A tomato heterotrimeric G protein γ-subunit suppresses auxin-induced root development while it facilitates ABA inhibition of seed germination.
Abstract
Heterotrimeric G proteins composed of α, β, and γ subunits are central signal transducers mediating the cellular response to multiple stimuli in most eukaryotes. Gγ subunits provide proper cellular localization and functional specificity to the heterotrimer complex. Plant Gγ subunits, divided into three structurally distinct types, are more diverse than their animal counterparts. Type B Gγ subunits, lacking a carboxyl-terminal isoprenylation motif, are found only in flowering plants. We present the functional characterization of type B Gγ subunit (SlGGB1) in tomato (Solanum lycopersicum). We show that SlGGB1 is the most abundant Gγ subunit in tomato and strongly interacts with the Gβ subunit. Importantly, the green fluorescent protein-SlGGB1 fusion protein as well as the carboxyl-terminal yellow fluorescent protein-SlGGB1/amino-terminal yellow fluorescent protein-Gβ heterodimer were localized in the plasma membrane, nucleus, and cytoplasm. RNA interference-mediated silencing of SlGGB1 resulted in smaller seeds, higher number of lateral roots, and pointy fruits. The silenced lines were hypersensitive to exogenous auxin, while levels of endogenous auxins were lower or similar to those of the wild type. SlGGB1-silenced plants also showed strong hyposensitivity to abscisic acid (ABA) during seed germination but not in other related assays. Transcriptome analysis of the transgenic seeds revealed abnormal expression of genes involved in ABA sensing, signaling, and response. We conclude that the type B Gγ subunit SlGGB1 mediates auxin and ABA signaling in tomato.
Heterotrimeric G proteins (G proteins) consisting of Gα, Gβ, and Gγ subunits are arguably the most important signaling mediators in eukaryotes. In animal systems, G proteins bind to seven-transmembrane-spanning G protein-coupled receptors (GPCRs) at the plasma membrane. Upon perception of a stimulus, GPCRs facilitate the replacement of GDP bound to the Gα subunit with GTP. This exchange results in the activation of the heterotrimer. The active GTP-bound Gα and the Gβγ dimer independently initiate specific signaling pathways. The intrinsic GTPase activity of Gα hydrolyzes GTP to GDP and thereby returns the heterotrimer to the steady-state mode (Gautam et al., 1998; McCudden et al., 2005).
Genetic approaches have revealed the involvement of G proteins in a wide variety of plant processes, including defense against pathogens (Llorente et al., 2005; Trusov et al., 2006, 2009, 2010; Trusov and Botella, 2012; Maruta et al., 2015), morphological development (Lease et al., 2001; Ullah et al., 2003; Chakravorty et al., 2011; Thung et al., 2012), cell proliferation (Ullah et al., 2001; Chen et al., 2006a), ion-channel regulation (Armstrong and Blatt, 1995; Chakravorty et al., 2012), stomatal control (Assmann, 1996; Zhang et al., 2008; Chakravorty et al., 2011), light perception (Warpeha et al., 1991, 2006, 2007; Okamoto et al., 2001; Jones et al., 2003; Ullah et al., 2003; Botto et al., 2009), early seedling development (Lapik and Kaufman, 2003), abiotic stresses (Booker et al., 2004; Joo et al., 2005; Misra et al., 2007; Bhardwaj et al., 2012), and responses to phytohormones including abscisic acid (ABA), GA, brassinosteroid, ethylene, jasmonic acid, and auxins (Ullah et al., 2002; Chen et al., 2004; Pandey and Assmann, 2004; Huang et al., 2006; Trusov et al., 2006; Wang et al., 2006; Okamoto et al., 2009).
During evolution, plant G proteins have acquired a number of unique characteristics not seen in animal G proteins (Chen et al., 2003, 2004; Jones and Assmann, 2004; Johnston and Siderovski, 2007; Temple and Jones, 2007; Chakravorty et al., 2011; Jones et al., 2011b; Urano et al., 2012, 2013; Urano and Jones, 2014). For example, in animals, activation of GPCRs catalyzes the exchange of GDP for GTP in the Gα subunit; however, in plants, this step seems to be spontaneous, without the need for accessory proteins (Jones et al., 2011a). Instead of GPCRs, plants can use alternative receptors such as ATRGS1 that keeps the plant G protein complex in its resting state. Upon binding of an agonist, the RGS undergoes phosphorylation and subsequent endocytosis, releasing the G protein complex, which spontaneously activates (i.e. loads with GTP), starting the signaling cycle (Jones et al., 2011b). A similar mechanism seems to exist in rice (Oryza sativa), where the COLD1 receptor serves as a GTPase-accelerating protein (Ma et al., 2015). Plant G proteins also have been proven to mediate responses from single-pass membrane receptors such as the BAK1 Interacting receptor-like kinase (BIR1), Nod factor receptors, and RECEPTOR-LIKE PROTEIN KINASE2 in Arabidopsis (Arabidopsis thaliana; Choudhury and Pandey, 2013; Liu et al., 2013; Ishida et al., 2014). In maize (Zea mays), the Gα subunit was functionally linked to FASCIATED EAR2, an ortholog of Arabidopsis CLAVATA2, receptor-like protein (Bommert et al., 2013).
While humans possess 23 Gα, six Gβ, and 12 Gγ subunits (Milligan and Kostenis, 2006), the Arabidopsis genome has only one Gα, one Gβ, and three Gγ genes (Ma et al., 1990; Weiss et al., 1994; Mason and Botella, 2000, 2001; Chakravorty et al., 2011). Recently it was demonstrated that a plant-specific group of Gα-like proteins, extra-large G proteins (Lee and Assmann, 1999; Ding et al., 2008), also form complexes with the canonical Gβγ dimer and initiate defense responses (Maruta et al., 2015) and, therefore, should be considered as G protein subunits. Diversity studies of plant Gγ subunits revealed three distinct types of these proteins, two of which were specific to plants (Trusov et al., 2012). Type A represents the canonical form of the Gγ subunits, which are structurally similar to their animal and fungal counterparts. These proteins are characterized by relatively small size, a conserved domain for coiled-coil interaction with Gβ, and a C-terminal isoprenylation motif, CaaX (where C is Cys, a is an aliphatic amino acid, and X is variable, with a preference for hydrophobic and aliphatic amino acids). Type B Gγ subunits are similar to type A Gγ subunits but lack the C-terminal CaaX motif, precluding the possibility of prenylation (Trusov et al., 2012). Finally, type C Gγ subunits have the conserved domain, a transmembrane domain, and a relatively long Cys-rich C-terminal end (Chakravorty et al., 2012; Trusov et al., 2012; Wolfenstetter et al., 2015). A similar classification has been reported for soybean (Glycine max) Gγ subunits (Choudhury et al., 2011). The roles of type A and type C Gγ subunits have been established in Arabidopsis and rice (Fan et al., 2006; Trusov et al., 2007; Huang et al., 2009; Chakravorty et al., 2011), while the functions of type B have not been studied yet.
In this study, we have identified four genes encoding Gγ subunits of heterotrimeric G proteins in tomato (Solanum lycopersicum) ‘MicroTom’. Relative expression levels were determined for all four genes. We demonstrate that the tomato type B Gγ subunit SlGGB1 interacts with the tomato Gβ subunit, but unlike any other known Gγ subunits, it localizes to the cytoplasm and nucleus in addition to the usual localization to the plasma membrane. Analyses of several RNA interference (RNAi) lines with significantly reduced levels of SlGGB1 revealed alterations in the development of lateral roots, fruits, and seeds. These transgenic lines also had altered responses to auxin and ABA. We conclude that the type B Gγ subunit SlGGB1 plays an important role in auxin signaling throughout plant development and is involved in ABA signaling during seed germination.
RESULTS
The Tomato Genome Contains Four Genes Encoding Heterotrimeric G Protein Gγ Subunits
BLAST searches of the tomato proteome (cv Heinz; ITAG release 2.40) using Arabidopsis Gγ subunits as queries identified four Gγ-like proteins. Of the four identified putative Gγ subunits, one belonged to the previously described type A, two to type B, and one to type C (Trusov et al., 2012). According to the nomenclature suggested by Trusov et al. (2012), we named these genes SlGGA1 (Solyc09g082940.2.1), SlGGB1 (Solyc12g096270.1.1), SlGGB2 (Solyc08g005950.2.1), and SlGGC1 (Solyc07g041980.2.1).
The type B subunit genes, SlGGB1 and SlGGB2, have open reading frames of 354 and 384 nucleotides encoding 117 and 127 amino acids, respectively, and consist of four exons and three introns. The proteins share 69% amino acid identity within the conserved central region responsible for the coiled-coil interaction with the Gβ subunit. In the conserved DPLL motif, both proteins have a substitution of Pro with Ala (DALL; Supplemental Fig. S1). In addition, both proteins have conserved residues important for interaction with the Gβ subunit (Supplemental Fig. S1; Temple and Jones, 2007). The most distinct feature of SlGGB1 and SlGGB2 is the lack of the C-terminal CaaX motif. The proteins end with RWI, the consensus sequence for all type B Gγ subunits in eudicots (Trusov et al., 2012).
SlGGB1 Is the Most Abundantly Expressed Gγ Gene in Tomato
We quantified the transcript levels of all four tomato Gγ subunit genes (SlGGA1, SlGGB1, SlGGB2, and SlGGC1) in several tissues using quantitative real-time PCR (RT-qPCR). Our results reveal similar expression profiles for SlGGA1 and SlGGB1, with SlGGB1 being the most abundant in almost all examined tissues, while SlGGC1 showed relatively low transcript levels and SlGGB2 levels were very low across all tested samples (Fig. 1A). SlGGB1 transcript levels were higher in reproductive tissues (i.e. flowers and fruits) compared with vegetative tissues (i.e. seedlings, leaves, and roots; Fig. 1A).
Figure 1.
SlGGB1 is the most abundantly expressed Gγ gene in tomato. A, Expression levels of SlGGA1, SlGGB1, SlGGB2, and SlGGC1 in the designated tissues. RNA was extracted from three biological replicates for each tissue and subjected to RT-qPCR. The expression values for the subunit genes were normalized with GAPDH expression. Values represent means of the three replicates with se. B, Histochemical analysis of SlGGB1 expression using the GUS reporter gene. Blue color corresponds to GUS activity. I, Flower buds; II, floral peduncle (AZ, abscission zone); III, opened flower; IV, pollen grains; V, calyx (sepals); VI, green fruit; VII, breaker fruit; VIII, mature fruit; IX, fresh seeds from ripe fruit; X, seed, 1 d after germination; XI, seed, 3 d after germination; and XII, longitudinal section of stem.
To further assess the SlGGB1 spatial and developmental expression patterns, we cloned 2 kb of the genomic DNA directly upstream of the SlGGB1 gene, including the 5′ untranslated region, and fused it to the GUS reporter gene (SlGGB1:GUS). This promoter construct was used to produce multiple independent transgenic lines in cv MicroTom. Preliminary analysis of all transgenic promoter lines showed that most lines had similar staining patterns, indicating that variability due to insertion events was insignificant. Three representative lines were selected for further analysis. In flower buds and open flowers, GUS staining was observed in the abscission zone peduncle, petals, anthers, and pollen grains but was nondetectable in sepals (Fig. 1B, I–IV). In immature and mature green fruits, GUS activity was detected in the area where the calyx is attached and in the pedicel (Fig. 1B, V–VII). GUS staining became more intense during the breaker stage, with staining visible in the funiculus, where the seed is attached to the columella, and in the vascular strands that attach the fruit to the calyx. GUS staining increased markedly during ripening, with very strong expression observed in the pericarp, spreading from the calyx through the columella, with intense staining also observed in the funiculus of each seed (Fig. 1B, VIII and IX). In germinating seeds, expression was confined to the micropyle region of the endosperm, but no GUS staining was detected in dry seeds (Fig. 1B, X and XI). Visible GUS expression was not detected in leaves and roots.
Tomato Type B Gγ Subunits Interact with Gβ
Gγ and Gβ subunits form an obligate functional dimer, and strong interaction between Gβ and all three Arabidopsis Gγ subunits has been demonstrated (Mason and Botella, 2000, 2001; McIntire, 2009; Chakravorty et al., 2012). Interaction between type B Gγ and Gβ subunits has been comprehensively demonstrated in rice and soybean (Kato et al., 2004; Choudhury et al., 2011). To confirm that SlGGB1 and SlGGB2 interact with the tomato Gβ subunit (SlGB1), we performed yeast (Saccharomyces cerevisiae) two-hybrid assays with SlGGB1 and SlGGB2 fused to the GAL4 activation domain (AD) and SlGB1 fused to the GAL4 binding domain (BD). When yeast was cotransformed with AD-SlGGB1 and BD-SlGB1, growth was observed on a medium lacking His, indicating interaction between both proteins (Fig. 2A). Yeast growth was also observed when AD-SlGGB2 and BD-SlGB1 were used in the assays. The canonical Arabidopsis Gγ2 subunit (AGG2) also showed strong interaction with SlGB1, serving as a positive control, while the empty pACT2 vector, a negative control, did not show any yeast growth (Fig. 2A).
Figure 2.
SlGGB1 interacts with the Gβ subunit. A, Yeast two-hybrid assays using pACT2 carrying SlGGB1 or SlGGB2 and pBridge vector carrying SlGB1. pACT2 with AGG2 and empty pACT2 were used as positive and negative controls, respectively. Growth on medium lacking His, Trp, and Leu (SC-LWH) indicates positive interaction, while growth on medium lacking His and Leu (SC-LW) indicates successful cotransformation of the yeast with two vectors. B, BiFC assessment of the interaction between SlGGB1 fused to cYFP and Arabidopsis AGB1 fused to nYFP in mesophyll protoplasts isolated from Arabidopsis. The interaction manifested as yellow fluorescence of the reconstituted YFP. The protoplasts were analyzed using confocal microscopy 16 to 24 h after transfection. The representative protoplasts were photographed with 510- to 550-nm (for YFP) and 640- to 700-nm (for chloroplasts) filters. Since focal planes for the plasma membrane and the nucleus differ, two photographs were taken for each protoplast and later stacked using Photoshop CS6 software. Bars = 20 μm.
We confirmed the SlGGB1 interacts with the Gβ subunit using bimolecular fluorescence complementation (BiFC) in Arabidopsis mesophyll protoplasts. The protoplasts were cotransfected with C-terminal yellow fluorescent protein (cYFP)-SlGGB1 and N-terminal yellow fluorescent protein (nYFP)-AGB1 as well as cYFP-AGG2 and nYFP-AGB1 as a positive control and with cYFP and nYFP-AGB1 as a negative control. Protoplasts coexpressing cYFP-SlGGB1 and nYFP-AGB1 showed strong fluorescence in the nucleus, with lower intensity observed in the cytoplasm and plasma membrane (Fig. 2B). The positive control coexpressing cYFP-AGG2 and nYFP-AGB1 showed strong fluorescence at the plasma membrane, with very weak fluorescence also apparent in the nucleus. No fluorescence was observed in negative controls (Fig. 2B).
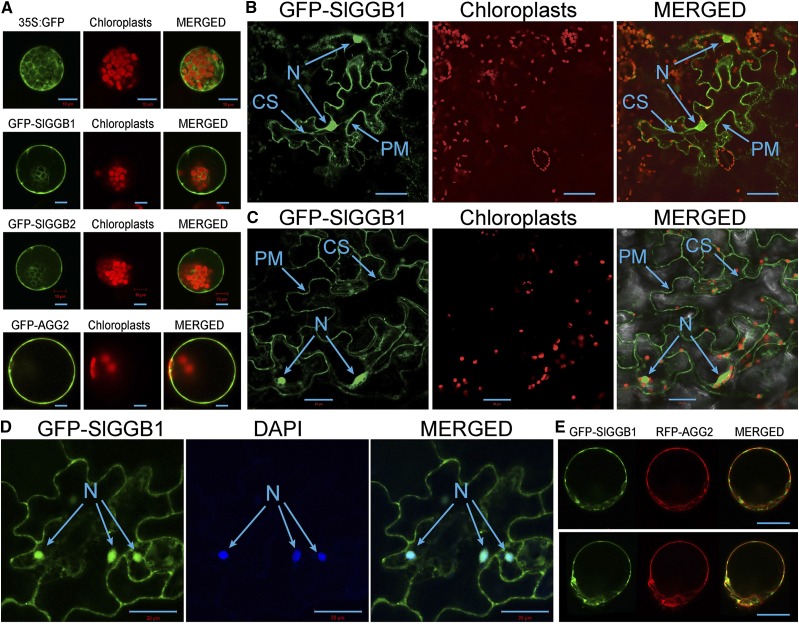
GFP-SlGGB1 Localizes to the Plasma Membrane Cytoplasm and the Nucleus
To establish the subcellular localization of the SlGGB1 and SlGGB2 subunits, we performed transient expression assays in tomato mesophyll protoplasts transfected with GFP-SlGGB1 and GFP-SlGGB2 fusion proteins under the control of the cauliflower mosaic virus 35S promoter. Confocal microscopy detected fluorescence in the plasma membrane, cytoplasm, and nucleus, similar to the pattern observed for free GFP (Fig. 3A). Under the same conditions, fluorescence produced by GFP fused to Arabidopsis AGG2 was localized at the plasma membrane (Fig. 3A). Transient expression of GFP-SlGGB1 in Nicotiana benthamiana leaves also yielded similar results (Fig. 3B). Furthermore, we tested the localization of the GFP-SlGGB1 protein in stably transformed Arabidopsis. Here, the GFP-SlGGB1 was also detected in the nucleus, cytoplasm, and plasma membrane (Fig. 3C). The nuclear localization was confirmed by 4′,6-diamino-2-phenylindole staining (Fig. 3D). To elucidate if GFP-SlGGB1 is located at the plasma membrane or just in peripheral cytoplasm, we produced mesophyll protoplasts from transgenic Arabidopsis plants expressing GFP-SlGGB1 and transfected them with the Arabidopsis Gγ subunit AGG2 fused to mCherry as a control. The plasma membrane localization of AGG2 was established previously (Adjobo-Hermans et al., 2006; Zeng et al., 2007). Both proteins were detected at the plasma membrane, although with a different pattern, as depicted by red and green colors (Fig. 3E, top). Analysis of ruptured protoplasts confirmed that both proteins remained attached to the plasma membrane (Fig. 3E, bottom). Our combined observations indicate that GFP-SlGGB1 is present at the plasma membrane, nucleus, and cytoplasm.
Figure 3.
SlGGB1 localizes to the nucleus, cytoplasm, and plasma membrane. A, Transient expression of unfused GFP, GFP-SlGGB1, GFP-SlGGB2, and GFP-AGG2 in mesophyll protoplasts isolated from tomato leaves. B, Transient expression of GFP-SlGGB1 in N. benthamiana leaves. C, Constitutive expression of GFP-SlGGB1 in Arabidopsis leaves. D, Constitutive expression of GFP-SlGGB1 in Arabidopsis leaves stained with 4′,6-diamino-phenylindole (DAPI). CS, Cytoplasmic strands; N, nucleus; PM, plasma membrane. Bars = 20 μm. E, Colocalization of GFP-SlGGB1 and RFP-AGG2 in mesophyll protoplasts (top); GFP-SlGGB1 and RFP-AGG2 were retained at the plasma membrane after protoplast rupture (bottom). Bars = 20 μm.
Silencing of SlGGB1 Results in Increased Lateral Root Formation and Auxin Sensitivity
To establish the physiological role of SlGGB1, we produced transgenic lines carrying RNAi constructs designed to silence the SlGGB1 gene. Several independent SlGGB1 RNAi lines were generated (hereafter referred to as slggb1), and the SlGGB1 expression levels were analyzed by RT-qPCR. Three transgenic lines with very low or undetectable SlGGB1 expression in T0 plants (slggb1-35, slggb1-36, and slggb1-50) were selected, and T3 homozygous lines were produced and used for further studies. RT-qPCR expression analysis was repeated on the homozygous lines, showing almost undetectable SlGGB1 transcript levels in slggb1-35 and slggb1-36, while in slggb1-50, SlGGB1 transcript levels were approximately 3% of those in wild-type plants (Fig. 4). To ensure that the silencing of SlGGB1 was not compensated by increased expression of the remaining γ genes that could potentially counteract the effects of the silencing, we determined SlGGA1, SlGGB2, and SlGGC1 expression levels in the transgenic lines. The expression levels of the second type B Gγ subunit, SlGGB2, in all three transgenic lines were reduced by about 50% compared with wild-type plants (P ≤ 0.05; Fig. 4). No alterations in transcript levels were detected for SlGGA1 and SlGGC1.
Figure 4.
Expression of all Gγ subunits in transgenic plants carrying SlGGB1 RNAi. The expression of SlGGB1 and SlGGB2 was down-regulated in slggb1 lines. Total RNA extracted from 3-week-old seedlings was subjected to RT-qPCR; the tomato GAPDH gene was used to normalized the expression values. Values represent average relative expression in three biological replicates, and error bars indicate se. Letters represent groups of statistically significant differences based on one-way ANOVA with Tukey’s multiple comparison method. WT, Wild type.
The formation of lateral roots is strongly affected in Arabidopsis mutants lacking Gβ or Gγ subunits (Ullah et al., 2003; Trusov et al., 2007), prompting us to evaluate the number of lateral roots in wild-type and transgenic tomato lines. All three SlGGB1-silenced lines showed a 2 to 2.5 times increase in lateral root numbers compared with the wild type, with high statistical significance (P ≤ 0.001; Fig. 5A). The enhanced lateral root formation observed in SlGGB1-silenced lines could be the result of increased lateral root primordium (LRP) formation, but it could also be due to an increased rate of cell elongation from an otherwise wild-type number of LRPs. To distinguish between these two scenarios, the total numbers of lateral roots as well as LRPs of 3-week-old slggb1 and wild-type seedlings were counted. The roots of slggb1 seedlings had approximately 2-fold more lateral roots + LRPs than wild-type roots (Fig. 5B). Since lateral root formation is under tight auxin control (Celenza et al., 1995), our observations imply that the down-regulation of SlGGB1 may result in either an increased auxin pool or an altered auxin sensitivity in roots.
Figure 5.
slggb1 lines have increased sensitivity to auxin. A, Average number of lateral roots on 3-week-old seedlings grown vertically on plates (1× Murashige and Skoog [MS] medium, 3% Suc, and 0.8% phytagel). B, Average number of lateral roots (LRs) and LRPs on 3-week-old seedlings. C, Auxin-induced lateral root development. Roots were excised from 3-d-old seedlings grown on MS medium and transferred to medium supplemented with the designated concentrations of NAA. The number of lateral roots was counted 5 d later. D, Cotyledons from 9-d-old seedlings were excised and transferred to MS medium supplemented with the designated concentrations of NAA. The cotyledons were incubated for 10 d at 26°C under 16/8 h of light/dark and photographed. The number of adventitious roots was counted for each cotyledon. Mean values were calculated from at least 10 measurements, and error bars represent se; asterisks indicate statistically significant differences between the wild type (WT) and transgenic lines (Student’s t test, *P < 0.05, **Student’s t test, P < 0.01, ***Student’s t test, P < 0.001). E, Representative cotyledons developing auxin-induced adventitious roots. The shaded squares indicate genotypes on the plate as designated in C and D.
The increase in lateral root formation observed in slggb1 plants prompted us to examine their auxin sensitivity by determining the effect of different auxin concentrations on lateral root and LRP formation. Germinated slggb1 seeds were grown on MS minimal medium for 3 d before excising the roots from the seedlings and transferring them to MS medium supplemented with various concentrations (0–1 μm) of naphthaleneacetic acid (NAA). The apical meristem was excised from seedlings to eliminate the flow of endogenous auxin from the shoot tip, as auxin synthesized in the apical region of the plant is translocated to the roots (Laskowski et al., 1995). Five days after incubation with NAA, the numbers of lateral roots and LRPs were counted. In the absence of auxin, the excised roots of slggb1 lines showed no significant differences in the number of lateral roots from wild-type excised roots (Fig. 5C). When the medium was supplemented with NAA, all genotypes, including the wild type, demonstrated substantial increases in lateral root and LRP formation, even at the lowest concentration of NAA, 0.1 μm (Fig. 5C). However, slggb1 lines produced significantly more lateral roots and LRPs than wild-type plants (Fig. 5C). Combined, these results indicate that slggb1 lines are more sensitive than the wild type to exogenous auxin.
To further assess the auxin response of slggb1 lines, we examined the effect of exogenous auxin on tissues lacking preexisting root primordia. Cotyledons from 9-d-old seedlings grown on MS medium were excised and transferred to MS medium supplemented with various concentrations (0–4 μm) of NAA. The treated slggb1 cotyledons developed adventitious roots starting from 0.05 μm NAA, while wild-type cotyledons produced the roots only at 0.1 μm NAA. Quantification revealed that slggb1 lines had significantly more adventitious roots formed compared with the wild type at concentrations of 0.05 and 0.1 μm NAA (Fig. 5D). On the plates supplemented with 0.05 and 0.1 μm NAA, the difference between the wild type and the transgenic lines was noticeable by eye (Fig. 5E). At higher concentrations (1 and 4 μm), the number of the roots was too high to quantify reliably. Considering the increased auxin sensitivity observed in slggb1 lines, we conclude that SlGGB1 might be a negative regulator of auxin signaling.
Silencing of SlGGB1 Affects Fruit and Seed Morphology
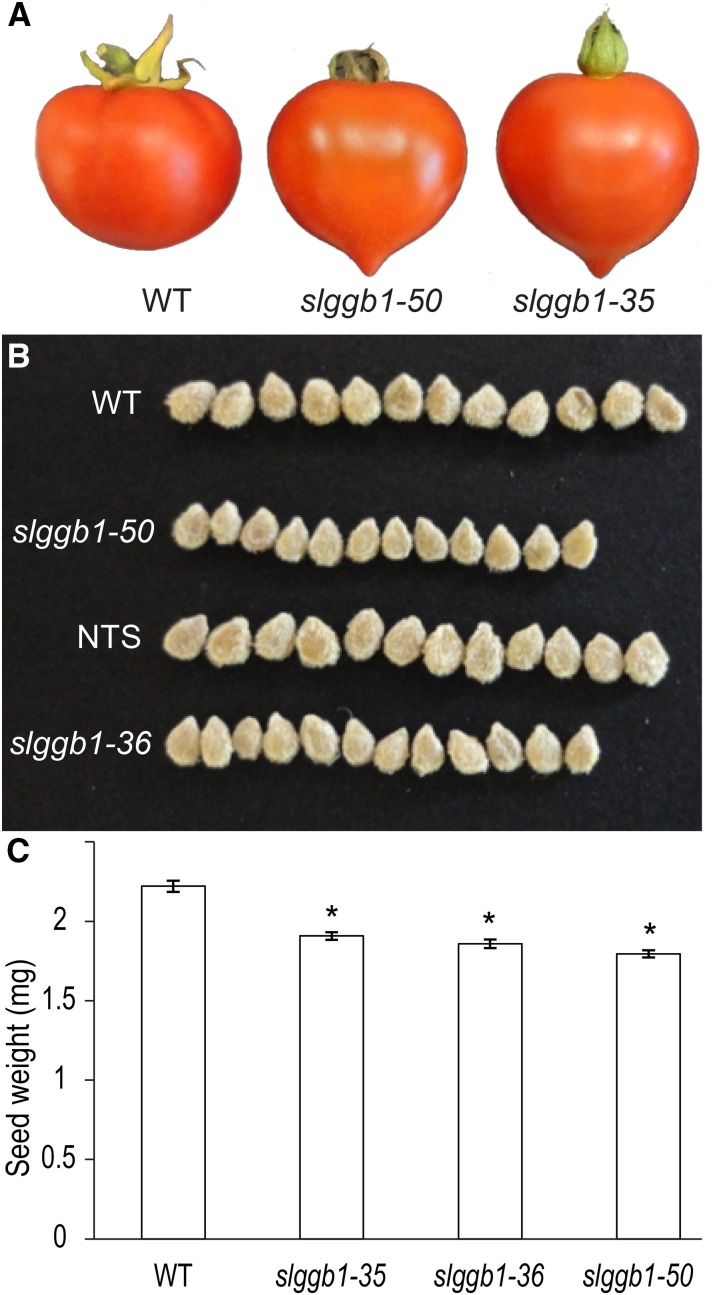
Fruit development is a complex process involving highly synchronized molecular, biochemical, and structural changes mediated by phytohormones such as auxin, GA, cytokinin, ABA, and ethylene (Gillaspy et al., 1993; Ozga and Reinecke, 2003). Fruits of slggb1 plants exhibited a pointy tip, giving them a heart-like shape, in contrast to the blunt tip of wild-type fruits (Fig. 6A). Previous studies have reported that the heart-like shape of tomato fruits can be a result of increased auxin sensitivity (de Jong et al., 2009). This is in accord with our findings observed in root development. On the other hand, the pointy tip and elevated auxin signal were also associated with parthenocarpy (seedless fruits; de Jong et al., 2009). Our slggb1 lines produced fruits with normal numbers of seeds, although they were smaller in appearance than wild-type seeds (Fig. 6B). Quantitative measurements revealed that seeds from the slggb1 plants were significantly lighter than wild-type seeds (Fig. 6C) and had smaller values for length and width (Table I; P < 0.001). The length-width ratio was similar for slggb1 and wild-type seeds, indicating that the seed shape was not altered. Noteworthy, the small size of slggb1 seeds was not reflected in viability or germination rates, as demonstrated in our germination experiments described below.
Figure 6.
slggb1 plants have heart-like fruits and small seeds. A, Ripe wild-type (WT) and slggb1 fruits displaying normal and heart-like shapes, respectively. B, Row of 12 representative seeds from fully ripened fruits of the designated genotypes. C, An average seed weight was calculated from 50 seeds per genotype. NTS, Non-transgenic segregant. Data sets are average values, and error bars represent se. Asterisks indicate values with significant differences from the wild type determined by Student’s t test (P < 0.05; n = 50). The experiment was repeated twice with similar results.
Table I. Quantification of seed length and width.
| Sample | Seed Length (n = 50) |
Seed Width (n = 5) |
Ratio, Seed Length to Width (n = 50) |
|||
|---|---|---|---|---|---|---|
| Mean ± se | P | Mean ± se | P | Mean ± se | P | |
| mm | ||||||
| Wild type | 3.06 ± 0.02 | 2.05 ± 0.03 | 1.51 ± 0.02 | |||
| slggb1-35 | 2.65 ± 0.02 | <0.001 | 1.81 ± 0.03 | <0.001 | 1.48 ± 0.03 | 0.3432 |
| slggb1-36 | 2.48 ± 0.03 | <0.001 | 1.68 ± 0.02 | <0.001 | 1.49 ± 0.04 | 0.4742 |
| slggb1-50 | 2.59 ± 0.03 | <0.001 | 1.71 ± 0.03 | <0.001 | 1.53 ± 0.03 | 0.5536 |
SlGGB1 Is Regulated by Auxin and Is Involved in the Regulation of Auxin-Responsive Genes, But Not in Auxin Biosynthesis
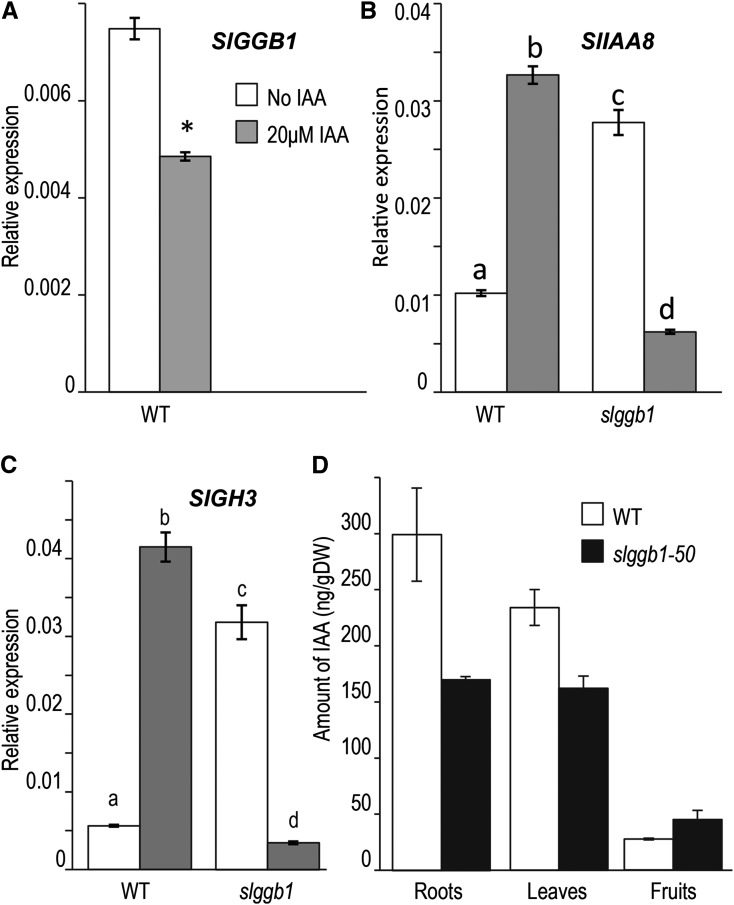
SlGGB1 expression in response to exogenous auxin was determined by RT-qPCR in wild-type seedling tissues. SlGGB1 gene expression was down-regulated by 40% 3 h after treatment with 20 μm indole-3-acetic acid (IAA; Fig. 7A). We also investigated the effect of SlGGB1 down-regulation on the expression of two early auxin-responsive genes: INDOLE-3-ACETIC ACID INDUCIBLE8 (IAA8; Abel et al., 1995) and GRETCHEN HAGEN3 (GH3; Hagen et al., 1984; Hagen and Guilfoyle, 1985). As expected, the tomato homologs of IAA8 and GH3 were strongly induced by auxin treatment in wild-type plants (Fig. 7, B and C). In contrast, in slggb1 lines, SlIAA8 and SlGH3 steady-state levels were elevated significantly compared with wild-type plants. Interestingly, however, the transcript levels of both genes in slggb1 lines decreased significantly after treatment with auxins (Fig. 7, B and C).
Figure 7.
SlGGB1 in an auxin-mediated network. Two-week-old wild-type (WT) and slggb1 seedlings were incubated with 20 μm IAA or with water for 3 h. Total RNA was extracted and subjected to RT-qPCR; the tomato GAPDH gene was used for normalization. A, Auxin treatment suppressed the expression of SlGGB1 in wild-type tomato seedlings. The asterisk signifies a statistically significant difference (P < 0.05). B and C, The expression pattern of IAA8 (B) and GH3 (C) genes was reversed in slggb1 seedlings. Values represent average relative expression in three biological replicates, and error bars indicate se. Letters represent groups of statistically significant differences based on one-way ANOVA with Tukey’s multiple comparison method. D, Levels of IAA. IAA was quantified in leaves and roots of 4-week-old and ripe fruits from mature wild-type and slggb1-50 plants. Values represent average values from two biological replicates, and error bars indicate se. DW, Dry weight.
The elevated expression of auxin-responsive genes together with the auxin hypersensitivity of slggb1 plants strongly indicate that SlGGB1 is a negative regulator of the auxin signaling pathway. Alternatively, down-regulation of SlGGB1 could increase endogenous auxin levels in the plant. Quantification of endogenous IAA levels in leaves and roots of 2-week-old plants as well as in ripe fruits revealed that they were either similar or lower in slggb1 plants compared with the wild type (Fig. 7D).
Silencing of SlGGB1 Decreases Sensitivity to Exogenous ABA during Seed Germination
The involvement of plant G proteins in ABA signaling has been well documented in Arabidopsis (Wang et al., 2001; Ullah et al., 2002; Chen et al., 2003, 2006b; Pandey and Assmann, 2004; Pandey et al., 2009; Chakravorty et al., 2011). Treatment of wild-type seeds with 10 μm ABA resulted in significant up-regulation of SlGGB1 expression, while no significant expression was detected in slggb1 seeds (Fig. 8A).
Figure 8.
SlGGB1 in an ABA-mediated network. A, ABA induces SlGGB1 expression in wild-type (WT) seeds. Wild-type seeds were imbibed in water or 10 μm ABA for 24 h. The tomato GAPDH gene was used for normalization. Average relative expression values from three biological replicates and se are shown. The asterisk signifies a statistically significant difference (P < 0.05). B to E, Germination rates of wild-type and slggb1 seeds plated on medium without ABA (B) or with 5 μm ABA (C), 10 μm ABA (D), and 50 μm ABA (E). Seeds were surface sterilized and plated on MS medium without Suc (one-half-strength MS medium and 0.8% phytagel) supplemented or not with ABA. Plates were kept in darkness at 26°C, and germination was monitored daily. The experiments were repeated at least three times with similar results. Values are averages from three replicates, and error bars indicate se. F, Three-day-old seedlings grown on MS medium (1× MS medium, 3% Suc, and 0.8% phytagel) were transferred to medium supplemented or not with ABA at the designated concentrations. The number of lateral roots was counted 10 d later. Plates were kept under a 16/8-h light/dark cycle at 26°C. The experiment was repeated at least three times with similar results. Bars represent average values from 15 seedlings, and error bars indicate se.
To establish if SlGGB1 plays a role in ABA signaling in tomato, we studied ABA-mediated germination inhibition in slggb1 and wild-type seeds. The seeds used in our assays were harvested on the same day and stored for 5 weeks before the test. Sterilized seeds were sown on MS medium devoid of Suc, supplemented with 0, 5, 10, or 50 μm ABA, and kept in darkness. Germination was judged by protrusion of the radicle, and counts were performed from day 3. Without the addition of ABA, the germination rate was similar in all slggb1 transgenic lines and the wild type (Fig. 8B), with almost 90% of seeds germinated by day 3 and 100% by day 6. The addition of 5 μm ABA to the medium resulted in strong inhibition of germination in wild-type plants, with only 20% of seeds germinating by day 6. In contrast, slggb1 seeds were clearly less affected, displaying 66% to 82% germination by day 6 (Fig. 8C). A similar trend was observed in the presence of 10 and 50 μm ABA, with wild-type seeds showing a stronger inhibition than slggb1 lines (Fig. 8, D and E).
We also tested the sensitivity of the slggb1 plants to ABA during lateral root formation. Five-day-old slggb1 and wild-type seedlings were transferred to MS medium with various concentrations of ABA (0.25–2 μm), and the number of lateral roots was counted after 10 d. We observed that lateral root formation was appreciably suppressed by ABA in all genotypes. The differences between wild-type and slggb1 plants were not significant (Fig. 8F).
Transcriptome Analysis of slggb1 Transgenic Seeds in Response to ABA Reveals Important Alterations in ABA-Related Gene Expression
In an effort to understand the physiological causes for the low ABA sensitivity exhibited by slggb1 seeds during germination, we performed a genome-wide transcriptome analysis of wild-type and slggb1 seeds (line slggb1-50) imbibed with and without exogenous ABA. At 24 h after imbibition in either water or 10 μm ABA, RNA was extracted from seeds and the complementary DNA (cDNA) was subjected to next-generation sequencing on an Illumina HiSeq 2000 platform. Three independent biological replicates were performed for each treatment.
Analysis of the expression patterns of slggb1 and wild-type seeds incubated with ABA for 24 h revealed a total of 54 genes with a 2-fold or greater difference in expression levels (Supplemental Table S1). All but two of the genes with altered expression were less responsive in slggb1 seeds compared with the wild type. Importantly, differential expression was observed in several late embryogenesis-abundant (LEA) genes, ABA-associated genes, osmotic stress-related genes, heat shock protein genes, and cold- or low temperature-inducible genes (Table II). Particularly interesting was the decreased expression of orthologs of the Arabidopsis PYR/PYL/RCAR-type ABA receptor PYL4 (Ma et al., 2009; Park et al., 2009) and MEDIATOR OF ABA-REGULATED DORMANCY1 (MARD1; He and Gan, 2004). Aside from ABA, GAs also have an essential role in seed germination (Metzger, 1983; Grappin et al., 2000). A GA biosynthetic gene, GIBBERELLIN 2-OXIDASE, as well as a transcription factor involved in GA response was less responsive to ABA treatment in slggb1 seeds (Table II). Finally, a number of genes not directly related to ABA but associated with seed storage and oil bodies such as vicilin and oleosin were identified. A complete list is provided in Supplemental Table S1.
Table II. Genes with decreased response to exogenous ABA in slggb1 seeds compared with the wild type.
| Gene Identifier | slggb1:Wild-Type Ratio | Gene Name/Description | Gene Function |
|---|---|---|---|
| ABA-responsive genes | |||
| Solyc01g095140.2 | −1.80899 | Late embryogenesis abundant protein | Response to desiccation |
| Solyc02g062770.1 | −1.79538 | Late embryogenesis abundant protein | Uncharacterized protein |
| Solyc05g053160.2 | −1.64584 | Late embryogenesis abundant protein9 | ABA-induced plasma membrane protein |
| Solyc06g048840.2 | −1.51523 | Late embryogenesis abundant protein | Uncharacterized protein |
| Solyc06g009000.1 | −1.88533 | Mediator of ABA-regulated dormancy1 | ABA-induced gene zinc-finger protein with Pro-rich N terminus |
| Solyc06g050500.2 | −1.93566 | ABA receptor PYL4 | Protein binding |
| Solyc12g010920.1 | −1.39275 | Vicilin-like protein | ABA-inducible storage protein |
| Solyc11g072380.1 | −1.75859 | Vicilin-like protein | Nutrient reservoir activity |
| Solyc07g061720.2 | −3.71946 | GA 2-oxidase | GA biosynthesis |
| Solyc06g034040.1 | −4.23294 | Oleosin | Storage of oil bodies |
| Solyc12g056720.1 | −1.93829 | 3-Ketoacyl-CoA synthase2 | Down-regulated by ABA |
| Solyc04g072470.2 | −2.86554 | Defensin-like protein | Induced by drought |
| Transcription factors | |||
| Solyc01g100200.2 | −2.49753 | GRAS family transcription factor | Repressor of GA response |
| Solyc04g017670.2 | −2.85787 | F-box family protein/Skp2-like | Protein ubiquitination |
| Solyc04g078300.2 | −2.35978 | MADS box transcription factor1 | DNA binding |
Analysis of the gene expression in seeds imbibed in water showed 19 genes with lower expression levels in slggb1 compared with the wild type and only two with higher levels (Supplemental Table S2). A number of the differentially expressed genes in water-imbibed seeds, such as xyloglucan endotransglucosylase, lipase, extensin, and lipid transfer protein, are involved in cell wall modification, seed storage, and fatty acid mobilization (Table III). Our analysis did not discover any overlaps in the set of differentially expressed genes between the water- and ABA-imbibed seeds.
Table III. Genes differentially expressed in response to water in slggb1 compared with wild-type seeds.
| Gene Identifier | slggb1/Wild-Type Ratio | Gene Name/Description | Gene Function |
|---|---|---|---|
| Solyc01g006400.2 | −3.19965 | Cys-rich extensin-like protein4 | Cell wall structural protein expressed in the endosperm |
| Solyc02g077020.2 | −3.54834 | Lipase-like protein | Storage lipid breakdown |
| Solyc02g067870.2 | −2.00572 | Chalcone isomerase | Increased fatty acid storage in developing embryo |
| Solyc04g071890.2 | −1.93209 | Peroxidase4 | Induced by wounding of the endosperm |
| Solyc06g073580.2 | −3.02537 | 1-Aminocyclopropane-1-carboxylate oxidase1 | Ethylene biosynthesis |
| Solyc06g073570.2 | −2.50084 | Cytochrome P450 | Oxidation |
| Solyc08g074480.1 | −2.34505 | Cortical cell-delineating protein | Plant lipid transfer protein and hydrophobic protein |
| Solyc08g014000.2 | −1.87166 | Lipoxygenase | Peroxidation of polyunsaturated fatty acid |
| Solyc08g080660.1 | −1.7881 | Osmotin-like protein | Pathogenesis related |
| Solyc09g092520.2 | −2.68984 | Xyloglucan endotransglucosylase | Cell wall-modifying enzymes/loosens cell wall |
| Solyc10g075110.1 | −1.95594 | Nonspecific lipid-transfer protein | Facilitates transfer of phospholipids and fatty acids |
| Solyc10g080730.1 | 1.50593 | Thioredoxin family protein | Cell redox homeostasis |
DISCUSSION
Although Gγ subunits were initially regarded as a passive partner in the Gβγ dimer whose only function was to anchor the dimer to the plasma membrane, they have now emerged as an important member of the heterotrimer, providing functional selectivity to Gβγ dimer signaling in plants and animals (Gautam et al., 1990; Trusov et al., 2007; Thung et al., 2013). Classically, Gγ subunits consist of three domains: a variable N terminus, a conserved region for coiled-coil interaction with Gβ, and a C-terminal isoprenylation motif, CaaX (Temple and Jones, 2007). In plants, three distinct structural types of Gγ subunits were identified (Choudhury et al., 2011; Trusov et al., 2012). Type A is represented by Gγ subunits with classical structure, type B subunits are very similar but lack the CaaX motif, and type C subunits are characterized by the presence of a Cys-rich tail (Trusov et al., 2012). Unlike types A and C Gγ subunits, whose functions were studied in Arabidopsis or rice, type B subunits had not been functionally characterized so far, perhaps due to the fact that there are not present in the model species Arabidopsis. In this work, we carried out molecular characterization and genetic studies on a type B Gγ subunit from tomato. Interestingly, in all tested tomato tissues, SlGGB1 expression levels were highest among all Gγ genes. This observation contrasts with previously reported expression patterns in soybean, where types A and C were considerably more abundant than type B (Choudhury et al., 2011). As mentioned above, Arabidopsis and other Brassicaceae species seem to have lost this type completely (Trusov et al., 2012; Arya et al., 2014). It is tempting, therefore, to hypothesize that type B Gγ subunits are functionally more important in asterid species (tomato) compared with rosids (soybean and Arabidopsis).
The Type B Gγ Subunit SlGGB1 Has a Unique Localization Pattern
Lack of the isoprenylation motif in canonical (type A) Gγ subunits results in the failure of plasma membrane targeting (Kino et al., 2005; Adjobo-Hermans et al., 2006; Zeng et al., 2007). We showed that GFP-SlGGB1 localizes to the nucleus, the plasma membrane, and the cytoplasm (Fig. 2). Moreover, when SlGGB1 and the Gβ subunit were coexpressed in the same cell (in our BiFC study), they formed a heterodimer that was most abundant in the nucleus, with the fluorescence intensity noticeably weaker in cytoplasm and at the plasma membrane. It could be argued that the use of the cauliflower mosaic virus 35S promoter and, hence, excessive expression could result in mislocalization to the nucleus. To evaluate this possibility, we also examined the localization of the Arabidopsis AGG2-AGB1 heterodimer in a parallel experiment. This heterodimer was predominantly observed at the plasma membrane, only weakly in the cytoplasm, and was barely detectable in the nucleus. While localization to the nucleus and the cytoplasm is not surprising considering that type B Gγ subunits are small proteins and do not have an isoprenylation motif, any conclusion about plasma membrane targeting requires special caution. Therefore, we studied GFP-SlGGB1 behavior in ruptured protoplasts, which allowed distinction between localization in the peripheral cytoplasm and the plasma membrane (Serna, 2005). Our analysis confirmed the plasma membrane location of GFP-SlGGB1. Plasma membrane localization was also reported for all three type B Gγ subunits from soybean (Choudhury et al., 2011) and RGG2, a single type B Gγ subunit from rice (Kato et al., 2004). It was hypothesized that the localization of RGG2 to the plasma membrane could be due to palmitoylation of the single Cys residue situated within the conserved central region (Kato et al., 2004). Another possibility is that the presence of positively charged aromatic amino acids at the SlGGB1 C terminus could result in the formation of an amphipathic α-helix able to anchor the protein to the plasma membrane (Prinz and Hinshaw, 2009; Trusov et al., 2012). Further studies are required to ascertain the structural characteristics and possible posttranslational modifications of the type B subunits. At this point, it is important to note that, in contrast to the majority of the known Gγ subunits (in plants, animals, or fungi), the type B subunits localize not only at the plasma membrane but in the cytoplasm and the nucleus. This unusual localization for Gγ subunits could be required to perform specific functions. As discussed below, SlGGB1 is involved in auxin and ABA signaling. Both hormones are perceived by intracellular receptors (Kepinski and Leyser, 2005; Ma et al., 2009; Park et al., 2009; Scherer, 2011); therefore, a cytosolic localization could allow G protein heterotrimers with a type B Gγ to contribute to signal propagation, but further studies are needed to confirm or deny such a speculative hypothesis.
SlGGB1 Attenuates Auxin Responses during Lateral Root Formation and Fruit Development
The histochemical analysis of SlGGB1 expression using the SlGGB1:GUS lines displayed some resemblance to that of the synthetic auxin-responsive promoter DR5 in DR5:GUS tomato fruits (Pattison and Catalá, 2012). At the same time, treatment with auxin significantly suppressed SlGGB1 expression in wild-type seedlings. These observations suggest that SlGGB1 function might be associated with auxin signaling. Compared with the wild type, the slggb1 lines with strongly reduced expression of SlGGB1 showed an increased number of lateral roots on standard medium and medium supplemented with NAA. These results are consistent with previous reports on Arabidopsis mutants that also displayed increased lateral root production as well as deregulation of a set of auxin-responsive genes in the presence of exogenous auxin (Ullah et al., 2003; Trusov et al., 2007). slggb1 lines were also more sensitive than the wild type to exogenous auxin in cotyledons producing adventitious roots.
The fruits of slggb1 plants have a characteristic pointy tip that is known to be a result of highly elevated auxin levels in flower buds (Pandolfini et al., 2002) or increased auxin sensitivity (de Jong et al., 2009; Bassa et al., 2012). We showed that the fruits of transgenic slggb1 and wild-type plants contain similar amounts of auxins; therefore, it is logical to assume that the phenotype is caused by enhanced sensitivity to auxin. The seeds of slggb1 were significantly smaller than those from wild-type plants. To the best of our knowledge, this phenotype was not linked to auxin signaling in tomatoes; however, the Arabidopsis mnt mutant, with dysfunctional AUXIN RESPONSE FACTOR (ARF) and hyposensitive to auxin, had larger seeds than the wild type (Schruff et al., 2006).
In tomato, several AUXIN/IAA and ARF genes were shown to be negative regulators of auxin signaling. Down-regulation of these genes in tomato results in distinct, easily quantifiable phenotype alterations. A number of reports have described that auxin sensitivity in tomato is composed of several independent pathways, which, however, have overlaps. Tomato lines expressing antisense transcripts for SlIAA9 were hypersensitive to auxin and displayed altered leaf morphology, fused petioles/leaves, fused flowers, and fused cotyledons; their fruits were parthenocarpic, their stems were elongated, and apical dominance was weakened (Wang et al., 2005). Transgenic lines with silencing of the SlIAA15 gene were hypersensitive to auxin and exhibited decreased plant height, reduced apical dominance, increased number of axillary shoots, increased lateral root formation, smaller number of flowers, and less efficient fruit set than wild-type plants (Deng et al., 2012). Notably, the number of trichomes on leaves and stems, as well as the density of epidermal cells, were decreased significantly in these transgenic lines (Deng et al., 2012). RNAi-mediated down-regulation of SlIAA27 also resulted in plants with increased sensitivity to auxin (Bassa et al., 2012). Their leaves contained less chlorophyll, hypocotyls and primary roots were elongated, the number of lateral roots was increased, fruits were significantly smaller and had pointy tips, seed number was decreased, and fertility was lowered (Bassa et al., 2012). RNAi silencing of SlARF7 caused parthenocarpy and heart-like fruits with pointy tips (de Jong et al., 2009). In short, it is clear that different transcriptional factors, such as IAAs and ARFs, control different pathways leading to plural auxin functions.
Our study revealed that slggb1 had heart-like pointy fruits very similar to those of SlARF7-silenced plants, but it was not parthenocarpic. On the other hand, similar to SlIAA27 down-regulated lines, slggb1 plants had more lateral roots than the wild type, but their fruits were fully fertile. Our results indicate that SlGGB1 does not exert its effect by controlling the activity of a specific transcription factor(s) but rather attenuates auxin-dependent signaling at a different level.
We also determined that G proteins are involved in the transcription regulation of auxin-inducible genes. The transcription pattern of auxin marker genes SlIAA8 and SlGH3 was reversed in slggb1 plants compared with the wild type. These genes were expressed without auxin in SlGGB1-deficient plants but down-regulated by IAA treatment. While the molecular mechanism of this reversion has yet to be established, the fact that IAA and ARF genes are deregulated is in agreement with the morphological alterations observed in the slggb1 plants.
SlGGB1 Regulates ABA Responses during Seed Germination and Modulates the Expression of ABA-Responsive Genes
The involvement of G proteins in ABA signaling is well documented in Arabidopsis (Wang et al., 2001; Ullah et al., 2002; Chen et al., 2003, 2006b; Pandey and Assmann, 2004; Chakravorty et al., 2011). Noteworthy, the sensitivity to ABA in G protein knockout mutants changes dramatically depending on the tissue and/or developmental process. For instance, Arabidopsis Gα-, Gβ-, and Gγ3-deficient mutants exhibited reduced sensitivity to ABA during stomatal opening but not in ABA-promoted stomatal closure. In contrast, the same mutants showed increased sensitivity to ABA during seed germination and postgermination development (Wang et al., 2001; Ullah et al., 2002; Lapik and Kaufman, 2003; Pandey et al., 2006). Interestingly, Gγ1 and Gγ2 have not been involved in ABA signaling (Trusov et al., 2007; Chakravorty et al., 2012). Mutants lacking the regulator of G protein signaling, RGS1, showed reduced sensitivity to ABA during germination (Chen et al., 2003, 2006b). GCR1, a putative GPCR, has also been implicated in the regulation of ABA signaling. gcr1 mutants were hypersensitive to ABA inhibition of root growth and stomatal responses but exhibited wild-type responses to ABA during seed germination, while overexpression of GCR1 reduced seed dormancy (Colucci et al., 2002; Chen et al., 2004).
Several lines of evidence in our work point to an important role for SlGGB1 in ABA control of seed germination. We found distinctively strong SlGGB1 promoter-driven GUS expression near the seed micropyle. This region is important for the regulation of seed germination, as the loosening of cell walls at the micropyle region of the endosperm enables radicle protrusion (Bewley, 1997). Moreover, in wild-type plants, ABA treatment induced SlGGB1 expression. Analysis of three independent SlGGB1-silenced transgenic lines revealed reduced sensitivity to ABA during seed germination, while postgermination development and response to ABA in lateral root production were similar in the transgenic and wild-type tomatoes.
Our transcriptome analysis of germinating seeds further substantiated the observed reduction in ABA sensitivity. A number of genes associated with ABA signaling were less affected in slggb1 seeds compared with the wild type in response to ABA, resembling the pattern found for previously reported ABA-hyposensitive mutants (Hoth et al., 2002; Kinoshita et al., 2010). In particular, four LEA genes were significantly less responsive to ABA in slggb1 seeds compared with the wild type. Similar behavior was observed for their Arabidopsis homologs in ABA-insensitive1 (abi1) and abi5 and growth insensitive to ABA3 mutants (Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2002; Kinoshita et al., 2010). Importantly, in Arabidopsis, these genes confer salt tolerance during germination (Jia et al., 2014) and acquisition of desiccation tolerance during seed maturation (Manfre et al., 2009). Decreased levels of these proteins in slggb1 seeds probably contributed to the increased germination rates observed in the presence of external ABA.
The finding that PYL4 expression is down-regulated in ABA-treated seeds is also very revealing. PYR/PYL/RCAR proteins have been identified recently as intracellular ABA receptors (Ma et al., 2009; Park et al., 2009). In the presence of ABA, the PYR/PYL/RCAR proteins form a complex with the protein phosphatase PP2C, which leads to the inhibition of PP2C activity. This, in turn, activates Snf1-related protein kinases (SnRKs), which target membrane proteins, ion channels, and transcription factors and facilitate the transcription of ABA-responsive genes (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009; Sheard and Zheng, 2009; Umezawa et al., 2010). Therefore, reduced PYR/PYL/RCAR expression would necessarily impair the ability of the seeds to perceive ABA. A gene encoding a zinc-finger protein, MARD1, also showed hyposensitivity to exogenous ABA in slggb1 seeds. Arabidopsis mard1 mutant seeds were insensitive to external ABA at the stage of radicle protrusion (He and Gan, 2004).
Despite the multiple lines of evidence presented here, the ABA-insensitive phenotype of slggb1 seeds might not be completely due to defects in ABA signaling, since we also observed down-regulation of a gibberellin 2-oxidase, an enzyme involved in GA biosynthesis, and a GRAS family transcription factor also involved in the GA response.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tomato (Solanum lycopersicum ‘MicroTom’) plants were grown on soil in the greenhouse under standard conditions with 16/8 of light/dark and a daily temperature of 26°C to 28°C. For in vitro culture, seeds were dry sterilized by incubation in a chamber of chlorine gas for approximately 4 h. Seeds were sown on one-half-strength MS medium supplemented with one-half-strength Gamborg’s vitamin mixture, 3% (w/v) Suc, and 0.8% (w/v) phytagel, pH 5.8. Transgenic seeds were selected on MS culture medium containing 150 mg L−1 kanamycin. After sowing, all seeds were kept in darkness for 4 d until germination and then transferred to light under 16/8 h of light/dark at 26°C. Germination was determined as an obvious protrusion of the radicle.
Plant Transformation
To generate RNAi SlGGB1 transgenic lines, the forward 5′-ACTCGAGTCTAGATACAAATCGATCTCCATTTCCTC-3′ primer including part of the 5′ untranslated region and reverse 5′-AGAATTCGGATCCACTTGGGAAGTGTATGAGTTACAAAA-3′ primer including part of the 3′ untranslated region were used to amplify the full-length SlGGB1 cDNA clone. This fragment was first cloned into pHannibal (Wesley et al., 2001) intermediate RNAi vector in the sense and antisense orientations under the control of cauliflower mosaic virus 35S and the OCS terminator. Later, the RNAi construct was cloned into pUQC247 binary vector. The promoter region of SlGGB1 was amplified from wild-type cv MicroTom genomic DNA using forward primer 5′-TTTGTGCATTTGACTTGCCAC-3′ and reverse primer 5′-ACTCGAGTAAAGCTTCAAAATTAGAGCTTG-3′. Restriction sites (underlined) were added at the ends of each primer for cloning purposes. The SlGGB1 promoter fragment was cloned into pGEM-T Easy vector (Promega), transferred using XhoI and SacI into pHannibal vector incorporated with GUS, and then transferred to pART27 binary vector (Gleave, 1992). Transgenic plants were generated via Agrobacterium tumefaceins-mediated transformation according to Dan et al. (2006), and all experiments were carried out using homozygous lines from F3 or later generations. Histochemical GUS analysis was carried out according to Wang et al. (2005).
Morphological and Physiological Characterization of SlGGB1
Plate Assays
Unless specified otherwise, the plate medium contained 1× MS medium with Gamborg’s vitamins, 3% (w/v) Suc, and 0.8% (w/v) phytagel (pH 5.8, adjusted with potassium hydroxide). For lateral root assay, sterilized seeds were sown to the medium, and plates were placed vertically under 16/8 h of light/dark at 26°C. The lateral roots were counted 3 weeks after germination using a dissecting microscope. The adventitious root assay from cotyledons was adapted from Wang et al. (2005).
Germination Assay
Seeds were sterilized by soaking in 10% (v/v) sodium hypochlorite for 15 min and then rinsed extensively with sterile distilled water. Fifty to 70 slggb1 or wild-type seeds were germinated per petri dish. The medium contained 1× MS medium with Gamborg’s vitamins and 0.8% (w/v) phytagel (pH adjusted to 5.8 by KOH before autoclaving). ABA and fluridone were filter sterilized (0.22-μm Millex-GS filter unit; Millipore) and added to the medium after autoclaving. Plates with seeds were placed at an optimal temperature of 26°C in continuous darkness. Germination assays were carried out in triplicate, and three different batches of seeds were tested.
Seed Weight, Length, and Width
Approximately 30 dry seeds per line were weighed. About 50 seeds per line were photographed next to a ruler. Length (measured at the widest part of the seeds) and width measurements of seeds were made using ImageJ software (http://www.nih.gov/).
Yeast Two-Hybrid Assay
Yeast work and in vitro binding were carried out as described (Mason and Botella, 2000) using tomato Gβ subunit (SlGB1). SlGB1 was amplified with the following primer pair: 5′-ATGTCAGTTGCGGAGCTGAAAGAG-3′ and 5′-GTCGACTCAGACCACACTTCTGTGT-3′. The amplified SlGB1 was fused to GAL4-BD in pBridge vector using EcoRI and SalI restriction sites incorporated during PCR. pACT2-AD-AGG2 from Mason and Botella (2000, 2001) was used as a positive control, and empty pACT2 was used as a negative control. Full-length constructs of SlGGB1 and SlGGB2 were amplified using the following primer pairs: for SlGGB1, 5′-TGGAGTCGTCGTCGTCATCAC-3′ and 5′-TCATATCCAGCGTTTGTTGCGTCTTG-3′; and for SlGGB2, 5′-ATGGATTCATTAATTATAATTAATG-3′ and 5′-TCAGATCCACCGTTTGTTACG-3′. The amplified full-length genes were cloned in frame into pACT2 using the terminal NcoI and BamHI restriction sites incorporated during PCR to produce pACT-AD-SlGGB1 and pACT-AD-SlGGB2. The yeast strain AH109 Saccharomyces cerevisiae was used for transformation following the Matchmaker Yeast Protocols (Clontech). Yeast cotransformed with two plasmid constructs was grown on SC synthetic complete medium lacking Leu and Trp. For interaction tests, SC synthetic complete medium lacking His, Leu, and Trp was used. All media were made according to the Clontech protocol.
Isolation of RNA and Transcription Analysis
Total RNA was from various tissues and isolated as described previously (Purnell and Botella, 2007). First-strand DNA synthesis was performed using the SuperScript III RT Kit (Invitrogen) according to the manufacturer’s instructions. RT-qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and the 7900HT Sequence Detection System (Applied Biosystems). The following primer pairs, designed using Primer Express software (Applied Biosystems), were used in RT-qPCR: for SlGGA, 5′-GGAAACAAGGCCAGATCCATT-3′ and 5′-GATGCGTCTTGTGCTCCTTCA-3′; for SlGGB1, 5′-GGATCCCTAACGAAGAAAATAC-3′ and 5′-CGTGAAGCTGGTGATGACGACGA-3′; and for SlGGC, 5′-TTTGTATGGAAAGCGTCGAGAAT-3′ and 5′-CCTTCAATGGATTTCAGTTCTTCCT-3′. Internal reference GAPDH, TIP41, and CAC genes were coamplified with the target gene (Expósito-Rodríguez et al., 2008). Primer sequences for auxin-responsive genes were extracted from the work of Chaabouni et al. (2009). Gene expression analysis was performed using SDS version 2.2.2 software (Applied Biosystems). The results shown are average values from three independently prepared RNA samples.
IAA Quantification
Leaves and roots from 4-week-old plants and ripe fruits from mature wild-type and slggb1-50 plants were harvested and frozen in liquid nitrogen. Frozen tissues were further crushed and freeze dried. The freeze-dried tissues were homogenized in methanol:water (1:1) overnight at 4°C, purified using C18 Sep-Pak cartridges (Waters), and analyzed using gas chromatography-mass spectrometry. Endogenous auxin levels were calculated based on the addition of 40 ng of [13C6]IAA, 4 ng of [13C1]indole butyric acid, and 4 ng of [2H4]4Cl-IAA per sample. The average weight of the plant samples was 0.6 g (se = 0.01).
BiFC Analysis
Full-length SlGGB1 and AtAGG2 were cloned into pKannibal-cEYFP using NcoI/BamHI and NcoI/HindIII sites, respectively. pKannibal-cEYFP was produced by cloning a PCR fragment obtained with primers cYFP-F-XhoI (5′-TTCTCGAGATGGGCGGCAGCGTGCAGCT-3′) and cYFP-R-NcoI (5′-AACCATGGATCTACACTTGTACAG-3′) into pKannibal-GFP (Maruta et al., 2015), substituting GFP with cYFP. The cYFP fragment was fused to N termini of the proteins, since the C terminus of AGGs was prenylated posttranslationally and could not be altered (Adjobo-Hermans et al., 2006; Zeng et al., 2007). pKannibal-nEYFP-AGB1 with Arabidopsis (Arabidopsis thaliana) Gβ subunit cDNA was described previously (Aranda-Sicilia et al., 2015). Mesophyll protoplasts were isolated from 3- to 4-week-old Arabidopsis plants and transfected with the constructs of interest, according to the established protocol (Yoo et al., 2007). Transfected protoplasts were incubated at room temperature with gentle rocking for 16 to 18 h. Fluorescence was studied with a confocal microscope (Zeiss LSM700) with the parameters described below.
Subcellular Localization
Full-length coding regions of SlGGB1 and SlGGB2 were amplified by PCR from tomato cDNA with the following primer pairs: for SlGGB1, 5′-TCCATGGAGTCGTCGTCGTCATCACCA-3′ and 5′-TGGATCCTCATATCCAGCGTTTGTTGCGTCT-3′; and for SlGGB2, 5′-TCCATGGATTCATTAATTATAATTAATGATG-3′ and 5′-TGGATCCTCAGATCCACCGTTTGTTACG-3′.
The fragments were cloned into pKannibal-GFP (Maruta et al., 2015) using NcoI/BamHI restriction sites. The Arabidopsis AGG2 coding region was cloned into pKannibal-GFP using NcoI/HindIII restriction sites. These vectors were used to transfect mesophyll protoplasts isolated from 3- to 4-week-old tomato plants according to the established protocol (Yoo et al., 2007). Transfected protoplasts were incubated at room temperature with gentle rocking for 16 to 18 h. Fluorescence was studied with a confocal microscope (Zeiss LSM700).
The GFP-SlGGB1 expression cassette from pKannibal-GFP-SlGGB1 was cloned into pART27 (Gleave, 1992) using NotI restriction sites. The obtained binary vector was introduced into Agrobacterium tumefaciens (GV3101) via electroporation. For transient expression in Nicotiana benthamiana, A. tumefaciens harboring the construct was grown in 2 mL of Luria-Bertani medium with rifampicin (PCCA) and spectinomycin (Sigma) overnight at 28°C. The bacteria were harvested and resuspended in 10 mm MgCl2 with 150 μm acetosyringone (3,5-dimethoxy-acetophenone [Fluka]) and 10 mm MES at pH 5.5, to give a final optical density at 600 nm of 0.2. Leaves of N. benthamiana grown for 2 to 3 weeks were infiltrated using a syringe without a needle. For fluorescence analysis, a Zeiss LSM700 confocal microscope was used.
Arabidopsis plants were transformed with pART27-GFP-SlGGB1 using the A. tumefaciens-mediated floral dip method (Bent, 2006). True leaves and cotyledons of the transgenic plants were analyzed for GFP fluorescence.
To compare the localization of tomato SlGGB1 and Arabidopsis AGG2, the latter was fused to red fluorescent protein (RFP), an mCherry variant. For that, RFP was amplified by PCR using the primers mCherryF-XhoI (5′-CTCGAGATGGTGAGCAAGGGCGAGGA-3′) and mCherryR-EcoRI (5′-AGAATTCCTTGTACAGCTCGTCCATGCCG-3′) and cloned into pKannibal (Wesley et al., 2001) using XhoI/EcoRI sites. Full-length AGG2 was amplified with AGG2F-EcoRI (5′-TGAATTCATGGAAGCGGGTAGCTCCAAT-3′) and AGG2R-HindIII (5′-TAAGCTTCAAAGAATGGAGCAGCCACATC-3′) and cloned into pKannibal-mCherry. Two plasmids, pKannibal-GFP-SlGGB1 and pKannibal-RFP-AGG2, were mixed and introduced into Arabidopsis mesophyll protoplasts as described above for BiFC analysis. Protoplasts were assayed with a confocal microscope (Zeiss LSM700).
Confocal Microscopy
Fluorescence analysis was performed on a laser scanning confocal microscope (Zeiss LSM700). Argon laser line excitation wavelengths and emission bandpass filter wavelengths for GFP, YFP, and mCherry were 480 to 510, 510 to 550, and 580 to 630 nm, respectively. Chlorophyll autofluorescence was recorded at 640 to 700 nm. Image acquisition parameters, laser power set at 2.0 and pinhole set at automatic minimum, were not changed throughout the study; detector gain was optimized for each experiment but did not change upon optimization. 10× and 40× objectives were used. Raw data were processed in ZEN 2011 SP3 (black edition) software, version 8.1.
RNA Sequencing Data Analysis
Seeds of wild-type and slggb1-50-silenced lines imbibed in 10 μm ABA solution and water were used in this experiment. Wild-type and slggb1-50 seeds imbibed in water were used as controls. After 24 h of imbibition, seeds from both the ABA-treated and control lines were collected and immediately snap frozen in liquid nitrogen. These samples were labeled WT Control, WT ABA, B50 Control, and B50 ABA. Each sample had three biological replicates. Total RNAs from samples were prepared using Trizol reagent (Invitrogen) and subsequently used for mRNA purification and library construction with the Truseq RNA Sample Prep Kit (Illumina) following the manufacturer’s instructions. The samples were then sequenced on an Illumina HiSeq 2000 (Illumina), generating 595,464,013 reads. Sequencing was completed by the Australian Research Genome Facility. The reads were submitted to mapping analysis against a reference genome sequence (ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG2.4_release/; Tomato Genome Consortium, 2012) using TopHat (Trapnell et al., 2009) with parameters of minimum and maximum intron length to be 42 and 22,729, respectively, according to the annotation release 2.4. The aligned reads were then assembled with Cufflinks (http://cufflinks.cbcb.umd.edu/; Trapnell et al., 2012) to assemble and reconstruct the tomato transcriptome for each treatment. The assembled transcriptomes of the different treatments and the original genome annotations were then merged using the Cuffmerge utility, which is included with the Cufflinks package. Furthermore, the reads and merged transcriptomes of different treatments were fed to Cuffdiff, an RNA sequencing analysis tool used for transcript and gene quantification, which calculates the expression levels and differential gene expression for the different treatment data sets and then tests the statistical significance of the observed changes.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence alignment of tomato Gγ subunits.
Supplemental Table S1. Differentially expressed genes in slggb1 seeds imbibed in water.
Supplemental Table S2. Differentially expressed genes in slggb1 seeds imbibed in water.
Supplementary Material
Glossary
- GPCR
G protein-coupled receptor
- ABA
abscisic acid
- RNAi
RNA interference
- RT-qPCR
quantitative real-time PCR
- BiFC
bimolecular fluorescence complementation
- LRP
lateral root primordium
- NAA
naphthaleneacetic acid
- MS
Murashige and Skoog
- IAA
indole-3-acetic acid
- cDNA
complementary DNA
Footnotes
Articles can be viewed without a subscription.
References
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Adjobo-Hermans MJ, Goedhart J, Gadella TW Jr (2006) Plant G protein heterotrimers require dual lipidation motifs of Galpha and Ggamma and do not dissociate upon activation. J Cell Sci 119: 5087–5097 [DOI] [PubMed] [Google Scholar]
- Aranda-Sicilia MN, Trusov Y, Maruta N, Chakravorty D, Zhang Y, Botella JR (2015) Heterotrimeric G proteins interact with defense-related receptor-like kinases in Arabidopsis. J Plant Physiol 188: 44–48 [DOI] [PubMed] [Google Scholar]
- Armstrong F, Blatt MR (1995) Evidence for K+ channel control in Vicia guard-cells coupled by G-proteins to a 7TMS receptor mimetic. Plant J 8: 187–198 [Google Scholar]
- Arya GC, Kumar R, Bisht NC (2014) Evolution, expression differentiation and interaction specificity of heterotrimeric G-protein subunit gene family in the mesohexaploid Brassica rapa. PLoS ONE 9: e105771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM. (1996) Guard cell G proteins. Trends Plant Sci 1: 73–74 [Google Scholar]
- Bassa C, Mila I, Bouzayen M, Audran-Delalande C (2012) Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol 53: 1583–1595 [DOI] [PubMed] [Google Scholar]
- Bent A. (2006) Arabidopsis thaliana floral dip transformation method. Methods Mol Biol 343: 87–103 [DOI] [PubMed] [Google Scholar]
- Bewley JD. (1997) Breaking down the walls: a role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci 2: 464–469 [Google Scholar]
- Bhardwaj D, Lakhanpaul S, Tuteja N (2012) Wide range of interacting partners of pea Gβ subunit of G-proteins suggests its multiple functions in cell signalling. Plant Physiol Biochem 58: 1–5 [DOI] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldshmidt A, Jackson D (2013) The maize Galpha gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558 [DOI] [PubMed] [Google Scholar]
- Booker FL, Burkey KO, Overmyer K, Jones AM (2004) Differential responses of G-protein Arabidopsis thaliana mutants to ozone. New Phytol 162: 633–641 [DOI] [PubMed] [Google Scholar]
- Botto JF, Ibarra S, Jones AM (2009) The heterotrimeric G-protein complex modulates light sensitivity in Arabidopsis thaliana seed germination. Photochem Photobiol 85: 949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL Jr, Grisafi PL, Fink GR (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, Frasse P, Latché A, Pech JC, Bouzayen M (2009) Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot 60: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D, Trusov Y, Botella JR (2012) Site-directed mutagenesis of the Arabidopsis heterotrimeric G protein β subunit suggests divergent mechanisms of effector activation between plant and animal G proteins. Planta 235: 615–627 [DOI] [PubMed] [Google Scholar]
- Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR (2011) An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J 67: 840–851 [DOI] [PubMed] [Google Scholar]
- Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM (2004) GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol 135: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, Ecker JR, Jones AM (2006a) RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot 57: 2697–2708 [DOI] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Chen Y, Ji F, Xie H, Liang J, Zhang J (2006b) The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol 140: 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury SR, Bisht NC, Thompson R, Todorov O, Pandey S (2011) Conventional and novel Gγ protein families constitute the heterotrimeric G-protein signaling network in soybean. PLoS ONE 6: e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury SR, Pandey S (2013) Specific subunits of heterotrimeric G proteins play important roles during nodulation in soybean. Plant Physiol 162: 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colucci G, Apone F, Alyeshmerni N, Chalmers D, Chrispeels MJ (2002) GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc Natl Acad Sci USA 99: 4736–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan Y, Yan H, Munyikwa T, Dong J, Zhang Y, Armstrong CL (2006) MicroTom: a high-throughput model transformation system for functional genomics. Plant Cell Rep 25: 432–441 [DOI] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57: 160–170 [DOI] [PubMed] [Google Scholar]
- Deng W, Yang Y, Ren Z, Audran-Delalande C, Mila I, Wang X, Song H, Hu Y, Bouzayen M, Li Z (2012) The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytol 194: 379–390 [DOI] [PubMed] [Google Scholar]
- Ding L, Pandey S, Assmann SM (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J 53: 248–263 [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam N, Downes GB, Yan K, Kisselev O (1998) The G-protein betagamma complex. Cell Signal 10: 447–455 [DOI] [PubMed] [Google Scholar]
- Gautam N, Northup J, Tamir H, Simon MI (1990) G protein diversity is increased by associations with a variety of gamma subunits. Proc Natl Acad Sci USA 87: 7973–7977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M (2000) Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta 210: 279–285 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ (1985) Rapid induction of selective transcription by auxins. Mol Cell Biol 5: 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Kleinschmidt A, Guilfoyle T (1984) Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta 162: 147–153 [DOI] [PubMed] [Google Scholar]
- He Y, Gan S (2004) A novel zinc-finger protein with a proline-rich domain mediates ABA-regulated seed dormancy in Arabidopsis. Plant Mol Biol 54: 1–9 [DOI] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Ishida T, Tabata R, Yamada M, Aida M, Mitsumasu K, Fujiwara M, Yamaguchi K, Shigenobu S, Higuchi M, Tsuji H, et al. (2014) Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep 15: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Qi S, Li H, Liu P, Li P, Wu C, Zheng C, Huang J (2014) Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. Biochem Biophys Res Commun 454: 505–511 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Siderovski DP (2007) Receptor-mediated activation of heterotrimeric G-proteins: current structural insights. Mol Pharmacol 72: 219–230 [DOI] [PubMed] [Google Scholar]
- Jones AM, Assmann SM (2004) Plants: the latest model system for G-protein research. EMBO Rep 5: 572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Ecker JR, Chen JG (2003) A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol 131: 1623–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Duffy JW, Machius M, Temple BRS, Dohlman HG, Jones AM (2011a) The crystal structure of a self-activating G protein alpha subunit reveals its distinct mechanism of signal initiation. Sci Signal 4: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Temple BRS, Jones AM, Dohlman HG (2011b) Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana. J Biol Chem 286: 13143–13150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV (2005) Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato C, Mizutani T, Tamaki H, Kumagai H, Kamiya T, Hirobe A, Fujisawa Y, Kato H, Iwasaki Y (2004) Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J 38: 320–331 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kino T, Kozasa T, Chrousos GP (2005) Statin-induced blockade of prenylation alters nucleocytoplasmic shuttling of GTP-binding proteins γ2 and β2 and enhances their suppressive effect on glucocorticoid receptor transcriptional activity. Eur J Clin Invest 35: 508–513 [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Berr A, Belin C, Chappuis R, Nishizawa NK, Lopez-Molina L (2010) Identification of growth insensitive to ABA3 (gia3), a recessive mutation affecting ABA signaling for the control of early post-germination growth in Arabidopsis thaliana. Plant Cell Physiol 51: 239–251 [DOI] [PubMed] [Google Scholar]
- Lapik YR, Kaufman LS (2003) The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein α-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15: 1578–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM (1995) Formation of lateral root meristems is a two-stage process. Development 121: 3303–3310 [DOI] [PubMed] [Google Scholar]
- Lease KA, Wen J, Li J, Doke JT, Liscum E, Walker JC (2001) A mutant Arabidopsis heterotrimeric G-protein β subunit affects leaf, flower, and fruit development. Plant Cell 13: 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Assmann SM (1999) Arabidopsis thaliana ‘extra-large GTP-binding protein’ (AtXLG1): a new class of G-protein. Plant Mol Biol 40: 55–64 [DOI] [PubMed] [Google Scholar]
- Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y (2013) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol 161: 2146–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sánchez-Rodriguez C, Jorda L, Molina A (2005) ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J 43: 165–180 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM (1990) Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc Natl Acad Sci USA 87: 3821–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, et al. (2015) COLD1 confers chilling tolerance in rice. Cell 160: 1209–1221 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Manfre AJ, LaHatte GA, Climer CR, Marcotte WR Jr (2009) Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana mutant atem6-1. Plant Cell Physiol 50: 243–253 [DOI] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Brenya E, Parekh U, Botella JR (2015) Membrane-localized extra-large G proteins and Gβγ dimer of the heterotrimeric G proteins form functional complexes engaged in plant immunity in Arabidopsis. Plant Physiol 167: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR (2000) Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein gamma-subunit cDNA. Proc Natl Acad Sci USA 97: 14784–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR (2001) Isolation of a novel G-protein gamma-subunit from Arabidopsis thaliana and its interaction with Gbeta. Biochim Biophys Acta 1520: 147–153 [DOI] [PubMed] [Google Scholar]
- McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS (2005) G-protein signaling: back to the future. Cell Mol Life Sci 62: 551–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire WE. (2009) Structural determinants involved in the formation and activation of G protein betagamma dimers. Neurosignals 17: 82–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger JD. (1983) Role of endogenous plant growth regulators in seed dormancy of Avena fatua. II. Gibberellins. Plant Physiol 73: 791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan G, Kostenis E (2006) Heterotrimeric G-proteins: a short history. Br J Pharmacol (Suppl 1) 147: S46–S55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Wu Y, Venkataraman G, Sopory SK, Tuteja N (2007) Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): role in salinity and heat stress and cross-talk with phospholipase C. Plant J 51: 656–669 [DOI] [PubMed] [Google Scholar]
- Okamoto H, Göbel C, Capper RG, Saunders N, Feussner I, Knight MR (2009) The alpha-subunit of the heterotrimeric G-protein affects jasmonate responses in Arabidopsis thaliana. J Exp Bot 60: 1991–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Matsui M, Deng XW (2001) Overexpression of the heterotrimeric G-protein α-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13: 1639–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM (2003) Hormonal interactions in fruit development. J Plant Growth Regul 22: 73–81 [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol 141: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandey S, Nelson DC, Assmann SM (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Pandolfini T, Rotino GL, Camerini S, Defez R, Spena A (2002) Optimisation of transgene action at the post-transcriptional level: high quality parthenocarpic fruits in industrial tomatoes. BMC Biotechnol 2: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison RJ, Catalá C (2012) Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J 70: 585–598 [DOI] [PubMed] [Google Scholar]
- Prinz WA, Hinshaw JE (2009) Membrane-bending proteins. Crit Rev Biochem Mol Biol 44: 278–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell MP, Botella JR (2007) Tobacco isoenzyme 1 of NAD(H)-dependent glutamate dehydrogenase catabolizes glutamate in vivo. Plant Physiol 143: 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer GFE. (2011) AUXIN-BINDING-PROTEIN1, the second auxin receptor: what is the significance of a two-receptor concept in plant signal transduction? J Exp Bot 62: 3339–3357 [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261 [DOI] [PubMed] [Google Scholar]
- Serna L. (2005) A simple method for discriminating between cell membrane and cytosolic proteins. New Phytol 165: 947–952 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Zheng N (2009) Signal advance for abscisic acid. Nature 462: 575–576 [DOI] [PubMed] [Google Scholar]
- Temple BRS, Jones AM (2007) The plant heterotrimeric G-protein complex. Annu Rev Plant Biol 58: 249–266 [DOI] [PubMed] [Google Scholar]
- Thung L, Chakravorty D, Trusov Y, Jones AM, Botella JR (2013) Signaling specificity provided by the Arabidopsis thaliana heterotrimeric G-protein γ subunits AGG1 and AGG2 is partially but not exclusively provided through transcriptional regulation. PLoS ONE 8: e58503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung L, Trusov Y, Chakravorty D, Botella JR (2012) Gγ1 + Gγ2 + Gγ3 = Gβ: the search for heterotrimeric G-protein γ subunits in Arabidopsis is over. J Plant Physiol 169: 542–545 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Botella JR (2012) New faces in plant innate immunity: heterotrimeric G proteins. J Plant Biochem Biotechnol 21: S40–S47 [Google Scholar]
- Trusov Y, Chakravorty D, Botella JR (2012) Diversity of heterotrimeric G-protein γ subunits in plants. BMC Res Notes 5: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Jorda L, Molina A, Botella JR (2010) G proteins and plant innate immunity. In Yalovsky S, Baluska F, Jones AM, eds, Integrated G Protein Signaling in Plants. Springer-Verlag, Berlin, pp 221–250 [Google Scholar]
- Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR (2006) Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol 140: 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen JG, Jones AM, Botella JR (2007) Heterotrimeric G protein gamma subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell 19: 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Sewelam N, Rookes JE, Kunkel M, Nowak E, Schenk PM, Botella JR (2009) Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J 58: 69–81 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM (2003) The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15: 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Wang S, Jones AM (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol 129: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Chen JG, Botella JR, Jones AM (2013) Heterotrimeric G protein signalling in the plant kingdom. Open Biol 3: 120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones AM (2014) Heterotrimeric G protein-coupled signaling in plants. Annu Rev Plant Biol 65: 365–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM (2012) G protein activation without a GEF in the plant kingdom. PLoS Genet 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech JC, Bouzayen M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu YY, Ma QB, Li D, Xu ZH, Chong K (2006) Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res 16: 916–922 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Warpeha KM, Hamm HE, Rasenick MM, Kaufman LS (1991) A blue-light-activated GTP-binding protein in the plasma membranes of etiolated peas. Proc Natl Acad Sci USA 88: 8925–8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KM, Lateef SS, Lapik Y, Anderson M, Lee BS, Kaufman LS (2006) G-protein-coupled receptor 1, G-protein Gα-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis. Plant Physiol 140: 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, Anderson MB, Kaufman LS (2007) The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol 143: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss CA, Garnaat CW, Mukai K, Hu Y, Ma H (1994) Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc Natl Acad Sci USA 91: 9554–9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wolfenstetter S, Chakravorty D, Kula R, Urano D, Trusov Y, Sheahan MB, McCurdy DW, Assmann SM, Jones AM, Botella JR (2015) Evidence for an unusual transmembrane configuration of AGG3, a class C Gγ subunit of Arabidopsis. Plant J 81: 388–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeng Q, Wang X, Running MP (2007) Dual lipid modification of Arabidopsis Gγ-subunits is required for efficient plasma membrane targeting. Plant Physiol 143: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, He SY, Assmann SM (2008) The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J 56: 984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.