demonstrate that the MultiSite Gateway-compatible inducible system confers stringent regulation of gene expression.
Figure 2.
Stringent regulation of gene expression conferred by the inducible system. A and B, A 20-h induction of pG10-90:XVE>>GUS with 1 μm 17-β-estradiol leads to strong and ubiquitous GUS expression throughout the plant, including the root tip. Note that the induced GUS expression appears high, as the representative root tip in B has experienced relatively short incubation time (90 min, room temperature) with its substrate 5-bromo-4-chloro-3-indolyl-β-glucuronic acid. C and D, Similar amounts of seeds obtained from plants transformed with pG10-90:XVE>>DTA were surface sterilized and spread on agar plates containing kanamycin (C) or kanamycin and 5 μm 17-β-estradiol (D). The images were taken 14 d after germination. E and F, pG10-90:XVE>>PLT5-YFP control seedlings grown for 15 d on Murashige and Skoog (MS) medium (E) or MS medium containing 5 μm 17-β-estradiol (F), the latter displaying ectopic somatic embryo-like structures (red arrowheads). Bars = 1 mm.
Cell Type Specificity of the Inducible System
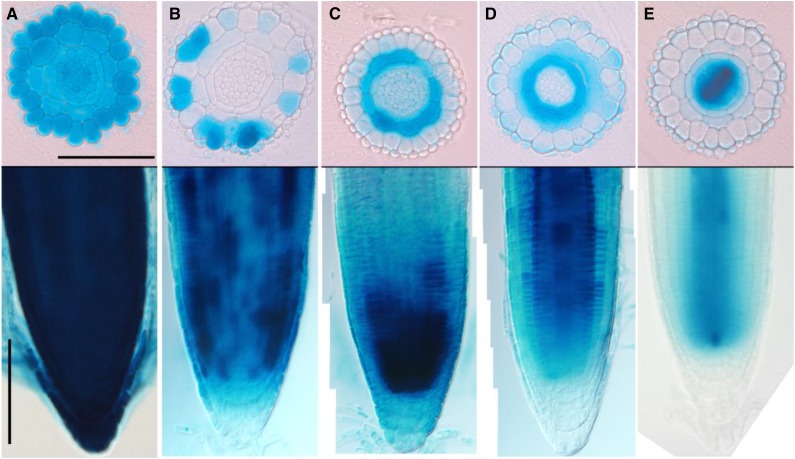
In order to test the cell type specificity of the system, we generated inducible GUS reporter lines specific for the different cell layers in the root (Fig. 3). We selected well-established promoters with cell type-restricted expression patterns to facilitate testing of the inducible promoters: G10-90 (Fig. 3A) for ubiquitous expression (Ishige et al., 1999), WEREWOLF (Fig. 3B) for nonhair cells in epidermis (Lee and Schiefelbein, 1999), CO2 (Fig. 3C) for the cortex (Heidstra et al., 2004), SCARECROW (Fig. 3D) for endodermis (Di Laurenzio et al., 1996), and WOODENLEG (Fig. 3E) for root meristem procambium (Mähönen et al., 2006). All four constructs showed characteristic, layer-specific GUS expression after 17-β-estradiol induction (Fig. 3). In some transformant lines, GUS expression was observed in unexpected cell types (Fig. 3; data not shown). This underlines the need to carefully select transformants, similar to when selecting standard transgenic lines.
Figure 3.
Tissue-specific induction of GUS enzyme after 17-β-estradiol treatment. Histological cross sections (top) of root meristems and bright-field images (bottom) of 4-d-old seedlings transferred for 24 h on 5 μm 17-β-estradiol are shown. A, pG1090:XVE>>GUS. B, pWEREWOLF:XVE>>GUS. C, pCO2:XVE>>GUS. D, pSCARECROW:XVE>>GUS. E, pWOODENLEG:XVE>>GUS. Bars = 100 μm.
Effect of 17-β-Estradiol on Root Growth
The use of chemical inducers in laboratory experimental procedures always raised concerns about any possible harmful effect on plant growth and physiology. High levels of ethanol used in some inducible systems (Roslan et al., 2001; Claassens et al., 2005; Craft et al., 2005; Samalova et al., 2005; Camargo et al., 2007) were shown to be variably toxic when used experimentally, while dexamethasone (Craft et al., 2005; Moore et al., 2006) or 17-β-estradiol (Zuo et al., 2000; Tornero et al., 2002; Vilarrasa-Blasi et al., 2014) have exhibited little or no toxicity when tested at physiological concentrations by different laboratories.
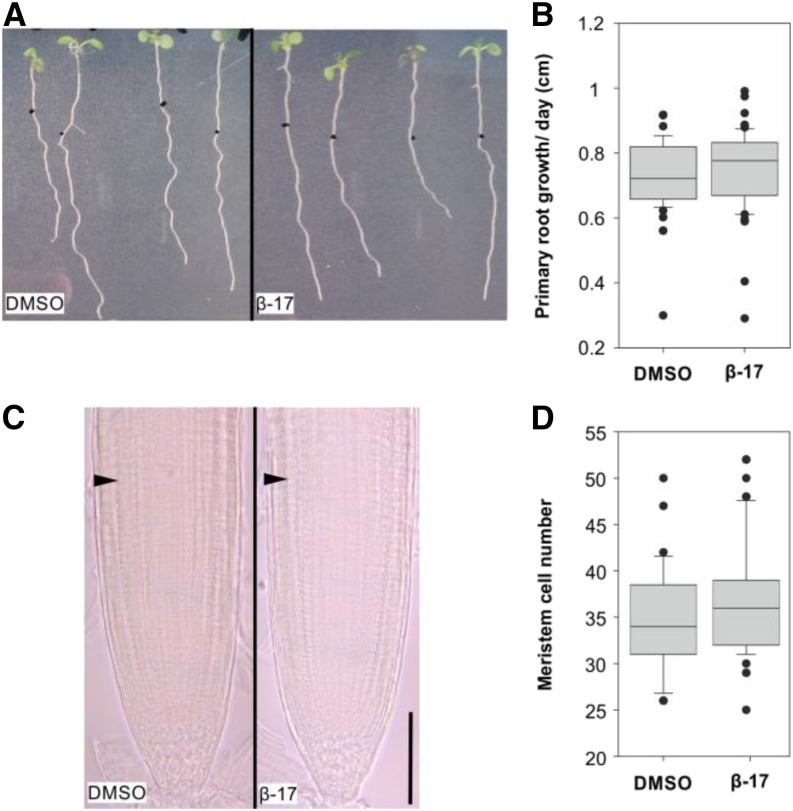
We decided to examine in detail whether 17-β-estradiol treatment shows any effect on plant growth. First, we focused on the various aspects of root growth and used 5 µm 17-β-estradiol, as at this concentration the system seems to be saturated but does not confer toxicity to plants (Zuo et al., 2000). Transferring 4-d-old Columbia-0 (Col-0) seedlings on 5 µm 17-β-estradiol for 4 d did not affect primary root growth (Fig. 4, A and B) or root meristem size (Fig. 4, C and D), thus confirming that 5 µm concentration is noninhibitory to axial growth of the root.
Figure 4.
Primary root growth is not affected by treatment with 17-β-estradiol. A, Arabidopsis seedlings grown for 4 d on MS medium were transferred to dimethyl sulfoxide (DMSO) solvent control (left) or 5 μm 17-β-estradiol (right) plates and incubated for 4 d. Black dots represent the positions of the root tips after transfer. B, Box-plot graph representing the primary root growth per day (cm) measured on 4-d-old seedlings transferred for 4 d on DMSO (n = 57) or 5 μm 17-β-estradiol (β-17; n = 58). P = 0.127 (Mann-Whitney U test). C, Root apical meristems of 4-d-old seedlings transferred for 4 d on DMSO (left) or 5 μm 17-β-estradiol (right). Arrowheads indicate the beginning of the transition zone. Bar = 100 μm. D, Box-plot graph representing the meristem size (cortex cell number) of 4-d-old seedlings transferred for 4 d on DMSO (n = 33) or 5 µm 17-β-estradiol (n = 53). P = 0.127 (Mann-Whitney U test).
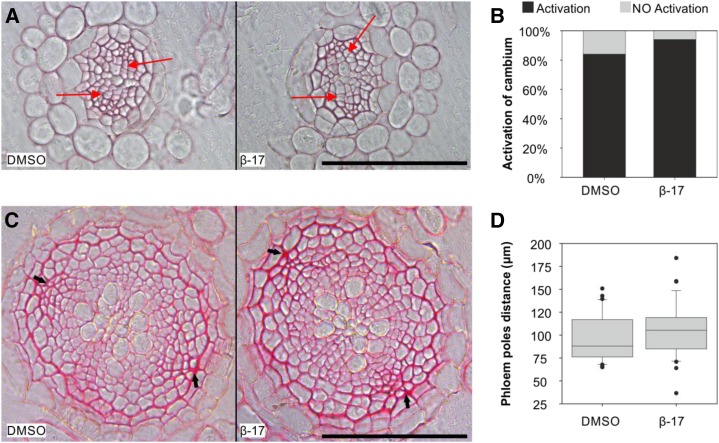
The secondary growth in Arabidopsis root is activated 5 d after germination (Matsumoto-Kitano et al., 2008; Nieminen et al., 2015), and it is characterized by the appearance of periclinal cell divisions in the root (pro)cambial and pericycle cells 5 mm below the hypocotyl (Fig. 5A). Treatment with 5 µm 17-β-estradiol had no apparent effect on the activation of secondary growth (Fig. 5, A and B). Next, we investigated the long-term effect of the inducer on secondary growth. Four-day-old Col-0 seedlings were transferred on mock (DMSO) or 5 µm 17-β-estradiol medium for 12 d, after which the distance between the primary phloem poles was measured. No significant difference in the width of the vasculature was observed (Fig. 5, C and D).
Figure 5.
Secondary root growth is not affected by treatment with 17-β-estradiol. A, Histological cross sections of roots grown for 4 d on MS medium and 4 d on DMSO solvent control (left) or 5 μm 17-β-estradiol (right). Red arrows mark a subset of periclinal cells divisions in the cambium. Bar = 100 μm. B, Percentage of plants that activated secondary growth. Four-day-old seedlings were grown for 4 d on DMSO (n = 51) or 5 μm 17-β-estradiol (n = 53). P = 0.1792 (Mann-Whitney U test). C, Histological cross sections of roots grown for 4 d on MS medium and transferred for 12 d on DMSO (left) or 5 μm 17-β-estradiol (right). Arrows indicate primary phloem poles. Bar = 100 μm. D, Box-plot graph representing the distance between the primary phloem poles (μm) measured on 4-d-old seedlings grown for 12 d on DMSO (n = 30) or 5 μm 17-β-estradiol (n = 39). P = 0.130 (Mann-Whitney U test).
Growth conditions vary between laboratories; consequently, this affects the results obtained (Massonnet et al., 2010). Therefore, a similar root growth assay to that performed in Helsinki was carried out in a different location (Purdue University) by a different scientist. Unlike at the University of Helsinki, 5 µm 17-β-estradiol slightly but significantly affected primary root growth at Purdue University, and the application of 8 µm or higher inhibited growth even more (Supplemental Fig. S1). These results together show the importance of carrying out the necessary chemical control experiments each time the XVE inducible system is used.
Effect of the XVE System on Root Growth
Next, we tested whether the XVE construct by itself or after induction affects root growth. Four-day-old Col-0, p35S:XVE>>GUS, pG1090:XVE>>GUS (two lines), and pG1090:XVE>>axr3-1-RFP were transferred on DMSO or 5 µm 17-β-estradiol plates, and their root growth was monitored for 4 d. None of the four XVE lines showed inhibition of root growth in the absence of the inducer (Fig. 6). In the presence of inducer, two-thirds of the ubiquitously induced GUS lines showed modest but significant reduction of root growth (Fig. 6). Since neither the XVE construct nor 5 µm 17-β-estradiol alone is inhibitory to root growth, these data indicate that either the strong and ubiquitous GUS overexpression or XVE activation can have an inhibitory effect on root growth. As a positive control for the functionality of the system, we detected a strong inhibition of primary root growth after constitutively inducing the dominant negative regulator of auxin signaling axr3-1 (Leyser et al., 1996), thus confirming results published previously (Fig. 6; Mähönen et al., 2014). In conclusion, the XVE system in general is not inhibitory for root growth; however, after induction, some lines exhibit growth inhibition. This highlights the importance of using appropriate controls while selecting lines for experiments monitoring growth.
Figure 6.
Effect of the XVE system on root growth. Col-0 and transgenic lines were grown for 4 d on one-half-strength GM plates and then grown for 4 d on DMSO solvent control or 5 μm 17-β-estradiol plates. The data represent the distance the primary root has grown per day. Asterisks indicate significant differences (Student’s t test or Mann-Whitney U test, P < 0.001).
Slow Diffusion of the 17-β-Estradiol from Root to Shoot
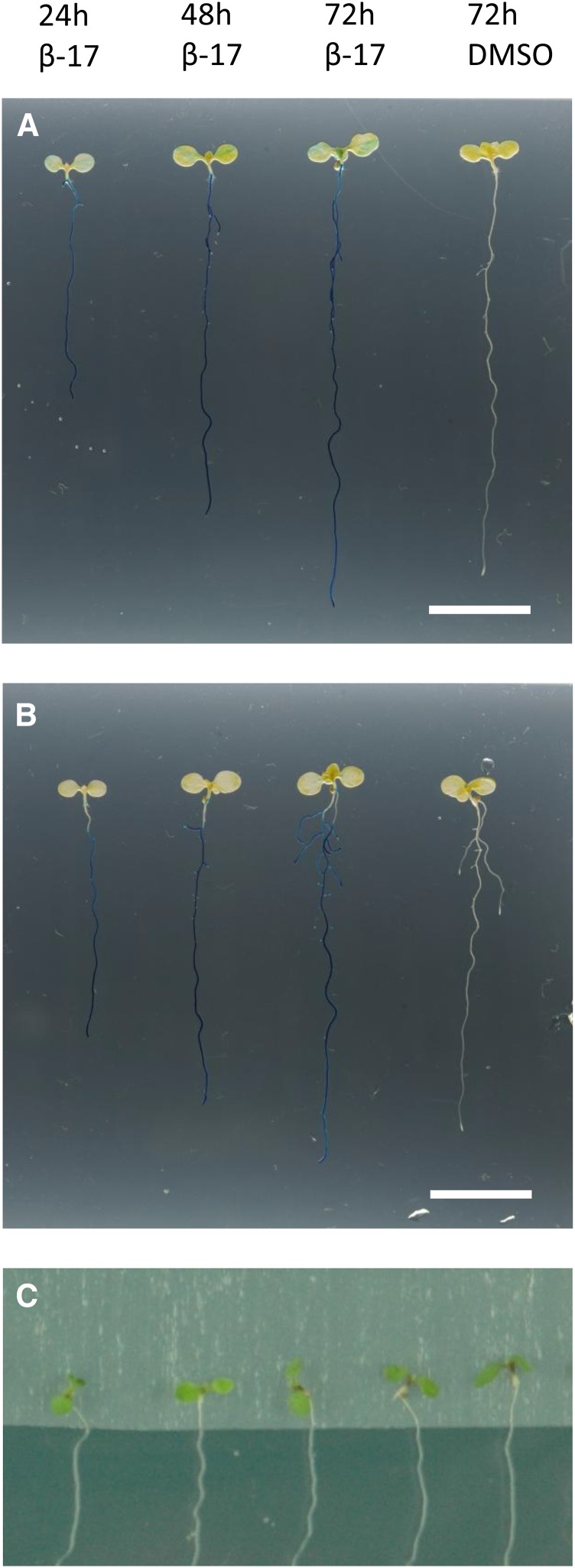
Arabidopsis seedlings are often examined on agar plates, where their growth can be monitored in well-controlled conditions. Germinated seedlings are typically in contact with the agar medium via the root system and part of the hypocotyl. Since 17-β-estradiol is often administered for the seedlings via the medium, the question is raised whether the shoot tissues will also receive the inducer. Therefore, we decided to investigate whether the 17-β-estradiol molecule diffuses or moves from the root to the shoot and activates the XVE inducible system there. We used the pG1090:XVE>>GUS #1 line, which, in the previous experiment, did not show inhibition of root growth after 17-β-estradiol treatment (Fig. 6). After transferring the seedlings onto 5 µm 17-β-estradiol plates, only a weak and occasional GUS signal was detected in the aerial parts (Fig. 7A). This weak signal appeared typically on one side of the cotyledon, suggesting that the cotyledon received the inducer through contact with the agar medium. To test that theory, a sterile sheet of Parafilm was placed between the aerial part of the seedling and the medium (Fig. 7C). In these conditions, no GUS staining was observed in the shoot (Fig. 7B). These data demonstrate that the 17-β-estradiol diffuses very slowly from root to shoot, thus confirming the findings by Brand et al. (2006).
Figure 7.
17-β-Estradiol diffuses slowly in Arabidopsis seedlings. A, Five-day-old pG1090:XVE>>GUS #1 seedlings incubated for the indicated hours on DMSO solvent control or 5 μm 17-β-estradiol (β-17) plates, without Parafilm underneath the shoot. Note a weak GUS staining in the cotyledons. B, Five-day-old pG1090:XVE>>GUS #1 seedlings transferred for the indicated hours on 5 μm 17-β-estradiol or DMSO control plates, with Parafilm underneath the shoot. C, Example of the induction method used in B, consisting of a Parafilm layer to avoid contact between the shoot and the induction medium. Ten or more plants were analyzed for each time point. Selected roots show the most representative seedling. Bars in A and B = 1 cm.
Construction of Reporter Protein Entry Clones
During the past two decades, the use of fluorescent proteins (FPs) revolutionized imaging studies in biology, opening the way to, for example, colocalization studies of different proteins in the same biological context. In addition, the histochemical reporter GUS (Jefferson et al., 1987) is still a valuable tool to analyze the expression pattern of interest whenever confocal microscopy studies are challenging.
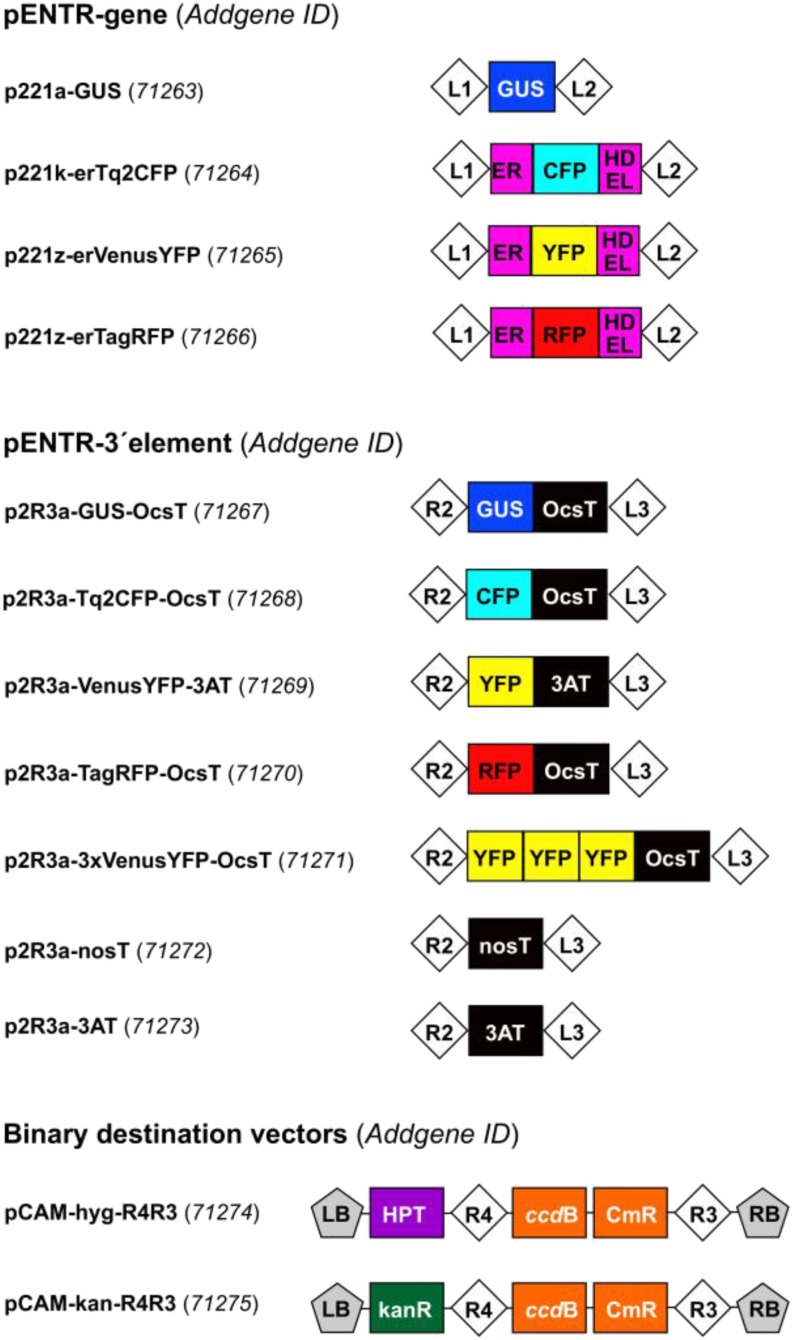
We developed two different sets of entry clones (pENTR-gene and pENTR-3′ element), compatible with the MultiSite Gateway system (Fig. 8). These reporter clones can be used to construct transcriptional or translational reporters of the genes of interest or to combine with the XVE inducible system. Three different fluorescent proteins compatible for colocalization studies were selected: TURQUOISE2 CYAN FLUORESCENT PROTEIN (Tq2CFP; Goedhart et al., 2012), VENUS YELLOW FLUORESCENT PROTEIN (VenusYFP; Nagai et al., 2002), and TAG RED FLUORESCENT PROTEIN (TagRFP; Merzlyak et al., 2007). The genes encoding three FPs and the GUS enzyme were inserted into two different MultiSite Gateway-compatible plasmids: (1) the pENTR-gene plasmid, where the fluorescent proteins (but not the GUS enzyme) are targeted into the endoplasmic reticulum (ER) and can be used to construct transcriptional reporters; and (2) the pENTR-3′ element plasmid, where the FPs and the GUS enzyme can be fused with a flexible Gly linker at the C-terminal end of the protein of interest. In addition, three repeats of VenusYFP were cloned into the pENTR-3′ element to allow expression analysis with enhanced sensitivity. To demonstrate the increased sensitivity, the fluorescence of PLETHORA2-1xYFP and PLETHORA-3xYFP was compared, and a clear increase in fluorescence intensity with the 3xYFP version was observed (Fig. 9, F–H). In addition to this, 3xYFP and 1xYFP fusions can be utilized to study protein cell-to-cell movement (Mähönen et al., 2014). We also generated pENTR-3′ clones with nos (Depicker et al., 1982) or 3AT (Fluhr et al., 1986) sequence, necessary to complete the transcriptional reporters by providing the poly(A) tail to the nascent mRNA (Fig. 8). We provide these two different terminators to lower the risk of cosuppression when several constructs are present in the same plant.
Figure 8.
MultiSite Gateway reporter entry clones and binary destination vectors. The plasmids can be obtained from Addgene (https://www.addgene.org/Ari_Pekka_Mahonen/) by using the Addgene identifier codes presented in parentheses. Abbreviations are as follows: L1, L2, R2, L3, R3, and R4, Gateway recombination sites attL1, attL2, attR2, attL3, attR3, and attR4, respectively; ER, Arabidopsis vacuolar basic chitinase ER targeting signal; HDEL, ER retention signal; OcsT, octaline synthase terminator; nosT, nopaline synthase poly(A) addition sequence; 3AT, T3A pea (Pisum sativum) ribulose-1,5-bisphosphate carboxylase 3A subunit terminator; LB and RB, left and right border of the T-DNA, respectively; ccdB, gene conferring negative selection (inhibits E. coli DNA gyrase); CmR, chloramphenicol resistance gene; kanR, kanamycin resistance gene. A letter after p221 and p2R3 refers to bacterial selection: A, ampicillin; K, kanamycin; Z, zeocin. Note that the maps are not drawn to scale.
Figure 9.
Examples of MultiSite Gateway-generated fluorescent reporter lines. A and B, DR5rev:erGFP/pARR5:erTagRFP. C and D, DR5rev:erTagRFP/pTCS:nGFP. In A to D, green signal shows GFP and magenta RFP fluorescence, respectively. E, pARR5:erVenusYFP. Images show the expression pattern in longitudinal (A, C, and E) or radial (B and D) optical sections. F and G, Representative images of the PLT2 protein fused to the single VenusYFP (F) or triple VenusYFP (G) expressed under the control of the endogenous PLT2 promoter. All seedlings were 5 d old. The magenta signal in E to G visualizes propidium iodide staining. Bars = 25 μm. H, Scatter graph of the fluorescence intensities represented in F and G. Twelve root meristems of three different transgenic lines were analyzed to generate the pPLT2:PLT2-3xYFP values, while 11 root meristems of two different transgenic lines were analyzed to generate the pPLT2:PLT2-YFP values. Central lines represent average values, and error bars indicate 95% confidence intervals.
To test these new entry clones, we developed two ARR5 fluorescent reporters in the Col-0 background by expressing VenusYFP and TagRFP under the control of 2,260 bp of the ARR5 promoter and a new DR5rev reporter designed with the TagRFP fluorescent protein (Fig. 9, A–E). The expression pattern of all three reporters recapitulated the published data (Bishopp et al., 2011a).
DISCUSSION
Selecting Nonleaky, Highly Inducible, Cell Type-Specific Inducible Lines
Temporally and spatially controllable gene expression is a prerequisite for modern plant molecular biology studies. Here, we present a MultiSite Gateway-based vector system which allows fast and easy combination of tissue-specific inducible promoters with genes of interest.
The control of inducibility in our system is stringent, as GUS expression, DTA-mediated lethality, and PLT5/AIL5-mediated phenotypes in the transgenic lines are 17-β-estradiol dependent. Comparable nonleakiness was observed also in the previous XVE versions (Zuo et al., 2000; Brand et al., 2006). Cell type-specific promoters can be easily cloned into the inducible system, where they provide cell type-specific expression, similar to the constitutive versions of these promoters. However, when generating inducible lines, the positional effect of the transgene needs to be taken into account, similar to when selecting standard transgenic lines. Therefore, we propose the following work flow when selecting transgenic lines: (1) screen for lines that show minimal expression of the gene to be induced in the absence of 17-β-estradiol; (2) transfer these nonleaky lines on plates containing 17-β-estradiol and select for lines that show strong and robust inducibility; and (3) if cell type-specific promoters are used, test the nonleaky, highly inducible lines for correct cell type-specific expression. These steps are important to follow, since we occasionally identify lines with gene expression in the absence of inducer or with expression in ectopic locations. These phenotypes are likely caused by endogenous enhancers near the transgenes interfering with the inducible system. Depending on the inducible promoter used, the frequency of the occurrence of these unwanted characteristics is variable. However, we have been able to isolate nonleaky, highly inducible, cell type-specific lines for each promoter tested when enough independent transgenic lines have been screened. To make the screening faster and easier, the gene to be induced is useful to be recombined with a pENTR-3′ clone containing a reporter gene (FPs or GUS; Fig. 8) to provide a visual method to monitor gene inducibility.
Toxicity of 17-β-Estradiol for Plants
The hormone estrogen is present in all vertebrates, but not in plants, which do not have a recognized estrogen receptor. Therefore, estrogen and its receptor were employed in the development of the plant XVE inducible system with the goal of minimizing interference with endogenous plant processes (Zuo et al., 2000). Previous work suggests that expression of the XVE system itself has no effect on plant growth or physiology (Zuo et al., 2000; Tornero et al., 2002). Apart from positional effects produced by the interruption of endogenous genes, we observed no growth inhibition in induced or uninduced XVE system transformants carrying no additional transgene. However, previous experiments suggest that estrogen concentrations greater than 5 µm can have a negative effect on growth (Zuo et al., 2000; Moore et al., 2006). In order to assess possible toxicity effects, we studied various aspects of root growth in the presence of 2 and 5 µm 17-β-estradiol. At 2 µm, no interference with growth was detected under any conditions. Furthermore, at the University of Helsinki, we did not observe any negative impacts of 5 µm 17-β-estradiol on plant growth. However, independent experiments at Purdue University and the University of Maryland showed slight, but significant, decreases in root growth at 5 µm and above. Efforts to isolate the underlying cause for this difference suggested that temperature and light levels, but not relative humidity, impact the 17-β-estradiol toxicity threshold (Supplemental Fig. S2). Even though these factors are among many others that can affect toxicity, these differences emphasize the need to establish inducer toxicity thresholds before analyzing responses to gene expression with 17-β-estradiol and the XVE system. The results also show the importance of optimal growth conditions, as toxic effects might be enhanced in response to stress. In conclusion, the XVE system and moderate concentrations of 17-β-estradiol are not toxic for Arabidopsis; however, the use of the inducer and the XVE system needs to be controlled properly in each experiment.
Diffusibility of the 17-β-Estradiol in Plant Tissue
Previously, it was shown that, due to the nonvolatile nature of 17-β-estradiol, the chemical can be applied to small regions on leaves and induce local expression of a reporter (Brand et al., 2006). Our results show that young seedlings placed on medium containing 17-β-estradiol show reporter expression mainly on tissues that are in contact with the medium, thus supporting the idea that the inducer diffuses slowly in plant tissue. The slow diffusibility of 17-β-estradiol makes it possible to separate the shoot from inducing medium with Parafilm when only root phenotypes are to be studied. However, when making such manipulations, care must be taken to avoid 17-β-estradiol contamination via tweezers or other implements. In experiments where the induction in shoot tissues is desired, the shoot can be pushed gently into the agar to mediate the diffusion of the inducer into the tissue. This approach was used when generating ectopic embryo-like structures in the shoot with PLT5/AIL5 induction (Fig. 2F). In conclusion, the low diffusibility of 17-β-estradiol is important to take into an account when conducting experiments.
MultiSite Gateway-Compatible Reporter Entry Clones
In this article, we also present a set of Gateway-compatible entry clones to provide reporters for the inducible system and for standard constitutive expression studies. Two different sets of entry clones, pENTR-gene and pENTR-3′ element, were generated. pENTR-gene vectors can be used to generate transcriptional reporters and pENTR-3′ element vectors to construct translational gene-reporter fusions. We provide entry clones containing genes encoding the histological reporter GUS as well as three different fluorescent reporters. These fluorescent proteins (Tq2CFP, VenusYFP, and TagRFP) were chosen because these color variants can be visualized simultaneously with confocal laser scanning microscopy (Shaner et al., 2005). Additionally, for each color variant, the brightest or one of the brightest versions of the FPs tested in plants was selected. The pENTR-3′ element reporters are especially useful when testing the cell type-specific inducibility of the XVE system, as explained above. However, when the gene to be induced does not allow C-terminal fusions, pENTR-3′ element containing only terminator (nos or 3AT) can be used.
A MultiSite Gateway-Compatible Vector Series for Plant Molecular Biology Studies
The MultiSite Gateway-compatible XVE inducible system and entry clones presented in Table I and Figure 8 have been widely used in our laboratories to generate published and unpublished lines. For example, we developed various fluorescent variants of the popular cytokinin and auxin signaling reporters ARR5:GUS (D’Agostino et al., 2000) and DR5rev:erGFP (Friml et al., 2003), and they showed expression patterns similar to the published versions (Bishopp et al., 2011a). Additionally, we have generated hundreds of inducible lines by using the MultiSite Gateway-compatible XVE system. Our inducible system has already been utilized in published studies to control PIN localization (Dhonukshe et al., 2010) and expression (Bishopp et al., 2011a), auxin signaling (Bishopp et al., 2011a; Mähönen et al., 2014) and conjugation (Xuan et al., 2015), cytokinin degradation (Bishopp et al., 2011b) and signaling (Bishopp et al., 2011a), PLT gene overexpression and RNA interference-mediated silencing (Mähönen et al., 2014), MEMBRANE-ASSOCIATED KINASE REGULATOR4 gene overexpression and artificial microRNA-mediated silencing (Xuan et al., 2015), expression of a microbe-associated molecular pattern (Oome et al., 2014), callose deposition (Bishopp et al., 2011b; Vatén et al., 2011), as well as transcription factors driving phloem development (Furuta et al., 2014). In most of these cases, tissue specificity and inducibility were essential, since constitutive or ubiquitous induction of those genes would lead to drastic or transient phenotypes, making it difficult to assess their primary role in the process.
CONCLUSION
Here, we describe a MultiSite Gateway-based inducible system that allows high-throughput cloning of cell type-specific inducible constructs. The simultaneous recombination of tissue-specific inducible promoters, genes, reporters, and plant selection markers guarantees numerous possibilities for generating inducible transgenic lines. The availability of three fluorescent protein variants, compatible for colocalization studies, can help to unravel the localization of different proteins or to study the signaling of different pathways. These tools will be readily available for the plant science community (for details, see below).
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Analysis
Arabidopsis (Arabidopsis thaliana) Col-0 seeds were grown according to Mähönen et al. (2014) and transferred on one-half-strength GM medium containing 5 µm 17-β-estradiol (Sigma) or an equal volume of DMSO (Sigma) as a control for the described times. 17-β-Estradiol was prepared as 20 mm stock solutions in DMSO and stored at −20°C. The lines DR5rev:erGFP (Friml et al., 2003), pTCSn:erGFP (Zürcher et al., 2013), pG1090:XVE>>axr3-1-RFP, pPLT2:PLT2-YFP, and pPLT2:PLT2-3xYFP (Mähönen et al., 2014) have been published before.
Construction of the Inducible Promoter Entry Clones
The first step to construct MultiSite Gateway-compatible inducible promoter entry clones was to remove the pNOS and HPT coding sequence between the XVE-e9T and nosT-pLexA units present in the original XVE vector, pER8 (Zuo et al., 2000). The pNOS:HPT unit was removed by amplifying first the XVE-e9T and nosT-pLexA units separately and then ligating them together, thus obtaining the XVE-e9T-nosT-pLexA core unit. The cloning procedure proceeded as follows. To obtain the 5′ part of the promoterless version p1R4-ML:XVE containing AscI-AgeI-KpnI-XhoI-SnaBI restriction endonuclease sites for cloning new promoters, XVE-e9T was amplified with primers MLF-attB4 and rbcTR-not1 (for details, see Supplemental Table S1). The 3′ end part, nosT-pLexA, was amplified with primers nostF-not1 and laxR-attB1 using pER8 as template. PCR products of XVE-e9T and nosT-pLexA were digested with NotI, purified, and then ligated together to obtain ML:XVE-e9T-nosT-pLexA. In the second step, the ligated XVE-e9T-nosT-pLexA unit was amplified with MLF-attB4 and laxR-attB1 primers having attB4 and attB1 overhangs, respectively. The amplified fragment was purified and recombined in a Gateway BP reaction with an ampicillin-resistant variant of the pDONRP4-P1R donor vector and then transformed into Escherichia coli. p1R4-pG1090:XVE, p1R4-pWOL:XVE, and p1R4-p6xUAS:XVE were amplified from existing inducible constructs based on the original pER8 vector and containing the above promoters. For cloning, the procedure followed the same protocol as for p1R4-ML:XVE, except that G1090F-attB4 primer was used instead of MLF-attB4 when cloning the 5′ part (for details, see Supplemental Table S1).
The majority of inducible promoter entry clones (Table I) were constructed as follows (for visualization, see Fig. 1). The 5′ regulatory regions of the desired genes were amplified with primers containing restriction endonuclease site overhangs (Supplemental Table S1). The PCR products were digested at the unique cutting sites located in the overhangs and ligated with T4 DNA ligase into one (or two) of the unique sites located in the p1R4-ML:XVE vector (Fig. 1). Alternatively, the digested promoter fragments were cloned by replacing existing promoters located in the p1R4-PROM:XVE vector with digestion and ligation. After identifying the correct plasmid clones with restriction enzyme cleavage site analysis, the 5′ and 3′ ends of the promoters were typically sequenced. The cloned inducible promoter entry clones were directly used in subsequent MultiSite LR Clonase reactions to obtain inducible overexpression constructs to be used in plant transformations (Fig. 1). The recombination reactions were carried out according to the MultiSite Gateway Three-Fragment Vector Construction Kit manual (Thermo Fisher Scientific; catalog no. 12537-023).
When cloning inducible constructs, vectors containing attR4 and attR3 sites can be used as binary destination vectors. These include, for example, the pXm43GW (where X refers to a letter indicating different plant selection markers) vector series by Karimi et al. (2005) or the two new destination vectors, pCAM-hyg-R4R3 (hygromycin resistant in plants) and pCAM-kan-R4R3 (kanamycin resistant in plants), developed in this study. pCAM-hyg-R4R3 was cloned by replacing 35S:HPT sequences of pCAMBIA1300 (CAMBIA) with the pNOS:HPT-attR4-ccdB-CmR-attR3 cassette from a modified pGII0227 (Hellens et al., 2000), with pGII0227-R4R3 as SmaI-PdmI fragment. pCAM-kan-R4R3 was cloned by replacing 35S:HPT sequences of pCAMBIA1300 (CAMBIA) with the pNOS:kanR-attR4-ccdB-CmR-attR3 cassette from a modified pGII0226 (Hellens et al., 2000), with pGII0226-R4R3 as SmaI-PdmI fragment.
Occasional Instability of the LexA Operator Copies in E. coli
We have occasionally noticed that copies of the LexA operator sequences disappear from the XVE entry clones when the plasmids are propagated in E. coli. LexA operator sequences are identical in these copies (Zuo et al., 2000) and therefore might increase the likelihood of recombination. These recombination events are rare in the XVE entry clones; however, we advise to confirm the presence of all LexA operator copies whenever new plasmid preparations have been prepared. Digestion of the XVE entry clones with PstI restriction endonucleases should generate a 527-bp band, if all the copies are present.
Construction of the pENTR-Gene Entry Clones
p221a-GUS was cloned by amplifying GUS (Jefferson et al., 1987)-encoding sequence with primers 221RH_GUS_FP (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGTATGTTACGTCCTGTAGAAACCCCAACCCG-3′) and 221RH_GUS_RP (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTCATTGTTTGCCTCCCTGCTGCGGT-3′) and recombining the purified gene product in a Gateway BP reaction with an ampicillin-resistant variant of the pDONR 221 donor vector.
p221z-erTagRFP was cloned by amplifying ER-targeting erTagRFP (Merzlyak et al., 2007) containing N-terminal signal peptide derived from an Arabidopsis vacuolar basic chitinase (Haseloff et al., 1997) with primers tagF-attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAACAATGAAGACTAATCTTTTTCTCTTTCTCATCT-3′) and tagR-attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTTAAAGCTCATCATGCTTGTGC-3′) and recombining the purified gene product in a Gateway BP reaction with a zeocin-resistant variant of the pDONR 221 donor vector.
p221z-erVenusYFP was cloned by amplifying ER-targeting erVenusYFP (Haseloff et al., 1997; Nagai et al., 2002) with primers VerF-attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAAGCTTAACAATGAAGACTAATCTTTTTCTC-3′) and VerR-attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTATTAAAGCTCATCATGCTTGTACAGC-3′) and recombining the purified gene product in a Gateway BP reaction with a zeocin-resistant variant of the pDONR 221 donor vector.
The entry clone p221k-erTq2CFP was generated in three steps. (1) Sequence encoding GFP and ER-targeting sequence was amplified with the primers GFP-F.attB1 (5′-AAAAAGCAGGCTATGAAGACTAATCTTTTTCTC-3′) and GFP-R.attB2 (5′-AGAAAGCTGGGTTAGGCCCGGGAAGCTCATCATG-3′) from a vector containing ER-targeting sequence, GFP, and EcoRI cutting site. (2) The resulting fragment was recombined in a BP reaction with kanamycin-resistant pDONOR/221 donor vector, producing a fragment with ER-targeting sequence, GFP, and EcoRI restriction site between them. (3) Finally, sequence encoding TURQUOISE2 CYAN FLUORESCENT PROTEIN was amplified from mT2-C1 plasmid (Goedhart et al., 2012) using the forward primer mTq2.EcoRI.F (5′-GCCGAATTCATGGTGAGCAAGGGCGAG-3′) and reverse primer mTq2.SmaI.Rnostop (5′-AGGCCCGGGAAGCTCATCATGCTTGTACAGCTCGTCCATGCC-3′). The 3′ sequence encoding ER retention signal together with the SmaI restriction site were included in the reverse primer sequence. The amplified Tq2 fragment and the entry clone (containing the GFP and ER-targeting sequence) were digested with the restriction enzymes EcoRI and SmaI, and the ligation reaction was set where the GFP fragment was replaced with Tq2 to produce the p221k-erTq2CFP entry clone.
The entry clone p221-gPLT5 was generated by amplifying PLT5 with primers PLT5.aatB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGTATGAAGAACAATAACAACAAATCTTC-3′) and PLT5.aatB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTTCCAACCCAAAAACCGGTGTGTGCA-3′) and recombined with BP reaction into the pDONORzero plasmid.
Construction of the pENTR-3′ Element Entry Clones
p2R3a-nosT was cloned by amplifying the nosT terminator with primers nosTF-attB2 (5′-GGGGACAGCTTTCTTGTACAAAGTGGGGTAGCGATCGTTCAAACATTTGGCA-3′) and nostR-attB3 (5′-GGGGACAACTTTGTATAATAAAGTTGAAGCTTCTGTCGAGGGGGGATCAATTC-3′) and recombining the purified gene product in a Gateway BP reaction with an ampicillin-resistant variant of the pDONR P2R-P3 donor vector.
p2R3a-3AT was generated by amplifying the 3AT terminator with primers 3ATF-attB2 (5′-GGGGACAGCTTTCTTGTACAAAGTGGAATAACAGGCCTCCCAGCTTTCG-3′) and 3ATR2-attB3 (5′-GGGGACAACTTTGTATAATAAAGTTGTGTTTAAACCCTCGACACAAAAAGCCTATACTGTACTTAAC-3′) and recombining the product in a BP reaction as explained above.
p2R3a-VenusYFP-3AT was generated by amplifying VenusYFP (Nagai et al., 2002) with primers glyVenF-attB2 (5′-GGGGACAGCTTTCTTGTACAAAGTGGGTGGTGGTGGCGCGGTG-3′) and 3ATR-attB3 (5′-GGGGACAACTTTGTATAATAAAGTTGCCCTCGACACAAAAAGCCTATACTGTAC-3′) and recombining the product in a BP reaction as explained above.
p2R3a-GUS-OcsT was generated by first amplifying the gene encoding GUS (Jefferson et al., 1987) with the primers GUSF-Hind3 (5′-TATTAAGCTTCGGGAGGTATGTTACGTCCTGTAGAAACCCCAAC-3′) and GUSR-Xho1 (5′-TATTCTCGAGTCATTGTTTGCCTCCCTGCTG-3′). The amplified fragment was then cloned into the HindIII and XhoI sites located upstream of the OcsT sequences of the vector p2R3a-AS-OcsT.
p2R3a-3xVenusYFP-OcsT was generated by first amplifying sequences containing three repeats of genes encoding VenusYFP (Nagai et al., 2002) with the primers 3XYF-Hind3 (5′-TATAAAGCTTCGCAGGCTGGATCCATGGT-3′) and 3XYR-Xho1 (5′-AAAACTCGAGTTAATCATGCTTGTACAGCTCGTCCA-3′). The amplified fragment was then cloned into the HindIII and XhoI sites located upstream of the OcsT sequences of the vector p2R3a-AS-OcsT.
The entry clone p2R3a-Tq2CFP-OcsT was generated by first amplifying the gene encoding TURQUOISE2 CYAN FLUORESCENT PROTEIN (Goedhart et al., 2012) with the primers HindIIITorqF (5′-TATATATTAAAGCTTCGGGTGGTGGTGGCGTGAGCAAGGGCGAGGAGCTG-3′) and XhoITorqR (5′-TATATAATCTCGAGTTACTTGTACAGCTCGTCCATGCCG-3′). The GUS gene of p2R3a-GUS-OcsT was replaced by the amplified product by digesting both the vector and the PCR product with HindIII and XhoI and ligating the fragments together.
The entry clone p2R3a-TagRFP-OcsT was constructed by amplifying the gene encoding TAG RED FLUORESCENT PROTEIN (Merzlyak et al., 2007) with the primers BamHI-4gly-TagRFP (5′-ATTAGGATCCGCGGTGGTGGTGGCAGCGAGCTGATTAAGGAGAA-3′) and TagRFP-XhoI (5′-TTAACTCGAGTCACTTGTGCCCCAGTTTGCTAGGGAGGTCGCAGT-3′). The amplified fragment was cloned into the BamHI and XhoI sites located upstream of the OcsT sequences of the vector p2R3a-AS-OcsT.
GUS Staining
Samples were fixed for 1 h in 90% (v/v) acetone in deionized water and washed for 30 min in ice-cold sodium phosphate buffer (0.05 m, pH 7.2) and an additional 30 min in ice-cold sodium phosphate buffer (0.05 m, pH 7.2) containing 1 mm ferrocyanide and 1 mm ferricyanide. Samples were vacuum infiltrated for 1 min with GUS staining solution (30 mm Na2HPO4, 20 mm NaH2PO4, 1.5 mm ferrocyanide, 1.5 mm ferricyanide, 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 0.1% [v/v] Triton X-100) and incubated from 30 min to overnight at 37°C in darkness. Multiple longitudinal images of the primary root were combined using the Photomerge option in the Adobe Photoshop program (Fig. 3).
Primary Growth Analysis
Primary root growth was calculated by measuring in ImageJ the distance between the position of the root tip immediately after transfer (marked with a black dot) and the position of the root tip after 4 d of treatment. Root meristem size was analyzed according to Perilli and Sabatini (2010), and multiple images were combined using the Photomerge option in the Adobe Photoshop program.
Secondary Growth Analysis
We analyzed the activation stage as in Figure 5 and considered secondary growth to be activated when the procambial cell files present between the primary phloem and xylem had divided into two layers of cell files on both sides of the xylem. We considered the presence of secondary xylem beside the primary xylem also as proof for the activation of secondary growth. Cambium growth was analyzed by measuring in ImageJ the distance between the primary phloem poles.
Histological Analysis
Histological analysis was carried out as described by Idänheimo et al. (2014) on root samples cut around 5 mm below the root-hypocotyl junction. Cross sections in Figure 5 were stained for 30 s in a solution of 0.05% (w/v) Ruthenium Red (Fluka Biochemika) in deionized water.
Microscopy
Histological cross sections were imaged using a Leica 2500 microscope (20× and 40× objectives). Primary roots were imaged using a Leica 2500 microscope (20× and 63× objectives) or an Olympus BX-61 microscope (20× and 40× objectives). The confocal images were acquired with the LAS AF Lite Leica Software using water as an imaging medium, with or without propidium iodide.
Statistical Analysis
Statistical analysis was carried out in SigmaPlot using the Mann-Whitney U statistical test. Five micromolar 17-β-estradiol toxicity experiments were repeated several times with the following results: at the University of Helsinki, four of six experiments showed no inhibition of primary root growth (Figs. 4B and 6), two of two experiments showed no decrease of root meristem size (Fig. 4D), three of three experiments showed no delay in the activation of secondary growth (Fig. 5B), and two of three experiments showed no decrease in cambium growth (Fig. 5D), while two of two experiments at Purdue University showed reduced growth (Supplemental Fig. S1).
Quantification of PLT2 Translational Reporters
The graph in Figure 9H was generated as described by Mähönen et al. (2014).
Distribution of the Plasmids
The cell type-specific XVE inducible entry clones (Table I) are distributed via our laboratory’s Web site: http://www.biocenter.helsinki.fi/bi/mahonen/index.htm. The maps and the sequences of the plasmids are available through this Web site. The reporter and terminator entry clones as well as the binary destination vectors (Fig. 8) are distributed through Addgene: https://www.addgene.org/Ari_Pekka_Mahonen/. The Addgene identifiers for each entry and destination vector are given in parentheses in Figure 8.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of 17-β-estradiol treatment on root growth (at Purdue University).
Supplemental Figure S2. Effects of growth conditions (temperature, light intensity, and relative humidity) on 17-β-estradiol-mediated root growth inhibition.
Supplemental Table S1. Primers to amplify and clone the inducible promoters.
Supplementary Material
Acknowledgments
We thank Nam-Hai Chua and Tatsuo Kakimoto for the pER8 vector, Tuomas Puukko, Anne-Maarit Bågman, and Mikko Herpola for technical assistance, and Tommi Anttonen and Ulla Pirvola for help with the Olympus BX-61 microscope.
Glossary
- Col-0
Columbia-0
- DMSO
dimethyl sulfoxide
- FPs
fluorescent proteins
- ER
endoplasmic reticulum
- MS
Murashige and Skoog
Footnotes
This work was supported by the Academy of Finland and the University of Helsinki (to R.S., X.W., S.R.Y., S.L., I.S., R.U., J.Z., M.W., and A.P.M.), by the Human Frontier Science Program (fellowship no. LT00558/2006–L to A.P.M.), by the Department of Energy, Basic Energy Sciences, grant no. DE-FG02-13ER16405, to A.M., by the Viikki Doctoral Programme in Molecular Biosciences (Ph.D. grant to R.S.), and by the Netherlands Genomic Initiative (Horizon grant no. 050–71–054 to M.G.).
Articles can be viewed without a subscription.
References
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, Marteaux B, Brunoud G, Chambrier P, Rozier F, Mirabet V, Legrand J, Lainé S, et al. (2014) Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505: 417–421 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y (2011a) A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol 21: 917–926 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Lehesranta S, Vatén A, Help H, El-Showk S, Scheres B, Helariutta K, Mähönen AP, Sakakibara H, Helariutta Y (2011b) Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr Biol 21: 927–932 [DOI] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Borghi L. (2010) Inducible gene expression systems for plants. Methods Mol Biol 655: 65–75 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Brand L, Hörler M, Nüesch E, Vassalli S, Barrell P, Yang W, Jefferson RA, Grossniklaus U, Curtis MD (2006) A versatile and reliable two-component system for tissue-specific gene induction in Arabidopsis. Plant Physiol 141: 1194–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo SR, Cançado GMA, Ulian EC, Menossi M (2007) Identification of genes responsive to the application of ethanol on sugarcane leaves. Plant Cell Rep 26: 2119–2128 [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martínez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Claassens MMJ, Verhees J, van der Plas LHW, van der Krol AR, Vreugdenhil D (2005) Ethanol breaks dormancy of the potato tuber apical bud. J Exp Bot 56: 2515–2525 [DOI] [PubMed] [Google Scholar]
- Collier RJ. (1967) Effect of diphtheria toxin on protein synthesis: inactivation of one of the transfer factors. J Mol Biol 25: 83–98 [DOI] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I (2005) New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J 41: 899–918 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CD, Galgoci BF, Irish VF (1995) Genetic ablation of petal and stamen primordia to elucidate cell interactions during floral development. Development 121: 2887–2895 [DOI] [PubMed] [Google Scholar]
- Depicker A, Stachel S, Dhaese P, Zambryski P, Goodman HM (1982) Nopaline synthase: transcript mapping and DNA sequence. J Mol Appl Genet 1: 561–573 [PubMed] [Google Scholar]
- De Rybel B, Möller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith RS, Borst JW, Weijers D (2013) A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell 24: 426–437 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Huang F, Galvan-Ampudia CS, Mähönen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, et al. (2010) Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R, Moses P, Morelli G, Coruzzi G, Chua NH (1986) Expression dynamics of the pea rbcS multigene family and organ distribution of the transcripts. EMBO J 5: 2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Furuta KM, Yadav SR, Lehesranta S, Belevich I, Miyashima S, Heo JO, Vatén A, Lindgren O, De Rybel B, Van Isterdael G, et al. (2014) Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science 345: 933–937 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, van Weeren L, Gadella TW Jr, Royant A (2012) Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun 3: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Harrison GS, Maxwell F, Long CJ, Rosen CA, Glode LM, Maxwell IH (1991) Activation of a diphtheria toxin A gene by expression of human immunodeficiency virus-1 Tat and Rev proteins in transfected cells. Hum Gene Ther 2: 53–60 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B (2004) Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18: 1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hématy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Höfte H (2007) A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol 17: 922–931 [DOI] [PubMed] [Google Scholar]
- Idänheimo N, Gauthier A, Salojärvi J, Siligato R, Brosché M, Kollist H, Mähönen AP, Kangasjärvi J, Wrzaczek M (2014) The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem Biophys Res Commun 445: 457–462 [DOI] [PubMed] [Google Scholar]
- Ishige F, Takaichi M, Foster R, Chua N, Oeda K (1999) A G-box motif (GCCACGTGCC) tetramer confers high-level constitutive expression in dicot and monocot plants. Plant J 18: 443–448 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AA, Hibberd JM, Gay C, Essah PA, Haseloff J, Tester M, Guiderdoni E (2005) Spatial control of transgene expression in rice (Oryza sativa L.) using the GAL4 enhancer trapping system. Plant J 41: 779–789 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Leyser HM, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G (2005) Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B (2014) PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GT, Meyer RC, Messerli G, Gronlund JT, et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152: 2142–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Kitano M, Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Václavíková K, Miyawaki K, Kakimoto T (2008) Cytokinins are central regulators of cambial activity. Proc Natl Acad Sci USA 105: 20027–20031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, et al. (2007) Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods 4: 555. [DOI] [PubMed] [Google Scholar]
- Moore I, Samalova M, Kurup S (2006) Transactivated and chemically inducible gene expression in plants. Plant J 45: 651–683 [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Nieminen K, Blomster T, Helariutta Y, Mähönen AP (2015) Vascular cambium development. The Arabidopsis Book 13: e0177. doi: 10.1199/tab.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812 [DOI] [PubMed] [Google Scholar]
- Oome S, Raaymakers TM, Cabral A, Samwel S, Böhm H, Albert I, Nürnberger T, Van den Ackerveken G (2014) Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc Natl Acad Sci USA 111: 16955–16960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye ZH (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Sabatini S (2010) Analysis of root meristem size development. Methods Mol Biol 655: 177–187 [DOI] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Nagel DH, Kang SE, Bonaldi K, Doherty CJ, Ravelo S, Galli M, Ecker JR, Kay SA (2014) A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Reports 8: 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo H, Demura T, Fukuda H (2004) Spatial and temporal tracing of vessel differentiation in young Arabidopsis seedlings by the expression of an immature tracheary element-specific promoter. Plant Cell Physiol 45: 1529–1536 [DOI] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, White MR, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, et al. (2001) Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J 28: 225–235 [DOI] [PubMed] [Google Scholar]
- Samalova M, Brzobohaty B, Moore I (2005) pOp6/LhGR: a stringently regulated and highly responsive dexamethasone-inducible gene expression system for tobacco. Plant J 41: 919–935 [DOI] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2: 905–909 [DOI] [PubMed] [Google Scholar]
- Tornero P, Chao RA, Luthin WN, Goff SA, Dangl JL (2002) Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell 14: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Sauer N (1995) The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Tsuwamoto R, Yokoi S, Takahata Y (2010) Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Mol Biol 73: 481–492 [DOI] [PubMed] [Google Scholar]
- Ursache R, Miyashima S, Chen Q, Vatén A, Nakajima K, Carlsbecker A, Zhao Y, Helariutta Y, Dettmer J (2014) Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development 141: 1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E (2009) APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA 106: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J, González-García MP, Frigola D, Fàbregas N, Alexiou KG, López-Bigas N, Rivas S, Jauneau A, Lohmann JU, Benfey PN, et al. (2014) Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev Cell 30: 36–47 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R (2003) Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol 133: 1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Audenaert D, Parizot B, Möller BK, Njo MF, De Rybel B, De Rop G, Van Isterdael G, Mähönen AP, Vanneste S, et al. (2015) Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr Biol 25: 1381–1388 [DOI] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B (2013) A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol 161: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.