A protein conserved in the green cell lineage binds quantitatively to PSII monomers at the stromal side of thylakoid membranes and facilitates supercomplex assembly during PSII repair.
Abstract
The remarkable capability of photosystem II (PSII) to oxidize water comes along with its vulnerability to oxidative damage. Accordingly, organisms harboring PSII have developed strategies to protect PSII from oxidative damage and to repair damaged PSII. Here, we report on the characterization of the THYLAKOID ENRICHED FRACTION30 (TEF30) protein in Chlamydomonas reinhardtii, which is conserved in the green lineage and induced by high light. Fractionation studies revealed that TEF30 is associated with the stromal side of thylakoid membranes. By using blue native/Deriphat-polyacrylamide gel electrophoresis, sucrose density gradients, and isolated PSII particles, we found TEF30 to quantitatively interact with monomeric PSII complexes. Electron microscopy images revealed significantly reduced thylakoid membrane stacking in TEF30-underexpressing cells when compared with control cells. Biophysical and immunological data point to an impaired PSII repair cycle in TEF30-underexpressing cells and a reduced ability to form PSII supercomplexes after high-light exposure. Taken together, our data suggest potential roles for TEF30 in facilitating the incorporation of a new D1 protein and/or the reintegration of CP43 into repaired PSII monomers, protecting repaired PSII monomers from undergoing repeated repair cycles or facilitating the migration of repaired PSII monomers back to stacked regions for supercomplex reassembly.
Oxygenic photosynthesis is essential for almost all life on Earth, as it provides the reduced carbon and the oxygen required for respiration. A key enzyme in oxygenic photosynthesis is PSII, which catalyzes the light-driven oxidation of water. The core of PSII in algae and land plants contains D1 (PsbA), D2 (PsbD), CP43 (PsbC), CP47 (PsbB), the α-subunit (PsbE) and β-subunit (PsbF) of cytochrome b559, as well as several intrinsic low-molecular-mass subunits. The core monomer is associated with the extrinsic oxygen-evolving complex (OEC) consisting of OEE1 (PSBO), OEE2 (PSBP), and OEE3 (PSBQ), which stabilize the inorganic Mn4O5Ca cluster required for water oxidation (for review, see Pagliano et al., 2013). PSII core monomers assemble into dimers to which, at both sides, light-harvesting proteins (LHCII) bind to form PSII supercomplexes. In land plants, each PSII dimer binds two each of the monomeric minor LHCII proteins CP24, CP26, and CP29 in addition to up to four major LHCII trimers (Caffarri et al., 2009; Kouřil et al., 2011). Biochemical evidence suggests that, in the thylakoid membrane, up to eight LHCII trimers can be present per PSII core dimer, presumably because of the existence of a pool of extra LHCII (Kouřil et al., 2013). In Chlamydomonas reinhardtii, lacking CP24, each PSII dimer binds two each of the CP26 and CP29 monomers as well as up to six major LHCII trimers (Tokutsu et al., 2012). The reaction center proteins D1 and D2 bind all the redox-active cofactors required for PSII electron transport (Umena et al., 2011). Light captured by the internal antenna proteins CP43 and CP47 and the outer antenna induces charge separation in PSII, which in turn enables the OEC to oxidize water and provide electrons to the electron transfer chain. In land plants and green algae, PSII supercomplexes are localized to stacked regions of the thylakoid membranes, while the synthesis of PSII cores is considered to take place in stroma lamellae.
A particular feature of PSII is its vulnerability to light, with the D1 protein being a target of light-induced damage and the damage being proportional to the photon flux density (PFD) applied (Tyystjärvi and Aro, 1996). To cope with this damage, an elaborate, highly conserved repair mechanism has evolved termed the PSII repair cycle, during which damaged PSII complexes are partially disassembled and the defective D1 protein is replaced by a de novo synthesized copy (for review, see Nixon et al., 2010; Komenda et al., 2012; Mulo et al., 2012; Nath et al., 2013a; Nickelsen and Rengstl, 2013; Tyystjärvi, 2013; Järvi et al., 2015). Photodamage occurs at all light intensities, but when the rate of damage exceeds the capacity for repair, photoinhibition is manifested as a decrease in the proportion of active PSII reaction centers (Aro et al., 1993). While PSII photodamage occurs in the supercomplexes in the stacked membrane regions, the replacement of damaged D1 takes place in stroma lamellae (Aro et al., 2005). Thus, the PSII repair cycle requires the lateral migration of PSII complexes, which is impaired by the macromolecular crowding in stacked thylakoid membranes (Kirchhoff, 2014). Lateral migration of damaged PSII complexes is facilitated by thylakoid membrane unfolding and PSII supercomplex disassembly. Both processes are enhanced by the phosphorylation of the PSII core subunits D1, D2, CP43, and PsbH, which is mainly mediated by the protein kinase STATE TRANSITION8 (STN8; Tikkanen et al., 2008; Fristedt et al., 2009; Herbstová et al., 2012; Nath et al., 2013b; Wunder et al., 2013). Efficient PSII supercomplex disassembly also requires the THYLAKOID FORMATION1 (THF1)/NON-YELLOW COLORING4 (NYC4)/Psb29 protein (Huang et al., 2013; Yamatani et al., 2013). After the migration of PSII monomers to unstacked thylakoid regions, PSII core subunits are dephosphorylated by the PSII core phosphatase PBCP (Samol et al., 2012), which is required for the efficient degradation of D1 (Koivuniemi et al., 1995; Rintamäki et al., 1996; Kato and Sakamoto, 2014). Degradation of D1 is subsequently realized by the membrane-integral FtsH protease (Lindahl et al., 2000; Silva et al., 2003) and by lumenal and stromal Deg proteases (Haussühl et al., 2001; Kapri-Pardes et al., 2007; Sun et al., 2010). Degradation is assisted by the THYLAKOID LUMEN PROTEIN18.3 (TLP18.3), presumably by its phosphatase activity and ability to interact with lumenal Deg1 (Sirpiö et al., 2007; Wu et al., 2011; Zienkiewicz et al., 2012). D1 proteolysis follows the partial disassembly of the PSII complex, during which CP43 and low-molecular-mass subunits are released to generate a CP43-free PSII monomer (Aro et al., 2005). Thereafter, a newly synthesized D1 copy is cotranslationally inserted from a plastidial 70S ribosome into the thylakoid membrane and processed by the CARBOXYL TERMINAL PEPTIDASE A (CTPA; Zhang et al., 1999, 2000; Che et al., 2013). In Arabidopsis (Arabidopsis thaliana), the D1 synthesis rate appears to be negatively regulated by the PROTEIN DISULFIDE ISOMERASE6 (PDI6; Wittenberg et al., 2014). Moreover, yet unknown steps during PSII repair require the stromal cyclophilin ROTAMASE CYP4 and stromal HEAT SHOCK PROTEIN70 (Schroda et al., 1999; Yokthongwattana et al., 2001; Cai et al., 2008). The PSII repair cycle is completed by the reassembly of the CP43 protein, ligation of the OEC, back migration of PSII to stacked membrane regions, and supercomplex formation. Except for CtpA, all mentioned factors appear to be specific for PSII repair, while many more auxiliary factors play roles in PSII de novo synthesis and repair (for review, see Järvi et al., 2015).
In this study, we report on the functional characterization of the THYLAKOID ENRICHED FRACTION30 (TEF30) protein in C. reinhardtii. In this organism, TEF30 was first identified in a proteomics study on isolated thylakoid membranes (Allmer et al., 2006). TEF30 attracted our attention because its abundance increased 1.7-fold in membrane-enriched fractions of C. reinhardtii cells that had been shifted from 41 to 145 µmol photons m−2 s−1 for 8 h (Mettler et al., 2014; Supplemental Fig. S1). The TEF30 ortholog in Arabidopsis M-ENRICHED THYLAKOID PROTEIN1 (MET1; where M stands for mesophyll cells) was functionally characterized only recently (Bhuiyan et al., 2015). Both MET1 and TEF30 interact quantitatively with monomeric PSII core particles at the stroma side of the thylakoid membranes and play a role in the assembly of PSII monomers and/or their migration to stacked membrane regions for supercomplex assembly. While MET1 appears to exert this function during PSII de novo biogenesis and during the PSII repair cycle in Arabidopsis, TEF30 appears to function exclusively during PSII repair in C. reinhardtii.
RESULTS
TEF30 Is an Abundant Chloroplast Protein Capable of Self-Oligomerization
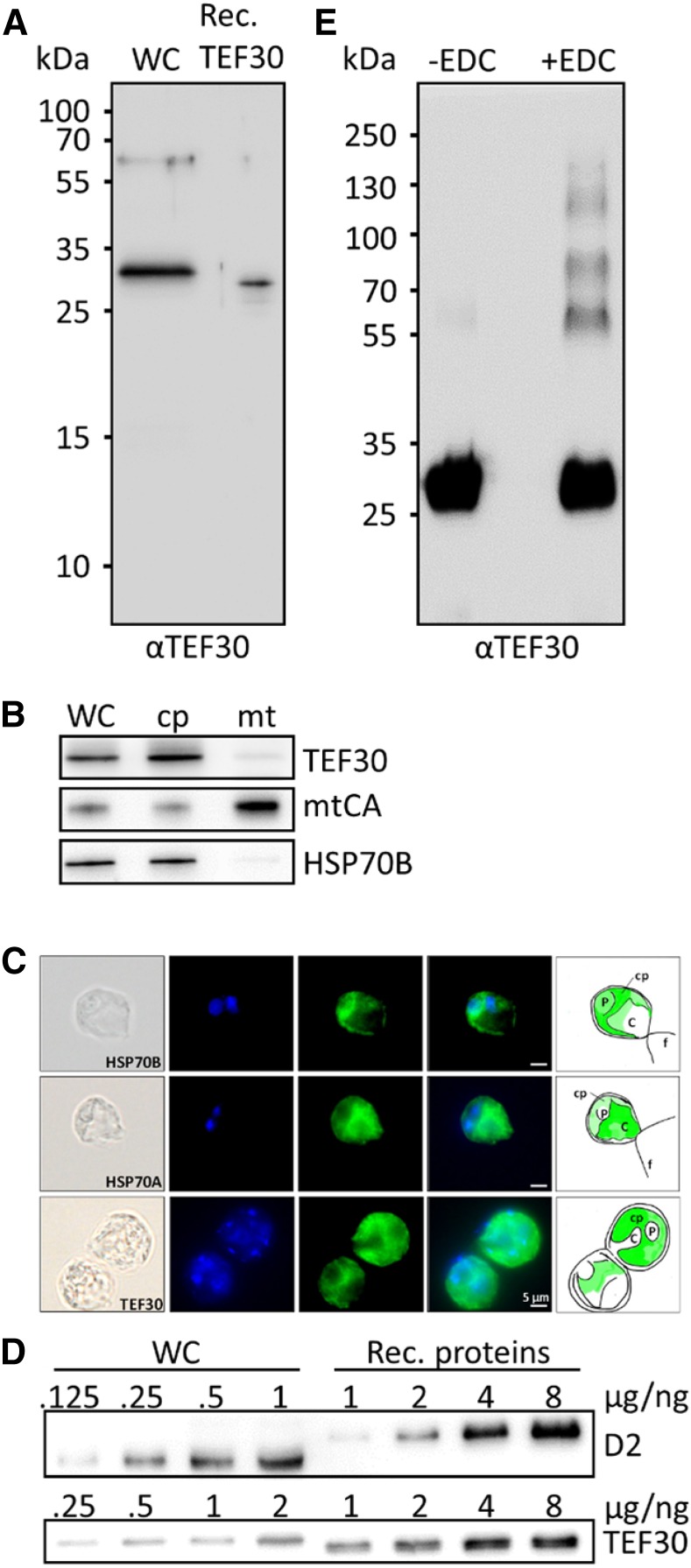
As shown in Supplemental Figure S2A, TEF30 proteins are well conserved in the green lineage. The N termini of TEF30 proteins are predicted to qualify as chloroplast transit peptides, and all TEF30 homologs contain PDZ and TPR domains known to mediate protein-protein interactions (Allan and Ratajczak, 2011; Ye and Zhang, 2013; Supplemental Fig. S2B). Following the 36-amino acid chloroplast transit peptide predicted by ChloroP (Emanuelsson et al., 1999), C. reinhardtii TEF30 (but not its homologs from Arabidopsis or moss) harbors sequence motifs characteristic for thylakoid TAT pathway targeting signals (i.e. an RRXXφ twin-Arg motif and an AXA processing site, with X standing for any amino acid and φ for Leu, Val, Phe, or Met; Albiniak et al., 2012). The region between both motifs is supposed to be hydrophobic; however, this is not the case in C. reinhardtii TEF30. The calculated molecular mass of C. reinhardtii TEF30 lacking the predicted 36-amino acids chloroplast transit peptide is 33.3 kD, while it is 30.4 kD if the putative TAT targeting signal was processed. As shown in Figure 1A, mature TEF30 on SDS gels with C. reinhardtii whole-cell protein extracts migrated slightly above recombinant TEF30, having a calculated molecular mass of 31.4 kD (in recombinant TEF30, the predicted chloroplast and TAT targeting signals were replaced with a hexa-His tag; Supplemental Fig. S2B). This indicates that the predicted chloroplast transit peptide is cleaved off in mature TEF30, while the predicted TAT signal is not. Perhaps the incomplete TAT signal mediates TEF30’s targeting to the thylakoid periphery? Mature TEF30 was detected by immunoblot analyses in isolated chloroplasts but hardly in isolated mitochondria (Fig. 1B). Moreover, immunofluorescence microscopy revealed TEF30 to be confined to the cup-shaped chloroplast (Fig. 1C), thus indicating that TEF30 is a chloroplast protein.
Figure 1.
Antibody characterization and analysis of TEF30 oligomer formation, cellular abundance, and intracellular localization. A, Immunodetection of TEF30 in cw15-325 whole-cell proteins (WC) corresponding to 2 µg of chlorophyll compared with 0.25 ng of recombinant (Rec.) TEF30 protein. B, Subcellular localization of TEF30 by immunoblotting. Ten or 3 µg (depending on the antiserum used) of protein from whole cells, chloroplasts (cp), and mitochondria (mt) isolated from strain cw15-302 was separated by SDS-PAGE and immunodetected with antisera against TEF30, mitochondrial carbonic anhydrase (mtCA), and stromal HSP70B. C, Microscopy images taken from strain cw15-325. Shown from left to right are differential interference contrast images, 4′,6-diamino-phenylindole (DAPI) staining, immunofluorescence, merge of DAPI and immunofluorescence, and a schematic drawing that combines information from the images. Antisera used for immunofluorescence were against HSP70B and HSP70A as markers for stroma and cytosol, respectively, and against TEF30. C, Cytosol; f, flagella; P, pyrenoid. Immunofluorescence signal is shown in green. D, One to 8 ng of recombinant proteins TEF30 (purified from E. coli) and D2 (purchased from Agrisera) was separated by SDS-PAGE together with cw15-325 whole-cell proteins corresponding to 0.125 to 2 µg of chlorophyll and immunodetected with antibodies against TEF30 and D2. E, Recombinant TEF30 protein was incubated with EDC/N-hydroxysuccinimide, separated on an SDS-polyacrylamide gel, and immunodetected with antibodies against TEF30.
The routine detection of TEF30 in shotgun proteomics approaches on C. reinhardtii indicates that it is a well-expressed protein (Hemme et al., 2014; Mettler et al., 2014; Ramundo et al., 2014; Schmollinger et al., 2014). To get an estimate of its cellular abundance, we separated C. reinhardtii whole-cell proteins next to recombinant TEF30 and D2 proteins on SDS gels and quantified the signals obtained from immunoblot analyses (Fig. 1D). These analyses revealed TEF30 and D2 to represent 0.05 ± 0.007 ng µg−1 chlorophyll (n = 4) and 0.59 ± 0.016 ng µg−1 chlorophyll (n = 3), respectively.
The PDZ and TPR domains in TEF30 are likely to mediate protein-protein interactions. To test whether TEF30 is capable of self-oligomerization, recombinant TEF30 was subjected to 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)-mediated zero-length cross-linking and separated by SDS-PAGE. As shown in Figure 1E, untreated recombinant TEF30 migrated with an apparent molecular mass of approximately 30 kD, while EDC-treated TEF30 also migrated with apparent masses of approximately 60, 90, 120, and 150 kD, thus indicating that TEF30 is indeed capable of self-oligomerization.
TEF30 Is Localized to the Stromal Side of Thylakoid Membranes
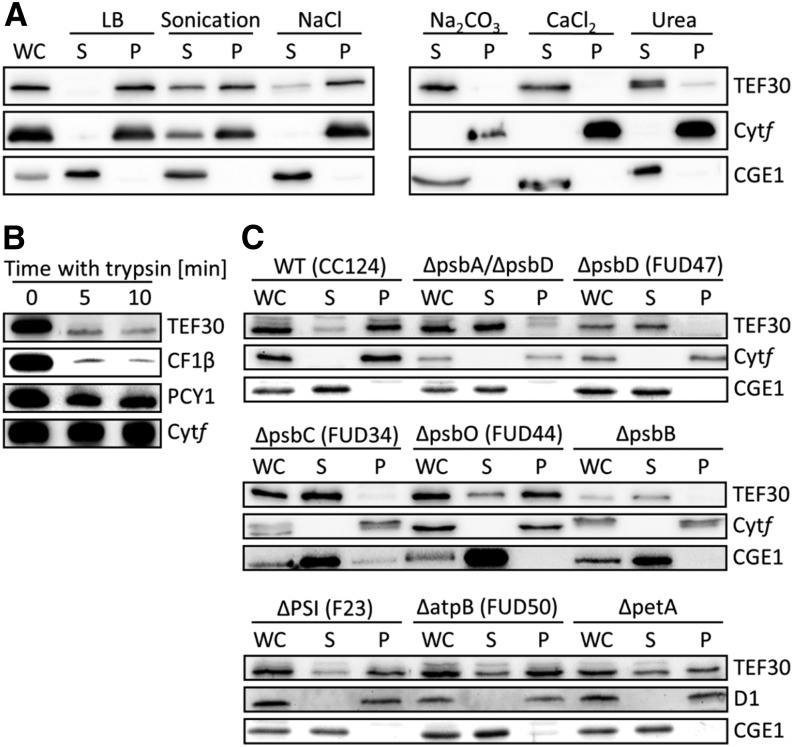
TEF30 was detected previously in C. reinhardtii thylakoid membrane fractions (Allmer et al., 2006). To verify TEF30’s thylakoid localization, C. reinhardtii cells were separated into soluble and membrane-enriched fractions by freezing/thawing, and fractions were analyzed by immunoblotting. As shown in Figure 2A, TEF30, like integral membrane protein cytochrome f, was only detected in the membrane-enriched pellet, while stromal CHLOROPLAST GrpE HOMOLOG1 (CGE1) was only found in the soluble fraction. Cell lysis by sonication had no effect on CGE1’s exclusive localization to the soluble fraction but resulted in a partial localization of TEF30 and cytochrome f to the soluble fraction, presumably because sonication created small thylakoid membrane vesicles that were not precipitated by centrifugation. Incubation of cells lysed by freezing/thawing in the presence of NaCl, Na2CO3, CaCl2, or urea had no effect on the exclusive localization of cytochrome f to the pellet and of CGE1 to the soluble fraction. However, NaCl led to a partial, and Na2CO3, CaCl2, and urea led to a virtually complete, shifting of TEF30 to the soluble fraction (Fig. 2A). Hence, TEF30 appears to be associated with thylakoid membranes rather than embedded into them, which is in agreement with the TMHMM algorithm (Sonnhammer et al., 1998) predicting TEF30 not to contain any transmembrane helices.
Figure 2.
Localization of TEF30 in thylakoids of C. reinhardtii wild-type and mutant cells. A, Salt treatments during cell fractionation. Whole cells (WC) of strain cw15-325 were resuspended in lysis buffer (LB) containing the indicated salts, subjected to freezing/thawing, and separated into soluble (S) and pellet (P) fractions. One sample resuspended in lysis buffer only was subjected to sonication instead of freezing/thawing. Proteins were separated by SDS-PAGE and immunodetected with antibodies against TEF30 and against integral membrane protein cytochrome f (Cytf) and stromal CGE1 as controls. B, Trypsin treatment of thylakoid membranes. Isolated thylakoids from strain cw15-325 were incubated on ice with trypsin, separated by SDS-PAGE, and immunodetected with antibodies against TEF30 and the peripheral CF1β subunit of the ATPase complex, lumenal plastocyanin (PCY1), and integral membrane protein cytochrome f as controls. C, Cell fractionation of various photosynthesis mutants. Whole cells of the wild type (WT) and thylakoid membrane protein mutants were separated into soluble and pellet fractions via freezing/thawing. Proteins were separated by SDS-PAGE and immunodetected with antibodies against TEF30 and against integral membrane protein cytochrome f or D1 and stromal CGE1 as controls.
To determine whether TEF30 is associated with the stromal or the lumenal side of thylakoid membranes, thylakoid membranes from cells lysed by freezing/thawing were purified by flotation on Suc step gradients and incubated with trypsin (Fig. 2B). While the beta-subunit of chloroplast coupling factor 1 (CF1β), situated at the stromal side of thylakoid membranes, was almost completely degraded by trypsin, lumenal plastocyanin was degraded only to a small extent and integral membrane protein cytochrome f was not degraded at all. TEF30 was degraded to a similar extent as CF1β, indicating that TEF30 is associated with the stromal side of thylakoid membranes.
To test whether TEF30’s association with thylakoid membranes was due to its interaction with one of the major thylakoid membrane protein complexes, we fractionated C. reinhardtii mutants devoid or depleted of PSII, PSI, cytochrome b6/f, or the ATP synthase by freezing/thawing into soluble and membrane-enriched fractions and analyzed the localization of TEF30 by immunoblotting. Although cell-walled strains were used (in contrast to the experiments shown in Figure 2A, which were done with a cell wall-deficient strain), TEF30 in wild-type cells was again mainly detected in membrane-enriched fractions (Fig. 2C). Also in PSI, ATP synthase, and cytochrome b6/f mutants, most of the TEF30 protein was detected in membrane-enriched fractions, suggesting that TEF30 was not associated with one of these complexes. In contrast, virtually all TEF30 shifted to soluble fractions in mutants affected in the core subunits of PSII (PsbA–PsbD), while TEF30 retained its location to membrane-enriched fractions in a mutant lacking the O subunit of the OEC of PSII. As this mutant still accumulates considerable amounts of PSII core subunits (de Vitry et al., 1989), our combined data indicate that TEF30 is associated with (a) PSII core subunit(s) exposed to the stromal side of the thylakoid membranes.
TEF30 Copurifies with PSII Particles
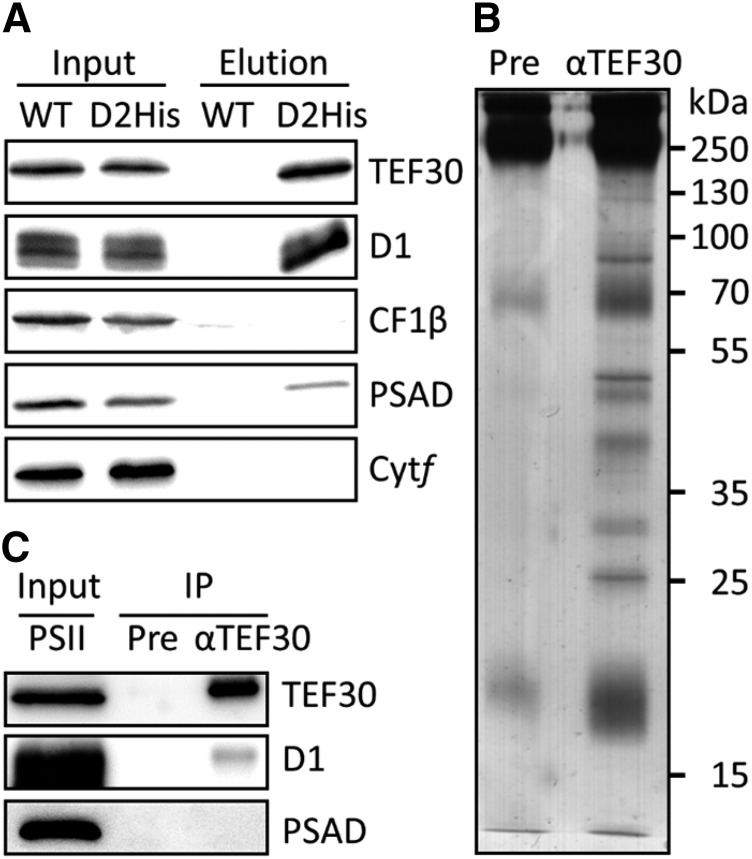
To verify the interaction of TEF30 with PSII, we took advantage of a C. reinhardtii strain (D2His) in which the D2 subunit was genetically engineered to contain a hexa-His tag at its C terminus (Sugiura et al., 1998). Detergent-solubilized thylakoid membranes prepared from wild-type and D2His cells were subjected to nickel-affinity chromatography, and bound proteins were eluted with imidazole. Immunoblot analyses revealed TEF30 and D1 to be amply present, the D-subunit of PSI (PSAD) to be present in small amounts, and CF1β and cytochrome f not to be present in eluates from the D2His strain (Fig. 3A). None of these proteins was detected in control eluates from the wild-type strain. To make sure that TEF30 was not interacting with PSI present as a contamination in the PSII preparation, we immunoprecipitated TEF30 from purified PSII particles derived from the D2His strain. As shown in Figure 3, B and C, several proteins coprecipitated with TEF30 but not with preimmune serum. Among the coprecipitating proteins was D1 but not PSAD, indicating that TEF30 interacts tightly with PSII particles and not with PSI.
Figure 3.
Copurification of TEF30 with PSII particles. A, Nickel-affinity purification of PSII particles. Detergent-solubilized thylakoid membranes from a C. reinhardtii strain harboring a hexa-His-tagged D2 protein (D2His) and from wild-type control strain cc124 (WT) were subjected to nickel-affinity purification. Thylakoid membrane input proteins corresponding to 1 µg of chlorophyll and 4% of the imidazole eluate were separated by SDS-PAGE and immunodetected with antibodies against TEF30 and proteins representative for the major thylakoid membrane complexes. B, Silver staining of proteins immunoprecipitated from hexa-His-tagged PSII particles with preimmune serum (Pre) and antibodies against TEF30. C, Immunodetection of proteins immunoprecipitated from PSII particles. Purified PSII particles equivalent to approximately 1 µg of chlorophyll (Input) and 20% of the immunoprecipitates obtained with preimmune serum and antibodies against TEF30 (IP) were separated by SDS-PAGE and immunodecorated with the antisera indicated.
TEF30 Comigrates with PSII Core Subunits on Suc Density Gradients and Blue Native Gels
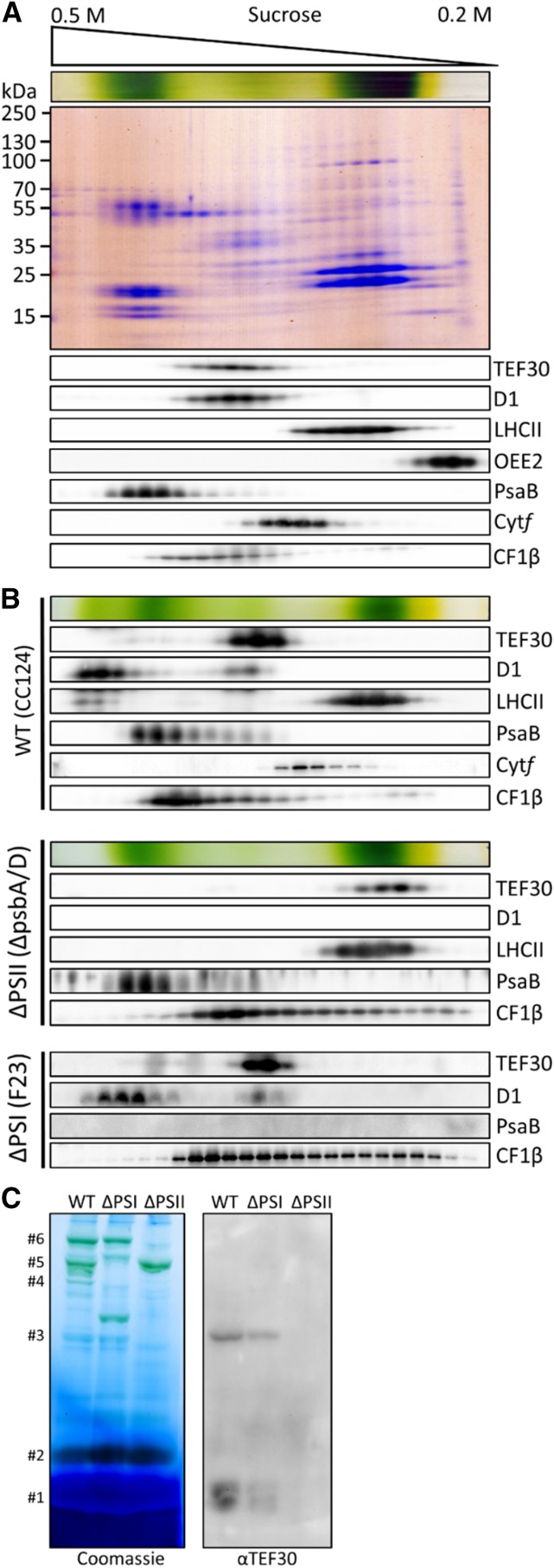
To further substantiate our finding that TEF30 interacts with PSII core particles, we isolated thylakoid membranes from cell wall-deficient C. reinhardtii cells by flotation on Suc step gradients and, following detergent solubilization, separated protein complexes on Suc density gradients. Immunoblot analyses of fractions taken from these gradients revealed that sample processing resulted in the separation of PSII cores from LHCII and the OEC. This was observed before when Suc density gradient centrifugation was employed to analyze PSII supercomplex compositions (Caffarri et al., 2009; Tokutsu et al., 2012) and, in our hands, for unknown reasons occurred predominantly with cells lacking cell walls. TEF30 clearly comigrated with monomeric PSII core complexes (Drop et al., 2014) and not with LHCII, the OEC, PSI, the cytochrome b6/f complex, or the ATP synthase (Fig. 4A).
Figure 4.
Comigration of TEF30 with PSII core subunits as revealed by Suc density gradient centrifugation and blue native (BN)-PAGE. A, Thylakoid membranes from strain cw15-325 (1 mg chlorophyll mL−1) were solubilized with 1% β-dodecyl maltoside (β-DDM) and separated on a linear 0.2 to 0.5 m Suc gradient at 205,000g for 16 h. Proteins were collected in 25 fractions, separated by SDS-PAGE, and detected with Colloidal Blue staining (top gel) or immunologically with the antisera indicated (bottom gels). B, Analysis of thylakoid membrane protein complexes by Suc density gradient centrifugation as described in A on wild-type (WT) strain cc124, PSI mutant F23, and PSII mutant ΔpsbA/D. C, Analysis of thylakoid membrane protein complexes by BN-PAGE. Thylakoids from wild-type strain cc124, PSI mutant F23, and PSII mutant nac2 were solubilized with 1% β-DDM, and proteins corresponding to 5 µg of chlorophyll were separated on a 5% to 15% polyacrylamide BN gradient gel, followed by Coomassie Blue staining (left gel) and immunodetection with antibodies against TEF30 (right gel). Protein bands were assigned to LHCII monomers (#1), LHCII trimers (#2), PSII monomers (#3), PSII dimers (#4), PSI supercomplexes (#5), and PSII supercomplexes (#6) according to Järvi et al. (2011).
Suc density gradients were also run with solubilized thylakoid membranes isolated from cell-walled cells of wild-type, PSII mutant, and PSI mutant strains. Here, sample processing left most PSII supercomplexes intact (Fig. 4B). On gradients run with wild-type thylakoids, TEF30 comigrated again quantitatively with monomeric PSII core complexes, and only traces of TEF30 comigrated with larger PSII assemblies that were still smaller than fully assembled PSII supercomplexes. TEF30 also did not comigrate with LHCII, PSI, the cytochrome b6/f complex, or the ATP synthase. While the quantitative comigration of TEF30 with monomeric PSII core complexes was not altered in gradients run with thylakoids from the F23 PSI mutant, TEF30 was found to migrate much less deeply into the gradient, to a position even above LHCII, in gradients run with thylakoids from the ΔpsbA/D PSII mutant (Fig. 4B). Solubilized thylakoids from cell-walled wild-type and mutant strains were also separated by BN-PAGE and analyzed by immunoblotting. As shown in Figure 4C, TEF30 was again found to comigrate quantitatively with PSII monomers, but not with PSII supercomplexes, in gel lanes containing solubilized thylakoids from wild-type and PSI mutant strains, while TEF30 was not detected in lanes with thylakoids from the nac2 PSII mutant strain. In summary, these results confirm that TEF30 is specifically associated with monomeric PSII core complexes.
PSII in TEF30-Artificial MicroRNA Cells Is More Susceptible to Photoinhibition Than PSII in Control Cells
To get insights into the function of TEF30 in C. reinhardtii chloroplasts, we generated strains constitutively expressing artificial microRNAs (TEF30-amiRNAs) targeting the coding region of TEF30 transcripts based on the pChlamiRNA2 vector (Molnar et al., 2009). Strains cw15-325 and cw15-302 were used as recipients, and in both we could routinely identify transformants in which TEF30 protein levels were reduced to less than 20% of wild-type levels (Supplemental Fig. S3). When comparing cells from control and TEF30-amiRNA strains, we observed no obvious morphological differences (as judged from light microscopy) and no differences in growth under heterotrophic, mixotrophic, or photoautotrophic conditions at various light and temperature regimes (Supplemental Fig. S4).
As we found membrane-localized TEF30 to accumulate in response to higher light intensities and to be associated with PSII core complexes (Figs. 2–4; Supplemental Fig. S1), we reasoned that TEF30 might play a role in the protection/repair of PSII from photoinhibition. To test this idea, we monitored PSII maximum quantum efficiency (Fv/Fm) in cells from control and TEF30-amiRNA strains grown at approximately 30 µmol photons m−2 s−1 that had been shifted to high light (HL) at a PFD of 1,800 µmol photons m−2 s−1 for 60 min and then allowed to recover at approximately 30 µmol photons m−2 s−1 for 3 h. As shown in Figure 5A, PSII maximum quantum efficiency declined more strongly and recovered more slowly in TEF30-amiRNA cells as compared with control cells. The same was observed by 77 K fluorescence emission spectroscopy (Supplemental Fig. S5). To test whether this apparent higher sensitivity of PSII to HL in TEF30-amiRNA cells was also reflected at the level of PSII subunit accumulation, we shifted cells from control and TEF30-amiRNA strains grown at approximately 30 µmol photons m−2 s−1 to HL of 800 µmol photons m−2 s−1 for 6 h and monitored the accumulation of subunits of all four major thylakoid membrane complexes, as well as of TEF30, the VESICLE-INDUCING PROTEIN IN PLASTIDS1 (VIPP1), and the LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 (LHCSR3), by immunoblotting (Fig. 5B). VIPP1 and LHCSR3 were monitored as controls because they are strongly induced by HL (Supplemental Fig. S1; Naumann et al., 2005; Nordhues et al., 2012). VIPP1 was suggested to play a role in the organization of lipid microdomains in thylakoid membranes (Rütgers and Schroda, 2013), whereas LHCSR3 was shown to be essential for the induction of nonphotochemical quenching (NPQ) in C. reinhardtii (Peers et al., 2009).
Figure 5.
PSII activity and levels of PSII core subunits are more sensitive to HL intensities or low temperatures in TEF30-amiRNA cells than in control cells. A, cw15-325 control (Con) and TEF30-amiRNA strains were photoinhibited (PI) for 60 min at 1,800 μmol photons m–2 s–1, then allowed to recover at 30 μmol photons m–2 s–1. PSII fluorescence was measured by pulse amplitude-modulated fluorometry. Values represent means from three biological replicates, and error bars indicate sd. B, Control and TEF30-amiRNA #1.48 cells in the cw15-325 strain background were exposed to high PFDs (HL) of approximately 800 µmol photons m−2 s−1 for 6 h. Whole-cell proteins corresponding to 0.2, 1, or 2 µg of chlorophyll (depending on the antiserum used) were separated on 10% or 14% SDS-polyacrylamide gels and analyzed by immunoblotting. C, Control and TEF30-amiRNA #2.48 cells in the cw15-325 strain background were grown at a PFD of approximately 30 μmol photons m–2 s–1 and then shifted from a growth temperature of 25°C to 10°C for 18 h. Samples were analyzed as in B.
While TEF30 was virtually absent in the TEF30-amiRNA strain used, it accumulated during HL exposure in cells of the control strain, thus confirming the proteomics data (Supplemental Fig. S1). The exposure to HL in neither strain affected the accumulation of cytochrome f, CF1β, LHCII, or the CP26/CP29 minor antenna. In contrast, levels of OEE1/2 proteins declined only in TEF30-amiRNA cells. Moreover, levels of D1, D2, and CP43 declined in control and TEF30-amiRNA cells, but to a much larger extent in the latter, indeed indicating a role of TEF30 in the protection/repair of PSII from photoinhibition. We noted elevated levels of PsaA in TEF30-amiRNA cells that declined during HL treatment to similar levels as did PsaA in control cells. This observation appears to be specific to the strain used here, as we did not detect elevated PsaA levels in other TEF30-amiRNA strains.
Significant photoinhibition of PSII is known to occur also at low light intensities under chilling conditions (Greer et al., 1988; Li et al., 2004). This is because temperature-dependent reactions during the PSII repair cycle occur with reduced pace at low temperatures, while photochemical reactions are temperature independent (Tyystjärvi, 2013). To test whether TEF30-amiRNA cells are also more prone to photoinhibition under chilling conditions, we shifted control and TEF30-amiRNA cells grown at approximately 30 µmol photons m−2 s−1 from 25°C to 10°C for 18 h and monitored the levels of TEF30 and of marker subunits of the four major thylakoid membrane complexes by immunoblotting. As shown in Figure 5C, chilling led to a slight increase in TEF30 protein levels in control cells, but in cells of neither strain did chilling affect the levels of PSAD, cytochrome f, and CF1β. However, chilling did induce a decline of D1 levels in TEF30-amiRNA cells, while it did not affect D1 levels in control cells, therefore corroborating the notion that TEF30 plays a role in the protection/repair of PSII from photoinhibition.
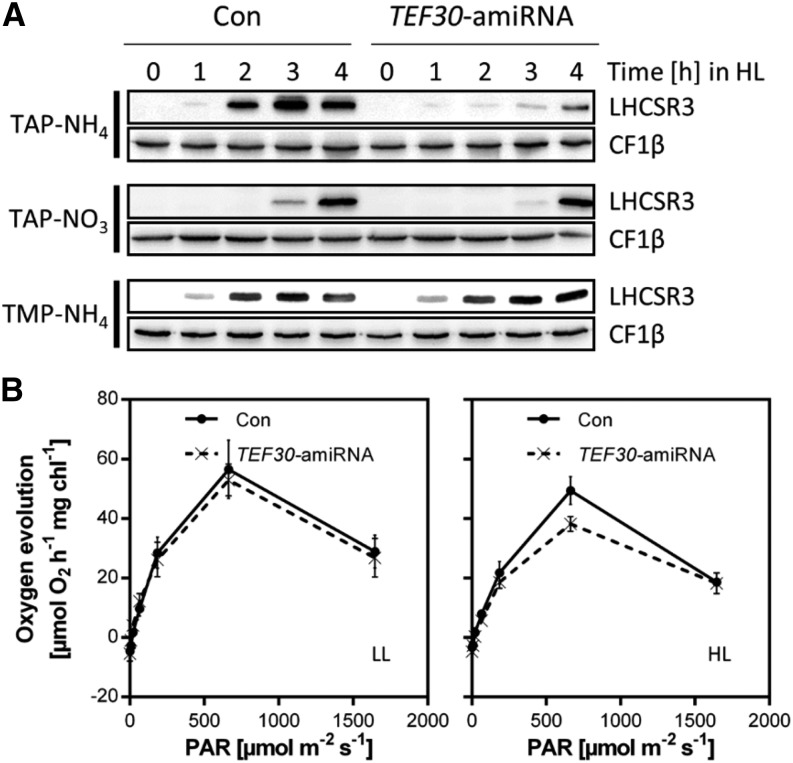
We made the interesting observation that the accumulation of the LHCSR3 protein in HL was impaired in TEF30-amiRNA cells when compared with control cells, while that of VIPP1 was unaffected (Fig. 5B). The reduced accumulation of the LHCSR3 protein in TEF30-amiRNA cells was only observed at time points later than 1 h of HL exposure and, as revealed by quantitative reverse transcription (qRT)-PCR, was due to a reduced induction of LHCSR3 gene expression (Fig. 6A; Supplemental Fig. S6A). Moreover, this effect was only observed in cells grown under mixotrophic conditions with ammonium as nitrogen source (TAP-NH4 medium), while it was not observed in cells grown mixotrophically on nitrate (TAP-NO3) or photoautotrophically on ammonium (TMP-NH4). Accordingly, the induction of NPQ was similar in photoautotrophically grown control and TEF30-amiRNA cells (Supplemental Fig. S6B).
Figure 6.
Comparison of LHCSR3 expression and oxygen evolution in control and TEF30-amiRNA cells. A, Time course of LHCSR3 accumulation at 600 μmol photons m–2 s–1 (HL) in cw15-325 control (Con) and TEF30-amiRNA cells grown in Tris-acetate-phosphate (TAP)-NH4, TAP-NO3, or Tris-minimal-phosphate (TMP)-NH4. Whole-cell proteins were separated by SDS-PAGE and immunodetected with antibodies against LHCSR3 and against CF1β as loading control. B, Oxygen evolution from thylakoid membranes isolated from cw15-325 control and TEF30-amiRNA cells grown in TAP-NH4 medium at LL of approximately 30 µmol photons m−2 s−1 or HL of approximately 800 µmol photons m−2 s−1 for 4 h. Values represent means from experiments on two independent control and TEF30-amiRNA lines, and error bars indicate sd. (Note that the same thylakoid preparation was used for all PFDs applied, thus explaining the decline in oxygen evolution at 1,600 µmol photons m−2 s−1.)
The induction of LHCSR3 gene expression was shown to correlate with the PFD applied and to be abolished in the presence of the PSII inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea; therefore, it appears to correlate with the rate of linear electron flow (LEF; Naumann et al., 2007; Petroutsos et al., 2011; Maruyama et al., 2014). Therefore, a smaller LEF in TEF30-amiRNA cells versus control cells grown mixotrophically in the presence of ammonium and exposed to HL could provide an explanation for the reduced induction of LHCSR3 gene expression in TEF30-amiRNA cells. This decrease in LEF might be a consequence of an increased fraction of photodamaged PSII in TEF30-amiRNA cells. To test this idea, we measured oxygen evolution as a direct measure for LEF rates on thylakoid membranes isolated from TEF30-amiRNA and control cells grown at low light (LL; approximately 30 µmol photons m−2 s−1) or HL (approximately 800 µmol photons m−2 s−1) for 4 h. As shown in Figure 6B, the oxygen evolution capacity at 600 µmol photons m−2 s−1 was indeed approximately 25% lower in HL-grown TEF30-amiRNA cells compared with control cells, while no difference was seen in LL-grown cells. These data therefore indicate a reduced LEF in HL-exposed TEF30-amiRNA cells versus control cells.
Neither PSII de Novo Synthesis nor PSII Stability, But Efficient Repair of Photodamaged PSII, Is Impaired in TEF30-amiRNA Cells
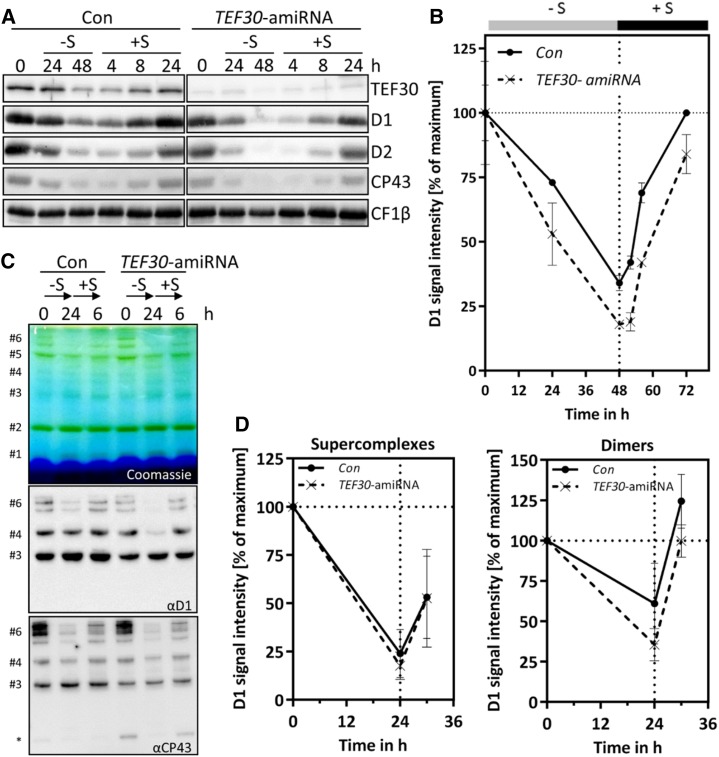
We observed that levels of the D1 protein of PSII were sometimes slightly lower in LL-grown TEF30-amiRNA cells than in control cells (Fig. 5, B and C). We envisaged two explanations for this finding, namely that TEF30 plays a role in D1 synthesis during de novo biogenesis of PSII reaction centers or that a role of TEF30 in counteracting photoinhibition leads to increased photodamage of PSII in TEF30-amiRNA cells also under LL conditions. To distinguish between these two possibilities, we induced PSII degradation in control and TEF30-amiRNA cells by sulfur starvation for 48 h and monitored the reaccumulation of PSII subunits through de novo synthesis upon sulfur repletion for another 24 h (Kosourov et al., 2005; Malnoë et al., 2014). As shown in Figure 7, A and B, levels of D1, D2, and CP43 declined faster during sulfur starvation in TEF30-amiRNA cells when compared with control cells, but recovery kinetics were the same in both strains. As PSII subunits reaccumulating during sulfur repletion may not necessarily be incorporated into PSII supercomplexes, we analyzed PSII supercomplexes by BN-PAGE during sulfur starvation (for 24 h) and sulfur repletion (for 6 h; Fig. 7, C and D). As judged from D1 and CP43 signals from immunoblots on native gels, PSII supercomplexes in TEF30-amiRNA cells declined and reaccumulated during sulfur starvation and repletion as fast as those in control cells. Interestingly, PSII dimers declined faster in TEF30-amiRNA cells during sulfur starvation, but during sulfur repletion they reaccumulated with the same kinetics as in control cells. Moreover, the levels of unassembled CP43 were higher in TEF30-amiRNA cells than in control cells under control conditions and during sulfur repletion (Fig. 7C).
Figure 7.
PSII de novo synthesis is not impaired in TEF30-amiRNA strains. A, TEF30-amiRNA and control (Con) cells in the cw15-325 background were grown in TAP-NH4 at approximately 30 µmol photons m−2 s−1 to the exponential phase. Cells were then harvested, washed, and resuspended in TAP medium lacking sulfur (–S). After 48 h under –S conditions, cells were shifted back to regular TAP medium (+S) for another 24 h. Whole-cell proteins from samples taken during this time course were separated by SDS-PAGE, and PSII polypeptide abundance was measured via immunoblotting using the antibodies indicated and CF1β as a loading control. B, D1 signal intensities from A were quantified using the FUSIONCapt Advance program. Signals were normalized to the initial D1 levels in each strain. Error bars represent sd; n = 2. C, Sulfur depletion was done as in A, but time in –S and +S conditions was reduced to 24 and 6 h, respectively. Thylakoids were solubilized with 1% α-dodecyl maltoside (α-DDM), and proteins corresponding to 5 µg of chlorophyll were separated on a 5% to 15% polyacrylamide BN gradient gel, followed by Coomassie Blue staining (top gel) and immunodetection with antibodies against D1 and CP43 (bottom gels). Protein bands were assigned as in Figure 4C; the asterisk designates free CP43. D, D1 signal intensities from protein bands #6 (PSII supercomplexes) and #4 (PSII dimers) from C were quantified using the FUSIONCapt Advance program. Signals were normalized to the initial D1 levels in each strain. Error bars represent sd; n = 4.
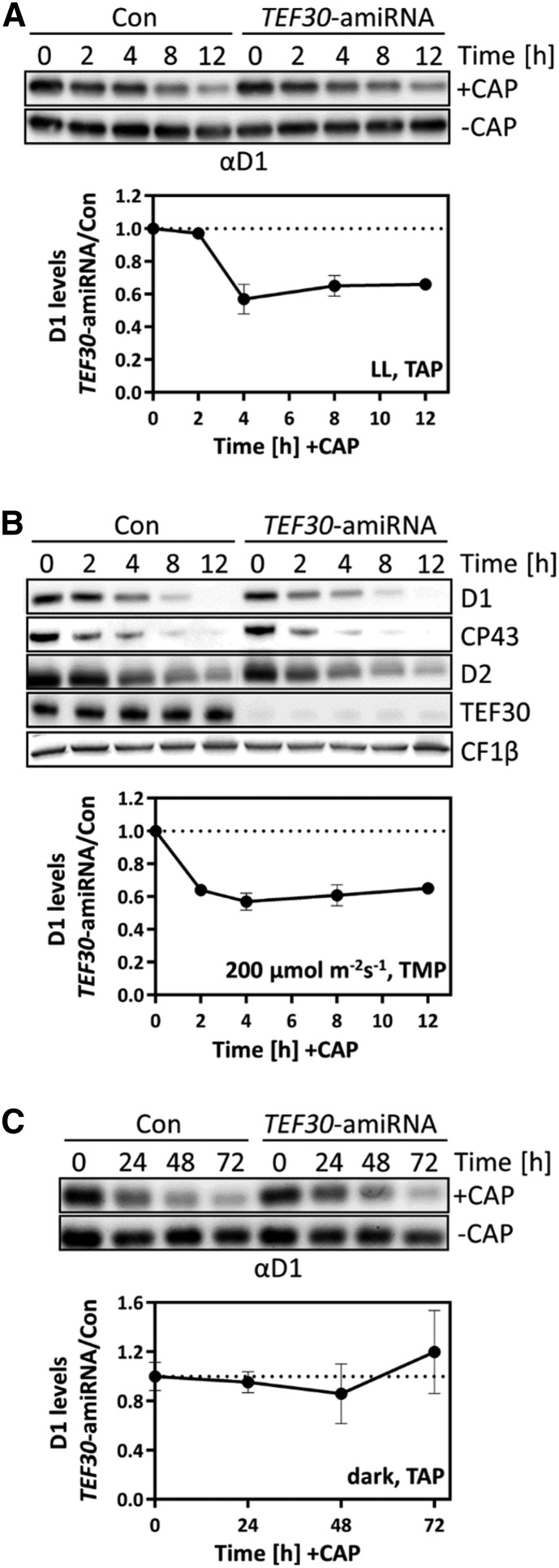
Although these data do not support a role of TEF30 in PSII de novo biosynthesis, it was still possible that the absence of TEF30 during PSII de novo synthesis rendered PSII more unstable. To test this, we grew TEF30-amiRNA and control cells under LL conditions, added chloramphenicol (CAP) to inhibit the de novo synthesis of plastid-encoded proteins, and left the cultures at LL, placed them in the dark, or exposed them to 200 µmol photons m−2 s−1 and monitored levels of PSII subunits. We found D1 levels to decline faster in TEF30-amiRNA cells than in control cells only if CAP-treated cultures were placed in the light (Fig. 8). Hence, PSII complexes assembled in TEF30-amiRNA cells are not generally more unstable than those assembled in control cells.
Figure 8.
PSII stability is not impaired in TEF30-amiRNA strains. A, TEF30-amiRNA and control (Con) cells in the cw15-325 background were grown at approximately 30 µmol photons m−2 s−1 in TAP medium in the presence or absence of 100 µg mL−1 CAP for 12 h. Proteins from whole cells harvested during the time course corresponding to 0.5 or 1 µg of chlorophyll (depending on the antiserum used) were analyzed by immunoblotting. D1 levels were quantified with the FUSIONCapt Advance program. D1 levels were normalized to the values before the addition of CAP (at 0 h), and the ratio between D1 levels from TEF30-amiRNA strains and controls was calculated. Values represent means from four independent replicates, and error bars indicate sd. B, TEF30-amiRNA and control cells in the cw15-325 background were grown in TMP minimal medium at 200 μmol photons m–2 s–1 in the presence of 100 µg mL−1 CAP for 12 h. Analyses were done as in A, with two biological replicates. C, TEF30-amiRNA and control cells in the cw15-325 background were grown in TAP medium in the dark in the presence of 100 µg mL−1 CAP for 72 h. Analyses were done as in A, with two biological replicates.
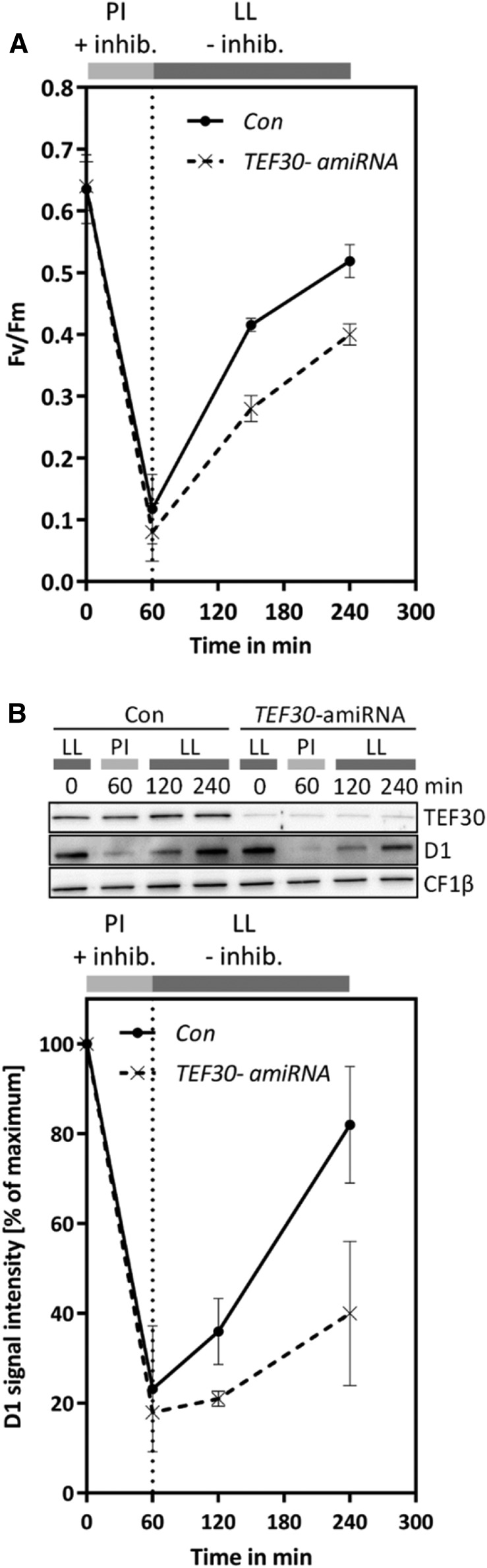
However, we still could not rule out the possibility that improper de novo assembly of PSII in TEF30-amiRNA cells increased the sensitivity of PSII to photodamage. To address this possibility, we exposed cells from control and TEF30-amiRNA strains to HL of 1,800 µmol photons m−2 s−1 in the presence of CAP for 1 h. CAP was added to prevent the repair of photodamaged PSII during HL exposure, and the exposure time of 1 h was chosen because, after 1 h of HL treatment, LHCSR3 levels were still comparable in both strains (Fig. 6A). After HL exposure, CAP was washed out to allow PSII repair to proceed. As there was no significant difference regarding the decline in Fv/Fm and D1 protein levels between both strains after HL exposure, we can conclude that PSII is equally light sensitive in TEF30-amiRNA and control cells (Fig. 9). However, Fv/Fm values and D1 protein levels recovered significantly faster in control than in TEF30-amiRNA cells, strongly suggesting that TEF30 plays a role in PSII repair from photodamage.
Figure 9.
Not PSII sensitivity to HL, but PSII repair, is impaired in TEF30-amiRNA strains. A, Cells of cw15-325 control (Con) and TEF30-amiRNA strains were photoinhibited (PI) for 60 min at 1,800 μmol photons m–2 s–1 in the presence of the chloroplast translation inhibitors CAP and lincomycin and allowed to recover at 30 μmol photons m–2 s–1 (LL) without inhibitors. PSII fluorescence was measured by pulse amplitude-modulated fluorometry. Values represent means from two biological replicates, and error bars indicate sd. B, Immunoblot analysis of whole-cell proteins collected from the experiments described in A. Results from a typical experiment are shown at the top, and means from quantified D1 signals (normalized relative to the initial D1 signal) are shown at the bottom. Values represent means from five biological replicates, and error bars indicate sd.
TEF30-amiRNA Cells Appear to Be Impaired in PSII Supercomplex Formation during PSII Repair
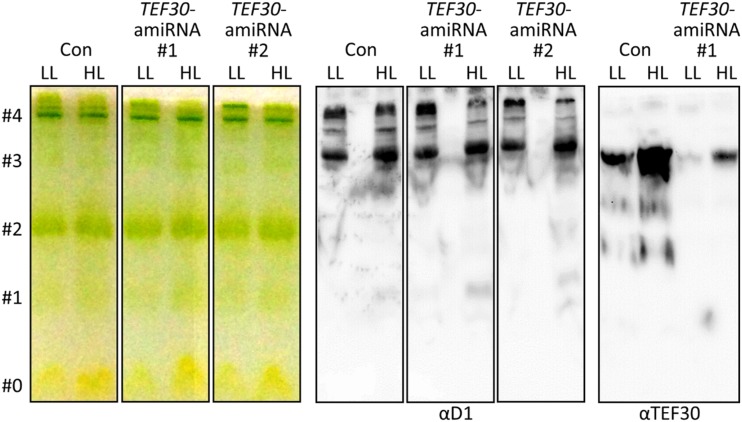
To get an idea at which step of the PSII repair cycle TEF30 might be involved, we analyzed the partitioning of the D1 protein into PSII core and supercomplexes in cells from control and TEF30-amiRNA cells by native Deriphat-PAGE (Formighieri et al., 2012). As shown in Figure 10, there was no difference in the partitioning of D1 in cells of both strains grown under LL conditions. However, much less D1 was found to accumulate in PSII supercomplexes than in core complexes in TEF30-amiRNA cells compared with control cells when cells had been exposed to HL of 800 µmol photons m−2 s−1 for 4 h. TEF30 in native extracts from both strains comigrated quantitatively with PSII monomers, again corroborating that TEF30 specifically interacts with PSII monomers but not with PSII supercomplexes. While TEF30 levels were clearly reduced in TEF30-amiRNA cells, HL inducibility and the comigration of TEF30 with monomers were retained in mutant cells. Overall, these data indicate a possible role of TEF30 in PSII supercomplex reassembly after repair.
Figure 10.
PSII supercomplex accumulation is impaired in TEF30-amiRNA strains exposed to HL. cw15-325 control (Con) and TEF30-amiRNA cells were grown at 30 µmol photons m−2 s−1 (LL) and exposed to 800 µmol photons m−2 s−1 (HL) for 4 h. Thylakoid membranes were isolated and solubilized with 1% α-DDM, and protein complexes were separated electrophoretically on a 7.5% native Deriphat-polyacrylamide gel. The gel was then photographed (left) and processed for immunoblotting using antibodies against TEF30 and the D1 protein (right). Bands were assigned to free pigments (#0), antenna monomers (#1), LHCII trimers (#2), PSI/II cores (#3), and PSI/II supercomplexes (#4) according to Formighieri et al. (2012).
Thylakoid Membrane Stacks in TEF30-amiRNA Cells Contain Fewer Lamellae Than Control Cells
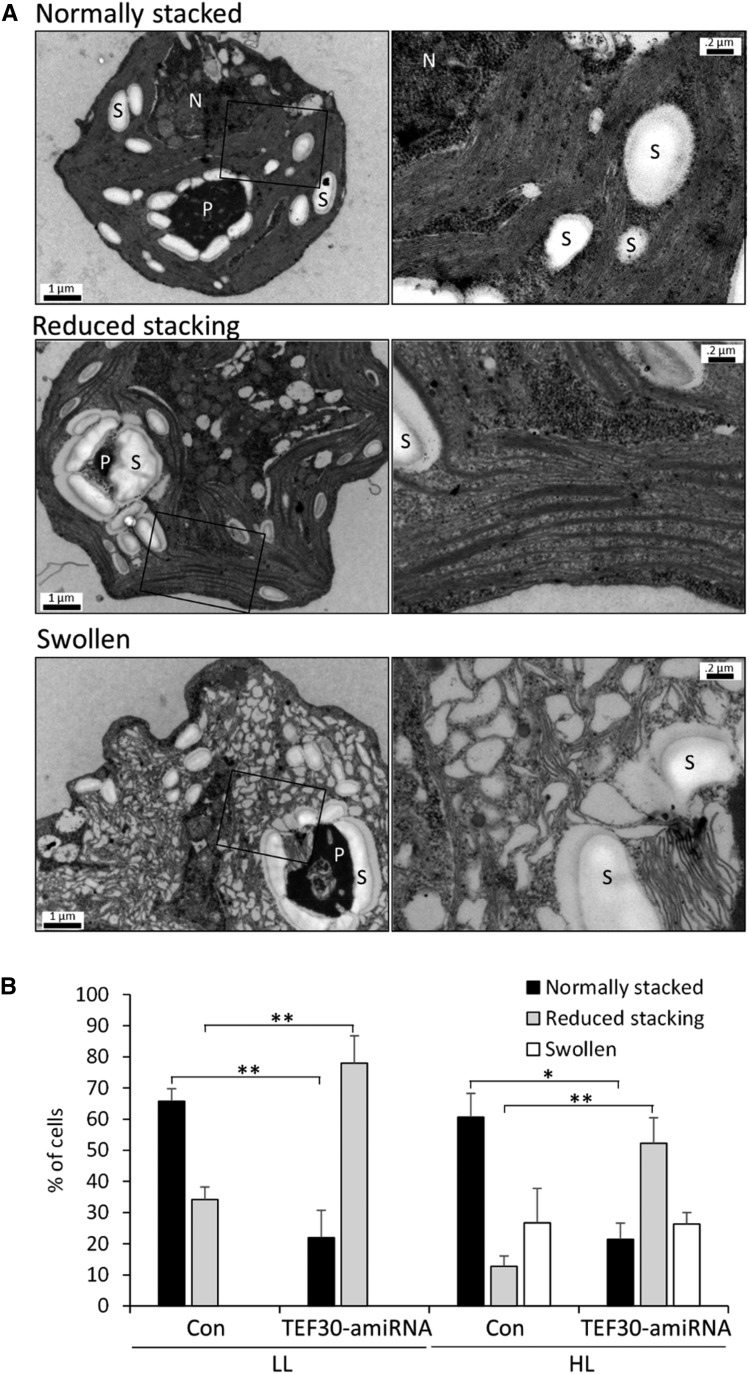
Finally, to investigate whether the depletion of TEF30 had any phenotypes on thylakoid membrane ultrastructure, we analyzed a total of 500 images by transmission electron microscopy from cells of two control and three TEF30-amiRNA strains grown at LL or at HL of approximately 800 µmol photons m−2 s−1 for 4 h. In approximately 66% of cw15-325 control cells grown in LL, we found stacks containing at least 10 thylakoid lamellae (Fig. 11; Nordhues et al., 2012). However, only approximately 22% of LL-grown TEF30-amiRNA cells contained stacks with more than 10 lamellae, whereas thylakoids were much more loosely organized in approximately 78% of the mutant cells (Fig. 11). While the fraction of cells containing thylakoid stacks with more than 10 lamellae remained unchanged in HL-exposed cells (approximately 61% in control cells and approximately 21% in TEF30-amiRNA cells), 26% of control and mutant cells contained swollen thylakoids. The fraction of cells with loosely organized thylakoids declined in control and mutant cells (to 13% and 53%, respectively). These data suggest that the depletion of TEF30 results in less pronounced thylakoid membrane stacking in cells grown in both LL and HL intensities.
Figure 11.
Cells of TEF30-amiRNA strains contain fewer thylakoid membranes per stack. A, Electron microscopy images of cw15-325 cells characteristic for the categories normally stacked, reduced stacking, and swollen. Overview images are shown on the left, and enlarged images of the region demarcated by the black box are shown on the right. N, Nucleus; P, pyrenoid; S, starch. Bars in overview images = 1 µm, and those in enlarged images = 0.2 µm. B, cw15-325 control (Con) and TEF30-amiRNA cells were grown in TAP medium at approximately 30 µmol photons m−2 s−1 (LL) and exposed to approximately 800 µmol photons m−2 s−1 (HL) for 4 h. Fifty electron micrographs each for two independent control strains and three independent TEF30-amiRNA strains were taken for each condition, and thylakoid phenotypes were assessed according to the categories shown in A. Values are given in percent and represent means from the two control and three TEF30-amiRNA strains. Error bars indicate sd. Significance was tested using Student’s t test (*, P < 0.05 and **, P < 0.01).
DISCUSSION
We report here on the functional characterization of the TEF30 protein in C. reinhardtii. As a protein present in the green algal and flowering and nonflowering plant lineages, but not detected in nonphotosynthetic organisms, TEF30 is one out of 597 GreenCut proteins. Moreover, because TEF30 is not present in cyanobacteria, Cyanidioschyzon merolae, Phaeodactylum tricornutum, or Thalassiosira pseudonana, it is part of the ViridiCut subcategory comprising 312 proteins (Supplemental Fig. S1A; Merchant et al., 2007; Heinnickel and Grossman, 2013). ViridiCut proteins are likely enriched in green lineage-specific functions, such as mechanisms of chlorophyll a/b protein regulation (Karpowicz et al., 2011), a prediction that, according to the results of this study, turned out to be correct.
TEF30 Is a Chloroplast Protein Associated with PSII Monomers at the Stromal Side of Thylakoid Membranes
Immunological data on isolated organelles and immunofluorescence data clearly indicate that TEF30 is localized in the chloroplast (Fig. 1, B and C). Cell fractionation and trypsin digestion experiments revealed TEF30 to be largely associated with the stromal side of thylakoid membranes, although no transmembrane helices were predicted in TEF30. Accordingly, fractionations done in the presence of (chaotropic) salts indicated that the membrane association of TEF30 is realized via electrostatic interactions (Fig. 2A). The following evidence indicates that TEF30’s association with thylakoid membranes is realized by its direct interaction with PSII monomers: (1) TEF30 was largely recovered in soluble fractions when mutant cells deficient in D1, D2, CP43, or CP47 proteins were fractionated into membrane and soluble fractions (Fig. 2C); (2) TEF30 coeluted with affinity-purified PSII particles (Fig. 3); (3) TEF30 comigrated quantitatively with PSII monomers on Suc density gradients, on BN gels, and on native Deriphat gels (Figs. 4 and 10); and (4) comigration of TEF30 with PSII monomers was abolished if thylakoid membrane proteins were derived from PSII mutants (Fig. 4, B and C).
With TEF30 and D2 representing approximately 0.05 and approximately 0.59 ng of protein per μg of chlorophyll (Fig. 1D) and exhibiting molecular masses of 33.3 and 39.45 kD, respectively, TEF30 on a molar basis is approximately 10 times less abundant than D2. Given that some TEF30 was also recovered in soluble fractions and that TEF30 was found to be able to form oligomers (Figs. 1E and 2, A and C), the stoichiometry of TEF30 (oligomers?) to PSII is likely below 1:10. PSII monomers are not the main active form of PSII and are mainly located to stroma thylakoids (Danielsson et al., 2006). Therefore, TEF30’s association with PSII monomers is in agreement with the calculated stoichiometries between TEF30 and D2. A localization of TEF30 to stroma lamellae was also indicated by the finding that TEF30 and CF1β with a known location to stroma lamellae shared the same sensitivity to trypsin digestion of isolated thylakoid membranes (Fig. 2B). Consistently, proteomics approaches on thylakoid membranes separated into grana and stroma lamellae identified the Arabidopsis thaliana TEF30 homolog (AT1G55480) exclusively in stroma lamellae (Tomizioli et al., 2014).
TEF30 Is Involved in PSII Repair
TEF30 shares its ability to associate with PSII components at the stroma side with a number of other stromal proteins, including Deg7 (Sun et al., 2010), THF1/Psb29/NYC4 (Wang et al., 2004; Huang et al., 2013), AtPDI6 (Wittenberg et al., 2014), LOW PSII ACCUMULATION3 (LPA3 Cai et al., 2010), and ROC4 (Lippuner et al., 1994; Cai et al., 2008). While Deg7, THF1/Psb29/NYC4, AtPDI6, and ROC4 play roles in the PSII repair cycle (Keren et al., 2005; Cai et al., 2008; Sun et al., 2010; Huang et al., 2013; Yamatani et al., 2013; Wittenberg et al., 2014), LPA3 appears to be required for coordinating the membrane insertion of de novo synthesized CP43 (Cai et al., 2010).
Of these possible functions of TEF30 (PSII de novo synthesis or repair), our data argue against a role of TEF30 in PSII de novo synthesis. This is because control and TEF30-amiRNA cells, in which PSII degradation was induced by sulfur starvation (Kosourov et al., 2005; Malnoë et al., 2014), reaccumulated de novo synthesized PSII core components, dimers, and supercomplexes with the same kinetics after sulfur repletion (Fig. 7). As judged from BN and native Deriphat gels, PSII (super)complexes in TEF30-underexpressing cells also appear to be correctly assembled (Figs. 7C and 10). This is supported by the observation that, in the presence of chloroplast protein synthesis inhibitors, PSII stability in the dark and PSII sensitivity to HL exposure were comparable with those of control cells (Figs. 8C and 9). Moreover, the oxygen evolution capacities of LL-grown cells were the same in control and TEF30-underexpressing strains (Fig. 6B).
Rather, our results point to a role of TEF30 in the PSII repair cycle: most importantly, PSII activity in the absence of protein synthesis inhibitors was more sensitive to HL and recovered more slowly in TEF30-underexpressing cells than in control cells (Fig. 5A; Supplemental Fig. S5). This was reflected by the accumulation kinetics of PSII core subunits, which after HL exposure or in LL under chilling conditions (known to enhance photoinhibition; Greer et al., 1988; Li et al., 2004) declined more pronouncedly in TEF30-underexpressing than in control cells, while minor and major antennae were unaffected (Fig. 5, B and C). Moreover, PSII stability in the presence of chloroplast protein synthesis inhibitors was impaired in TEF30-underexpressing cells only when they were grown in the light but not when they were grown in the dark (Fig. 8). Finally, like TEF30-underexpressing cells, mutants in factors known to be involved in PSII repair hardly display any phenotypes if not grown under high/fluctuating light (Supplemental Fig. S4; Keren et al., 2005; Sirpiö et al., 2007; Cai et al., 2008; Sun et al., 2010).
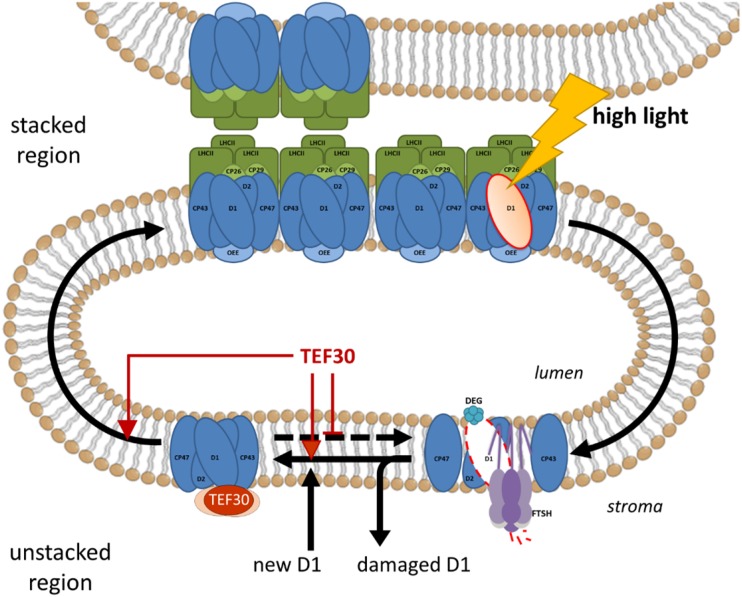
At which step of the PSII repair cycle may TEF30 play a role? Native Deriphat gels indicated a strongly reduced accumulation of PSII supercomplexes in TEF30-underexpressing versus control cells following an exposure to HL for 4 h (Fig. 10). This observation, combined with the finding that TEF30 did not comigrate with PSII-LHCII supercomplexes on Suc density gradients, Deriphat-PAGE, and BN-PAGE (Figs. 4 and 10), points to a role of TEF30 in directing repaired PSII monomers from stroma-exposed regions back to stacked regions for PSII dimer/supercomplex reassembly and/or in stabilizing/protecting repaired PSII monomers (Fig. 12). In Arabidopsis, grana margins (when compared with grana cores and stroma membranes) were shown to represent the membrane regions in which PSII core subunits are mainly dephosphorylated, D1 levels are lowest, and FtsH levels are highest and, therefore, were identified as the main thylakoid subdomain for proteolytic degradation of D1 (Tietz et al., 2015). Hence, if unprotected PSII monomers in TEF30-underexpressing cells linger too long in such proteolytically active domains after repair, they may undergo further cycles of D1 degradation and replacement, thus explaining the inefficient PSII repair in TEF30-underexpressing cells. However, TEF30 may also facilitate complex reassembly (e.g. of temporally released CP43) or the incorporation of de novo synthesized D1 during repair (Fig. 12). In the absence of de novo D1 synthesis during PSII repair, PSII core monomer complexes completely disappeared from stroma membranes (Aro et al., 2005). Therefore, PSII monomers whose reassembly is delayed during repair may be more prone to proteolytic attack. It is important to note that TEF30 apparently facilitates the PSII repair cycle but is not essential for it. The PSII repair cycle exists in all organisms capable of performing oxygenic photosynthesis, but TEF30 is found only in the Viridiplantae. Therefore, its role in facilitating PSII repair may have evolved to help overcome constraints to the PSII repair cycle that are specific to the Viridiplantae.
Figure 12.
Model for TEF30 function during PSII repair. When PSII is damaged (e.g. at HL intensities), the PSII complex enters the repair cycle. For this, PSII supercomplexes are disassembled and PSII monomers migrate from stacked into stroma-exposed membrane regions. After PSII monomers are partially disassembled to generate CP43-less forms, damaged D1 is degraded by FtsH and Deg proteases and a newly synthesized D1 copy is inserted into PSII monomers. At this step, TEF30 may facilitate incorporation of the new D1 protein and/or reassembly of CP43, bind to repaired PSII monomers to protect them from undergoing repeated repair cycles, or facilitate the migration of repaired PSII monomers back to stacked regions for supercomplex reassembly. These functions are not mutually exclusive.
Reduced Accumulation of LHCSR3 and Reduced Thylakoid Membrane Stacking May Be Pleiotrophic Effects of Impaired PSII Repair in TEF30-Underexpressing Cells
Upon HL exposure, the LHCSR3 protein accumulated to lower levels in TEF30-underexpressing cells than in control cells (Fig. 5B). LHCSR3 becomes activated by an increased acidification of the thylakoid lumen and mediates the dissipation of excess light energy by a protonation-induced interaction with PSII supercomplexes that is dependent on the R-subunit of PSII (PSBR) (Bonente et al., 2011; Tokutsu and Minagawa, 2013; Xue et al., 2015). The reduced accumulation of LHCSR3 in TEF30-underexpressing cells was observed only when cells were exposed for more than 1 h to HL and was realized at the level of gene expression (Fig. 6A; Supplemental Fig. S6A). Moreover, the effect was only observed in cells grown mixotrophically with ammonium as nitrogen source and not in cells grown under photoautotrophic conditions with ammonium as nitrogen source or grown mixotrophically with nitrate as nitrogen source (Fig. 6A). Hence, it appear as if the impaired induction of the LHCSR3 gene in TEF30-underexpressing cells is limited to conditions in which sinks for products of the light reactions are limiting (nitrate reduction and carbon fixation are highly ATP and NADPH demanding). LHCSR3 gene expression was shown to correlate with the PFD applied and thus with LEF rates (Naumann et al., 2007; Peers et al., 2009; Petroutsos et al., 2011; Maruyama et al., 2014). Accordingly, in cells grown mixotrophically in the presence of ammonium and exposed to HL for more than 1 h, LEF rates are supposed to be lower if TEF30 levels are low. This indeed appeared to be the case, as the oxygen evolution capacity was approximately 25% lower in HL-exposed TEF30-underexpressing cells than in control cells but was unaltered in LL-grown cells (Fig. 6B). Therefore, we propose that the reduced accumulation of LHCSR3 in TEF30-underexpressing cells grown under apparently sink-limited conditions is a consequence of increased PSII photoinhibition and thus of reduced LEF under such conditions rather than the cause for increased photoinhibition. In support of this idea, blocking the Calvin-Benson cycle was shown to enhance photoinhibition in C. reinhardtii (Takahashi and Murata, 2005).
Using transmission electron microscopy, we observed that approximately 66% of the cells of TEF30-underexpressing strains contained fewer than 10 lamellae in thylakoid membrane stacks, whereas this was true for only approximately 22% of the cells in control strains (Fig. 11). Therefore, the thylakoid organization in control cells under our growth conditions (LL of approximately 30 µmol photons m−2 s−1) resembles that of shade leaves, with many thylakoids per stack, while that of TEF30-underexpressing cells resembles the organization found in sun leaves, with fewer lamellae per stack (Guillot-Salomon et al., 1978; Lichtenthaler et al., 1981). One possible explanation for this phenotype is that TEF30 directly affects thylakoid stacking and impaired PSII repair is a consequence of this. The number of membrane layers in grana stacks was shown to correlate with the abundance of the CURVATURE THYLAKOID1 (CURT1) protein, which is located to grana margins and induces membrane curvature (Armbruster et al., 2013). Hence, TEF30 might positively modulate the function of CURT1, leading to reduced membrane stacking in TEF30-underexpressing cells. An argument against this idea is that reduced thylakoid membrane stacking was shown to improve PSII repair by facilitating the lateral migration of damaged PSII supercomplexes and, therefore, increasing their accessibility to the FtsH protease (Tikkanen et al., 2008; Fristedt et al., 2009; Kirchhoff et al., 2013; Nath et al., 2013b). Indeed, reduced membrane stacking may account for the faster degradation rates of PSII dimers and supercomplexes upon sulfur starvation in TEF30-underexpressing cells (Fig. 7) but conflicts with their impaired capacity for PSII repair. A second, more plausible explanation for the reduced thylakoid membrane stacking in TEF30-underexpressing cells is that it represents a countermeasure of these cells to improve PSII repair (e.g. by reducing CURT1 function at the levels of CURT1 protein accumulation or phosphorylation state or by increasing the phosphorylation state of PSII core subunits; Tikkanen et al., 2008; Fristedt et al., 2009; Goral et al., 2010; Samol et al., 2012; Armbruster et al., 2013; Kirchhoff et al., 2013; Tietz et al., 2015).
Did an Additional Function for TEF30 in Supercomplex Assembly of de Novo Synthesized PSII Evolve in Land Plants?
The Arabidopsis TEF30 homolog (AT1G55480) was first described by Ishikawa et al. (2005), who termed it ZKT, for protein containing a PDZ domain, a K-box region, and a TPR region, and proposed a role for the protein in wounding responses. Very recently, the same protein was named MET1 (Bhuiyan et al., 2015). Our results in C. reinhardtii TEF30 are largely congruent with those from the excellent study by Bhuiyan et al. (2015) with respect to the inducibility of TEF30/MET1 by HL, the interaction of the proteins with the stroma side of PSII complexes in thylakoid membranes, the lack of obvious phenotypes in mutants under standard growth conditions, as well as the increased sensitivity of PSII and the reduced accumulation of PSII supercomplexes in TEF30/MET1-depleted mutants compared with controls upon HL exposure. Therefore we agree with Bhuiyan et al. (2015) that a function of TEF30/MET1 in wounding is unlikely.
However, there are also differences between the traits observed for TEF30/MET1 in C. reinhardtii and Arabidopsis, which led to slightly different conclusions regarding the function of this protein. By immunoblot analysis of Arabidopsis thylakoid membrane protein complexes separated by BN-PAGE, Bhuiyan et al. (2015) found MET1 to comigrate with several PSII (sub)complexes, including dimers, monomers, CP43-less monomers, reaction centers, and unassembled proteins. With the yeast two-hybrid system, they also found MET1 to directly interact with stromal loops of CP43 and CP47. Based on these data, they concluded that MET1 is involved in the folding and/or stabilization of these stroma-exposed CP43/CP47 domains, thereby ensuring their correct configuration upon assembly of the PSII core during de novo PSII biogenesis and during the PSII repair cycle. By analyzing C. reinhardtii thylakoid membrane protein complexes on Suc density gradients, BN-PAGE, and Deriphat-PAGE, we observed TEF30 to quantitatively interact with PSII monomers and observed only traces of TEF30 to comigrate with larger PSII assemblies (Suc gradients; Fig. 4B) or smaller ones (Deriphat-PAGE; Fig. 10). Moreover, based on the finding that PSII supercomplex formation was only impaired in TEF30-underexpressing cells pushed toward PSII repair by HL exposure (Fig. 10) and not in cells pushed toward de novo PSII synthesis during recovery from sulfur starvation (Fig. 7), we concluded that TEF30 is rather involved in PSII repair and not in the assembly of de novo synthesized PSII complexes. Whether these differences are due to experimental setups or MET1 has acquired an additional role in PSII de novo assembly during land plant evolution remains to be elucidated. The latter idea may be supported by the finding that PSII de novo synthesis and repair appear to be spatially separated in C. reinhardtii (Uniacke and Zerges, 2007), while both processes appear to be only temporally separated during cell differentiation in land plants (Nickelsen and Rengstl, 2013).
MATERIALS AND METHODS
Chlamydomonas reinhardtii Strains and Culture Conditions
C. reinhardtii strains cw15-325 (cwd, mt+, arg7−) and cw15-302 (cwd, mt+, arg7−, nit−), kindly provided by R. Matagne (University of Liège, Belgium), were used as recipient strains for transformation with plasmid pCB412 (containing only the ARG7 gene; Schroda et al., 1999) to generate control strains or with pMS632 (containing the ARG7 gene and the TEF30-amiRNA construct) to down-regulate TEF30. Transformations were carried out using 1 µg of HindIII-linearized plasmids and vortexing with glass beads (Kindle, 1990). Photosynthesis mutants used were F23 (lacking PsaA; Girard et al., 1980), FUD34 (lacking CP43; Rochaix et al., 1989), FUD44 (lacking OEE1; Mayfield et al., 1987), ΔbB/ΔpsbB (lacking CP47; Minai et al., 2006), FUD47 and nac2 (both lacking D2; Erickson et al., 1986; Kuchka et al., 1989), FUD50 (lacking CF1β; Woessner et al., 1986), ΔpetA (lacking cytochrome f; Rimbault et al., 2000), and the double mutant ΔpsbA/ΔpsbD (lacking D1 and D2). Strain cc124 (mt−) was employed as a wild-type control for experiments with photosynthesis mutants. Strain D2His (Sugiura et al., 1998) was a gift from J. Minagawa (National Institute for Basic Biology, Japan). Unless specified otherwise, all strains were grown mixotrophically in TAP medium (Harris et al., 2008) supplemented with 7.5 mm NH4Cl and a revised micronutrient composition (Kropat et al., 2011) on a rotatory shaker at 25°C and approximately 30 μmol photons m−2 s−1. Minimal medium (TMP) was prepared in the same way but lacking acetate. Cultures were diluted on the day before experiments were conducted and grown to midlog phase. Cell densities were determined using a Z2 Coulter Counter (Beckman Coulter).
Phylogenetic Analyses
Multiple sequence alignments were performed with protein sequences from C. reinhardtii (Cre01.g031100.t1.2), Ostreococcus lucimarinus (XP_001416302), Micromonas pusilla (XP_003059792), Volvox carteri (XP_002955399), Physcomitrella patens (Phpat.019G052300.1), Selaginella moellendorffii (XP_002988351), Zea mays (XP_008652074), Oryza sativa (AP004670.4), Arabidopsis (Arabidopsis thaliana; AT1G55480.1), Medicago truncatula (XP_003629926), and Glycine max (XP_001235451) using the ClustalW2.1 program (Larkin et al., 2007). Transit peptide sequences were predicted with ChloroP (Emanuelsson et al., 1999) or TargetP (Emanuelsson et al., 2000). The phylogenetic tree was inferred by the neighbor-joining method using MEGA5.1 (Tamura et al., 2011), and statistical support was assessed with 1,000 bootstrap replicates.
Cloning, Expression, and Purification of Recombinant TEF30
The TEF30 coding sequence was amplified from C. reinhardtii complementary DNA with primers 5′-TACTGGATCCGCCGAATACATCGAGCTGGA-3′ and 5′-GCAACATGTTCAACAAGGAGAAGTAAGCTTACTCA-3′. The resulting 817-bp PCR product was digested with BamHI and HindIII and cloned in frame into the PetDuet vector (Novagen) cleaved with the same enzymes, yielding pMS649. TEF30 was overexpressed in Escherichia coli BL21 and purified by nickel-nitrilotriacetic acid agarose according to the manufacturer’s instructions (Qiagen). Imidazole was largely removed by three successive runs of concentration/dilution with KMH buffer (20 mm HEPES-KOH, pH 7.2, 80 mm KCl, and 2.5 mm MgCl2) using Amicon Ultra-4 tubes (Millipore). After that, proteins were supplemented with 10% (v/v) glycerol and 1 mm dithiothreitol. For antibody generation, TEF30 was purified under denaturing conditions with guanidine-HCl and eluted with 200 mm imidazole in 6 m urea. One milligram of purified protein was used for a 3-month rabbit immunization protocol (SeqLab) for antibody generation.
Protein Analyses and Thylakoid Membrane Isolation
Protein extractions, SDS-PAGE, semidry blotting, and immunodetections were carried out as described before (Liu et al., 2005; Schulz-Raffelt et al., 2007). Proteins on SDS-polyacrylamide gels were loaded on the basis of chlorophyll concentrations or protein concentrations determined according to Lowry et al. (1951). Immunodetection was done using enhanced chemiluminescence and the FUSION-FX7 Advance imaging system (PEQLAB). Antisera described previously were against mitochondrial carbonic anhydrase (Agrisera AS11 1737), HSP70B (Schroda et al., 1999), HSP70A (Schulz-Raffelt et al., 2007), VIPP1 (Liu et al., 2005), LHCSR3 (Naumann et al., 2007), CF1β (Lemaire and Wollman, 1989), PSAD (Naumann et al., 2005), LHCII (Vallon et al., 1986), CP43 (Vallon et al., 1985), cytochrome f (Pierre and Popot, 1993), D1 (Agrisera AS05 084), OEE1 (Agrisera AS06 142-33), OEE2 (de Vitry et al., 1989), PsaA (Agrisera AS06 172), PsaB (Agrisera AS10 695), CP26 (Agrisera AS09 407), CP29 (Agrisera AS06 117), D2 (Agrisera AS06 146), and CGE1 (Schroda et al., 2001). Densitometric quantification of the bands after immunodetection was calculated by the FUSIONCapt Advance program for image capture and analysis (PEQLAB). Thylakoids were isolated as described previously (Chua and Bennoun, 1975). Briefly, cells were lysed using a BioNebulizer (Glas-Col) with an operating N2 pressure of 1.5 to 2 bar, and lysates were centrifuged through a Suc step gradient (0.6, 1.3, and 1.8 m Suc) at 40,000 rpm for 1 h. Thylakoids floating between the 1.8 and 1.3 m transition were collected, diluted in a buffer containing 5 mm HEPES-KOH, pH 7.5, and 10 mm EDTA, and pelleted by centrifugation at 40,000 rpm for 15 min.
EDC Cross-Linking of Recombinant TEF30
A total of 1 μg of recombinant TEF30 was incubated in KMH buffer with 2.5 mm EDC and a double concentration of N-hydroxysuccinimide. The mixture was incubated for 30 min at 25°C. The cross-linking reaction was stopped by the addition of 0.2 m ammonium acetate, and samples equivalent to 100 ng of TEF30 were analyzed by immunoblotting.
Cell Fractionations
Isolation of chloroplasts was performed as described previously (Zerges and Rochaix, 1998). Mitochondria were isolated according to Eriksson et al. (1995) but using a BioNebulizer (Glas-Col) for cell disruption. For the separation of soluble and membrane-enriched fractions, 2 × 107 cells were harvested by a 2-min centrifugation at 1,300g and 4°C and resuspended in lysis buffer (10 mm Tris-HCl, pH 8, 1 mm EDTA, and 0.25× protease inhibitor cocktail [Roche]). An aliquot was taken for whole-cell proteins. The remainder was frozen in liquid nitrogen and thawed at 25°C. This cycle was repeated two times. Disrupted cells were centrifuged for 30 min at 18,000g and 4°C. The supernatant was collected as the soluble protein fraction. Pellets were resuspended by brief sonication with the same volume of lysis buffer. When indicated, a final concentration of 250 mm NaCl, 200 mm Na2CO3, 1 m CaCl2, or 6 m urea was added to the lysis buffer before freezing/thawing the cells. For localization experiments, isolated thylakoids were incubated on ice with 0.1 mg mL−1 trypsin. After the addition of 0.25× proteinase inhibitor cocktail (Roche), proteins were precipitated with 80% (v/v) acetone. Pellets were resuspended in Laemmli buffer for SDS-PAGE (Laemmli, 1970).
Vector Construction and Screening for TEF30-Underexpressing Transformants
The artificial microRNA targeting TEF30 was designed according to Molnar et al. (2009) using the WMD2 Web platform (Ossowski et al., 2008; current version at http://wmd3.weigelworld.org). The resulting oligonucleotides, 5′-ctagtAAGTTCGCTCGCGGTGGCGAAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTTCGTTACCGCGAGCGAACTTg-3′ and 5′-ctagcAAGTTCGCTCGCGGTAACGAAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTTCGCCACCGCGAGCGAACTTa-3′ (lowercase sequences form the SpeI recognition site and the hairpin/loop, uppercase sequences the amiRNA duplex), were annealed by a gradual temperature reduction from 100°C to 60°C (0.3°C min−1) in a thermocycler and ligated into pChlamiRNA2 (Molnar et al., 2009), yielding pMS632. Correct constructs were screened as described by Molnar et al. (2009) and sequenced. After transformation of this construct into C. reinhardtii cells, 50 Arg-prototrophic transformants of each recipient strain (cw15-325 or cw15-302) were screened by immunoblotting.
Cold Stress, HL, Photoinhibition, and Sulfur Starvation Experiments
For cold stress, 150 mL of 2 × 106 cells grown at 25°C were incubated under continuous agitation in a water bath at 10°C and approximately 30 μmol photons m−2 s−1 for 18 h. HL experiments were performed with 150 mL of 2 × 106 cells in 400-mL beakers placed onto spots on a rotary shaker that were illuminated (approximately 800 μmol photons m−2 s−1) by Osram HLX 250-W 64663 Xenophot bulbs. A glass plate nonpermissive to infrared and UV irradiation was placed between light sources and beakers. The position of the beakers was altered in regular intervals, and the cell culture temperature was maintained at 25°C. The same setup was used for photoinhibition, but cultures were exposed to approximately 1,800 μmol photons m−2 s−1. For recovery after photoinhibition, cultures were placed on a rotatory shaker at 25°C and approximately 30 μmol photons m–2 s–1. When indicated, 500 μg mL−1 lincomycin and/or 100 μg mL−1 chloramphenicol were added before photoinhibition or at the beginning of the recovery phase. To wash out the inhibitors, cells were centrifuged for 2 min at 956g and 25°C and cell pellets were resuspended in fresh TAP-NH4 medium. PFDs were determined with a light meter (LI-COR LI-250A). Sulfur starvation experiments were performed in TAP-NH4 according to Malnoë et al. (2014). To follow the de novo synthesis of PSII, cells were centrifuged and resuspended in TAP-NH4 medium and kept in the light (approximately 30 μmol photons m−2 s–1) for 24 h.
Isolation of PSII Particles and Immunoprecipitation
Isolation of PSII particles was performed with approximately 2 L of 3 × 106 cells mL−1 of the D2His strain according to Cullen et al. (2007) with a ProBond resin column (Invitrogen). After their elution, PSII particles were mixed with an equal volume of 20 mm MES, pH 6.3, 15 mm NaCl, 20% (w/v) polyethylene glycol 8000, 10 mm ascorbic acid, and 10 mm EDTA and then centrifuged at 11,000g for 10 min. The pellet was resuspended in 20 mm MES, pH 6.3, 15 mm NaCl, 10 mm ascorbic acid, 10% (w/v) polyethylene glycol, and 10 mm CaCl2 and then centrifuged as before to decrease the imidazole concentration. The pellet containing isolated PSII particles was resuspended in 250 µL of buffer A (20 mm MES, pH 6.3, 15 mm NaCl, 333 mm Suc, and 10 mm CaCl2). The same procedure was followed with cc124 cells as a control. The coupling of antibodies against TEF30 and of preimmune serum to protein A-Sepharose beads for immunoprecipitation was carried out as described previously (Schroda et al., 2001; Liu et al., 2005). Coupled beads were washed twice in buffer A and incubated with 250 µL of isolated PSII particles for 1 h at 4°C. Beads were then washed three times in buffer B (buffer A with the addition of 0.1% [v/v] Triton X-100) and two times in 10 mm Tris-HCl, pH 7.5. Immunoprecipitated proteins were collect by boiling the beads in 2% (w/v) SDS and 12% (w/v) Suc for 40 s. After the addition of 0.1 m dithiothreitol, eluates were subjected to SDS-PAGE.
BN-PAGE, Deriphat-PAGE, and Suc Density Centrifugation
BN-PAGE was performed with isolated thylakoids, crude membrane fractions, or whole-cell proteins as described (Schägger and von Jagow, 1991; Schägger et al., 1994). Native Deriphat-PAGE was performed according to Peter and Thornber (1991) with a 4% (w/v) acrylamide stacking gel poured on the 7.5% (w/v) acrylamide running gel. Thylakoid membranes (0.25 mg chlorophyll mL−1) were solubilized for 20 min on ice in 5 mm HEPES-KOH, pH 7.5, and 1 mm EDTA containing 1% (w/v) α-DDM. For Suc density centrifugation, isolated thylakoids were solubilized with 1% (w/v) β-DDM and loaded onto a linear 0.2 to 0.5 m Suc gradient containing 10 mm HEPES-KOH, pH 7.5, and 0.05% (w/v) β-DDM. On the bottom of the gradient, a 2 m Suc cushion was present. The gradient was centrifuged at 205,000g for 16 h at 4°C, and 25 to 26 fractions were collected from bottom to top.
Immunofluorescence Staining and Transmission Electron Microscopy
Immunofluorescence microscopy was performed with control transformants in the cw15-325 strain background. Cells were fixed and stained as described previously (Uniacke et al., 2011). Primary antibodies were against TEF30, HSP70B, and HSP70A, used in 1:300, 1:8,000, and 1:4,000 dilutions, respectively. As a secondary antibody, we used a fluorescein isothiocyanate-labeled goat anti-rabbit antibody (Sigma-Aldrich) in a 1:500 dilution. After incubation with the secondary antibody, slides were washed in phosphate-buffered saline, and a drop of mounting solution containing DAPI (Vectashield; Vector Laboratories) was applied at the center of each slide. Images were obtained with an Olympus BX53 microscope with a violet filter for DAPI and a green filter for fluorescein isothiocyanate and an Olympus DP26 color camera. For transmission electron microscopy, cells were collected, fixed, and embedded as described previously (Nordhues et al., 2012). Images were analyzed with a JEM-2100 (JEOL) or EM 902A (Carl Zeiss) transmission electron microscope (both operated at 80 kV). Micrographs were taken using a 4,080- × 4,080-pixel or a 1,350- × 1,040-pixel CCD camera (UltraScan 4000 or Erlangshen ES500W, respectively; Gatan) and the Gatan Digital Micrograph software (version 1.70.16).
RNA Extraction and qRT-PCR
A total of 2 × 107 cells were collected for RNA extraction with the RNeasy Plant Mini kit (Qiagen) following the manufacturer’s instructions. DNA contaminations were removed with DNase (RNase-free Turbo DNase; Ambion). Complementary DNA synthesis was done with the murine leukemia virus reverse transcriptase (Promega), deoxynucleotide triphosphate, and oligo(dT)18 primers. qRT-PCR was performed using the StepOnePlus RT-PCR system (Applied Biosystems) and the Maxima SYBR Green kit from Fermentas as described previously (Strenkert et al., 2011). Primers for LHCSR3.1 were the same as described by Peers et al. (2009).
Biophysical Measurements
Fluorescence induction measurements were performed at 25°C with a DeepGreen Fluorometer (JBeamBio). Fv/Fm values were determined after incubating cells in the dark for 1 min according to the equation Fv = Fm − F0, where Fv is the variable PSII fluorescence, F0 represents the fluorescence when the electron acceptor of PSII is fully oxidized and Fm is the maximum PSII fluorescence. Fm was measured during a 250-ms saturating flash of light or in the presence of 10 mm 3-(3,4-dichlorophenyl)-1,1-dimethylurea. Low-temperature fluorescence emission spectra were determined with a laboratory-built setup. Samples were submerged in liquid nitrogen and excited with a light-emitting diode source (LS-450; Ocean Optics; blue light-emitting diode, 450 nm). The emission spectra were measured by a CCD spectrophotometer (QE6500; Ocean Optics). The device was calibrated by the manufacturer, and calibration was verified using standard band-pass filters. For the fluorescence quenching measurements, cells were grown mixotrophically in TAP-NH4 medium at 50 µmol photons m−2 s−1, harvested in the exponential phase, and resuspended in TMP-NH4 at a concentration of 2 × 107 cells mL−1. Cells were agitated and exposed to HL intensities of 500 µmol photons m−2 s−1 provided by a WH1200 Phyto (JBeamBio) to induce the accumulation of LHCSR3. The NPQ response was measured using a chlorophyll fluorescence imaging system (Johnson et al., 2009). Samples were incubated in the dark under strong agitation for 15 min before each measurement. NPQ was calculated as (Fm − Fm′)/Fm′, where Fm′ is the maximum PSII fluorescence in light (after 10 s at 700 μmol photons m−2 s−1). Fm and Fm′ were measured after exposure to a brief saturating pulse of 2,100 μmol photons m−2 s−1 for 250 ms. Oxygen evolution was measured with a Clark-type oxygen electrode in the presence of 0.5 mm 2,6-dichlorobenzoquinone and 2 mm potassium hexacyanoferrate (III) as electron acceptors. The measurements were carried out with crude membrane fractions in TMP-NH4 medium at 25°C.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. TEF30 protein accumulates in C. reinhardtii cells exposed to HL.
Supplemental Figure S2. Phylogenetic tree and alignment of TEF30 homologs.
Supplemental Figure S3. Screening of TEF30-amiRNA transformants.
Supplemental Figure S4. TEF30-underexpressing transformants exhibit no obvious growth phenotype under a variety of growth conditions.
Supplemental Figure S5. PSII fluorescence is more affected by HL in TEF30-amiRNA cells than in control cells.
Supplemental Figure S6. Reduced induction of LHCSR3 expression in TEF30-amiRNA cells is realized at the transcriptional level, and NPQ is not affected under photoautotrophic conditions.
Supplementary Material
Acknowledgments
We thank Jun Minagawa for providing the D2His strain, Michael Hippler for providing antisera against LHCSR3 and PSAD, and Fabrice Rappaport for helping with the biophysical measurements.
Glossary
- OEC
oxygen-evolving complex
- PFD
photon flux density
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- BN
blue native
- Fv/Fm
PSII maximum quantum efficiency
- HL
high light
- NPQ
nonphotochemical quenching
- qRT
quantitative reverse transcription
- TAP
Tris-acetate-phosphate
- LEF
linear electron flow
- LL
low light
- CAP
chloramphenicol
- α-DDM
α-dodecyl maltoside
- β-DDM
β-dodecyl maltoside
- DAPI
4′,6-diamino-phenylindole
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Schr 617/9–1), the Deutscher Akademischer Austauschdienst/Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant no. 290020/2010–7), and the Forschungsschwerpunkt BioComp.
Articles can be viewed without a subscription.
References
- Albiniak AM, Baglieri J, Robinson C (2012) Targeting of lumenal proteins across the thylakoid membrane. J Exp Bot 63: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Allan RK, Ratajczak T (2011) Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16: 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmer J, Naumann B, Markert C, Zhang M, Hippler M (2006) Mass spectrometric genomic data mining: novel insights into bioenergetic pathways in Chlamydomonas reinhardtii. Proteomics 6: 6207–6220 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Labs M, Pribil M, Viola S, Xu W, Scharfenberg M, Hertle AP, Rojahn U, Jensen PE, Rappaport F, et al. (2013) Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25: 2661–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro EM, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamäki E (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56: 347–356 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Bhuiyan NH, Friso G, Poliakov A, Ponnala L, van Wijk KJ (2015) MET1 is a thylakoid-associated TPR protein involved in photosystem II supercomplex formation and repair in Arabidopsis. Plant Cell 27: 262–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G, Ballottari M, Truong TB, Morosinotto T, Ahn TK, Fleming GR, Niyogi KK, Bassi R (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol 9: e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarri S, Kouril R, Kereïche S, Boekema EJ, Croce R (2009) Functional architecture of higher plant photosystem II supercomplexes. EMBO J 28: 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Ma J, Chi W, Zou M, Guo J, Lu C, Zhang L (2010) Cooperation of LPA3 and LPA2 is essential for photosystem II assembly in Arabidopsis. Plant Physiol 154: 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cai W, Ma J, Guo J, Zhang L (2008) Function of ROC4 in the efficient repair of photodamaged photosystem II in Arabidopsis. Photochem Photobiol 84: 1343–1348 [DOI] [PubMed] [Google Scholar]
- Che Y, Fu A, Hou X, McDonald K, Buchanan BB, Huang W, Luan S (2013) C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc Natl Acad Sci USA 110: 16247–16252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua NH, Bennoun P (1975) Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc Natl Acad Sci USA 72: 2175–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen M, Ray N, Husain S, Nugent J, Nield J, Purton S (2007) A highly active histidine-tagged Chlamydomonas reinhardtii photosystem II preparation for structural and biophysical analysis. Photochem Photobiol Sci 6: 1177–1183 [DOI] [PubMed] [Google Scholar]
- Danielsson R, Suorsa M, Paakkarinen V, Albertsson PA, Styring S, Aro EM, Mamedov F (2006) Dimeric and monomeric organization of photosystem II: distribution of five distinct complexes in the different domains of the thylakoid membrane. J Biol Chem 281: 14241–14249 [DOI] [PubMed] [Google Scholar]
- de Vitry C, Olive J, Drapier D, Recouvreur M, Wollman FA (1989) Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J Cell Biol 109: 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drop B, Webber-Birungi M, Yadav SK, Filipowicz-Szymanska A, Fusetti F, Boekema EJ, Croce R (2014) Light-harvesting complex II (LHCII) and its supramolecular organization in Chlamydomonas reinhardtii. Biochim Biophys Acta 1837: 63–72 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JM, Rahire M, Malnoë P, Girard-Bascou J, Pierre Y, Bennoun P, Rochaix JD (1986) Lack of the D2 protein in a Chlamydomonas reinhardtii psbD mutant affects photosystem II stability and D1 expression. EMBO J 5: 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Gardestrom P, Samuelsson G (1995) Isolation, purification, and characterization of mitochondria from Chlamydomonas reinhardtii. Plant Physiol 107: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formighieri C, Ceol M, Bonente G, Rochaix JD, Bassi R (2012) Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Mol Plant 5: 1242–1262 [DOI] [PubMed] [Google Scholar]
- Fristedt R, Willig A, Granath P, Crèvecoeur M, Rochaix JD, Vener AV (2009) Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21: 3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J, Chua NH, Bennoun P, Schmidt G, Delosme M (1980) Studies on mutants deficient in the photosystem I reaction centers in Chlamydomonas reinhardtii. Curr Genet 2: 215–221 [DOI] [PubMed] [Google Scholar]
- Goral TK, Johnson MP, Brain AP, Kirchhoff H, Ruban AV, Mullineaux CW (2010) Visualizing the mobility and distribution of chlorophyll proteins in higher plant thylakoid membranes: effects of photoinhibition and protein phosphorylation. Plant J 62: 948–959 [DOI] [PubMed] [Google Scholar]
- Greer DH, Laing WA, Kipnis T (1988) Photoinhibition of photosynthesis in intact kiwifruit (Actinidia deliciosa) leaves: effect of temperature. Planta 174: 152–158 [DOI] [PubMed] [Google Scholar]
- Guillot-Salomon T, Tuquet C, De Lubac M, Hallais MF, Signol M (1978) Comparative analysis of ultrastructure and lipid composition of plastids from sun and shade plants [author’s translation]. Cytobiologie 17: 442–452 [PubMed] [Google Scholar]
- Harris EH, Stern DB, Witman GB, editors (2008) The Chlamydomonas Sourcebook, Ed 2, Vol 1, Introduction to Chlamydomonas and Its Laboratory Use. Elsevier/Academic Press, San Diego [Google Scholar]
- Haussühl K, Andersson B, Adamska I (2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J 20: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinnickel ML, Grossman AR (2013) The GreenCut: re-evaluation of physiological role of previously studied proteins and potential novel protein functions. Photosynth Res 116: 427–436 [DOI] [PubMed] [Google Scholar]
- Hemme D, Veyel D, Mühlhaus T, Sommer F, Jüppner J, Unger AK, Sandmann M, Fehrle I, Schönfelder S, Steup M, et al. (2014) Systems-wide analysis of acclimation responses to long-term heat stress and recovery in the photosynthetic model organism Chlamydomonas reinhardtii. Plant Cell 26: 4270–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbstová M, Tietz S, Kinzel C, Turkina MV, Kirchhoff H (2012) Architectural switch in plant photosynthetic membranes induced by light stress. Proc Natl Acad Sci USA 109: 20130–20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Chen Q, Zhu Y, Hu F, Zhang L, Ma Z, He Z, Huang J (2013) Arabidopsis thylakoid formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol Plant 6: 1673–1691 [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Tanaka H, Kato C, Iwasaki Y, Asahi T (2005) Molecular characterization of the ZKT gene encoding a protein with PDZ, K-Box, and TPR motifs in Arabidopsis. Biosci Biotechnol Biochem 69: 972–978 [DOI] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Aro EM (2015) Photosystem II repair in plant chloroplasts: regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim Biophys Acta 1847: 900–909 [DOI] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro EM (2011) Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem J 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Johnson X, Vandystadt G, Bujaldon S, Wollman FA, Dubois R, Roussel P, Alric J, Béal D (2009) A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth Res 102: 85–93 [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E, Naveh L, Adam Z (2007) The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz SJ, Prochnik SE, Grossman AR, Merchant SS (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem 286: 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2014) Phosphorylation of photosystem II core proteins prevents undesirable cleavage of D1 and contributes to the fine-tuned repair of photosystem II. Plant J 79: 312–321 [DOI] [PubMed] [Google Scholar]
- Keren N, Ohkawa H, Welsh EA, Liberton M, Pakrasi HB (2005) Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17: 2768–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H. (2014) Diffusion of molecules and macromolecules in thylakoid membranes. Biochim Biophys Acta 1837: 495–502 [DOI] [PubMed] [Google Scholar]
- Kirchhoff H, Sharpe RM, Herbstova M, Yarbrough R, Edwards GE (2013) Differential mobility of pigment-protein complexes in granal and agranal thylakoid membranes of C3 and C4 plants. Plant Physiol 161: 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivuniemi A, Aro EM, Andersson B (1995) Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry 34: 16022–16029 [DOI] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ (2012) Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15: 245–251 [DOI] [PubMed] [Google Scholar]
- Kosourov S, Makarova V, Fedorov AS, Tsygankov A, Seibert M, Ghirardi ML (2005) The effect of sulfur re-addition on H2 photoproduction by sulfur-deprived green algae. Photosynth Res 85: 295–305 [DOI] [PubMed] [Google Scholar]
- Kouřil R, Oostergetel GT, Boekema EJ (2011) Fine structure of granal thylakoid membrane organization using cryo electron tomography. Biochim Biophys Acta 1807: 368–374 [DOI] [PubMed] [Google Scholar]
- Kouřil R, Wientjes E, Bultema JB, Croce R, Boekema EJ (2013) High-light vs. low-light: effect of light acclimation on photosystem II composition and organization in Arabidopsis thaliana. Biochim Biophys Acta 1827: 411–419 [DOI] [PubMed] [Google Scholar]
- Kropat J, Hong-Hermesdorf A, Casero D, Ent P, Castruita M, Pellegrini M, Merchant SS, Malasarn D (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J 66: 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka MR, Goldschmidt-Clermont M, van Dillewijn J, Rochaix JD (1989) Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell 58: 869–876 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lemaire C, Wollman FA (1989) The chloroplast ATP synthase in Chlamydomonas reinhardtii. I. Characterization of its nine constitutive subunits. J Biol Chem 264: 10228–10234 [PubMed] [Google Scholar]
- Li XG, Duan W, Meng QW, Zou Q, Zhao SJ (2004) The function of chloroplastic NAD(P)H dehydrogenase in tobacco during chilling stress under low irradiance. Plant Cell Physiol 45: 103–108 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Buschmann C, Döll M, Fietz HJ, Bach T, Kozel U, Meier D, Rahmsdorf U (1981) Photosynthetic activity, chloroplast ultrastructure, and leaf characteristics of high-light and low-light plants and of sun and shade leaves. Photosynth Res 2: 115–141 [DOI] [PubMed] [Google Scholar]
- Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, Andersson B (2000) The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V, Chou IT, Scott SV, Ettinger WF, Theg SM, Gasser CS (1994) Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J Biol Chem 269: 7863–7868 [PubMed] [Google Scholar]
- Liu C, Willmund F, Whitelegge JP, Hawat S, Knapp B, Lodha M, Schroda M (2005) J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol Biol Cell 16: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Malnoë A, Wang F, Girard-Bascou J, Wollman FA, de Vitry C (2014) Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions. Plant Cell 26: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Tokutsu R, Minagawa J (2014) Transcriptional regulation of the stress-responsive light harvesting complex genes in Chlamydomonas reinhardtii. Plant Cell Physiol 55: 1304–1310 [DOI] [PubMed] [Google Scholar]
- Mayfield SP, Bennoun P, Rochaix JD (1987) Expression of the nuclear encoded OEE1 protein is required for oxygen evolution and stability of photosystem II particles in Chlamydomonas reinhardtii. EMBO J 6: 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler T, Mühlhaus T, Hemme D, Schöttler MA, Rupprecht J, Idoine A, Veyel D, Pal SK, Yaneva-Roder L, Winck FV, et al. (2014) Systems analysis of the response of photosynthesis, metabolism, and growth to an increase in irradiance in the photosynthetic model organism Chlamydomonas reinhardtii. Plant Cell 26: 2310–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L, Wostrikoff K, Wollman FA, Choquet Y (2006) Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Mulo P, Sakurai I, Aro EM (2012) Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta 1817: 247–257 [DOI] [PubMed] [Google Scholar]
- Nath K, Jajoo A, Poudyal RS, Timilsina R, Park YS, Aro EM, Nam HG, Lee CH (2013a) Towards a critical understanding of the photosystem II repair mechanism and its regulation during stress conditions. FEBS Lett 587: 3372–3381 [DOI] [PubMed] [Google Scholar]
- Nath K, Poudyal RS, Eom JS, Park YS, Zulfugarov IS, Mishra SR, Tovuu A, Ryoo N, Yoon HS, Nam HG, et al. (2013b) Loss-of-function of OsSTN8 suppresses the photosystem II core protein phosphorylation and interferes with the photosystem II repair mechanism in rice (Oryza sativa). Plant J 76: 675–686 [DOI] [PubMed] [Google Scholar]
- Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Naumann B, Stauber EJ, Busch A, Sommer F, Hippler M (2005) N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J Biol Chem 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B (2013) Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol 64: 609–635 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot (Lond) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordhues A, Schöttler MA, Unger AK, Geimer S, Schönfelder S, Schmollinger S, Rütgers M, Finazzi G, Soppa B, Sommer F, et al. (2012) Evidence for a role of VIPP1 in the structural organization of the photosynthetic apparatus in Chlamydomonas. Plant Cell 24: 637–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Pagliano C, Saracco G, Barber J (2013) Structural, functional and auxiliary proteins of photosystem II. Photosynth Res 116: 167–188 [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Peter GF, Thornber JP (1991) Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-proteins. J Biol Chem 266: 16745–16754 [PubMed] [Google Scholar]
- Petroutsos D, Busch A, Janssen I, Trompelt K, Bergner SV, Weinl S, Holtkamp M, Karst U, Kudla J, Hippler M (2011) The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23: 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre Y, Popot JL (1993) Identification of two 4-kDa miniproteins in the cytochrome b6f complex from Chlamydomonas reinhardtii. C R Acad Sci III 316: 1404–1409 [PubMed] [Google Scholar]
- Ramundo S, Casero D, Mühlhaus T, Hemme D, Sommer F, Crèvecoeur M, Rahire M, Schroda M, Rusch J, Goodenough U, et al. (2014) Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell 26: 2201–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimbault B, Esposito D, Drapier D, Choquet Y, Stern D, Wollman FA (2000) Identification of the initiation codon for the atpB gene in Chlamydomonas chloroplasts excludes translation of a precursor form of the beta subunit of the ATP synthase. Mol Gen Genet 264: 486–491 [DOI] [PubMed] [Google Scholar]
- Rintamäki E, Kettunen R, Aro EM (1996) Differential D1 dephosphorylation in functional and photodamaged photosystem II centers: dephosphorylation is a prerequisite for degradation of damaged D1. J Biol Chem 271: 14870–14875 [DOI] [PubMed] [Google Scholar]
- Rochaix JD, Kuchka M, Mayfield S, Schirmer-Rahire M, Girard-Bascou J, Bennoun P (1989) Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J 8: 1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rütgers M, Schroda M (2013) A role of VIPP1 as a dynamic structure within thylakoid centers as sites of photosystem biogenesis? Plant Signal Behav 8: e27037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samol I, Shapiguzov A, Ingelsson B, Fucile G, Crèvecoeur M, Vener AV, Rochaix JD, Goldschmidt-Clermont M (2012) Identification of a photosystem II phosphatase involved in light acclimation in Arabidopsis. Plant Cell 24: 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Schmollinger S, Mühlhaus T, Boyle NR, Blaby IK, Casero D, Mettler T, Moseley JL, Kropat J, Sommer F, Strenkert D, et al. (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Whitelegge JP, Beck CF, Wollman FA (2001) The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing. Plant Cell 13: 2823–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman FA, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Raffelt M, Lodha M, Schroda M (2007) Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas. Plant J 52: 286–295 [DOI] [PubMed] [Google Scholar]
- Silva P, Thompson E, Bailey S, Kruse O, Mullineaux CW, Robinson C, Mann NH, Nixon PJ (2003) FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. Plant Cell 15: 2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S, Allahverdiyeva Y, Suorsa M, Paakkarinen V, Vainonen J, Battchikova N, Aro EM (2007) TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem J 406: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6: 175–182 [PubMed] [Google Scholar]
- Strenkert D, Schmollinger S, Sommer F, Schulz-Raffelt M, Schroda M (2011) Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell 23: 2285–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M, Inoue Y, Minagawa J (1998) Rapid and discrete isolation of oxygen-evolving His-tagged photosystem II core complex from Chlamydomonas reinhardtii by Ni2+ affinity column chromatography. FEBS Lett 426: 140–144 [DOI] [PubMed] [Google Scholar]
- Sun X, Fu T, Chen N, Guo J, Ma J, Zou M, Lu C, Zhang L (2010) The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol 152: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Murata N (2005) Interruption of the Calvin cycle inhibits the repair of photosystem II from photodamage. Biochim Biophys Acta 1708: 352–361 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietz S, Puthiyaveetil S, Enlow HM, Yarbrough R, Wood M, Semchonok DA, Lowry T, Li Z, Jahns P, Boekema EJ, et al. (2015) Functional implications of photosystem II crystal formation in photosynthetic membranes. J Biol Chem 290: 14091–14106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Nurmi M, Kangasjärvi S, Aro EM (2008) Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochim Biophys Acta 1777: 1432–1437 [DOI] [PubMed] [Google Scholar]
- Tokutsu R, Kato N, Bui KH, Ishikawa T, Minagawa J (2012) Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J Biol Chem 287: 31574–31581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Minagawa J (2013) Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110: 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizioli M, Lazar C, Brugière S, Burger T, Salvi D, Gatto L, Moyet L, Breckels LM, Hesse AM, Lilley KS, et al. (2014) Deciphering thylakoid sub-compartments using a mass spectrometry-based approach. Mol Cell Proteomics 13: 2147–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjärvi E. (2013) Photoinhibition of photosystem II. Int Rev Cell Mol Biol 300: 243–303 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi E, Aro EM (1996) The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc Natl Acad Sci USA 93: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Uniacke J, Colón-Ramos D, Zerges W (2011) FISH and immunofluorescence staining in Chlamydomonas. Methods Mol Biol 714: 15–29 [DOI] [PubMed] [Google Scholar]
- Uniacke J, Zerges W (2007) Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell 19: 3640–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O, Wollman FA, Olive J (1985) Distribution of intrinsic and extrinsic subunits of the PSII protein complex between appressed and non-appressed regions of the thylakoid membrane: an immunocytochemical study. FEBS Lett 183: 245–250 [Google Scholar]
- Vallon O, Wollman FA, Olive J (1986) Lateral distribution of the main protein complexes of the photosynthetic apparatus in Chlamydomonas reinhardtii and in spinach: an immunocytochemical study using intact thylakoid membranes and PS II enriched membrane preparation. Photobiochem Photobiophys 12: 203–220 [Google Scholar]
- Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL (2004) Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol 136: 3594–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg G, Levitan A, Klein T, Dangoor I, Keren N, Danon A (2014) Knockdown of the Arabidopsis thaliana chloroplast protein disulfide isomerase 6 results in reduced levels of photoinhibition and increased D1 synthesis in high light. Plant J 78: 1003–1013 [DOI] [PubMed] [Google Scholar]
- Woessner JP, Gillham NW, Boynton JE (1986) The sequence of the chloroplast atpB gene and its flanking regions in Chlamydomonas reinhardtii. Gene 44: 17–28 [DOI] [PubMed] [Google Scholar]
- Wu HY, Liu MS, Lin TP, Cheng YS (2011) Structural and functional assays of AtTLP18.3 identify its novel acid phosphatase activity in thylakoid lumen. Plant Physiol 157: 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder T, Xu W, Liu Q, Wanner G, Leister D, Pribil M (2013) The major thylakoid protein kinases STN7 and STN8 revisited: effects of altered STN8 levels and regulatory specificities of the STN kinases. Front Plant Sci 4: 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Tokutsu R, Bergner SV, Scholz M, Minagawa J, Hippler M (2015) PHOTOSYSTEM II SUBUNIT R is required for efficient binding of LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 to photosystem II-light-harvesting supercomplexes in Chlamydomonas reinhardtii. Plant Physiol 167: 1566–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamatani H, Sato Y, Masuda Y, Kato Y, Morita R, Fukunaga K, Nagamura Y, Nishimura M, Sakamoto W, Tanaka A, et al. (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll: protein complexes during leaf senescence. Plant J 74: 652–662 [DOI] [PubMed] [Google Scholar]
- Ye F, Zhang M (2013) Structures and target recognition modes of PDZ domains: recurring themes and emerging pictures. Biochem J 455: 1–14 [DOI] [PubMed] [Google Scholar]
- Yokthongwattana K, Chrost B, Behrman S, Casper-Lindley C, Melis A (2001) Photosystem II damage and repair cycle in the green alga Dunaliella salina: involvement of a chloroplast-localized HSP70. Plant Cell Physiol 42: 1389–1397 [DOI] [PubMed] [Google Scholar]
- Zerges W, Rochaix JD (1998) Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol 140: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (1999) Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274: 16062–16067 [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (2000) Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12: 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz M, Ferenc A, Wasilewska W, Romanowska E (2012) High light stimulates Deg1-dependent cleavage of the minor LHCII antenna proteins CP26 and CP29 and the PsbS protein in Arabidopsis thaliana. Planta 235: 279–288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.