Light-inducible in vitro transcription from an Arabidopsis gene observed only from chromatin but not from naked DNA defines higher order of DNA structure.
Abstract
In vitro transcription is an essential tool to study the molecular mechanisms of transcription. For over a decade, we have developed an in vitro transcription system from tobacco (Nicotiana tabacum)-cultured cells (BY-2), and this system supported the basic activities of the three RNA polymerases (Pol I, Pol II, and Pol III). However, it was not suitable to study photosynthetic genes, because BY-2 cells have lost their photosynthetic activity. Therefore, Arabidopsis (Arabidopsis thaliana) in vitro transcription systems were developed from green and etiolated suspension cells. Sufficient in vitro Pol II activity was detected after the minor modification of the nuclear soluble extracts preparation method; removal of vacuoles from protoplasts and L-ascorbic acid supplementation in the extraction buffer were particularly effective. Surprisingly, all four Arabidopsis Rubisco small subunit (rbcS-1A, rbcS-1B, rbcS-2B, and rbcS-3B) gene members were in vitro transcribed from the naked DNA templates without any light-dependent manner. However, clear light-inducible transcriptions were observed using chromatin template of rbcS-1A gene, which was prepared with a human nucleosome assembly protein 1 (hNAP1) and HeLa histones. This suggested that a key determinant of light-dependency through the rbcS gene transcription was a higher order of DNA structure (i.e. chromatin).
In plants, the molecular mechanisms of light-dependent gene regulation remain largely unknown. Generally, transcription is the major regulation step during gene expression in the eukaryotic genome. In plants, most genes are mainly regulated at the level of transcription, and the expressions of many genes depend on surrounding light conditions relating to photosynthesis, photomorphogenesis, phototropism, circadian rhythms, gametogenesis, etc. Therefore, light-dependent transcription has been studied intensively. Three distinct classes of photoreceptors have been discovered: phytochromes, blue-light receptors (cryptochromes, phototropins, and ZTL / FKF1 / LKP2 family), and photoreceptors for UV-B (Banerjee and Batschauer, 2005). These photoreceptors affect downstream gene expression mediated by the specific binding of transcription factors to cis-regulatory elements in promoters. Light regulatory elements are essential cis-regulatory elements found in photo-responsible genes, such as the conserved modular array 5 (CMA5) on the Rubisco small subunit (rbcS) genes (López-Ochoa et al., 2007). Our knowledge of transacting factors that associate with light regulatory elements is quite limited, mainly because plant materials are inconvenient to use for biochemical research. Therefore, to understand transcription regulation comprehensively, we need to develop a plant-specific in vitro transcription system.
The eukaryotic (human cell) in vitro transcription system was reported by Weil et al. (1979) and Manley et al. (1980). Subsequently, Dignam et al. (1983) established finely tuned systems. Nowadays, in vitro transcription systems are indispensable for molecular biology; however, this technique has not been used routinely in plants, because plant materials are not particularly suitable for biochemical procedures. Although there were several early attempts to develop a plant in vitro transcription system (for review, see Sugiura, 1997), their applications have not been fully expanded. After that several practically useful in vitro transcription systems were established: Pol I-dependent system from broccoli (Brassica oleracea) by extensively purified Pol I holoenzyme (Saez-Vasquez and Pikaard, 1997; Saez-Vasquez et al., 2003), Pol II-dependent system by rice (Oryza sativa) whole cell extract (Zhu et al., 2002), and Pol IV and Pol V-dependent system from Arabidopsis (Arabidopsis thaliana) by immunopurification (Haag et al., 2012). And our group also established a practical in vitro transcription system from tobacco (Nicotiana tabacum) BY-2 cultured cells (Fan and Sugiura, 1995; Nagata et al., 1992; Yukawa and Sugiura, 2004).
The BY-2 cells have several advantages for biochemical study. They can be easily maintained in artificial medium, they grow rapidly, protoplasts are easy to prepare, and they have large nuclei and fewer plastids. The BY-2 in vitro transcription system was useful for basal studies of the three major RNA polymerase activities: Pol I (e.g. to transcribe clustered rRNAs genes; Fan et al., 1995), Pol II (e.g. to transcribe β-1, 3-glucanase gene, and light-harvesting chlorophyll a/b-binding protein (lhcb1) genes; Fan and Sugiura, 1995; Hasegawa et al., 2003b), and Pol III (e.g. to transcribe tRNA genes, U3 snoRNA genes, U6 snRNA gene, 7SL RNA genes, SINE retroposon, and 5S rRNA genes; Akama et al., 1998; Arnaud et al., 2001; Cloix et al., 2002; Hasegawa et al., 2003a; Mathieu et al., 2003; Yukawa et al., 2005; Dieci et al., 2006; Yukawa et al., 2011). However, the BY-2 cells are nonphotosynthetic. Thus, transcriptions from photosynthesis-related genes must be suppressed in the BY-2 system. However, a lhcb1 gene and rbcS genes were efficiently transcribed using the BY-2 nuclear extracts (Hasegawa et al., 2003b). To overcome this deficiency, we needed to develop another in vitro transcription system using Arabidopsis cultured cells.
The Arabidopsis MM2d suspension cell culture was established from accession Landsberg erecta as a rapidly growing cell culture permitting study of the cell cycle. The MM2d cell culture can be efficiently synchronized by controlling Suc in the medium or by applying an aphidicolin. MM2d cells were derived from MM1 green cell culture by being cultured in continuous darkness for more than 2 years. They show faster growth than MM1 but retain the ability to develop chloroplasts under light conditions (Menges and Murray, 2002). Thus, MM2d cells might be the best material from which to prepare an in vitro transcription system.
Here, we describe the preparation of a nuclear extract from MM2d cells cultured under different light conditions and show regulation of photo-responsive gene transcription from the rbcS gene using chromatin templates.
RESULTS
Preparation of Nuclear Extracts from Etiolated Arabidopsis Cells
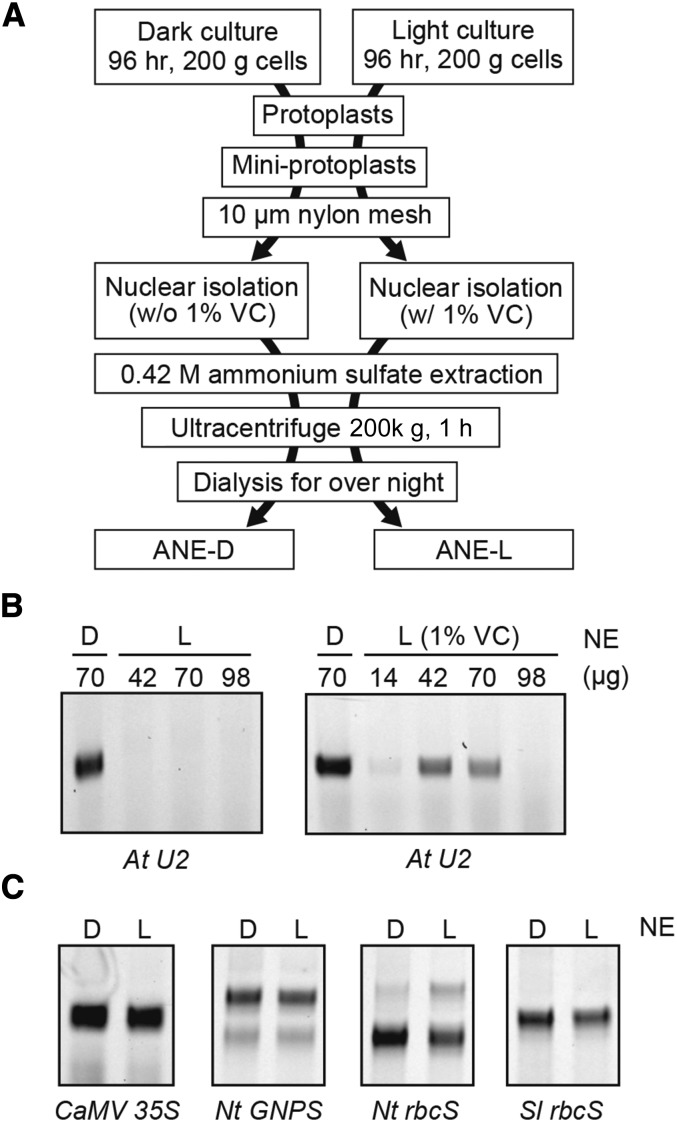
Initially, we prepared nuclear soluble extracts from etiolated Arabidopsis MM2d cell culture by the same procedure used for tobacco cultured cells (BY-2). Unfortunately, the method did not work for MM2d cells; no transcription was observed (data not shown). We then evaluated evacuolation (removal of vacuoles) from MM2d protoplasts (Igarashi et al., 2000; Hamada et al., 2013) before nuclei isolation. Nuclease-rich vacuoles could be efficiently separated from protoplasts using Percoll-mediated centrifugation (see “Experimental Procedures”), and nuclei were isolated from the resulting vacuole-less mini-protoplasts. As shown in Figure 1, the evacuolation step was so effective, because a substantial in vitro transcription signal (detected as the reverse transcription product from the labeled primer) was observed from a highly expressed Arabidopsis U2 small nuclear RNA (At U2) gene using the obtained Arabidopsis MM2d nuclear extracts (ANE-D).
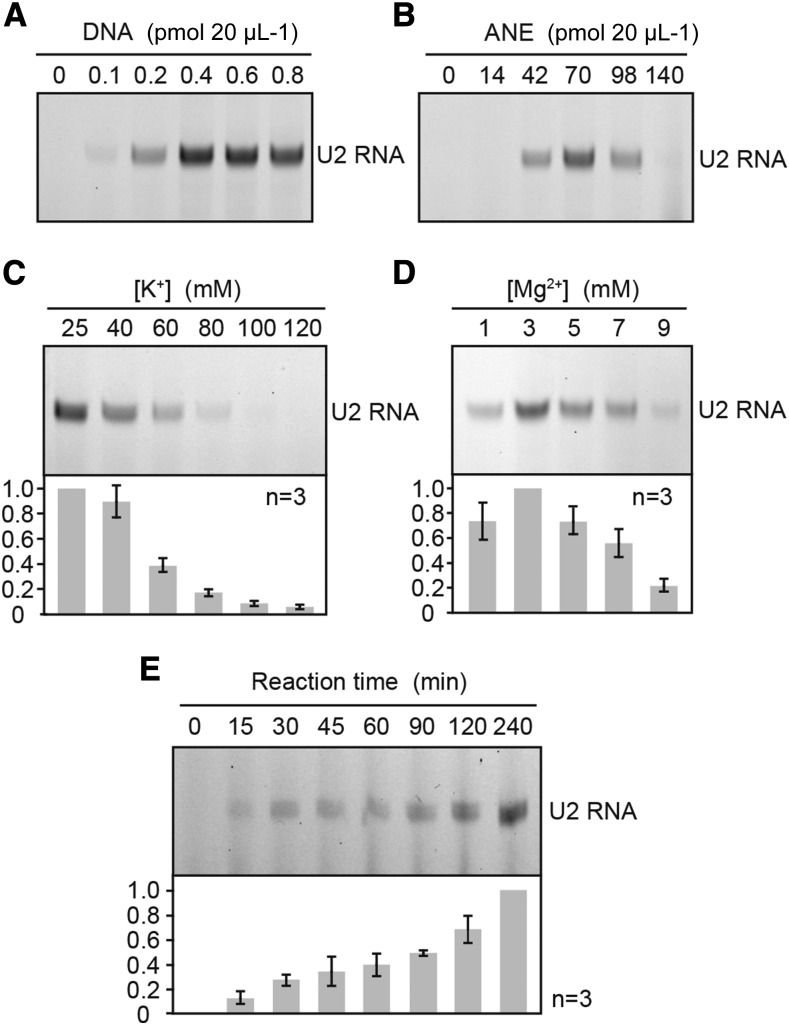
Figure 1.
Determination of optimal conditions for transcription in ANE. A and B, Effect of the amount of template DNA and ANE-D. In vitro transcription reactions were carried out in the presence of (A) 0 to 0.8 pmol of a circular plasmid containing At U2 gene containing plasmid (pAtU2.2) and (B) 0 to 140 µg / 20 µL ANE-D and 0.4 pmol pAtU2.2 for 90 min at 28°C. C and D, Optimal concentrations of K+ and Mg2+ for transcription. Incubations were carried out in the presence of 0.4 pmol of pAtU2.2 and either at various K+ (C) or Mg2+ (D) for 90 min at 28°C. Three independent assays were performed for each construct, and error bars indicate se. E, Time course of transcription in ANE-D. Incubations were carried out in the presence of 0.4 pmol of pAtU2.2 for 0 to 240 min at 28°C. Three independent assays were performed for each construct, and error bars indicate se. RNA products were analyzed for an 8% (w/v) polyacrylamide / 8 m urea gel.
Optimization of the reaction conditions for in vitro transcription was performed using the At U2 gene. Transcription signals were seen from 0.1 pmol DNA in a 20-μL reaction that used circular plasmid DNA. Transcription increased with rising amounts of DNA and became saturated by 0.4 pmol (Fig. 1A). The optimal amount of nuclear extract was determined as approximately 70 μg/20-μL reaction (Fig. 1B). Interestingly, a lower potassium concentration was required (25 mM; Fig. 1C), which was the lowest possible concentration in this reaction because nuclear extract contained 100 mm KOAc. The optimum magnesium ion concentration was 3 mM, and transcription gradually decreased with increasing Mg2+ concentrations (Fig. 1D). Transcription level increased stably up to 4 h of incubation (Fig. 1E); however, we did not measure the levels of the above-mentioned variables for longer incubation times.
Arabidopsis in vitro transcription levels were compared with the tobacco system. For this purpose, At U2 gene, the tobacco β-1,3-glucanase (Nt GNPS) gene, and tobacco and tomato (Solanum lycopersicum) rbcS (Nt rbcS and Sl rbcS) genes were selected, and transcriptions were detected by primer extension. As a result, the genes showed almost equivalent signal strength in the Arabidopsis system to those in the tobacco system (Fig. 2A). All transcriptions from ANE-D were RNA Pol II-dependent, because they were sensitive to a low concentration (1 μg mL−1) of α-amanitin (Fig. 2B). Interestingly, in both systems, rbcS genes transcription was observed.
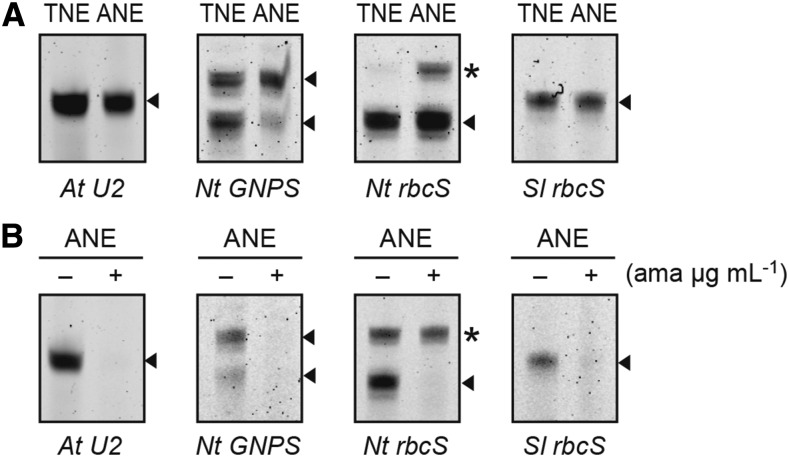
Figure 2.
In vitro transcription of Pol II-dependent genes with ANE-D. A, In vitro transcription assay from At U2, Nt GNPS gene, and Nt rbcS and Sl rbcS genes were compared with tobacco nuclear extracts (TNE) and ANE-D. At U2 gene was used as a positive control for Pol II activity. Expected extended products of reverse transcription from labeled primer are indicated by arrowheads, and α-amanitin tolerant signals are indicated by asterisks. B, Reaction conditions when α-amanitin (1 µg mL−1) was added to the indicated concentrations. Reactions were performed with (+) or without (-) α-amanitin. RNA products were analyzed for an 8% (w/v) polyacrylamide / 8 m urea gel.
Although the transcript profiles were similar in both plant systems, two obvious differences were observed. First, the Nt GNPS gene has two major transcription start sites on its DNA, and the lower-migrating band represented the main transcription signal used in Arabidopsis. Second, two clear extended products were shown from the Nt rbcS gene in the Arabidopsis system, but the lower-migrating signal was not Pol II-dependent, because it tolerated a low concentration of α-amanitin.
RNA Pol III Transcription with the Arabidopsis System
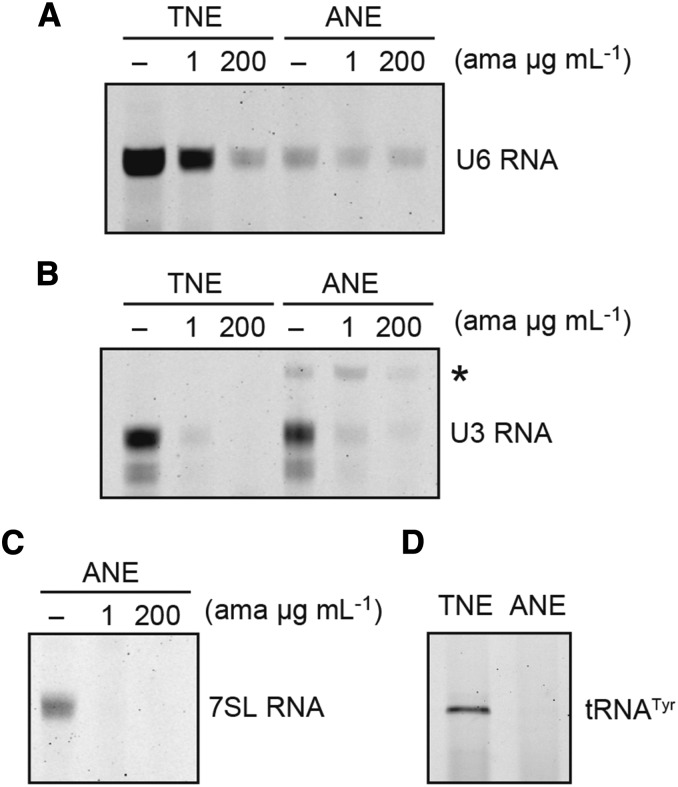
The tobacco nuclear extracts (TNE) exhibit substantial RNA Pol III transcription; therefore, we checked for Pol III transcription in the ANE-D. As shown in Figure 3A, the At U6 gene was efficiently transcribed by TNE, which showed clear Pol III dependency, because it could tolerate a low concentration of α-amanitin (1 μg mL−1) and was sensitive to a high concentration of α-amanitin (200 μg mL−1). By contrast, using ANE-D, less transcription was observed from the At U6 gene, and this transcription was not completely inhibited by 200 μg mL−1 α-amanitin; therefore, this low level of transcription was not Pol III dependent. However, as trace amounts of transcription were detected without α-amanitin, an additional two Pol III-dependent genes were tested: Arabidopsis U3 snoRNA and 7SL RNA genes (Fig. 3, B and C). Generally, U3 snoRNA genes are Pol II dependent; however, in plants, they are exceptionally transcribed by Pol III (Kiss et al., 1991). Both of the above genes were transcribed in ANE-D but were Pol II dependent and completely inhibited by a low concentration of α-amanitin. All the above genes belong to the plant U6 snRNA gene type promoter, which had Upstream Sequence Element (USE) and TATA-like sequence as conserved cis-sequences. These cis-elements have shown partial compatibility with Pol II and Pol III (Yukawa and Sugiura, 2004); we further tested a tRNA gene that shows strict specificity for Pol III. The tobacco nuclear tRNATyr gene was not transcribed in the Arabidopsis system at all (Fig. 3D); therefore, we concluded that the ANE-D did not include any Pol III activity.
Figure 3.
In vitro transcription of Pol III-dependent genes with ANE-D. A and B, In vitro transcription assay from At U6 gene and At U3 gene compared with TNE and ANE-D. Reactions were carried out for Pol II and Pol III with α-amanitin (1 µg mL−1 and 200 µg mL−1) or without (-); α-amanitin tolerant signal is indicated by an asterisk. C, In vitro transcription assay from At 7SL gene with ANE-D. Reaction conditions were the same as (B). D, In vitro transcription assay from tobacco tRNATyr gene with TNE and ANE-D. RNA products were analyzed for an 8% (w/v) polyacrylamide/8 m urea gel.
Nuclear Extracts from Green MM2d Cell Culture
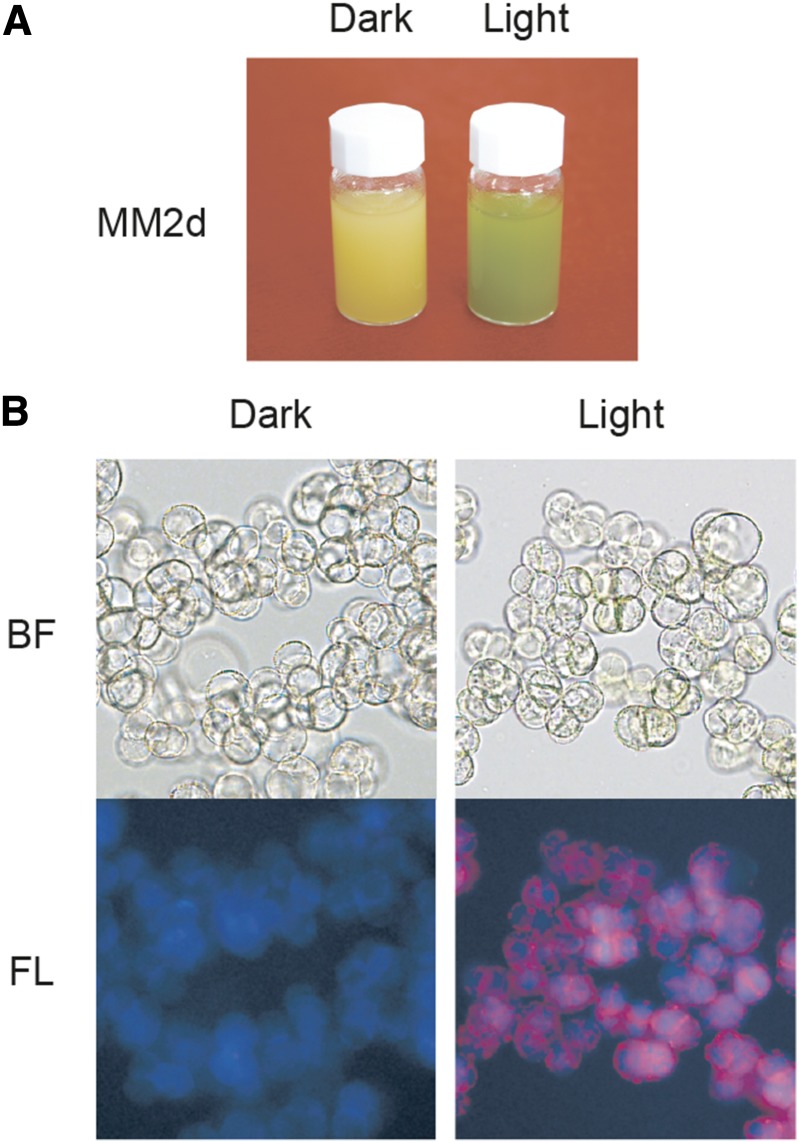
The MM2d cells were maintained under continuous darkness, but they retained their greening ability when transferred into the light (Menges and Murray, 2002). In addition to light, auxin / cytokinin addition to the medium also causes cell greening. As shown in Figure 4A, color differences were observed between cultures at 0.9 μm 2,4-dichlorophenoxyacetic acid (2,4-D) / no kinetin / dark (left; creamy yellow) and at no 2,4-D / 25 nm kinetin / light / 3% (v/v) CO2 flow (right; green). Cell shapes were similar, but only green cells formed chloroplasts (Fig. 4B).
Figure 4.
Characterization of Arabidopsis MM2d cultured cells. A, MM2d cells growing under dark condition / 0.9 µM 2,4-D / without kinetin, and continuous light conditions (150 µmol photons m−2 sec−1) / without 2,4-D / 25 nm kinetin / aerating 3% (v/v) CO2 flow in air. B, Microscope photos of MM2d dark and light conditions. BF, brightfield; FL, autofluorescence.
Unfortunately, the nuclear extraction method for the MM2d dark culture mentioned above did not work for the green cells (Fig. 5B, left). De novo transcription from the At U2 gene was not detected using the obtained light nuclear extract. We then modified the preparation method. The most effective change was 1% (w/v) L-ascorbic acid supplementation during nuclear isolation processes (Fig. 5B, right). Substantial transcription was detected by 42–70 µg (in 20 μL reaction) using modified green nuclear extract (ANE-L). We then compared the transcriptions of several nuclear genes using the two Arabidopsis nuclear extracts derived from dark (ANE-D) and light (ANE-L). Cauliflower mosaic virus (CaMV) 35S promoter, Nt GNPS, Nt rbcS, and Sl rbcS were all equally transcribed with ANE-D and ANE-L (Fig. 5C).
Figure 5.
Preparation of improved ANE-D and ANE-L. A, Preparation scheme of ANE-D and ANE-L. B, Effect of amount of ANE-L (L) for in vitro transcription of At U2 gene. Positive control of transcription was ANE-D (D). Effect of 1% (w/v) l-ascorbic acid sodium (VC) addition in preparation of nuclear extract is shown at right. C, In vitro transcription assay from CaMV 35S promoter, Nt GNPS gene, and Nt and Sl rbcS genes with ANE-D (70 µg) and ANE-L (42 µg) in 20 µL reactions.
Transcription from Photo-Dependent Genes
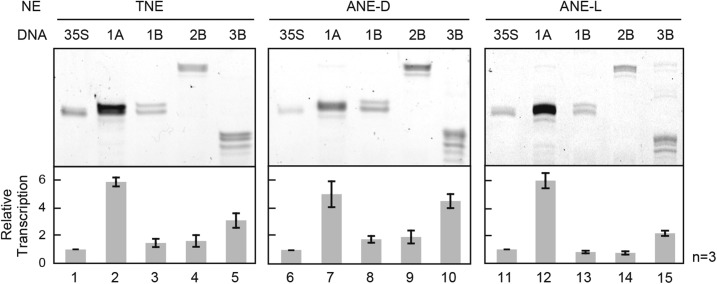
We then compared the relative transcription levels of four Arabidopsis rbcS genes (rbcS-1A, rbcS-1B, rbcS-2B, and rbcS-3B) with ANE-D, ANE-L, and TNE by the routine in vitro procedure as described above. The Arabidopsis rbcS gene family is classified into two subfamilies on the basis of linkage and sequence similarities: a single copy of type A (rbcS-1A) and the three copies of type B (rbcS-1B, rbcS-2B, and rbcS-3B; Krebbers et al., 1988). According to previous studies, 1A, 2B, and 3B genes are highly light responsive, but the 1B gene shows low photo-responsiveness (Dedonder et al., 1993).
In this assay, the transcription levels generated from rbcS genes in vitro were monitored by comparing them with those from CaMV 35S promoter as a nonlight-dependent standard (Fig. 6). Consequently, no significant differences were observed among three different in vitro systems. In particular, in all systems, the strongest transcription levels were observed from the 1A gene, being 5 to 6 times stronger than those from the 35S promoter. By contrast, the 1B and 2B genes showed weak transcription, and all the type B genes showed reverse light-inducibility: stronger expression in the dark and weaker in the light.
Figure 6.
In vitro transcription of four wild-type Arabidopsis rbcS genes. The relative transcription levels of four Arabidopsis rbcS genes (rbcS-1A, 1B, 2B, and 3B) were compared with in vitro transcription of TNE, ANE-D, and ANE-L, respectively. The CaMV 35S promoter was used as a control. Three independent assays were performed for each construct, and error bars indicate se.
Transcription from Chromatin Templates
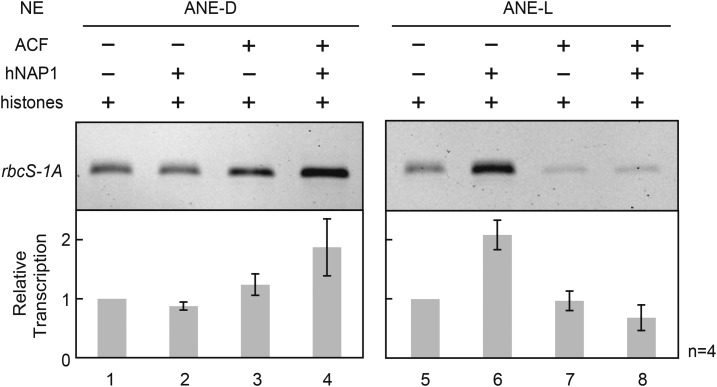
Clear photo-dependency of in vitro transcription could not be observed from Arabidopsis rbcS genes in both ANE-D and ANE-L; therefore, we hypothesized that light-dependent transcription might be largely controlled by chromatin. To confirm this, chromatin templates were in vitro prepared using histone chaperones, hNAP1 (human nucleosome assembly protein 1), and ACF (ATP-utilizing chromatin assembly and remodeling factor) complex, and histones and subjected to in vitro transcription reaction using ANE-D and ANE-L. An Arabidopsis rbcS-1A gene was chosen for this assay, because it showed the highest level and photo-dependent transcription among four members in vivo. Commercially available hNAP1 is highly similar to the four candidates in the Arabidopsis genome (NAP1;1, NAP1;2, NAP1;3, and NAP1;4; see Supplemental Fig. S2).
As shown in Figure 7, convincing light dependency of transcription was observed from rbcS-1A chromatin template prepared by hNAP1 and HeLa histones. The relative transcription levels with ANE-L were twice those with ANE-D. The up-regulation of rbcS-1A gene transcription by green nuclear extract had a rather minor effect, but it was the first observed and quite significant feature in this sort of assay. By contrast, the relative transcription levels from rbcS-1A chromatin template prepared only with ACF, or using hNAP1 and ACF, showed opposite effects to right photo-inducibility; they were higher with ANE-D than those with ANE-L. At least in this in vitro assay, human ACF had no effect on light-dependent transcription regulation.
Figure 7.
Effect of chromatin templates on light-dependent in vitro transcription from Arabidopsis rbcS-1A gene. Chromatin templates prepared with different combinations of human histones and histone chaperons (hNAP1 and ACF complex). They were subjected to in vitro transcription reactions using ANE-D and ANE-L. Control was HeLa histones alone (lanes 1, 5); the chromatin assembly reactions were carried out without ACF complex (lanes 2, 6) as well as hNAP1 (lanes 3, 7). Four independent assays were performed for each construct, and error bars indicate se.
DISCUSSION
In Vitro Transcription System from Arabidopsis Etiolated Cell Culture
Arabidopsis is a powerful model plant, because it was the first plant species whose whole genome was determined. This tiny Brassicaceae plant contains almost the minimum genome size (approximately 125 Mbp) in plants, has a short generation time, and can be easily transformed. Abundant postgenomic data and mutant lines are currently available. However, the small plant size would be a disadvantage for biochemical research. In particular, the molecular mechanisms of gene expression in this plant are still unclear compared with the other plants and kingdoms (e.g. animals, fungi, and bacteria). However, the use of plant cell culture could overcome this limitation. Arabidopsis MM2d cells grow rapidly and retain their greening ability (Menges and Murray, 2002). Therefore, we decided to prepare transcriptionally active nuclear soluble extract both from etiolated and greening cell cultures to study their photo-inducible gene expression.
Consequently, to obtain transcriptionally active extracts, two modifications of the procedure were required. First was evacuolation after protoplast preparation, and the second was 1% (w/v) L-ascorbic acid supplementation during nuclei isolation. Plant vacuoles include high protease and nuclease activities; therefore, they often degrade extracted soluble proteins. Evacuolation effect has already been shown by Frohnmeyer et al. (1994) when they established a light-responsive in vitro transcription system from parsley cell culture. As for Arabidopsis cells, they are likely to include more hydrolytic enzymes than tobacco BY-2 cells. Actually, whole cell extracts prepared from Arabidopsis cell culture for in vitro translation have shown high protease activities (Murota et al., 2011). The second was 1% (w/v) L-ascorbic acid supplementation during nuclei isolation. In general, plant materials also accumulate polyphenolic substances that are easily oxidized and turned into chemically active forms, which then damage many compounds in intact cells. It is reasonable to assume that such photo-reactive polyphenols are present in larger amounts in green cells than in etiolated cells. Therefore, we hypothesized that 1% (w/v) L-ascorbic acid would prevent such oxidization.
In plants, in vitro transcription has never been generally applied, which is a critical difference compared with other organisms. Several attempts to establish plant-specific in vitro transcription systems have not been successful because of the difficulties of working with plant materials. However, in this study, we established such systems from Arabidopsis, which were achieved by minor modifications of the method used to prepare the tobacco system. Previously, a report of Arabidopsis in vitro transcription system was done by Haag et al. (2012) in which they established a fine system to dissect RNA Pol IV and Pol V molecular functions. The transcription elongation activities by Pol IV and Pol V as well as Pol II have been shown using affinity-purified fractions and a tripartite or bipartite RNA extension template (DNA-RNA hybrid) mimicking RNA Pol transcription bubble. While our system is prepared by nuclear extraction and de novo transcripts are yielded from double-strand DNA templates. Therefore, this is the first report, to our knowledge, of the establishment of a complete in vitro Pol II-dependent transcription system in Arabidopsis. The activities of Arabidopsis nuclear extracts were comparable with those from tobacco cells and support transcription initiation and elongation, and probably also termination.
Differences between Arabidopsis and tobacco
Two differences were observed between our Arabidopsis and tobacco systems. First, we concluded that the Arabidopsis nuclear extracts did not include any Pol III activity, which could be explained as follows. Some transcription from Pol III-dependent U6 snRNA, U3 snoRNA, and 7SL RNA genes was observed in the Arabidopsis system; however, they were Pol II-dependent transcriptions, because all transcriptions were inhibited by a low concentration (1 μg mL−1) of α-amanitin (Fig. 3, A–C). No transcripts were observed from the tRNA gene (Fig. 3D). According to Willis (1993), Pol III-dependent (class III) genes are classified into four types by their promoter organization. Type 1 and 2 are 5S rRNA and tRNA genes, respectively; both types have cis-regulatory sequences within the transcribed region. Type 3 genes have upstream promoters only, namely USE and TATA-like sequence (TATA). The U6 snRNA and U3 snoRNA belong to this type. In general, U3 snoRNA genes in eukaryotes are transcribed by Pol II, but plants are an exception (Kiss et al., 1991). Type 4 has a mixed promoter organization of the above three types, and the 7SL RNA gene is just this type. Pol II-dependent (class II) and spliceosomal U1, U2, U4, and U5 snRNA genes also have USE and TATA and show similarity to class III type 3 and 4 genes. However, the gap length between the USE and TATA sequences in the DNA are 10 bp shorter than those of type 3 to 4 genes, and this has been considered a critical determinant of the two Pols’ specificities (Waibel and Filipowicz, 1990). Here, we observed slight Pol II transcriptions that occurred in a leaky manner from class III type 3 to 4 genes (Fig. 3). Thus, the Pol specificity of plant U snRNA genes is uncertain and should be further studied by means of tobacco and Arabidopsis in vitro systems. We do not know why the Arabidopsis systems lack Pol III activity. It is likely related to the MM2d cells; therefore, we should try cultures of other Arabidopsis cells to obtain Pol III-dependent transcription in vitro.
Second, species specificities of transcription were observed between the Arabidopsis and tobacco systems. When a Nt GNPS gene was transcribed, two equally strong transcription start sites were observed with the tobacco system. However, in the Arabidopsis system, the site producing the upper (lower migrating) band on Figure 2A was much stronger. When the Nt rbcS gene was transcribed with Arabidopsis system, additional nonspecific transcription was observed, because it was not inhibited by α-amanitin (Fig. 2B). These differences suggested that the fine dissection of plant gene regulation in vitro requires a species-specific assay system. Thus, our Arabidopsis in vitro system is ideal for studying Arabidopsis genes.
In Vitro Transcription System from Arabidopsis Green Cell Culture
One significant plant-specific characteristic must be their photosynthetic ability. In this context, we were interested in photo-inducible gene transcription regulation. The particular aim of this study was to develop an in vitro assay for photo-inducible gene transcription and to determine what factors control it under light conditions. As mentioned above, the plant light-dependent in vitro transcription system was already been developed from parsley cells (Frohnmeyer et al., 1994), but no report has been made for two decades using the parsley system. Therefore, we believe that another light-dependent in vitro transcription system had to be developed from Arabidopsis cell cultures growing under light condition.
In Arabidopsis, the rbcS gene family comprises four members: rbcS-1A, rbcS-1B, rbcS-2B, and rbcS-3B. In this report, these four members were differentially transcribed with three kinds of nuclear extracts: TNE, ANE-D, and ANE-L; however, each gene showed similar properties in all three systems (Fig. 6). Particularly, rbcS-1A showed the strongest in vitro transcription, and the other B-type genes were suboptimal. In particular, rbcS-1B was remarkably weak. These results were quite surprising for two reasons. First, even in the two dark systems (TNE and ANE-D), all rbcS genes were unexpectedly and highly transcribed, and their levels were almost equal to those in the light system (ANE-L). One plausible explanation for this result is that some repressive factors have been removed from our original in vitro transcription systems. They were in an artificial state using naked DNA as templates for transcription; therefore, they might be free from the inhibitory properties by chromatin. Thus, it is possible that major repression of rbcS under dark conditions is not performed by a specific transcription repressor that can bind to DNA directly. Second, each rbcS gene had a similar transcription profile among all the systems. Dedonder et al. (1993) and Yoon et al. (2001) reported that rbcS-1A is the major form and is highly expressed, whereas rbcS-1B, rbcS-2B, and rbcS-3B are less transcribed. In addition, rbcS-1A, rbcS-2B, and rbcS-3B are controlled under phytochrome induction, although rbcS-1B is not photo-inducible and shows the weakest transcription among them. These properties corresponded well with our in vitro results. This character of rbcS-1B must be linked to a defect of the CMA5 module in the promoter region. CMA5 was characterized as a minimal photo-regulatory element module found in Arabidopsis rbcS genes and comprises an I-box-associated module (IbAM5), an I-box, and a G-box (López-Ochoa et al., 2007). It has been suggested that the lack of a G-box on rbcS-1B causes lack of photo-responsiveness. As mentioned above, rbcS-1B showed less transcription in vitro; therefore, this light-responsive cis-regulatory element should be studied further using our in vitro systems.
Photo-Inducible In Vitro Transcription with Chromatin Template
The eukaryotic genome is packaged into a structure known as chromatin. Chromatin is a dynamic structure consisting essentially of DNA and core histones and is a key determinant of nuclear processes, including DNA repair, replication, recombination, and transcription. Generally, nucleosomes impede the accessibility of other proteins to the DNA sequences; therefore, they function predominantly in transcriptional repression. To test the hypothesis that repression of photo-inducible gene transcription could be related to chromatin structure, in vitro transcriptions were performed from chromatin templates. As shown in Figure 7, the chromatin templates comprising the rbcS-1A plasmid prepared with HeLa histones and hNAP1 showed the expected result: lower transcription in the dark (ANE-D) and higher transcription in the light (ANE-L). This result was convincing, because histones are highly conserved among eukaryotic organisms, and NAP1 is also highly conserved from yeast (Saccharomyces cerevisiae) to humans and plants (Yoon et al., 1995; Dong et al., 2003; Park and Luger, 2006). In Arabidopsis, the NAP1 family comprises four gene members: Nap1;1, Nap1;2, Nap1;3, and Nap1;4. Except for C-terminal truncated NAP1;4, the three Arabidopsis NAP1 proteins are highly similar to human NAP1 (Supplemental Fig. S2). Especially the Arabidopsis NAP1;1, NAP1;2, and NAP1;3 have a C-terminal CaaX box motif, which is needed for protein farnesyl transferase recognition, and Nap1;1 gene can rescue delay mitosis and elongated bud phenotype of yeast Nap1 mutant (Galichet and Gruissem, 2006). However, the contribution of Arabidopsis NAP1s in the assembly of chromatin remains unclear, because triple mutants of ubiquitously expressed Nap1;1, Nap1;2, and Nap1;3 exhibit a normal phenotype (Liu et al., 2009a) in contrast to embryo lethality of Drosophila melanogaster Nap1 knockout inactivation (Lankenau et al., 2003). In terms of their functions, a nucleotide excision repair function (Liu et al., 2009a), cell proliferation and cell expansion during leaf development (Galichet and Gruissem, 2006), response to abscisic acid in seedling growth (Liu et al., 2009b), and somatic homologous recombination (Gao et al., 2012) were suggested. Our study indicated that at least hNAP1 and HeLa histones contribute to promote chromatin assembly with DNA containing plant-specific genes. Furthermore, the chromatin template drives light-dependent transcription in vitro. In the next stage of this study, we will further confirm whether Arabidopsis’ four NAP1s contribute to chromatin assembly using the in vitro transcription systems described here.
By contrast, the opposite effect was observed from a chromatin template prepared using the ACF complex as well as the hNAP1: both are prominent histone chaperones in humans. The ACF complex is an ATP-dependent factor that mediates energy-dependent chromatin assembly, translocates the core histone onto the DNA, and assembles periodically arrayed nucleosome in conjunction with NAP1 activity (Ito et al., 1997). The ACF complex comprises only two factors: ISWI (imitation switch) ATPase and ACF1 (Ito et al., 1999). ISWI is the catalytic component and the ACF1 enhances and modulates the activity of ISWI during chromatin assembly and nucleosome sliding (Tyler et al., 1996; Eberharter et al., 2001). Two Arabidopsis ISWI proteins, CHROMATIN REMODELING 11 and 17, were identified (Knizewski et al., 2008; Li et al., 2014) that showed high similarity to human ISWI (Supplemental Fig. S3). Double mutation of these two genes resulted in the loss of evenly spaced nucleosome location in the gene body (Li et al., 2014). A SLIDE domain located in the C-terminal region of ISWI could associate directly with a DDT (DNA binding homeobox and different transcription factors) domain of its counterparts. There are at least twelve DDT domain-containing proteins in Arabidopsis; however, the ACF1 ortholog is missing (Dong et al., 2013). Thus, the human ACF complex could not form a plant-specific ordered periodic array of nucleosomes and promote plant transcription depending on light conditions. Indeed, in vitro transcription with a chromatin template that was prepared only with hNAP1 showed an intermediate effect with extracts grown under light conditions. Therefore, a real plant-specific, ACF-like activity must exist and should be identified using our Arabidopsis in vitro transcription system.
DNA methylation, which is an epigenetic mark, strongly influences chromatin structure and affects gene expression, including transcription and gene silencing. In plants, the addition of a methyl group to a cytosine occurs on CHH and CHG sites as well as CG sites (for review, see Law and Jacobsen, 2010). In this study, however, we did not investigate this aspect, because the relationship between methylation levels and photo-inducible transcription is poorly understood. In addition, the control of position-specific methylation on a DNA template is technically difficult. Therefore, we just used plasmid DNA prepared from Escherichia coli strain XL1-blue for chromatin template. However, DNA methylation is critical to comprehend photo-inducible gene expression.
In conclusion, naked DNA, including the rbcS-1A gene, did not show photo-dependency in the newly developed Arabidopsis in vitro transcription system. Therefore, major repression under dark conditions by certain repressive transcription factors is not likely to occur. The nucleosome can be considered a hindrance to transcription, which must be allowed access of Pol II and transcription factors to the DNA sequences. In this context, key repression of photo-dependent transcription was probably caused by a special chromatin status. Further development of chromatin-dependent in vitro transcription systems will reveal the mechanism of transcription regulation in plants as well as its photo-responsiveness.
MATERIALS AND METHODS
Plasmids for In Vitro Transcription Assays
Plasmids pSI01, pSI02, pSI03, and pSI04 include the promoter regions from four Arabidopsis (Arabidopsis thaliana) rbcS genes: rbcS-1A, rbcS-1B, rbcS-2B, and rbcS-3B, respectively (Schwarte and Tiedemann, 2011). Plasmids pAt7SL1, pAtU2.2, pAtU3CP, and pBAU6-28 include Arabidopsis 7SL RNA, U2 snRNA, U3 snoRNA, and U6 snRNA gene promoters, respectively (Fan et al., 1995; Yukawa et al., 2005; Yukawa et al., 2013). Plasmids pNS10, pNtYI, and pGNPS include tobacco (Nicotiana tabacum) promoters from rbcS (Ntss23), tRNATyr, and β-1,3-glucanase genes, respectively (Mazur and Chui, 1985; Stange and Beier, 1986; Fan and Sugiura, 1995; Yukawa et al., 2001). Plasmids pYY0847 and pYI0805 contain just the CaMV 35S promoter and the CaMV 35S promoter fused to part of the coding region of Arabidopsis elongation factor 1Bα1 (Héricourt and Jupin, 1999), respectively. Plasmid pRBC-1657 includes the promoter from the tomato (Solanum lycopersicum) rbcS gene (Fan and Sugiura, 1995; for details, see Supplemental Fig. S1 and Supplemental Table S1).
All plasmids were constructed in the pBluescript II KS+ vector except for pYI0805 and pNtY1 (pUC19), and DNAs were prepared by transforming Escherichia coli strain XL1-Blue.
Preparation of Chromatin Templates for In Vitro Transcription Assays
Chromatin templates, including the Arabidopsis rbcS-1A gene fragment, were prepared using a Chromatin Assembly kit (Active Motif). Briefly, 7 μg of recombinant hNAP1 protein and 2.7 μg of HeLa core histones were mixed with high salt buffer and incubated on ice for 15 min to assemble the complex. The complex was then mixed with low salt buffer including an ATP regeneration system, 0.625 μg recombinant ACF complex, and 0.4 pmol circular pSI01 DNA and incubated at 27°C for 4 h. In addition, control chromatin assembly reactions without hNAP1 or ACF were also carried out. The resulting four types of chromatin templates were directly subjected to the in vitro transcription assay.
Preparation of Nuclear Extracts
TNE preparations have been described previously (Fan and Sugiura, 1995; Yukawa et al., 1997; Yukawa and Sugiura, 2004). ANEs were prepared as follows. An Arabidopsis MM2d cell culture was maintained by weekly subculturing in Murashige and Skoog (MS) medium supplemented with 3% (w/v) Suc, 200 mg L−1 KH2PO4, 100 mg L−1 myo-inositol, 1 mg L−1 thiamine-HCl, and 0.9 µM 2,4-D under dark conditions on a rotary shaker (120 rpm) at 26°C, the same as the tobacco BY-2 cell culture (Wu et al., 2012). The MM2d green cell culture was grown in MS medium supplemented with 1% (w/v) Suc, 200 mg L−1 KH2PO4, 100 mg L−1 myo-inositol, 1 mg L−1 thiamine-HCl, and 25 nm kinetin under continuous light (150 μmol photons m−2 sec−1) on a rotary shaker (120 rpm) at 24°C aerated with 3% (v/v) CO2 flow in air.
A 15-mL aliquot of a 7-d-old culture in the dark and a 40-mL aliquot of a 7-d-old culture in the light were transferred separately into 250 mL of fresh media in a 1-L flask. Approximately 200 g of MM2d cells was harvested at a midlog phase (96 h after inoculation) from a 2-L cell culture by filtration through one layer of Miracloth (Merck Millipore). The cells were then digested in 600 mL of enzyme solution [2.5% (w/v) Sumizyme C and 0.4% (w/v) Sumizyme AP2 (Shin Nihon Chemical, Japan), 0.9× MS medium, 3% (w/v) Suc, and 0.4 m mannitol (pH 5.5)] at 30°C for 3 h with gentle agitation. To remove vacuoles from the MM2d protoplasts, protoplast pellets were suspended in Percoll solution [24% (v/v) Percoll (GE Healthcare), 6% (w/v) Suc, 0.5 m sorbitol, 89 mm mannitol, 30 mm MgSO4, and 4 mm HEPES (pH 7.5)] and centrifuged at 22,500g for 45 min at 20°C. Evacuolated miniprotoplasts were collected from the bottom layer and washed with 100 mL ice-cold 0.6 m sorbitol and centrifuged at 3,100g for 5 min at 2°C. The miniprotoplasts were suspended in 150 mL ice-cold nuclear isolation buffer [18% (w/v) Ficoll PM400 (GE Healthcare), 15 mm HEPES-KOH (pH 7.9), 28 mm 2-mercaptoethanol, 4 mm MgSO4, 3 mm DTT, 1 mm EGTA, 1 mm NaF, 0.5 mm EDTA, 1% (w/v) l-ascorbic acid sodium, 0.5 mm phenylmethylsulfonyl fluoride (PMSF), 0.5 mm benzamidine, 0.5 mm spermidine, 0.15 mm spermine, and 400 μL protease inhibitor cocktail for plant cell and tissue extracts (Sigma-Aldrich)]. The miniprotoplast suspension was vacuum filtered once through a 10-μm pore Nytal HD-10 nylon mesh (Sefar, Switzerland) and twice through two layers of nylon mesh. The homogenate was centrifuged at 3,100g for 10 min at 2°C, and the nuclear pellet was washed with 40 mL of nuclear isolation buffer and centrifuged. Purified nuclei were suspended in an equal amount of nuclear extraction buffer (2×) [40% (v/v) glycerol, 50 mm HEPES-KOH (pH 7.9), 8 mm MgSO4, 2 mm NaF, 5 mm DTT, and 0.8 mm EGTA] and lysed by the drop-wise addition of 4 m ammonium sulfate to a final concentration of 0.42 M; the highly viscous mixture was gently rotated by a tube rotator at 2°C for 30 min. The nuclear lysate was centrifuged at 200,000g for 1 h at 2°C, and the supernatant was precipitated in dialysis tubing (Wako Chemical, Japan) with 70% (w/v) ammonium sulfate-saturated D buffer [20% (v/v) glycerol, 20 mm HEPES-KOH (pH 7.9), 100 mm KOAc, 4 mM MgSO4, 2 mm DTT, 0.5 mm PMSF, 0.5 mm benzamidine, 0.2 mm EGTA, 0.1 mm EDTA] at 4°C overnight. The precipitate was centrifuged at 15,000g for 15 min at 2°C, and the pellet was dissolved with equal volume of D buffer and immediately dialyzed three times each against 500 mL of D buffer on ice for 1 h. The dialyzed extract was then centrifuged at 15,000g for 5 min at 2°C. The supernatant was aliquoted, frozen in liquid nitrogen, and stored at −80°C. Approximately 2 mL of nuclear extracts containing approximately 10 μg μL−1 protein was obtained from one preparation.
In Vitro Transcription Assay with TNEs and ANEs
In vitro transcription of Arabidopsis rbcS genes was carried out as previously described. Briefly, reactions were performed in a 20-μL volume containing 0.4 pmol circular plasmid DNA, approximately 40 μg protein from the nuclear extract, 30 mm HEPES-KOH (pH 7.9), 3 mm MgSO4, 40 mm KOAc, 0.1 mm EGTA, 2 mm DTT, 10% (v/v) glycerol, 1 mm each of ATP, CTP, UTP, and GTP, and 8 U RNase inhibitor (Takara Bio, Japan).
After incubation at 28°C for 90 min, the reactions were extracted once with phenol / chloroform / isoamylalcohol (25:24:1, v/v) and with chloroform / isoamylalcohol (24:1, v/v) and precipitated with 0.2 volume of 10 m ammonium acetate 10 μg Glyco Blue (Life Technologies) and 2.2 volumes of absolute ethanol. Precipitates were collected by centrifugation, and the resulting pellets were washed with 80% (v/v) ethanol and dried. The precipitates were dissolved in 10 μL of reverse transcription buffer (Toyobo) with 0.2 pmol 5′-fluorescein labeled primers (compare with Supplemental Table S2), denatured at 70°C for 4 min, and then annealed at 60°C for 8 min. After addition of 10 μL reverse transcriptase cocktail [1 mm each of dNTPs, 80 μm actinomycin D, and 10 U ReverTraAce (Toyobo) in 1× reverse transcription buffer], the mixture was incubated at 50°C for 1 h, and then the reaction was terminated by adding 20 μL loading buffer (deionized formamide with blue dextran). After heating at 96°C for 2 min, the extended products were resolved through an 8% (w/v) polyacrylamide gel (19:1) with 7 m urea and 1× TBE. Fluorescent signals were detected by a Typhoon9400 analyzer (GE Healthcare) and quantified using Image Quant TL (GE Healthcare).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank database under the following accession numbers: At rbcS-1A, At1g67090; At rbcS-1B, At5g38430; At rbcS-2B, At5g38420; At rbcS-3B, At5g38410; At U2, At3g57645; At U3, x52630; At U6, x52538; At 7SL, At4g02970; Nt rbcS, x02353; Nt GNPS, x53600; NtY1, x04781; and Sl rbcS, x05986.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic representation of the plasmids used in this study.
Supplemental Figure S2. Alignment of Arabidopsis and human NAP1 amino acid sequences.
Supplemental Figure S3. Alignment of Arabidopsis and human ISWI amino acid sequences.
Supplemental Table S1. List of the clones used in this study.
Supplemental Table S2. List of the primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. James A. H. Murray (University of Cambridge) for providing the Arabidopsis MM2d cell culture and Maki Yukawa and Dr. Akihiko Moriyama (Nagoya City University) for advice and discussion.
Glossary
- ANE
Arabidopsis MM2d nuclear extract
- ANE-D
Arabidopsis nuclear extract derived from dark
- ANE-L
Arabidopsis nuclear extract derived from light
- CaMV
Cauliflower mosaic virus
- 2,4-D
2,4-dichlorophenoxyacetic acid
- MS
Murashige and Skoog
- Pol
polymerase
- TNE
tobacco nuclear extract
- USE
Upstream Sequence Element
References
- Akama K, Yukawa Y, Sugiura M, Small I (1998) Plant cytosolic tRNAHis possesses an exceptional C54 in the canonical TΨC loop. Nucleic Acids Res 26: 2708–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud P, Yukawa Y, Lavie L, Pélissier T, Sugiura M, Deragon JM (2001) Analysis of the SINE S1 Pol III promoter from Brassica; impact of methylation and influence of external sequences. Plant J 26: 295–305 [DOI] [PubMed] [Google Scholar]
- Banerjee R, Batschauer A (2005) Plant blue-light receptors. Planta 220: 498–502 [DOI] [PubMed] [Google Scholar]
- Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillier C, Espagnol MC, Picard G, Tourmente S (2002) Analysis of the 5S RNA pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature 5S RNA. Genome Res 12: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedonder A, Rethy R, Fredericq H, Van Montagu M, Krebbers E (1993) Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol 101: 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Yukawa Y, Alzapiedi M, Guffanti E, Ferrari R, Sugiura M, Ottonello S (2006) Distinct modes of TATA box utilization by the RNA polymerase III transcription machineries from budding yeast and higher plants. Gene 379: 12–25 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A, Zhu Y, Yu Y, Cao K, Sun C, Shen WH (2003) Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta 216: 561–570 [DOI] [PubMed] [Google Scholar]
- Dong J, Gao Z, Liu S, Li G, Yang Z, Huang H, Xu L (2013) SLIDE, the protein interacting domain of Imitation Switch remodelers, binds DDT-domain proteins of different subfamilies in chromatin remodeling complexes. J Integr Plant Biol 55: 928–937 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Ferrari S, Längst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J 20: 3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Sugiura M (1995) A plant basal in vitro system supporting accurate transcription of both RNA polymerase II- and III-dependent genes: supplement of green leaf component(s) drives accurate transcription of a light-responsive rbcS gene. EMBO J 14: 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Yakura K, Miyanishi M, Sugita M, Sugiura M (1995) In vitro transcription of plant RNA polymerase I-dependent rRNA genes is species-specific. Plant J 8: 295–298 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H, Hahlbrock K, Schäfer E (1994) A light-responsive in vitro transcription system from evacuolated parsley protoplasts. Plant J 5: 437–449 [DOI] [PubMed] [Google Scholar]
- Galichet A, Gruissem W (2006) Developmentally controlled farnesylation modulates AtNAP1;1 function in cell proliferation and cell expansion during Arabidopsis leaf development. Plant Physiol 142: 1412–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhu Y, Zhou W, Molinier J, Dong A, Shen WH (2012) NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell 24: 1437–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS (2012) In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell 48: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Nagasaki-Takeuchi N, Kato T, Fujiwara M, Sonobe S, Fukao Y, Hashimoto T (2013) Purification and characterization of novel microtubule-associated proteins from Arabidopsis cell suspension cultures. Plant Physiol 163: 1804–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Yukawa Y, Obokata J, Sugiura M (2003a) A tRNALeu-like sequence located immediately upstream of an Arabidopsis clock-regulated gene is transcriptionally active: efficient transcription by an RNA polymerase III-dependent in vitro transcription system. Gene 307: 133–139 [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Yukawa Y, Sugiura M (2003b) In vitro analysis of transcription initiation and termination from the Lhcb1 gene family in Nicotiana sylvestris: detection of transcription termination sites. Plant J 33: 1063–1072 [DOI] [PubMed] [Google Scholar]
- Héricourt F, Jupin I (1999) Molecular cloning and characterization of the Arabidopsis thaliana alpha-subunit of elongation factor 1B. FEBS Lett 464: 148–152 [DOI] [PubMed] [Google Scholar]
- Igarashi H, Orii H, Mori H, Shimmen T, Sonobe S (2000) Isolation of a novel 190 kDa protein from tobacco BY-2 cells: possible involvement in the interaction between actin filaments and microtubules. Plant Cell Physiol 41: 920–931 [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155 [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev 13: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Marshallsay C, Filipowicz W (1991) Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell 65: 517–526 [DOI] [PubMed] [Google Scholar]
- Knizewski L, Ginalski K, Jerzmanowski A (2008) Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci 13: 557–565 [DOI] [PubMed] [Google Scholar]
- Krebbers E, Seurinck J, Herdies L, Cashmore AR, Timko MP (1988) Four genes in two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol Biol 11: 745–759 [DOI] [PubMed] [Google Scholar]
- Lankenau S, Barnickel T, Marhold J, Lyko F, Mechler BM, Lankenau DH (2003) Knockout targeting of the Drosophila nap1 gene and examination of DNA repair tracts in the recombination products. Genetics 163: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Liu S, Wang J, He J, Huang H, Zhang Y, Xu L (2014) ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J 78: 706–714 [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhu Y, Gao J, Yu F, Dong A, Shen WH (2009a) Molecular and reverse genetic characterization of NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in transcription and nucleotide excision repair in Arabidopsis thaliana. Plant J 59: 27–38 [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Gao J, Dong AW, Shen WH (2009b) A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3-2 mutant. Mol Plant 2: 688–699 [DOI] [PubMed] [Google Scholar]
- López-Ochoa L, Acevedo-Hernández G, Martínez-Hernández A, Argüello-Astorga G, Herrera-Estrella L (2007) Structural relationships between diverse cis-acting elements are critical for the functional properties of a rbcS minimal light regulatory unit. J Exp Bot 58: 4397–4406 [DOI] [PubMed] [Google Scholar]
- Manley JL, Fire A, Cano A, Sharp PA, Gefter ML (1980) DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci USA 77: 3855–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Yukawa Y, Prieto JL, Vaillant I, Sugiura M, Tourmente S (2003) Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana. Nucleic Acids Res 31: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur BJ, Chui CF (1985) Sequence of a genomic DNA clone for the small subunit of ribulose bis-phosphate carboxylase-oxygenase from tobacco. Nucleic Acids Res 13: 2373–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Murota K, Hagiwara-Komoda Y, Komoda K, Onouchi H, Ishikawa M, Naito S (2011) Arabidopsis cell-free extract, ACE, a new in vitro translation system derived from Arabidopsis callus cultures. Plant Cell Physiol 52: 1443–1453 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Park YJ, Luger K (2006) The structure of nucleosome assembly protein 1. Proc Natl Acad Sci USA 103: 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J, Albert AC, Earley K, Pikaard CS (2003) Purification and transcriptional analysis of RNA polymerase I holoenzymes from broccoli (Brassica oleracea) and frog (Xenopus laevis). Methods Enzymol 370: 121–138 [DOI] [PubMed] [Google Scholar]
- Saez-Vasquez J, Pikaard CS (1997) Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc Natl Acad Sci USA 94: 11869–11874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarte S, Tiedemann R (2011) A gene duplication/loss event in the ribulose-1,5-bisphosphate-carboxylase/oxygenase (rubisco) small subunit gene family among accessions of Arabidopsis thaliana. Mol Biol Evol 28: 1861–1876 [DOI] [PubMed] [Google Scholar]
- Stange N, Beier H (1986) A gene for the major cytoplasmic tRNATyr from Nicotiana rustica contains a 13 nucleotides long intron. Nucleic Acids Res 14: 8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. (1997) Plant in vitro transcription systems. Annu Rev Plant Physiol Plant Mol Biol 48: 383–398 [DOI] [PubMed] [Google Scholar]
- Tyler JK, Bulger M, Kamakaka RT, Kobayashi R, Kadonaga JT (1996) The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol 16: 6149–6159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waibel F, Filipowicz W (1990) RNA-polymerase specificity of transcription of Arabidopsis U snRNA genes determined by promoter element spacing. Nature 346: 199–202 [DOI] [PubMed] [Google Scholar]
- Weil PA, Luse DS, Segall J, Roeder RG (1979) Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell 18: 469–484 [DOI] [PubMed] [Google Scholar]
- Willis IM. (1993) RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem 212: 1–11 [DOI] [PubMed] [Google Scholar]
- Wu J, Okada T, Fukushima T, Tsudzuki T, Sugiura M, Yukawa Y (2012) A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis. RNA Biol 9: 302–313 [DOI] [PubMed] [Google Scholar]
- Yoon HW, Kim MC, Lee SY, Hwang I, Bahk JD, Hong JC, Ishimi Y, Cho MJ (1995) Molecular cloning and functional characterization of a cDNA encoding nucleosome assembly protein 1 (NAP-1) from soybean. Mol Gen Genet 249: 465–473 [DOI] [PubMed] [Google Scholar]
- Yoon M, Putterill JJ, Ross GS, Laing WA (2001) Determination of the relative expression levels of rubisco small subunit genes in Arabidopsis by rapid amplification of cDNA ends. Anal Biochem 291: 237–244 [DOI] [PubMed] [Google Scholar]
- Yukawa Y, Akama K, Noguchi K, Komiya M, Sugiura M (2013) The context of transcription start site regions is crucial for transcription of a plant tRNALys(UUU) gene group both in vitro and in vivo. Gene 512: 286–293 [DOI] [PubMed] [Google Scholar]
- Yukawa Y, Dieci G, Alzapiedi M, Hiraga A, Hirai K, Yamamoto YY, Sugiura M (2011) A common sequence motif involved in selection of transcription start sites of Arabidopsis and budding yeast tRNA genes. Genomics 97: 166–172 [DOI] [PubMed] [Google Scholar]
- Yukawa Y, Fan H, Akama K, Beier H, Gross HJ, Sugiura M (2001) A tobacco nuclear extract supporting transcription, processing, splicing and modification of plant intron-containing tRNA precursors. Plant J 28: 583–594 [DOI] [PubMed] [Google Scholar]
- Yukawa Y, Felis M, Englert M, Stojanov M, Matousěk J, Beier H, Sugiura M (2005) Plant 7SL RNA genes belong to type 4 of RNA polymerase III- dependent genes that are composed of mixed promoters. Plant J 43: 97–106 [DOI] [PubMed] [Google Scholar]
- Yukawa Y, Sugita M, Sugiura M (1997) Efficient in vitro transcription of plant nuclear tRNASer genes in a nuclear extract from tobacco cultured cells. Plant J 12: 965–970 [DOI] [PubMed] [Google Scholar]
- Yukawa Y, Sugiura M (2004) In vitro transcription systems from BY-2 cells. In Nagata T, Hasezawa S, Inzé D, eds, Tobacco BY-2 cells. Springer-Verlag, Berlin, Heidelberg, pp 265–282 [Google Scholar]
- Zhu Q, Ordiz MI, Dabi T, Beachy RN, Lamb C (2002) Rice TATA binding protein interacts functionally with transcription factor IIB and the RF2a bZIP transcriptional activator in an enhanced plant in vitro transcription system. Plant Cell 14: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.