The rice brassinosteroid receptor protein OsBRI1 disrupts the interaction between the tetratricopeptide repeat and kinase domains of the complementary brassinosteroid kinase via phosphorylation; this activates brasssinosteroid signaling by increasing the binding between the kinase domain and the brassinosteroid suppressor BSU1.
Abstract
Many plant receptor kinases transduce signals through receptor-like cytoplasmic kinases (RLCKs); however, the molecular mechanisms that create an effective on-off switch are unknown. The receptor kinase BR INSENSITIVE1 (BRI1) transduces brassinosteroid (BR) signal by phosphorylating members of the BR-signaling kinase (BSK) family of RLCKs, which contain a kinase domain and a C-terminal tetratricopeptide repeat (TPR) domain. Here, we show that the BR signaling function of BSKs is conserved in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) and that the TPR domain of BSKs functions as a “phospho-switchable” autoregulatory domain to control BSKs’ activity. Genetic studies revealed that OsBSK3 is a positive regulator of BR signaling in rice, while in vivo and in vitro assays demonstrated that OsBRI1 interacts directly with and phosphorylates OsBSK3. The TPR domain of OsBSK3, which interacts directly with the protein’s kinase domain, serves as an autoinhibitory domain to prevent OsBSK3 from interacting with bri1-SUPPRESSOR1 (BSU1). Phosphorylation of OsBSK3 by OsBRI1 disrupts the interaction between its TPR and kinase domains, thereby increasing the binding between OsBSK3’s kinase domain and BSU1. Our results not only demonstrate that OsBSK3 plays a conserved role in regulating BR signaling in rice, but also provide insight into the molecular mechanism by which BSK family proteins are inhibited under basal conditions but switched on by the upstream receptor kinase BRI1.
Brassinosteroids (BRs) are a group of natural polyhydroxy steroids that regulate diverse physiological processes in plants, including growth promotion, skotomorphogenesis, organ boundary formation, stomata development, sex determination, vascular differentiation, male fertility, seed germination, flowering, senescence, and resistance to various abiotic and biotic stresses (Gomes, 2011; Hartwig et al., 2011; Kim et al., 2012; Gendron et al., 2012; Kutschera and Wang, 2012). Over the past decade, using Arabidopsis (Arabidopsis thaliana) as a model plant, genetic, biochemical, molecular, and proteomic studies have revealed details about the BR signal transduction mechanisms, from perception of the signal by the receptor kinase BR INSENSITIVE1 (BRI1) to downstream transcriptional regulation. The BR signaling pathway in Arabidopsis represents one of the best-characterized receptor kinase pathways in plants and therefore serves as a model for understanding receptor kinase signaling in general and for understanding BR signaling in crops.
BRs are recognized by a membrane-localized Leu-rich repeat receptor-like kinase, BRI1, and its coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) (Li and Chory, 1997; Santiago et al., 2013; Sun et al., 2013). BR binding promotes the association of BRI1 with BAK1 and enables transphosphorylation between the cytoplasmic kinase domains of the two receptors (Li et al., 2002; Nam and Li, 2002; Wang et al., 2005, 2008). BRI1 then phosphorylates two membrane-localized receptor-like cytoplasmic kinases (RLCKs), BR SIGNALING KINASES (BSKs) and CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1), leading to activation of the protein phosphatase bri1-SUPPRESSOR1 (BSU1) (Tang et al., 2008; Kim et al., 2009, 2011). BSU1 dephosphorylates and inhibits the GSK3/Shaggy-like kinase BR INSENSITIVE2 (BIN2) (Kim et al., 2009). In the absence of BRs, BIN2 phosphorylates BRASSINAZOLE RESISTANT1 (BZR1) family transcription factors, preventing them from regulating the transcription of downstream targets (He et al., 2002, 2005; Vert and Chory, 2006). BR signaling inhibits BIN2 and allows BZR1 to be dephosphorylated by PROTEIN PHOSPHATASE 2A (Tang et al., 2011). Together with their binding partners, dephosphorylated BZR1 family transcription factors bind to BR response element or E-box cis-element and regulate the expression of many BR-responsive genes (He et al., 2005; Yin et al., 2005; Sun et al., 2010).
The interaction with a membrane-localized receptor-like kinase is not unique for BSKs and CDG1; it has also been observed for a number of other RLCKs, including M-LOCUS PROTEIN KINASE (Kakita et al., 2007), BOTRYTIS-INDUCED KINASE1 (Lu et al., 2010), CAST AWAY (CST) (Burr et al., 2011), OsRLCK185 (Yamaguchi et al., 2013), and AVRPPHB SUSCEPTIBLE-LIKE1 (Zhang et al., 2010). Therefore, interacting with a receptor-like kinase to regulate cellular signaling pathways might represent a general function of RLCKs. While many RLCKs regulate cellular responses by phosphorylating one or more substrates directly, there are many RLCKs encoded in the Arabidopsis genome (e.g. BSKs) that contain a kinase domain that lacks conserved amino acids believed to be essential for ATP binding or phosphoryl transfer (Castells and Casacuberta, 2007; Roux et al., 2014). The molecular mechanisms by which these atypical kinases switch signaling pathways on and off to regulate cellular functions remain to be explored.
In this study, we demonstrate that OsBSK3 plays a conserved role in transducing BR signal from OsBRI1 to downstream components in rice (Oryza sativa). Similar to Arabidopsis BSKs (AtBSKs), OsBSK3 contains a kinase domain and a C-terminal tetratricopeptide repeat (TPR) domain. Our data show that the TPR domain of OsBSK3 interacts directly with the protein’s kinase domain and that it serves as an autoinhibitory domain to prevent OsBSK3 from interacting with BSU1. Phosphorylation of OsBSK3 by OsBRI1 is critical for the regulation of BR signaling because it prevents binding between the TPR and kinase domains of OsBSK3, thus enabling binding between the kinase domain of OsBSK3 and BSU1.
RESULTS
Rice BSK3 Positively Regulates BR Signaling in Arabidopsis
Previously, it was shown that AtBSKs mediate BR signal transduction from the membrane receptor AtBRI1 to the downstream phosphatase AtBSU1 (Kim et al., 2009). To investigate whether a similar mechanism exists in rice, a BLAST search against the rice genome was conducted using twelve AtBSK protein sequences. Four AtBSK orthologs were identified in rice. Based on their sequence homology to AtBSK3, the orthologs were named OsBSK1 (Os03g04050), OsBSK2 (Os10g42110), OsBSK3 (Os04g58750), and OsBSK4 (Os03g61010). Similar to AtBSKs, these OsBSKs were found to contain an N-terminal kinase domain and a C-terminal TPR domain. In addition, several distinctive features of the kinase domain in AtBSKs were found to be conserved in the OsBSKs, including the lack of a classical GXGXXG motif (Gly-rich loop), the presence of an Ala gatekeeper, and amino acid substitutions within the conserved HRD and DFG motifs (Supplemental Fig. S1).
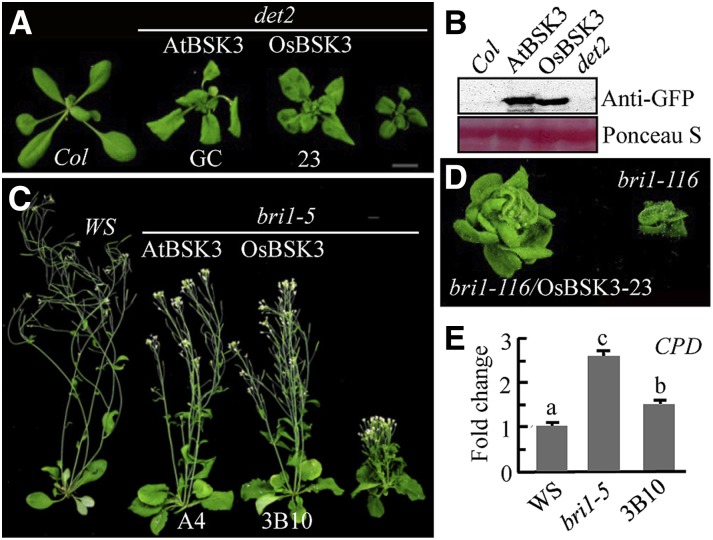
In preliminary semiquantitative RT-PCR analyses, we found that the expression of OsBSK3 in rice root and leaf tissues was up-regulated by BR treatment (Supplemental Fig. S2); therefore, we selected OsBSK3 for further analysis. When overexpressed in Arabidopsis, OsBSK3 rescued the dwarf phenotypes of the BR synthesis mutant det2, the weak BR signaling mutant bri1-5, and the strong BR signaling mutant bri1-116 (Fig. 1, A–D). Compared with bri1-5 control plants, expression of the BR synthesis gene CPD was greatly reduced in the transgenic plants overexpressing OsBSK3 (Fig. 1E), indicating that overexpression of OsBSK3 activates BR signaling in Arabidopsis.
Figure 1.
OsBSK3 overexpression rescued the mutant phenotypes of det2, bri1-5, and bri1-116 plants. A, C, and D, Phenotype of light-grown 4-week-old det2 (A), 7-week-old bri1-5 (C), and 8-week-old bri1-116 (D) mutant plants overexpressing AtBSK3 or OsBSK3 with a C-terminal YFP tag. B, Expression levels of OsBSK3-YFP and AtBSK3-YFP in the transgenic plants shown in (A). Ponceau S staining of the Rubisco large subunit was used as an equal loading control. E, Quantitative real-time RT-PCR analysis of the expression of CPD in 1-week-old wild-type (Ws), bri1-5, and OsBSK3-YFP-overexpressing bri1-5 (3B10) plants. A one-way ANOVA was performed; statistically significant differences are indicated by different lowercase letters (P < 0.05).
Membrane Localization Is Required for the Regulation of BR Signaling by OsBSK3
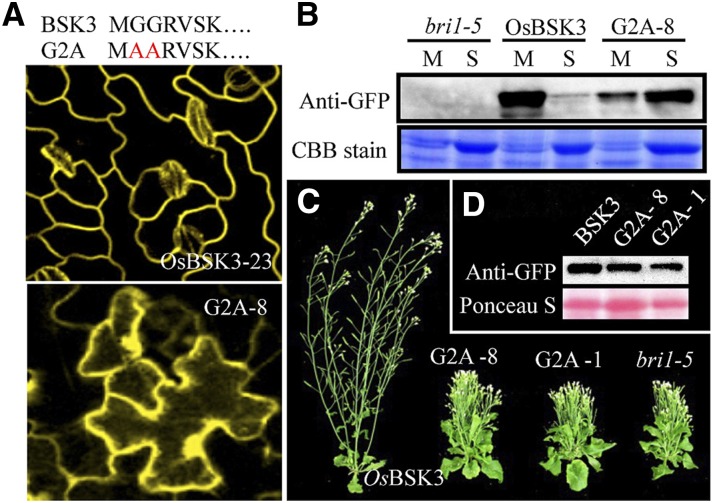
An analysis by confocal microscopy of transgenic plants overexpressing OsBSK3 with a C-terminal yellow fluorescent protein (OsBSK3-YFP) or GFP tag (OsBSK3-GFP) showed that OsBSK3 was localized to the plasma membrane in both Arabidopsis and rice seedlings (Figs. 2A; Supplemental Fig. 3). Sequence analysis revealed that OsBSK3 does not possess a transmembrane domain; however, it contains a potential myristoylation site at its N terminus, which serves as a membrane localization signal (Thompson and Okuyama, 2000). To examine whether this myristoylation site is critical for the membrane localization and biological function of OsBSK3, we produced transgenic Arabidopsis plants expressing a mutant version of OsBSK3 with a disrupted myristoylation site (the second and third Gly residues were substituted with Ala; G2A) and YFP fused to the C terminus. As predicted, at an expression level below that of OsBSK3-YFP, G2A-YFP was detected throughout the transgenic cells by confocal microscopy (Fig. 2A). Consistent with these observations, OsBSK3-YFP was detected only in the membrane fraction by immunoblotting, while G2A-YFP was mostly detected in the soluble fraction in the transgenic seedlings (Fig. 2B). Myristoylation-mediated membrane localization is critical for the biological function of OsBSK3. When overexpressed in bri1-5 plants, G2A was unable to rescue the semi-dwarf phenotype of the mutants (Fig. 2, C and D).
Figure 2.
Membrane localization is crucial for the regulation of BR signaling in Arabidopsis by OsBSK3. A, The upper panel shows the G2A mutation. Red letters show the mutated amino acid. The middle and lower panels show the YFP signal detected by confocal microscopy in 1-week-old light-grown OsBSK3-YFP- and G2A-YFP-overexpressing bri1-5 leaves. B, 100 µg of soluble (S) or microsomal (M) proteins from OsBSK3-YFP- and G2A-YFP-overexpressing bri1-5 mutant plants were separated by SDS-PAGE and immunoblotted using anti-YFP antibodies. Coomassie Brilliant Blue (CBB) staining of the Rubisco large subunit was used as an equal loading control. C, Seven-week-old bri1-5 mutant plants overexpressing OsBSK3-YFP or G2A-YFP. G2A-1 and -8 are two independent transgenic lines. D, OsBSK3-YFP and G2A-YFP expression in the 3-week-old transgenic plants shown in C. Ponceau S staining of the Rubisco large subunit was used as an equal loading control.
OsBSK3 Is a Direct Substrate of OsBRI1
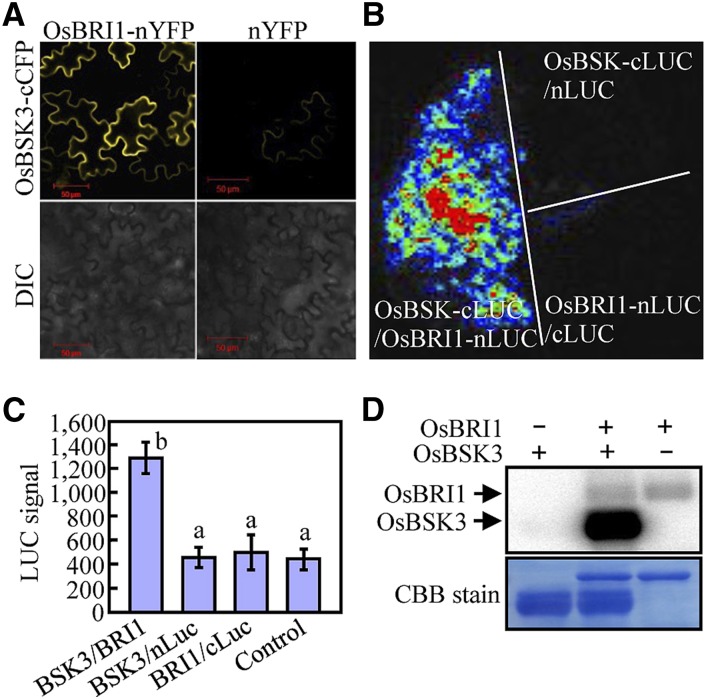
To test whether OsBSK3 interacts directly with the rice BR receptor OsBRI1, a bimolecular fluorescence complementation (BiFC) assay was performed. As shown in Figure 3A, tobacco (Nicotiana benthamiana) epidermal cells coexpressing OsBSK3 fused to the C-terminal half of cyan fluorescent protein (OsBSK3-cCFP) and full-length OsBRI1 fused to the N-terminal half of YFP (OsBRI1-nYFP) showed strong fluorescence at the plasma membrane, whereas tobacco cells coexpressing OsBSK3-cCFP and nYFP showed very weak fluorescence (Fig. 3A). The in vivo interaction of OsBRI1 with OsBSK3 was further confirmed using a split luciferase complementation assay. A strong luciferase signal was detected in tobacco leaf epidermal cells coexpressing OsBSK3 fused with the C-terminal half of luciferase (OsBSK3-cLuc) and full-length OsBRI1 fused with the N-terminal half of luciferase (OsBRI1-nLuc), but not in cells coexpressing OsBSK3-cLuc and the nLuc empty vector or OsBRI1-nLuc and the cLuc empty vector (Fig. 3, B and C).
Figure 3.
OsBSK3 is a direct substrate of OsBRI1. A and B, BiFC (A) and firefly luciferase complementation assays (B) revealed the direct interaction of OsBSK3 with full-length OsBRI1 in 4-week-old tobacco leaf epidermal cells. The leaves were coinfiltrated with Agrobacterium tumefaciens cells containing the indicated vector pairs. Images were collected 36 to 48 h after infiltration. DIC, differential interference contrast microscopy. C, Quantitation of the complemented luciferase activity in tobacco leaves cotransformed with the indicated vector pairs. The experiment was repeated at least three times with consistent results; representative results are shown. A one-way ANOVA was performed; statistically significant differences are indicated by different lowercase letters (P < 0.05). D, An in vitro assay for the phosphorylation of full-length OsBSK3 by OsBRI1.
An in vitro kinase assay showed that OsBSK3 could be phosphorylated by OsBRI1 (Fig. 3D). To elucidate the effect of phosphorylation by OsBRI1 on the biological function of OsBSK3, lambda phosphatase-treated recombinant OsBSK3 (see the “Materials and Methods” section) was phosphorylated in vitro by OsBRI1, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC/MS/MS). Mass spectrometry did not identify any phosphopeptides in the lambda phosphatase-treated OsBSK3 protein; however, Ser-213 and Ser-215 were found to be phosphorylated in OsBSK3 that had been coincubated with OsBRI1, suggesting that these residues are specific OsBRI1 phosphorylation sites (Table I; Supplemental Fig. S4). To investigate whether these two sites are phosphorylated in vivo, OsBSK3 was immunoprecipitated with anti-myc antibodies from 2-week-old rice seedlings overexpressing OsBSK3 with a C-terminal myc tag, digested with trypsin, and analyzed by LC/MS/MS. Our results indicate that the peptide SYSTNLAFTPPEYMR, which contained Ser-213 and Ser-215, was indeed phosphorylated in vivo and that the phosphorylation site was either Ser-213 or Ser-215 (Table I; Supplemental Fig. S5A).
Table I. The phosphorylated peptides identified from OsBSK3 and AtBSK3 by LC/MS/MS.
| Peptidea | Measured [M+H]+ | Calculated [M+H]+ | Score | P-Value | Identified Sites |
|---|---|---|---|---|---|
| Identified in vitro phosphopeptides from OsBSK3 by CID | |||||
| DGKpSYSTNLAFTPPEYM*R | 2172.9446 | 2172.9308 | 22.7 | 6.7E-06 | S213 |
| DGKSYpSTNLAFTPPEYM*R | 2172.9389 | 2172.9308 | 34.8 | 2.1E-09 | S215 |
| Identified in vivo phosphopeptides from OsBSK3 by HCD | |||||
| pSYpSTNLAFTPPEYMR | 1856.7945 | 1856.7925 | 48 | 2.0E-11 | S213 or S215 |
| Identified in vivo phosphopeptides from AtBSK3 by CID | |||||
| pSYpSTNLAFTPPEYLR | 1838.8317 | 1838.8361 | 24.2 | 6.6E-06 | S210 or S212 |
Met sulfoxide is denoted as M*; phosphoseryl residues are denoted as pS.
Ser-215 Phosphorylation Is Critical for the Biological Function of OsBSK3
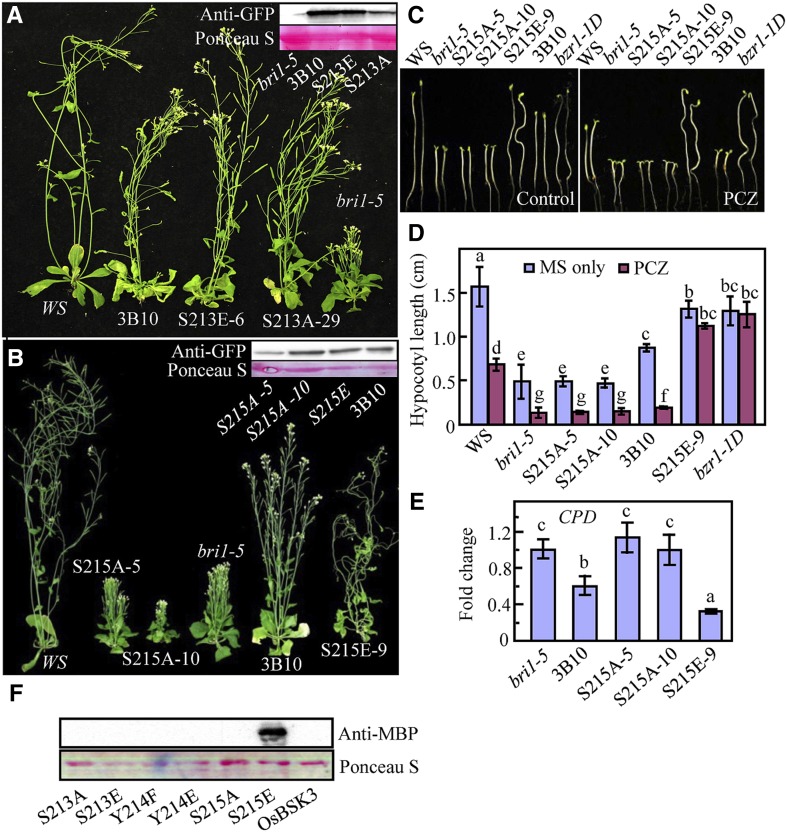
To determine the physiological significance of the OsBRI1 phosphorylation sites in OsBSK3, we generated multiple versions of OsBSK3 by site-directed mutagenesis (S213A and S215A) to eliminate phosphorylation at these residues. We also substituted Ser-213 or Ser-215 with Glu (generating S213E- and S215E-substituted OsBSK3, respectively) to mimic constitutive OsBRI1 phosphorylated forms of OsBSK3. These mutant forms of OsBSK3 were overexpressed in bri1-5 plants and examined for their ability to rescue the dwarf phenotype of the mutant. Surprisingly, replacement of Ser-213 with either Ala or Glu had little effect on the ability of OsBSK3 to rescue the dwarf phenotype of the bri1-5 mutant plants (Fig. 4A), suggesting that the phosphorylation of Ser-213 does not contribute to the regulatory role of OsBSK3 in BR signaling. In comparison, when the S215A-substituted version of OsBSK3 was expressed at a level similar to that of wild-type OsBSK3, no rescue of the bri1-5 phenotype was observed. Furthermore, S215A had a dominant-negative effect: Plants overexpressing the S215A version of OsBSK3 were smaller than the nontransgenic controls when grown in the light under the same conditions, and the severity of dwarfism was closely related to the level of expression of the mutant protein (Fig. 4B). When overexpressed, S215E-substituted OsBSK3 significantly rescued the dwarf phenotype of the bri1-5 mutant (Fig. 4B). The leaves of bri1-5 mutant plants overexpressing S215E-substituted OsBSK3 were similar in length to those of wild-type (Wassilewskija [Ws]) control plants (Supplemental Fig. S6). Meanwhile, the leaves, stems, and siliques of the S215E mutant-overexpressing plants were curled and twisted (Supplemental Fig. S6).
Figure 4.
Functional analysis of the OsBSK3 phosphorylation sites in Arabidopsis. A and B, Eight-week-old wild type (Ws), bri1-5 mutant, and bri1-5 mutant overexpressing wild type or a mutated form (S213A, S213E, S215A, and S215E) of OsBSK3. Immunoblotting for the expression of various OsBSK3 forms was conducted using anti-GFP antibodies since the OsBSK3s were produced with a C-terminal YFP tag. Ponceau S staining for the Rubisco large subunit was used as an equal loading control. C, The S215E transgenic plants were insensitive to PCZ-inhibited hypocotyl elongation. Young seedlings of wild type (Ws), bri1-5, or bri1-5 overexpressing various OsBSK3 forms were grown in the dark for 5 d on regular medium or medium containing 0.25 μm PCZ. D, A quantitative analysis of the hypocotyls of the seedlings shown in C. Each value in the graph represents the average of at least 20 individual seedlings. Error bars represent the se. E, Quantitative real-time RT-PCR analysis of the CPD expression levels in the seedlings shown in B. Error bars represent ± sd. F, A gel overlay analysis of the interaction between AtBSU1 and the various OsBSK3 mutant proteins. GST-tagged OsBSK3 proteins were immunoblotted onto a nitrocellulose membrane and probed with MBP-AtBSU1 and horseradish peroxidase-labeled anti-MBP antibodies. Total protein was visualized by Ponceau S staining. A one-way ANOVA was performed in D and E; statistically significant differences are indicated by different lowercase letters (P < 0.05).
Similarly, a peptide containing Ser-210 (equivalent to Ser-213 of OsBSK3) and Ser-212 (equivalent to Ser-215 of OsBSK3) from AtBSK3 was found to be phosphorylated in vivo (Table I; Supplemental Fig. S5B). Meanwhile, S212A-substituted AtBSK3 lost its ability to rescue the mutant phenotype of bri1-5 plants, and the substitution of Ser-212 with Glu enhanced the ability of AtBSK3 to rescue the dwarf phenotype of bri1-5 mutant plants compared with wild-type AtBSK3 under overexpression conditions (Supplemental Fig. S7). Taken together, these data suggest that the mechanism in which the phosphorylation of BSK3 regulates BR signaling is conserved between rice and Arabidopsis.
When grown in the dark, the etiolated hypocotyls of wild-type and S215E-substituted OsBSK3-expressing plants were significantly longer than those of bri1-5 plants, whereas the hypocotyls of S215A-substituted OsBSK3-expressing plants were similar to those of nontransgenic mutant control plants (Fig. 4, C and D). When grown in the presence of the BR biosynthesis inhibitor propiconazole (PCZ; 0.25 μm), the growth-promoting effect of wild-type OsBSK3 became less distinguishable, whereas the etiolated hypocotyls of the S215E-overexpressing bri1-5 mutant plants were wavy, twisted, and nearly insensitive to PCZ treatment (Fig. 4, C and D). Similar hypocotyl phenotypes were observed in bzr1-1D, a mutant that exhibits constitutively active BR signaling, suggesting that the S215E substitution produced constitutively active BR signaling. Consistent with these growth phenotypes, quantitative RT-PCR (qRT-PCR) showed that the expression levels of CPD were decreased in bri1-5 plants overexpressing wild-type or S215E-substituted OsBSK3, while they were unchanged in bri1-5 plants overexpressing S215A-substituted OsBSK3 (Fig. 4E).
It was previously reported that the phosphorylation of AtBSK1 by AtBRI1 increases the binding affinity of AtBSK1 for the downstream signaling component AtBSU1 (Kim et al., 2009). Although a sequence homology search showed that the rice genome contains several AtBSU1 orthologs, evidence showing that the BSU1 orthologs in rice regulate BR signaling in a manner similar to AtBSU1 is lacking. Therefore, we used AtBSU1 to investigate whether the identified OsBRI1 phosphorylation sites in OsBSK3 contribute to BSU1 binding. Wild-type and several mutated versions of OsBSK3 (S213A, S213E, Y214F, Y214E, S215A, and S215E) were purified from Escherichia coli using a GST tag, separated by SDS-PAGE, blotted onto nitrocellulose membranes, and incubated with purified MBP-tagged AtBSU1 (MBP-BSU1); the overlay signal was detected using horseradish peroxidase-conjugated anti-MBP antibodies. As shown in Figure 4F, S215E-substituted OsBSK3 exhibited strong affinity for AtBSU1, whereas no interaction was detected between AtBSU1 and the other forms of OsBSK3. The stronger interaction of S215E-substituted OsBSK3 with AtBSU1 was also confirmed using an in vivo BiFC assay (Supplemental Fig. S8).
The TPR Domain of OsBSK3 Negatively Regulates Its Function
Using transgenic plants, we discovered that overexpressed OsBSK3 carrying a C-terminal YFP tag rescued the bri1-5 mutant phenotype better than tag-free OsBSK3 (Supplemental Fig. S9). Given that the C terminus of OsBSK3 contains a TPR domain, which is believed to be involved in protein-protein interactions (Allan and Ratajczak, 2011), we tested the function of the TPR domain by overexpressing the kinase domain (amino acids 1–390) of OsBSK3 (N390) in wild-type plants and in various BR biosynthesis and signaling mutants. As shown in Supplemental Figure S10, overexpressing N390 partially rescued the semi-dwarf phenotype of the bri1-301 mutant, while wild-type seedlings overexpressing N390 and OsBSK3 showed a bzr1-1D mutant-like axillary branch kink phenotype (Supplemental Fig. S11) caused by BR-regulated organ fusion (Gendron et al., 2012). In addition, both sets of transgenic plants showed hypersensitivity to 24-epibrassinolide (eBL)-stimulated hypocotyl elongation and reduced sensitivity to brassinazole (BRZ; a BR biosynthesis inhibitor)-inhibited hypocotyl elongation. In comparison, the observed axillary branch kink phenotype and hypocotyl responses to exogenously applied eBL and BRZ were dramatically enhanced in N390-overexpressing plants, suggesting that BR signaling is hyperactivated in Arabidopsis plants overexpressing N390 compared with plants overexpressing full-length OsBSK3 (Supplemental Fig. S11). Consistent with these observations, N390 was able to rescue the dwarf phenotypes of bri1-5 and bri1-116 better than full-length OsBSK3 in plants expressing the proteins at similar levels (Fig. 5, A and B). qRT-PCR revealed that CPD expression was greatly reduced in the N390- and OsBSK3-overexpressing bri1-5 mutant plants (Fig. 5C), suggesting that the rescue of the mutant phenotype was due to the activation of BR signaling. We also overexpressed the TPR domain of OsBSK3 in wild-type Arabidopsis plants and analyzed their response to exogenous eBL. As shown, overexpression of the TPR domain reduced the sensitivity of the transgenic plants to eBL-stimulated hypocotyl elongation in the light (Fig. 5D).
Figure 5.
The TPR domain of OsBSK3 negatively regulates OsBSK3 function. A and B, Seven- to eight-week-old bri1-5 (A) and bri1-116 mutant (B) plants overexpressing full-length OsBSK3 or the kinase domain of OsBSK3 (N390). The upper panels in A and B show the expression levels of OsBSK3 and N390 in the transgenic plants. C, An analysis of the CPD expression level in the 2-week-old seedlings shown in A by qRT-PCR. D, Overexpression of the TPR domain of OsBSK3 reduced the BR sensitivity of the transgenic Arabidopsis plants. Plants were grown in half-strength Murashige and Skoog medium with or without the indicated concentration of eBL at 22°C under LD conditions for 1 week. Representative results from three independent experiments are shown. A one-way ANOVA was performed in C and D. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
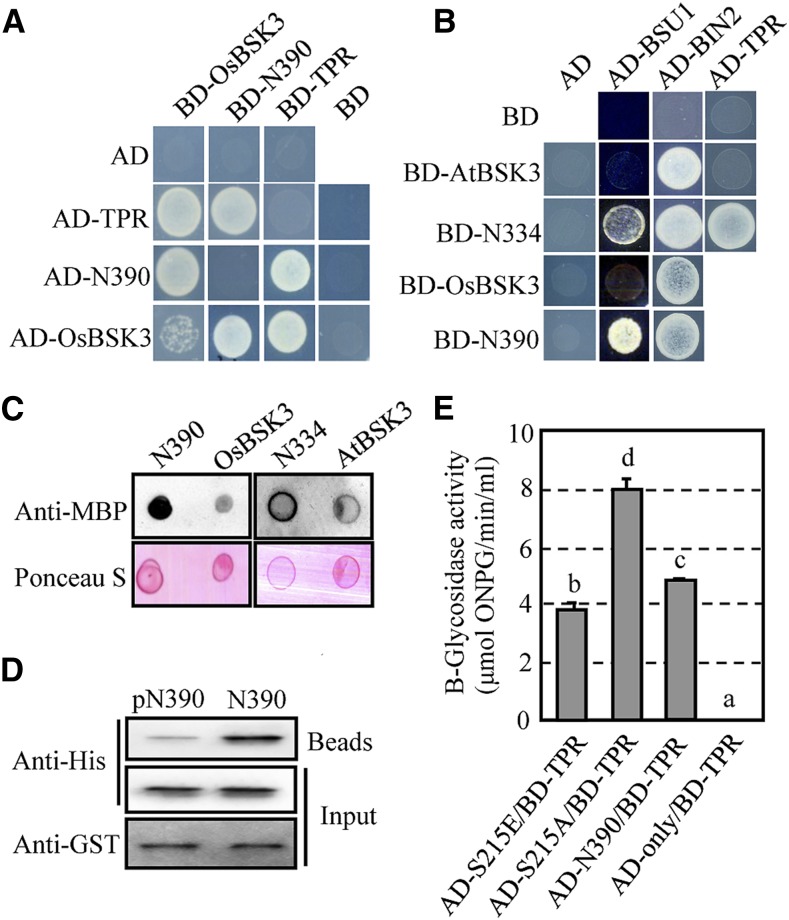
The strong ability of N390 to rescue the phenotypes of the BR signaling mutants and the reduced responses to eBL in plants overexpressing the TPR domain suggests that the TPR domain of OsBSK3 serves as an inhibitory domain. Thus, we next assessed whether the TPR and kinase domains of OsBSK3 (N390) could interact with each other directly by a yeast two-hybrid assay. In agreement with previous findings (Sreeramulu et al., 2013), OsBSK3 was able to interact with itself, but the interaction was relatively weak (Fig. 6A). When OsBSK3 was fragmented, the N390 or TPR domain (amino acids 391–502) interacted strongly with OsBSK3. Furthermore, although neither the N390 nor the TPR domain was able to bind to itself, they interacted strongly with each other (Fig. 6A; Supplemental Fig. S12A).
Figure 6.
The TPR domain of BSK3 prevents it from interacting with AtBSU1. A, Yeast two-hybrid assays of the interaction between full-length OsBSK3, the kinase domain of OsBSK3 (N390), and the TPR domain of OsBSK3 (TPR). B, Yeast two-hybrid assays of the interaction between full-length AtBSK3, full-length OsBSK3, the kinase domain of AtBSK3 (N334), and N390 with AtBSU1, AtBIN2, and the TPR domain from OsBSK3. C, AtBSU1 showed increased binding with the kinase domains of OsBSK3 and AtBSK3. The recombinant 6His-tagged kinase domain and full-length versions of OsBSK3 (N390 and OsBSK3) or AtBSK3 (N334 and AtBSK3) were dot blotted onto nitrocellulose membranes and then incubated with MBP-AtBSU1 and horseradish peroxidase-labeled anti-MBP antibodies. Total protein was visualized by Ponceau S staining. D, OsBRI1 phosphorylation prevents the TPR and kinase domains of OsBSK3 from binding. GST-TPR on Glutathione Sepharose 4B beads was used to pull down 6His-tagged N390, which had been incubated with MBP-tagged OsBRI1 kinase domain in the presence (pN390) or absence (N390) of ATP. The proteins were blotted onto nitrocellulose membranes and detected using anti-His or -GST antibodies. E, Ser-215 phosphorylation reduced the binding of the kinase and TPR domains of OsBSK3. Shown are quantitative β-glycosidase activity assays of the interaction between the TPR domain and wild type or Ser-215-substituted mutant form of N390. A one-way ANOVA was performed. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
Besides BRI1 and BSU1, BSK family proteins are able to bind directly with BIN2 (Sreeramulu et al., 2013). To further investigate the effect of the TPR domain of OsBSK3 or AtBSK3 on the protein’s physiological function, the ability of AtBSU1, AtBIN2, full-length OsBSK3, N390, full-length AtBSK3, and the kinase domain of AtBSK3 (N334) to interact was examined. AtBSU1 did not interact with full-length OsBSK3 or AtBSK3 in a yeast two-hybrid assay, whereas a strong interaction between AtBSU1 and N390 or N334 was detected (Fig. 6B). In comparison, AtBIN2 interacted with both the full length and the kinase domain of OsBSK3 or AtBSK3. These data suggest that the TPR domain prevented both OsBSK3 and AtBSK3 from binding with AtBSU1, but not AtBIN2. Our yeast two-hybrid data were further validated using an in vitro overlay assay, in which the kinase domain or full-length version of OsBSK3 (N390 and OsBSK3, respectively) or AtBSK3 (N334 and AtBSK3, respectively) carrying a 6His-tag were dot blotted onto a nitrocellulose membrane at a 1:1 molar ratio and incubated with MBP-AtBSU1. As shown in Figure 6C, MBP-AtBSU1 bound more strongly with the kinase domains of AtBSK3 and OsBSK3 than with the full-length versions of the proteins. This result was confirmed by a split luciferase assay (Supplemental Fig. S12B).
If OsBSK3’s TPR domain interacts with its kinase domain to prevent the protein from binding to the downstream target BSU1, could the phosphorylation of OsBSK3 by OsBRI1 prevent its TPR and kinase domains from binding? To test this hypothesis, a GST-TPR fusion protein was used to pull down N390-6His, which was preincubated with the OsBRI1 kinase domain in the presence or absence of ATP. Our findings indicate that unphosphorylated N390 bound the TPR domain much more strongly than phosphorylated N390 (Fig. 6D). Furthermore, quantitative yeast two-hybrid and split luciferase assays showed that the binding of the kinase and TPR domains of OsBSK3 was reduced when the S215E-substituted version of the protein was used and increased when the S215A-substituted version of the protein was used (Fig. 6E; Supplemental Fig. S12C).
OsBSK3 Regulates BR Signaling in Rice
To test whether OsBSK3 regulates BR signaling in rice, we overexpressed both full-length and the kinase domain of OsBSK3 in rice and measured the lamina joint angle and root length, which are typically regulated by BR signaling, in the transgenic plants. Seedlings overexpressing both full-length and the kinase domain of OsBSK3 showed a significantly increased leaf bending angle (Fig. 7, A and B). However, at a similar expression level, increased leaf bending and root growth inhibition were observed in BL-treated seedlings overexpressing N390, but not from seedlings overexpressing full-length OsBSK3 (Fig. 7, C and D). We also analyzed the expression levels of the BR biosynthesis genes DWARF and D2, which were previously shown to be feedback regulated by BR signaling in rice, in plants overexpressing N390 or OsBSK3. As shown in Figure 7E, the expression of DWARF and D2 was reduced in both types of transgenic plants; however, it was lower in the N390-overexpressing seedlings than in the OsBSK3-overexpressing seedlings. Taken together, these results suggest that BR signaling was activated in the OsBSK3- and N390-overexpressing rice seedlings. Similar to our finding in Arabidopsis, the kinase domain of OsBSK3 was more efficient at activating BR signaling than full-length OsBSK3.
Figure 7.
OsBSK3 regulates the BR response in rice. A, The lamina joint angle of the second leaves from 2-week-old rice seedlings overexpressing OsBSK3-myc. The 33 and 38 are two independent transgenic lines. B, Overexpression of the kinase domain of OsBSK3 (N390) enhanced the bending of the lamina joint in the transgenic rice seedlings. The upper panel shows the lamina joints of the second leaves from 2-week-old seedlings overexpressing C-terminal myc tag-fused OsBSK3 (BSK3) or kinase domain of OsBSK3 (N390). The lower panel shows the expression levels of OsBSK3-myc and N390-myc in the rice seedlings shown above using anti-myc antibodies. C, Quantitative analysis of BR-stimulated leaf lamina joint bending in OsBSK3- and N390-overexpressing rice seedlings. A total of 10 μL of the indicated concentration of eBL was applied to the lamina joint region of 10-d-old light-grown rice seedlings. The leaf bending angle was measured 3 d after eBL application. D, Quantitative analysis of BR-inhibited root elongation in OsBSK3- and N390-overexpressing rice seedlings. Seedlings were grown in Hoagland medium with or without 1 μm eBL for 1 week. Relative growth was calculated by dividing the root length in eBL by the root length under control conditions. E, Quantitative real-time RT-PCR analysis of DWARF and D2 expression in 2-week-old rice seedlings overexpressing OsBSK3 or N390. F, Left: qRT-PCR analysis of the expression of OsBSKs in 2-week-old wild-type and OsBSK3 CS transgenic rice seedlings. Right: Phenotypes of the seedlings used in the RT-PCR analysis. G, Internode length of fully grown wild-type and CS plants. H, Quantitative real-time RT-PCR analysis of DWARF expression in 2-week-old CS seedlings. I, Seeds from N390-overexpressing and CS transgenic rice plants. J, Weights of the seeds shown in I. A one-way ANOVA was performed in all experiments. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
While screening the transgenic rice seedlings, we identified several OsBSK3 cosuppression (CS) lines. qRT-PCR revealed that the expression of endogenous OsBSK3 in the CS plants was almost undetectable. Furthermore, the transcript levels of OsBSK1, OsBSK2, and OsBSK4 were all significantly reduced (Fig. 7F). The CS seedlings were all smaller in size and had a reduced leaf bending angle (Fig. 7F). Similar to a weak OsBRI1 loss-of-function mutant, when the plants were full grown, the elongation of the second internode was greatly reduced in the CS plants (Fig. 7G). Consistent with these phenotypes, the expression level of the BR biosynthesis gene OsDWARF was increased in the CS lines (Fig. 7H). Moreover, when grown in the field, the seeds of the N390-overexpressing plants were longer (not wider or thicker) and heavier, while the seeds from the CS plants were smaller and lighter, than seeds harvested from wild-type plants, which were grown alongside the transgenic plants (Fig. 7, I and J). These data strongly suggest that OsBSK3 expression is critical for the activation of BR signaling in rice.
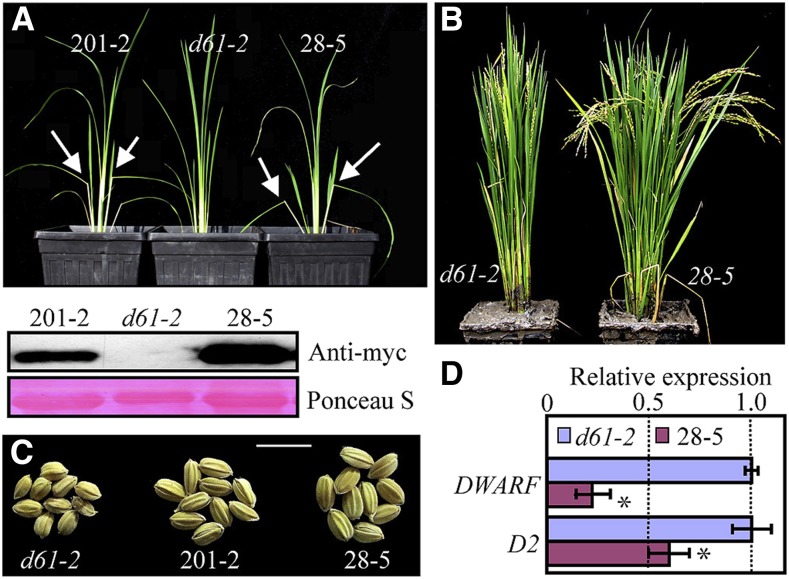
We also overexpressed wild-type or S215E-substituted N390 (N390-S215E) in the weak OsBRI1 mutant d61-2. The overexpression of N390 in d61-2 did not alter the mutant phenotype of the plants. However, overexpression of N390-S215E significantly increased the leaf bending angle in young d61-2 mutant plants, and the leaves of the full-grown transgenic plants were less erect (Fig. 8, A and B). The seeds of the N390-S215E-overexpressing plants were much larger than those of the d61-2 control plants (Fig. 8C). qRT-PCR revealed that the expression of DWARF and D2 was significantly reduced in the N390-S215E-overexpressing d61-2 mutant plants, suggesting that BR signaling was activated (Fig. 8D). Taken together, our results suggest that, similar to our observation in Arabidopsis, the ability of the various forms of OsBSK3 to activate BR signaling in rice was as follows: N390-S215E > N390 > full-length OsBSK3.
Figure 8.
Partial rescue of the d61-2 mutant phenotype via overexpression of the S215E-substituted kinase domain of OsBSK3. A, The upper panel shows 5-week-old d61-2 mutant plants or d61-2 plants overexpressing C-terminal myc tagged, S215E-substituted kinase domain of OsBSK3 (N390-S215E); 201-2 and 28-5 are two independent transgenic lines. Arrow, exaggerated lamina joint bending in the transgenic seedlings. The lower panel shows the expression levels of N390-S215E protein in the two independent transgenic lines shown in the upper panel. B, Full-grown d61-2 mutant and transgenic d61-2 plants overexpressing N390-S215E (28-5). C, Seeds of d61-2 mutant plants and d61-2 mutant plants overexpressing N390-S215E grown in the field. D, The expression levels of DWARF and D2 in 1-month-old d61-2 mutant plants and d61-2 plants overexpressing N390-S215E (28-5). Error bars represent ± se. Statistically significant differences are indicated by asterisks (P < 0.05).
DISCUSSION
As major growth-promoting hormones, BRs play important roles in regulating yield-related traits in rice plants, including leaf angle, tillering, plant height, and grain filling (Hong et al., 2004; Vriet et al., 2012). Compared with the elaborate BR signaling mechanism discovered in Arabidopsis, BR signaling in rice is not well understood. However, the severe growth-inhibiting phenotypes caused by the mutation of BRI1 orthologs in rice (Nakamura et al., 2006), tomato (Solanum lycopersicum; Koka et al., 2000), pea (Pisum sativum; Nomura et al., 2003), and barley (Hordeum vulgare; Gruszka et al., 2011) suggest that the BRI1-mediated BR signaling pathway is conserved across the plant kingdom. In addition to OsBRI1, orthologs of other Arabidopsis BR signaling components, such as OsBAK1 (Li et al., 2009), OsGSK2 (Tong et al., 2012), and OsBZR1 (Bai et al., 2007), have been found to regulate BR signaling in rice; however, the molecular mechanisms leading from the activation of OsBRI1 by BRs to the regulation of gene expression are mostly unknown. In this study, we identified four BSK orthologs in the rice genome by a sequence homology search. Overexpression of the kinase domain of OsBSK3 in rice plants reduced the expression of the BR biosynthesis genes DWARF and D2 and increased the sensitivity of the transgenic seedlings to eBL-induced leaf bending and eBL-inhibited root elongation. Consistent with our overexpression data, simultaneous knockdown of the four OsBSKs reduced the leaf bending angle and increased DWARF expression in rice. Taken together, our results suggest that OsBSK3 positively regulates BR signaling in rice.
Despite the fact that OsBSK3 does not contain a transmembrane domain, subcellular localization studies showed that it is localized to the plasma membrane by an N-terminal myristoylation site. This localization pattern is critical for the activation of BR signaling by OsBSK3; the mutation of this myristoylation site not only changed the localization of OsBSK3 from the plasma membrane to the cytoplasm, it also impaired the ability of OsBSK3 to rescue the mutant phenotype of bri1-5. Similar observations have been reported for other RLCKs, including AtBSK1 (Shi et al., 2013), AtLIP1, AtLIP2 (Liu et al., 2013), CST (Burr et al., 2011), and TPK1b (Abuqamar et al., 2008), indicating that lipidation-mediated membrane localization is a general feature of these membrane-associated RLCKs. Correlated with its plasma membrane localization pattern, BiFC and split luciferase assays showed that OsBSK3 interacted directly with, and was phosphorylated by, the membrane-localized BR receptor OsBRI1. Together, these results suggest that the role of OsBSK3 in regulating BR signaling is similar to that of AtBSKs, which transduce BR signals from BRI1 to downstream signaling component BSU1. This hypothesis is further supported by the observation that S215E-substituted OsBSK3, which mimicked the constitutively phosphorylated form of OsBSK3, bound BSU1 more strongly than wild-type OsBSK3.
Even though phosphorylation by AtBRI1 increases AtBSK1 binding to the downstream phosphatase BSU1 (Kim et al., 2009), direct evidence showing that the phosphorylation of BSKs by BRI1 activates BR signaling in vivo is lacking. By mass spectrometry, we identified Ser-213 and Ser-215 of OsBSK3 as OsBRI1 phosphorylation sites. Site-directed mutagenesis revealed that inhibiting the phosphorylation of OsBSK3 at Ser-215 by OsBRI1 prevented OsBSK3 from rescuing the mutant phenotype of bri1-5 plants, while substituting Ser-215 with Glu not only rescued the bri1-5 mutant phenotype, it also made the transgenic plants insensitive to 0.25 μm PCZ-inhibited hypocotyl elongation. Similarly, the phosphorylation of AtBSK3 at Ser-212 (equivalent to Ser-215 of OsBSK3) activated BR signaling in Arabidopsis. This conservative Ser residue was previously shown to be a major AtBRI1 phosphorylation site in AtBSK1 (Ser-230) and AtCDG1 (Ser-234); it is also important for the activation of AtBSU1 by AtBSK1 and AtCDG1 (Kim et al., 2009, 2011). Together, these results suggest that the mechanism by which BRI1 regulates BR signaling through the phosphorylation of BSKs is conserved in both Arabidopsis and rice. Although this idea was mostly tested using transgenic Arabidopsis plants, the partial rescue of the d61-2 mutant phenotype by overexpression of the S215E-substituted form of the OsBSK3 kinase domain (but not the wild-type form) suggests that a similar mechanism functions in rice plants.
With 72% homology to AtBSK3, OsBSK3 contains all of the key features of AtBSKs. A protein structure analysis of AtBSK8 suggested that AtBSKs are constitutively inactive protein kinases (Grütter et al., 2013). However, it was reported that AtBSK1 might contain weak Mn2+-dependent kinase activity (Shi et al., 2013). We also detected a weak OsBSK3 autophosphorylation signal during our in vitro kinase assay. The autoradiographic signal was stronger in the presence of Mn2+, and it was increased by OsBRI1 phosphorylation (Supplemental Fig. S13). However, the signal was so weak that we had to expose the dried gel under a storage phosphor screen for 1 week in order to obtain a clear image. Therefore, we cannot confidently claim that OsBSK3 is an active kinase based on this result. To further investigate whether the kinase activity of OsBSK3 is an essential part of its BR signaling function, we mutated two invariable amino acids that are considered to be critical for the kinase activity of OsBSK3 to create two mutant forms (K89E and K89E D183A) of the kinase by site-directed mutagenesis (Grütter et al., 2013). Surprisingly, when overexpressed, these “kinase-dead” forms of OsBSK3 were unable to rescue the semi-dwarf phenotype of bri1-5 mutant plants (Supplemental Fig. S14), suggesting that the kinase activity of OsBSK3 is required for its ability to transduce BR signals. As a binding partner-dependent activation mechanism has been shown to regulate AtRRK1 family RLCKs (Dorjgotov et al., 2009), whether a similar mechanism exists for BSK family proteins should be examined in a future study.
Besides BSKs, AtCDG1 family RLCKs positively regulate BR signaling (Kim et al., 2011). Similar to AtBSKs, AtCDG1 can interact directly with AtBRI1 and activate the downstream component AtBSU1 upon BRI1 phosphorylation. However, the overexpression of AtCDG1 in an AtBSK3 T-DNA insertion mutant could not rescue its BR-insensitive phenotype, suggesting that AtCDG1 regulates BR signaling differently from AtBSK3 (Kim et al., 2011). Here, we found that plants overexpressing S215E-substituted OsBSK3 had cdg1-D-like curled and twisted stems, leaves, and siliques (Muto et al., 2004). As cdg1-D is a gain-of-function mutant in which CDG1 is overexpressed due to the insertion of four 35S enhancers into its promoter sequence, the similar phenotypes of the CDG1- and S215E-overexpressing plants suggest that OsBSK3 and CDG1 regulate plant development via similar mechanisms.
A common feature of BSK family RLCKs is the presence of an N-terminal kinase domain and a C-terminal TPR domain; however, the role of the TPR domain in regulating the biological function of BSKs is unknown. Although it is generally accepted that the TPR domain acts mostly as a scaffold to promote the formation of multiprotein complexes (Allan and Ratajczak, 2011), our study shows that the TPR domain of both AtBSK3 and OsBSK3 acts as an autoinhibitory domain that binds to the kinase domain of BSK3 and prevents it from binding to and activating BSU1. Our in vitro pull-down and quantitative yeast two-hybrid assay results suggest that when OsBSK3 was phosphorylated by OsBRI1, the binding affinity of its TPR domain for its kinase domain was greatly reduced. A protein overlay assay showed that phosphorylation at Ser-215 greatly increased the binding of OsBSK3 to BSU1. Taken together, our results suggest that the binding of OsBSK3 with AtBSU1 is regulated by BRI1-dependent phosphorylation.
Based on our results, we propose a working model for BSKs in which the TPR domain binds with the kinase domain to prevent BSKs from binding to and activating BSU1 when BR signaling is turned off. The phosphorylation of BSKs by BR-activated BRI1 frees the kinase domain of BSKs from the TPR domain and increases the binding affinity of BSKs for BSU1, promoting the dephosphorylation of BIN2 by BSU1 via a still unknown mechanism.
MATERIALS AND METHODS
Plant Materials and Growth
The Arabidopsis (Arabidopsis thaliana) bri1-5 mutant is of the Ws ecotype, while the det2 and bri1-116 mutants are of the Columbia ecotype. For the observation of growth phenotypes, Arabidopsis seedlings were grown in a greenhouse at 22°C under long-day conditions (16 h of light:8 h of dark) at a light intensity around 100 μmol·m−2·s−1. For the hypocotyl growth assays, Arabidopsis seedlings were grown on agar medium containing half-strength Murashige and Skoog salts, 1% Suc, and the indicated concentration of eBL in the light or the BR biosynthesis inhibitor PCZ or BRZ in the dark at 22°C for 1 week before taking pictures for measurement using ImageJ software. The average ratio and sd were calculated based on values collected from at least three biological repeats, with at least 15 plants per experiment. The experiments included in this study were performed at least three times with similar results. Representative data from one repetition are shown in the figures.
Oryza sativa ssp japonica cultivar Dongjin was used for all transgenic experiments in rice. OsBRI1 mutant d61-2 is of the Taichung 65 background. For the BR-induced lamina joint bending assay, rice seeds were germinated in double-distilled water at 28°C in the dark for 4 d. The seedlings were then transferred to soil and grown for 10 additional days under long-day condition at 28°C before treatment with different concentrations of eBL at the leaf lamina joint to stimulate bending. The seedlings were allowed to grow three additional days before taking photos; the leaf bending angle for each seedling was calculated using ImageJ software. The average ratio and sd were calculated based on values collected from at least three biological repeats with 10 to 15 plants per experiment.
Overexpression of OsBSK3 in Arabidopsis and Rice
Full-length wild-type and mutated OsBSK3 or amino acids 1 to 390 of the wild-type OsBSK3 coding sequence (N390) without the stop codon were cloned into the binary vectors pEarlygate 101 (p101), pEarlygate 100 (p100), pMDC83, or pCAMBIA-1390 and then introduced into Agrobacterium tumefaciens strain GV3101 or EHA105. The constructs were transformed into Arabidopsis by the GV3101-mediated floral dip method. Multiple independent T3 homozygous transgenic lines were used for the phenotype analysis shown in this study; the reported results were consistent in these independent lines. Transgenic rice plants were generated by EHA105-mediated callus transformation (Yang et al., 2004). T2 homozygous transgenic rice seedlings were used for the phenotype analysis, unless indicated otherwise.
Gene Expression Analysis by Quantitative Real-Time RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized using M-MLV Reverse Transcriptase (Takara Bio) according to the manufacturer’s instructions. A SYBR Premix Ex Taq kit (Tli RNase H Plus; Takara Bio) was used for the real-time quantitative PCR analysis of gene expression with an ABI PRISM 7500 real-time system. The primer pairs used were as follows: 5′-TTGCTCAACTCAAGGAAGAG-3′ and 5′-TGATGTTAGCCACTCGTAGC-3′ for CPD (At5g05690); 5′-CAAATCCAAAACCCTAGAAACCGAA-3′ and 5′-ATCTCCCGTAGGACCTGCACTG-3′ for UBC30 (At5g56150); 5′-AGCTGCCTGGCACTAGGCTCTACAGATCAC-3′ and 5′-ATGTTGTCGGAGATGAGCTCGTCGGTGAGC-3′ for D2 (Os01g10040); 5′-CAAGCAGGGACCCAGTTTCAT-3′ and 5′-CCAGGATGTCCAGCATCGACT-3′ for DWARF (Os03g40540); 5′-ACACCTCCAGAGTATTTGAGAAATG-3′ and 5′-AACTAGAATCTGATGGATTGCTGTC-3′ for OsBSK1 (Os03g04050); 5′-CACCCTTTCCAAGCATCTCT-3′ and 5′-TATAACTTTTCCCATCGCGG-3′ for OsBSK2 (Os10g42110); 5′-CGCGGATCCCACCGCAAAGCCTGAGGTGGCCAG-3′ and 5′-CACCATGGGCGGGCGCGTGTCCAAG-3′ for OsBSK3 (Os04g58750); 5′-ACTGGGAGACAAAGCCATTGAG-3′ and 5′-GCCTTCTAAGCAAGAGTCCATCA-3′ for OsBSK4 (Os03g61010); 5′-AGCAACTGGGATGATATGGA-3′ and 5′-CAGGGCGATGTAGGAAAGC-3′ for OsACTIN1 (Os03g50885). The expression level of CPD was normalized against that of UBC30 in Arabidopsis, while the expression levels of DWARF and D2 were normalized against that of OsACTIN1 in rice. The relative gene expression level is presented as a ratio to the expression level in the control sample. The average ratio and sd were calculated based on values collected from at least three biological repeats.
Fractionation of Membrane Proteins
Microsomal protein was prepared according to Tang (2012), with slight modifications. In brief, Arabidopsis seedlings were ground in buffer H (25 mm HEPES, 10% glycerol, 0.33 m Suc, 0.6% polyvinylpyrrolidone, 5 mm ascorbic acid, 5 mm EDTA, 25 mm NaF, 1 mm sodium molybdate, 2 mm imidazole, 1 mm activated sodium vanadate, 5 mm DTT, 1 µm E-64, 1 µm bestatin, 1 µm pepstatin, 2 µm leupeptin, and 1 mm PMSF, pH 7.5) at 4°C. The homogenate was filtered through two layers of Miracloth and centrifuged at 10,000g for 15 min to pellet cell debris and organelles. The supernatant was then spun at 60,000g for 60 min to pellet microsomes. The supernatant (soluble proteins) and microsome pellet (membrane proteins) were boiled in 2× SDS extraction buffer (125 mm Tris-HCl, pH 6.8, 2% β-mercaptoethanol, 4% SDS, 20% glycerol, and 0.25% bromophenol blue) for 5 min, separated by 10% SDS-PAGE, transferred to a nitrocellulose membrane (EMD Millipore), and detected with anti-GFP antibodies.
In Vivo Protein-Protein Interaction Assays
Yeast two-hybrid and BiFC assays were performed according to Gampala et al. (2007). The full-length coding sequences of OsBSK3 and OsBRI1 (Os01g52050) were cloned in-frame into the BiFC expression vectors pX-NYFP and pX-CCFP. The vectors were then transformed into A. tumefaciens strain GV3101 and coinfiltrated into 4-week-old tobacco (Nicotiana benthamiana) leaves. At 36 to 48 h after infiltration, YFP fluorescence was observed by confocal microscopy (Zeiss LSM 510).
For the split luciferase assay, the full-length coding sequence of OsBRI1 was cloned into pCAMBIA-NLuc (Chen et al., 2008), with the N-terminal luciferase sequence fused to the C terminus of OsBRI1. The C-terminal luciferase sequence was amplified by PCR from pCAMBIA-CLuc (Chen et al., 2008) and added to the C terminus of the full-length OsBSK3 coding sequence in pENTR/SD/D-TOPO (Invitrogen), followed by subcloning into pEarlygate 100 using LR Clonase (Invitrogen). The resulting OsBRI1-NLuc- and OsBSK3-CLuc-expressing vectors were introduced to A. tumefaciens strain GV3101 and infiltrated into tobacco leaves as described previously (Gampala et al., 2007). At 36 to 48 h after infiltration, the tobacco leaves were sprayed with 2.5 mg/mL luciferin (Gold Biotechnology) and kept in the dark for 6 min to quench chloroplast fluorescence. Images showing luciferase bioluminescence were taken using a low-light cooled CCD imaging apparatus (Andor Technology) with the temperature of the camera precooled to −96°C (exposure time, 500 s; 1 × 1 binning).
Relative firefly luciferase activity was detected using a Centro LB 960 microplate luminometer (Berthold Technologies). At 36 to 48 h after infiltration, discs were punched from the tobacco leaves (0.3 cm in diameter) and placed into 96-well plates. The leaf discs were kept in the dark for 6 min to quench the fluorescence signal; 50 μL of 2.5 mg/mL luciferin (Gold Biotechnology) was then injected and the bioluminescence signal was recorded immediately for 10 s. The average ratio and sd were calculated based on values collected from at least three biological repeats, with five to six independent measurements per experiment.
Site-Directed Mutagenesis
The full-length coding sequence of OsBSK3 (without the stop codon) was cloned into pENTR/SD/D-TOPO (Invitrogen) and used as the template for all site-directed mutagenesis experiments. Site-directed mutagenesis was achieved using PCR, which was performed with 18 cycles in a 10 μL reaction mixture. The PCR product was digested overnight using DpnI (Takara Bio), and 1 μL of the digested product was used to transform Escherichia coli strain DH5α. Following the verification of each mutation by sequencing, the mutated forms of OsBSK3 were incorporated into a Gateway-compatible destination vector (pEarlygate 101, pGEX4T-1, gc-pGADT7, or gc-pCAMBIA-1390) using LR Clonase (Invitrogen).
Protein Purification and in Vitro Protein-Protein Interaction Assays
GST-, MBP-, and 6His-tagged recombinant proteins were expressed in E. coli and purified by standard protocols using glutathione Sepharose 4B beads (GE Healthcare), amylose agarose beads (New England Biolabs), or Ni-NTA resin (Qiagen). The purified recombinant proteins were concentrated by ultrafiltration using Centricon centrifuge tubes (EMD Millipore) with a 10-kD Mr cutoff.
For the dot blot assays, around 1 μmol each of recombinant 6His-OsBSK3 and 6His-N390 was dot blotted onto a nitrocellulose membrane. The membrane was allowed to air dry for 15 min in a cold room and then incubated in PBS containing 5% nonfat milk. Following incubation with 1 μg/mL of MBP-AtBSU1 followed by peroxidase-labeled anti-MBP antibodies, the overlay signal was detected using SuperSignal West Dura Chemiluminescence Reagent (Pierce). For the in vitro pull down assays, 2 μg of 6His-N390 was incubated overnight with 1 μg of MBP-tagged kinase domain of OsBRI1 in the presence or absence of 0.1 mm ATP. Glutathione Sepharose 4B beads bound GST-TPR were added to the reaction, and the mixture was rotated at 4°C for 2 h. The beads were then washed three times with PBS, and the proteins were eluted by boiling in 2× SDS sample buffer for 5 min. After SDS-PAGE, the proteins were transferred to a nitrocellulose membrane and the pull down signal was detected using anti-6His or -GST antibodies.
In Vitro Kinase Assays and Identification of the Phosphorylation Sites by Mass Spectrometry
In vitro phosphorylation assays were performed in a mixture containing 25 mm Tris-HCl, pH 7.5, 10 mm MgCl2 or 10 mm MnCl2, 100 mm NaCl, 1 mm DTT, 100 μm ATP, 5 to 10 μCi of 32P-γATP, 0.5 to 1 μg of GST-OsBRI1KD, and 2 to 5 μg of GST-OsBSK3. The mixtures were incubated at 30°C for 3 h with constant shaking. The reactions were stopped by the addition of 6× SDS sample buffer and incubation at 65°C for 10 min. Protein phosphorylation was analyzed by SDS-PAGE and autoradiography.
To identify the OsBRI1 phosphorylation sites on OsBSK3, GST-OsBSK3 attached to Glutathione Sepharose 4B beads was first incubated with lambda phosphatase for 6 h to generate hypophosphorylated OsBSK3 because it was reported that purified recombinant proteins can be phosphorylated by anonymous bacterial kinases when expressed in E. coli cells or during the protein purification process (Wu et al., 2012). After incubation, the Sepharose beads were washed extensively to remove the lambda phosphatase and eluted with 10 mm glutathione. Next, 2.5 μg of lambda phosphatase-treated GST-OsBSK3 was incubated with 5 μg of GST-OsBRI1KD overnight and then separated by SDS-PAGE. The Coomassie Brilliant Blue-stained gel containing GST-OsBSK3 was cut into 1- to 2-mm pieces and in-gel digested. The extracted peptides were freeze-dried using a SpeedVac, resuspended in 0.1% formic acid (FA), and separated by reverse phase liquid chromatography with an Easy-Spray PepMap column (75 µm × 15 cm; Thermo Fisher Scientific) using a nanoACQUITY Ultra Performance liquid chromatography system (Waters). Liquid chromatography was conducted using buffer A (2% acetonitrile and 0.1% FA) and buffer B (98% acetonitrile and 0.1% FA). A linear gradient from 2 to 30% B followed by washing with 50% B at a flow rate of 300 nL/min was used for peptide separation. MS/MS was conducted with an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) using the top six data-dependent acquisition method. The sequence included one survey scan in FT mode in the Orbitrap with a mass resolution of 30,000 followed by six collision-induced dissociation (CID) scans in LTQ, focusing on the first six most intense peptide ion signals whose m/z values were not on the dynamically updated exclusion list and whose intensities exceeded a threshold of 1,000 counts. The CID-normalized collision energy was set to 30. The analytical peak lists were generated from the raw data using PAVA software (Guan et al., 2011). The MS/MS data were searched against the proteome sequences for rice and Arabidopsis from Swiss-Prot using the Protein Prospector search engine (http://prospector.ucsf.edu/prospector/mshome.htm). The mass tolerance for precursor ions and fragment ions was set to 20 ppm and 0.7 D, respectively. The maximum number of missed cleavages was set at 2. Ser, Thr, and Tyr phosphorylation; Cys carbamidomethylation; and Met oxidation were included in the search as variable modifications.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsBSK1 (NM_001055405.1), OsBSK2 (NM_001071987.1), OsBSK3 (NM_001060843.2), OsBSK4 (NM_001058289.2), OsBRI1 (NM_001050612.1), D2 (NM_001048832.1), DWARF (NM_001057158.1), and OsACTIN (NM_001057621.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Four BSK orthologs are present in the rice genome.
Supplemental Figure S2. Semiquantitative RT-PCR analysis of the expression of the OsBSKs in rice seedlings.
Supplemental Figure S3. Subcellular localization of OsBSK3 in rice root.
Supplemental Figure S4. The in vitro OsBRI1-phosphorylated peptides identified by mass spectrometry from OsBSK3.
Supplemental Figure S5. In vivo-phosphorylated peptides identified by mass spectrometry from immunoprecipitated OsBSK3 or AtBSK3.
Supplemental Figure S6. Phenotypes of S215E-overexpressing bri1-5 mutant lines.
Supplemental Figure S7. The phosphorylation of AtBSK3 at Ser-212 activates BR signaling in Arabidopsis.
Supplemental Figure S8. BiFC assays showed that AtBSU1 interacted more strongly with the S215E form of OsBSK3.
Supplemental Figure S9. OsBSK3 with YFP fused to its C terminus was more efficient at suppressing the dwarf phenotype of bri1-5 mutant plants.
Supplemental Figure S10. Overexpression of the kinase domain of OsBSK3 partially suppressed the semi-dwarf phenotype of bri1-301 mutant plants.
Supplemental Figure S11. BR signaling was hyperactivated in N390-overexpressing Arabidopsis plants.
Supplemental Figure S12. Quantitative analysis of the interaction between the kinase and TPR domains of OsBSK3 and AtBSU1.
Supplemental Figure S13. Assay for OsBSK3 autophosphorylation.
Supplemental Figure S14. Overexpression of the K89E or K89E D183A forms of OsBSK3 could not rescue bri1-5 mutant plants.
Supplementary Material
Acknowledgments
We thank Tomoaki Sakamoto (Nagoya University) for providing the d61-2 rice mutant.
Glossary
- BiFC
bimolecular fluorescence complementation
- LC/MS/MS
liquid chromatography-tandem mass spectrometry
- Ws
Wassilewskija
- PCZ
propiconazole
- eBL
epibrassinolide
- BRZ
brassinazole
- CS
cosuppression
- FA
formic acid
References
- Abuqamar S, Chai MF, Luo H, Song F, Mengiste T (2008) Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20: 1964–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan RK, Ratajczak T (2011) Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16: 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr CA, Leslie ME, Orlowski SK, Chen I, Wright CE, Daniels MJ, Liljegren SJ (2011) CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol 156: 1837–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells E, Casacuberta JM (2007) Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J Exp Bot 58: 3503–3511 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorjgotov D, Jurca ME, Fodor-Dunai C, Szűcs A, Otvös K, Klement E, Bíró J, Fehér A (2009) Plant Rho-type (Rop) GTPase-dependent activation of receptor-like cytoplasmic kinases in vitro. FEBS Lett 583: 1175–1182 [DOI] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, et al. (2007) An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell 13: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Liu JS, Fan M, Bai MY, Wenkel S, Springer PS, Barton MK, Wang ZY (2012) Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci USA 109: 21152–21157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MMA (2011). Physiological effects related to brassinosteroid application in plants. In S Hayat, A Ahmad, eds, Brassinosteroids: A Class of Plant Hormone. Springer, Dordrecht, The Netherlands, pp 193–242 [Google Scholar]
- Gruszka D, Szarejko I, Maluszynski M (2011) New allele of HvBRI1 gene encoding brassinosteroid receptor in barley. J Appl Genet 52: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grütter C, Sreeramulu S, Sessa G, Rauh D (2013) Structural characterization of the RLCK family member BSK8: a pseudokinase with an unprecedented architecture. J Mol Biol 425: 4455–4467 [DOI] [PubMed] [Google Scholar]
- Guan SH, Price JC, Prusiner SB, Ghaemmaghami S, Burlingame AL (2011) A data processing pipeline for mammalian proteome dynamics studies using stable isotope metabolic labeling. Mol Cell Proteomics. 10: M111.010728. [DOI] [PMC free article] [PubMed]
- Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DPV, Choe S, Johal GS, Schulz B (2011) Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA 108: 19814–19819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka U, Matsuoka M (2004) Brassinosteroids and rice architecture. J Pestic Sci 29: 184–188 [Google Scholar]
- Kakita M, Murase K, Iwano M, Matsumoto T, Watanabe M, Shiba H, Isogai A, Takayama S (2007) Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka CV, Cerny RE, Gardner RG, Noguchi T, Fujioka S, Takatsuto S, Yoshida S, Clouse SD (2000) A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol 122: 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U, Wang ZY (2012) Brassinosteroid action in flowering plants: a Darwinian perspective. J Exp Bot 63: 3511–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7: 791–806 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhong S, Guo X, Hao L, Wei X, Huang Q, Hou Y, Shi J, Wang C, Gu H, Qu LJ (2013) Membrane-bound RLCKs LIP1 and LIP2 are essential male factors controlling male-female attraction in Arabidopsis. Curr Biol 23: 993–998 [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Yabe N, Asami T, Hasunuma K, Yamamoto KT (2004) Overexpression of constitutive differential growth 1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in Arabidopsis. Plant Physiol 136: 3124–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. (2006) The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol 140: 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nomura T, Bishop GJ, Kaneta T, Reid JB, Chory J, Yokota T (2003) The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J 36: 291–300 [DOI] [PubMed] [Google Scholar]
- Roux F, Noël L, Rivas S, Roby D (2014) ZRK atypical kinases: emerging signaling components of plant immunity. New Phytol 203: 713–716 [DOI] [PubMed] [Google Scholar]
- Santiago J, Henzler C, Hothorn M (2013) Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892 [DOI] [PubMed] [Google Scholar]
- Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D (2013) BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, Sessa G (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J 74: 905–919 [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Han Z, Tang J, Hu Z, Chai C, Zhou B, Chai J (2013) Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res 23: 1326–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. (2012) Quantitative analysis of plasma membrane proteome using two-dimensional difference gel electrophoresis. Methods Mol Biol 876: 67–82 [DOI] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GA Jr, Okuyama H (2000) Lipid-linked proteins of plants. Prog Lipid Res 39: 19–39 [DOI] [PubMed] [Google Scholar]
- Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C (2012) DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24: 2562–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J (2006) Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100 [DOI] [PubMed] [Google Scholar]
- Vriet C, Russinova E, Reuzeau C (2012) Boosting crop yields with plant steroids. Plant Cell 24: 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wu X, Oh MH, Kim HS, Schwartz D, Imai BS, Yau PM, Clouse SD, Huber SC (2012) Transphosphorylation of E. coli proteins during production of recombinant protein kinases provides a robust system to characterize kinase specificity. Front Plant Sci 3: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, Yoshimura S, Hayashi N, Uchihashi K, Ishihama N, Kishi-Kaboshi M, Takahashi A, Tsuge S, et al. (2013) A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13: 347–357 [DOI] [PubMed] [Google Scholar]
- Yang Y, Peng H, Huang H, Wu J, Jia S, Huang D, Lu T (2004) Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci 167: 281–288 [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.