Characterization of a maize tandem gene array reveals three kaurene synthases with varied roles in gibberellin and phytoalexin metabolism.
Abstract
While most commonly associated with its role in gibberellin phytohormone biosynthesis, ent-kaurene also serves as an intermediate in more specialized diterpenoid metabolism, as exemplified by the more than 800 known derived natural products. Among these are the maize kauralexins. However, no ent-kaurene synthases (KSs) have been identified from maize. The maize gibberellin-deficient dwarf-5 (d5) mutant has been associated with a loss of KS activity. The relevant genetic lesion has been previously mapped, and was found here to correlate with the location of the KS-like gene ZmKSL3. Intriguingly, this forms part of a tandem array with two other terpene synthases (TPSs). Although one of these, ZmTPS1, has been previously reported to encode a sesquiterpene synthase, and both ZmTPS1 and that encoded by the third gene, ZmKSL5, have lost the N-terminal γ-domain prototypically associated with KS(L)s, all three genes fall within the KS(L) or TPS-e subfamily. Here it is reported that all three genes encode enzymes that are targeted to the plastid in planta, where diterpenoid biosynthesis is initiated, and which all readily catalyze the production of ent-kaurene. Consistent with the closer phylogenetic relationship of ZmKSL3 with previously identified KSs from cereals, only transcription of this gene is affected in d5 plants. On the other hand, the expression of all three of these genes is inducible, suggesting a role in more specialized metabolism, such as that of the kauralexins. Thus, these results clarify not only gibberellin phytohormone, but also diterpenoid phytoalexin biosynthesis in this important cereal crop plant.
Gibberellins (GAs) are diterpenoid hormones with a variety of roles in vascular plant growth and development (Sun, 2011). GA biosynthesis begins with sequential cyclization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate, first to ent-copalyl diphosphate (ent-CPP) and then to ent-kaurene, catalyzed by CPP synthases (CPSs) and ent-kaurene synthases (KSs), respectively. This is followed by oxidative reactions catalyzed by cytochrome P450 mono-oxygenases and 2-oxoglutarate-dependent dioxygenases that lead to the production of bioactive GAs (Yamaguchi, 2008; Hedden and Thomas, 2012). A key step is the ring contraction that transforms the 6-6-6-5 ring structure of ent-kaurene to the 6-5-6-5 arrangement that characterizes the GAs (Peters, 2013).
Notably, the GAs are the ancestral members of the labdane-related diterpenoids, a superfamily of over 7000 natural products united by the common occurrence of a labda-13E-en-8-yl+ diphosphate intermediate in the initial cyclization reaction catalyzed by class II diterpene cyclases such as CPSs (Peters, 2010). While such natural products can be found in many types of organisms, they are particularly prevalent in plants, where the CPS and KS required for GA biosynthesis serve as a biosynthetic reservoir (Zi et al., 2014). Indeed, this is true for not only this superfamily, but also the entire class of terpenoid natural products more generally, as a bifunctional CPS/KS, such as those still found in the early diverging bryophytes (Hayashi et al., 2006), seems to have served as the ancestral plant terpene synthase (TPS). The TPSs form moderate-sized gene families in plants, although the vast majority has lost the N-terminal γ-domain that has been retained in and characterizes plant KSs (Chen et al., 2011). Notably, the TPSs involved in more specialized labdane-related diterpenoid biosynthesis seem to be those most closely related to KSs, and these have been termed KS-like (KSLs), and together the KSs and KSLs make up the KS(L)/TPS-e subfamily.
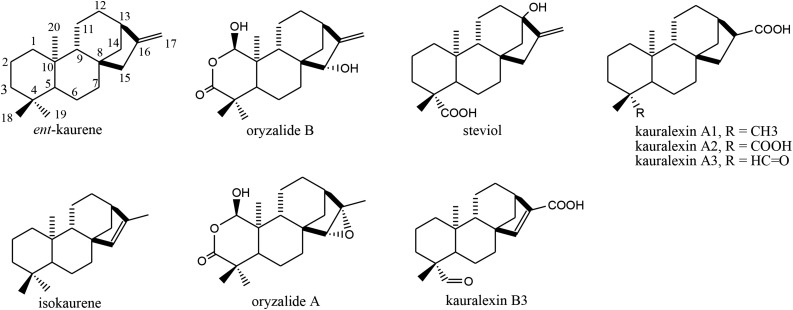
The proliferation of labdane-related diterpenoids from GA biosynthesis can perhaps be most readily appreciated by noting that more than 800 of these natural products derived from ent-kaurene, with retention of the signature 6-6-6-5 hydrocarbon ring structure, are already known (Dictionary of Natural Products, http://dnp.chemnetbase.com). However, the biosynthesis of only a few of these has been studied. This includes the steviosides from Stevia rebaudiana (Brandle and Telmer, 2007), the orzyalides and related diterpenoids from rice (Oryza sativa), and the kauralexins from maize (Zea mays; Schmelz et al., 2014). Some of these seem to be derived from the carbon-carbon double bond (C = C) isomer ent-(iso)kaur-15-ene, rather than the ent-kaur-16-ene relevant to GA metabolism. In particular, while the steviosides and oryzalide B series of diterpenoids contain the exocyclic C = C of ent-kaurene, the oryzalide A and kauralexin B series contain the endocyclic C = C characterizing ent-isokaurene. By contrast, as the kauralexin A series no longer has a C = C, these may be derived from ent-kaurene (Fig. 1).
Figure 1.
Natural products derived from ent-(iso)kaurene.
Two closely related KSs relevant to stevioside biosynthesis have been identified (Richman et al., 1999), and extensive investigation of the rice KS(L) gene family has been reported (Schmelz et al., 2014; Tezuka et al., 2015). This includes identification of OsKS1, required for GA production (Sakamoto et al., 2004; Margis-Pinheiro et al., 2005), as well as a separate ent-isokaurene synthase, OsKSL6 (Kanno et al., 2006; Xu et al., 2007). Interestingly, no KS specific to more specialized metabolism has yet been identified. Potentially, this is because KS seems to be constitutively expressed, as flux to ent-kaurene is controlled by CPS, at least in Arabidopsis thaliana, where over-expression of the native AtCPS increases ent-kaurene levels > 1000-fold, with only a minimal (< 2-fold) additional increase observed from over-expression of AtKS as well (Fleet et al., 2003).
There has been extensive investigation of GAs in maize, including early isolation of various GA-deficient mutants (Phinney, 1956). Among these are anther ear-1 (an1) and dwarf-5 (d5), whose growth deficiencies can be rescued by application of ent-kaurene (Katsumi et al., 1964). An1 was later found to encode the ent-CPP producing ZmCPS1 (Bensen et al., 1995), suggesting that the separate D5 would encode ZmKS. However, despite previously reported mapping studies (Beavis et al., 1991), and the availability of the maize genome sequence (Schnable et al., 2009), the relevant gene for D5 has not yet been identified.
Beyond GAs, maize also was reported some time ago to exhibit an increased capacity for production of various labdane-related diterpenes, including both ent-kaurene and ent-isokaurene, in response to fungal infection (Mellon and West, 1979). More recently, transcription of An2, encoding the ent-CPP producing ZmCPS2, was found to be induced by fungal infection (Harris et al., 2005). This eventually led to identification of the maize kauralexins as antifungal phytoalexins (Schmelz et al., 2011), and the role of An2 in such biosynthesis has just been verified (Vaughan et al., 2014). However, while substantial characterization of other maize TPSs has been reported (Degenhardt et al., 2009), this has not yet been extended to the KS(L)/TPS-e subfamily.
Here the previously mapped region for d5 is reported to contain a tandem array of three TPSs, ZmTPS1, ZmKSL3, and ZmKSL5. All three are targeted to plastids in planta, and catalyze the production of ent-kaurene from ent-CPP. However, only the expression of ZmKSL3 is abolished in d5 plants, indicating that this serves nonredundantly as the KS for gibberellin metabolism in maize, consistent with the closer phylogenetic relationship of this with previously identified KSs from cereals. On the other hand, transcription of all three genes is inducible, suggesting that these may play a role in biosynthesis of the kauralexin A series of phytoalexins.
RESULTS
The d5 Region Contains a Tripartite Array of KS(L)s
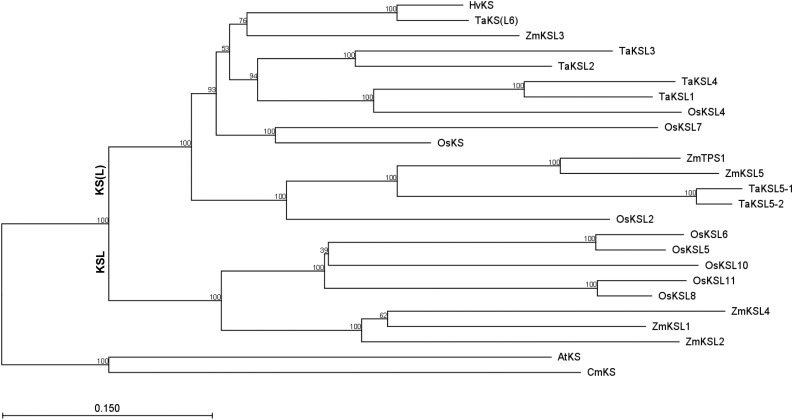
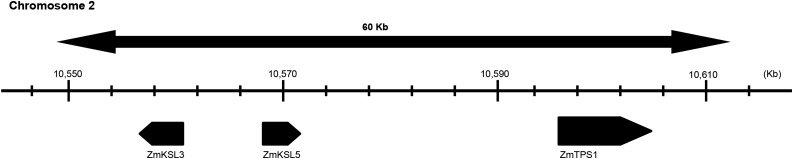
It has been previously reported that maize contains four KS(L)s, termed ZmKSL1 to ZmKSL4 (Schmelz et al., 2014). Phylogenetic analysis reveals the presence of two distinct clades within the cereal KS(L)s, with one clade containing only KSLs involved in more specialized metabolism, while the other contains both KSLs and KSs with known roles in GA biosynthesis. Notably, of ZmKSL1 to ZmKSL4, only ZmKSL3 falls within this latter clade (Fig. 2). Moreover, ZmKSL3 falls within the region on chromosome 2 that quantitative trait loci analysis previously indicated contains the genetic lesion responsible for the dwarfism associated with d5 (Beavis et al., 1991). Closer examination of this region of the maize genome revealed the presence of two other TPSs within 60 kb of ZmKSL3 (Fig. 3). One of these corresponds to ZmTPS1, which has been previously reported to be a sesquiterpene synthase (Schnee et al., 2002), while the other is closely related. Although both of these resemble other TPSs in lacking the N-terminal γ-domain prototypically associated with KS(L)s, phylogenetic analysis demonstrated that both fall within the KS(L)/TPS-e subfamily, and the third gene is then termed ZmKSL5 (Fig. 2). ZmTPS1 and ZmKSL5 fall into the same phylogenetic clade as ZmKSL3. Thus, together these form a tandem array of KS(L) TPSs. Notably, ZmTPS1 and ZmKSL5 further group with two previously characterized homeologs from wheat (Triticum aestivum), TaKSL5-1 and -2, which also no longer have the γ-domain (Zhou et al., 2012). Intriguingly, these react not only with the sesquiterpene precursor (E,E)-farnesyl diphosphate, producing (E)-nerolidol (similar to the activity originally reported for ZmTPS1), but also with ent-CPP, producing a 7:3 mixture of ent-kaurene and ent-beyerene (Hillwig et al., 2011). Thus, it seemed possible that ZmTPS1 and ZmKSL5 also might exhibit KS activity.
Figure 2.
Phylogenetic tree of cereal KS(L)s (AtKS and CmKS from dicots included as designated outgroup). This tree was constructed with neighbor joining method with the CLC Sequence Viewer 7.0 software package (CLC bio). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.
Figure 3.
A 60-kb tandem gene array on maize chromosome 2 consists of ZmTPS1, ZmKSL3, and ZmKSL5. Gene position was based on the Mo17 genomic sequence data from Phytozome 10.0.
For the KS activity exhibited by these enzymes to be relevant, they should be targeted to plastids, where diterpenoid biosynthesis is initiated (Zi et al., 2014). Notably, a high probability for such chloroplast targeting was predicted by a number of available algorithms for not only ZmKSL3, but also ZmTPS1 and ZmKSL5 (Supplemental Table S1). The predicted signal peptide sequences further enabled construction of pseudo-mature open reading frames for recombinant expression.
Recombinant ZmTPS1, ZmKSL3, and ZmKSL5 Exhibit KS Activity
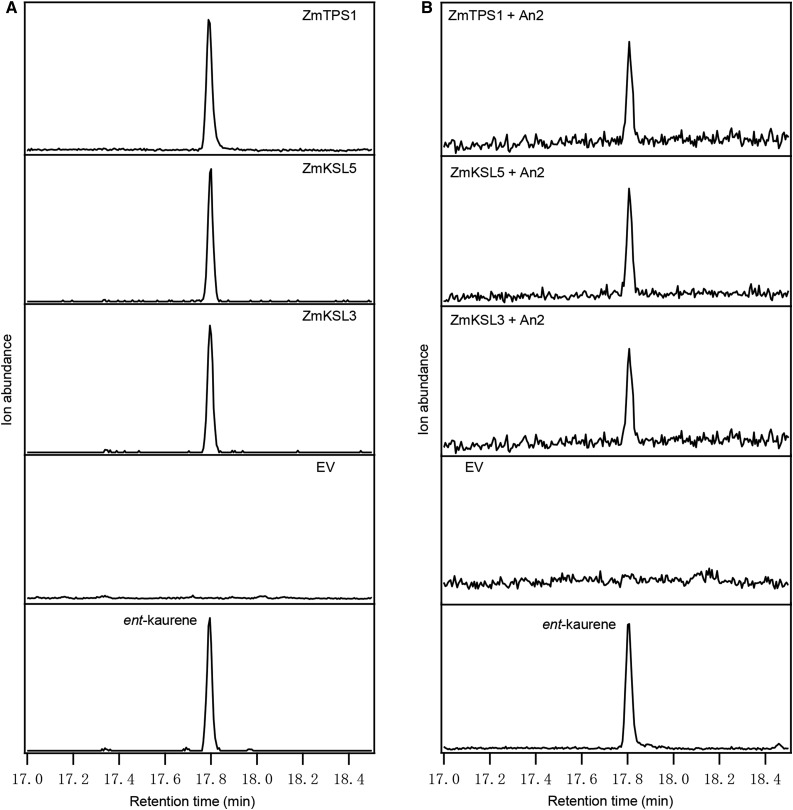
Biochemical characterization of ZmTPS1, ZmKSL5, and ZmKSL3 was carried out using a previously described modular metabolic engineering system (Cyr et al., 2007). Briefly, each of these was expressed as pseudo-mature enzymes in Escherichia coli also engineered to produce ent-CPP, leading to formation of ent-kaurene and demonstrating that all three exhibit KS activity (Fig. 4A). When these further were expressed in E. coli engineered to produce (E,E)-farnesyl diphosphate instead, no sesquiterpene product(s) were detected for ZmKSL3 or ZmKSL5, although ZmTPS1 did produce a few sesquiterpenes (Supplemental Fig. S1), consistent with the previously reported characterization (Schnee et al., 2002). These recombinant results encouraged further examination of biochemical function in planta.
Figure 4.
Characterization of KS activity for ZmTPS1, ZmKSL3, and ZmKSL5. A, GC-MS analysis (272 m/z extracted ion chromatographs) of the enzymatic products of ZmTPS1, ZmKSL5, and ZmKSL3. B, Production of ent-kaurene by ZmTPS1, ZmKSL5, and ZmKSL3 transiently expressed in N. benthamiana with An2 (ZmCPS2). GC-MS analysis (272 m/z extracted ion chromatographs) was carried out using leaf extract after infiltration for 5 d. Agrobacterium GV3101 harboring P19 was coinfiltrated in all treatment. Authentic ent-kaurene (retention time, 17.80 min.) made by OsKS was used for retention time and mass spectra comparison. EV, Empty vector control.
ZmTPS1, ZmKSL5, and ZmKSL3 Targeting and in Planta KS Activity
Nicotiana benthamiana has been reported to be a good host for plant metabolic engineering, particularly including diterpene production, with transient expression of the relevant TPSs (Brückner and Tissier, 2013). In an initial trial, transient expression of ZmTPS1, ZmKSL3, or ZmKSL5 under control of the 35S promoter in N. benthamiana did not lead to the detection of any new terpenes by GC-MS analysis. Given the low endogenous levels of ent-CPP in N. benthamiana, it was reasoned that coexpression of ent-CPS might be required to detect KS activity. Coexpression of An2/ZmCPS2 with either ZmTPS1, ZmKSL3, or ZmKSL5 resulted in detection of ent-kaurene in the transformed tobacco tissue (Fig. 4B). These results not only provide further evidence for KS activity for all three enzymes, but also indicate that these are in fact targeted to the plastid along with ZmCPS2. Further evidence for plastid targeting was obtained by fusing ZmKSL3, ZmTPS1, and ZmKSL5 to eGFP (enhanced green fluorescence protein). These constructs also were transiently expressed in N. benthamiana, with confocal laser scanning microscopy revealing their localization to the chloroplasts (Supplemental Fig. S2). Accordingly, ZmTPS1, ZmKSL3, and ZmKSL5 all exhibit KS activity in planta and could plausibly have a role in gibberellin biosynthesis.
ZmKSL3 Corresponds to d5
To determine which of the ZmTPS1, ZmKSL3, and/or ZmKSL5 genes are involved in GA biosynthesis, their expression was analyzed in d5 maize seedlings, whose GA-deficient dwarf phenotype was confirmed by the ability of exogenous GA3 to restore internode growth and plant height (Supplemental Fig. S3). Expression of all three genes can be detected in the parental line. However, ZmKSL3 was not expressed in the d5 mutant, while ZmKSL5 and ZmTPS1 expression was still observed (Fig. 5A). By contrast, analysis of the available Uniform Mu mutants for ZmKSL5 and ZmTPS1 demonstrated that, despite significant reduction in expression of the relevant gene, in both cases no dwarf phenotype was observed (Supplemental Fig. S4). In addition, cDNA for both ZmTPS1 and ZmKSL3 were cloned from d5 plants, and found to contain no significant changes relative to those cloned from the B73 line (above), with the encoded enzymes further exhibiting the expected KS activity (Supplemental Fig. S5). Thus, ZmKSL3 clearly corresponds to the d5 mutation and encodes the KS relevant to maize GA metabolism.
Figure 5.
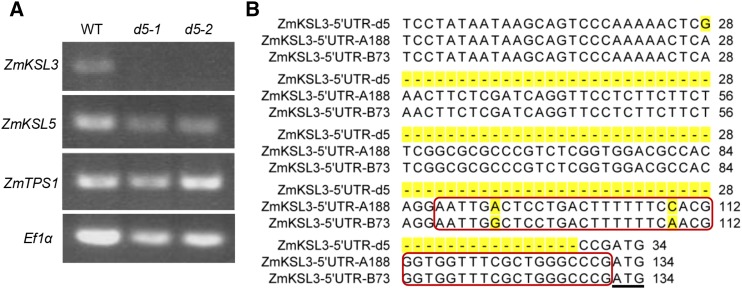
Only ZmKSL3 is not expressed in d5 mutant. A, RT-PCR analysis of expression of ZmTPS1, ZmKSL3, and ZmKSL5 in the A188 parental/wild-type (WT) line, relative to two d5 mutant maize plants (d5-1 and d5-2), reveals that only ZmKSL3 expression is abrogated in d5 plants. B, 100-bp loss close to the initial codon (underlined) was observed in genomic region corresponding to 5′UTR of ZmKSL5 in d5, in which one CAAT box (red box) was predicted. Detailed alignment and sequence information are located in the Supplemental Data.
The genetic lesion underlying the loss of ZmKS(L3)/D5 expression and dwarf phenotype of d5 was determined by sequencing the relevant genomic region. While there were no significant differences in the protein-coding region of ZmKSL3 between d5 and B73, there was a substantial deletion and several mutations in the 5′ untranslated region close to the start codon in d5. The corresponding ZmKS(L3)/D5 region from B73 and the parental line A188 contains a CAAT box, loss of which presumably underlies the loss of ZmKS(L3)/D5 expression in d5 (Fig. 5B).
Inducible Expression Suggests Role in More Specialized Metabolism
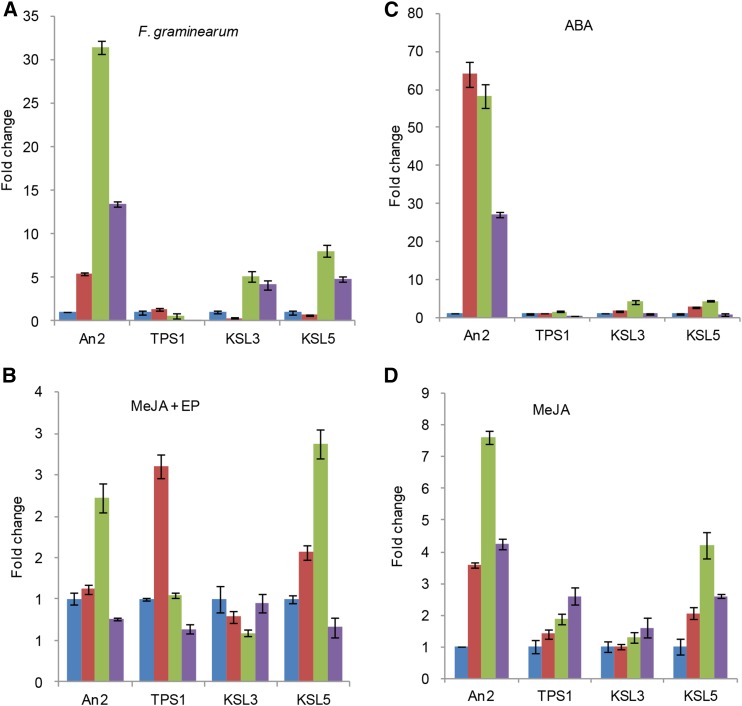
Given the potential use of ent-kaurene as a precursor in biosynthesis of the kauralexin A series of antifungal phytoalexins (Fig. 1), we examined the expression of ZmTPS1, ZmKS(L3)/D5, and ZmKSL5 in response to fungal infection. In particular, the accumulation of phytoalexins in response to infection is typified by transcriptional induction of the relevant biosynthetic genes (Ahuja et al., 2012). The kauralexins were reported to be constitutively present at low levels in seedlings, with dramatic increases induced by biotic and abiotic stress, suggesting constitutive and inducible expression of the relevant biosynthetic genes (Schmelz et al., 2011; Vaughan et al., 2014). Previously reported microarray analysis indicates that all three of these KS(L)/TPS-e genes were constitutively expressed in B73 seedlings, with ZmKS(L3)/D5 specifically expressed in actively growing tissues, such as the meristem, internode, tassel, embryo, and young seeds (Supplemental Fig. S6; and Sekhon et al., 2011). Such constitutive expression was verified here in young seedlings of both Mo17 and B73 cultivars (Supplemental Fig. S7). More interestingly, after combining mechanical wounding with inoculation of the maize fungal pathogen Fusarium graminearum, all three of these genes exhibited higher transcript abundance (Fig. 6A). There was some variation in the timing and amount of increase, with ZmKSL5 exhibiting the largest increase 24 h postinfection (hpi), similar to what is observed with An2/ZmCPS2, which has a known role in kauralexin biosynthesis (Vaughan et al., 2014). By contrast, ZmTPS1 expression is only slightly induced at 12 hpi, and is actually lower at 24 hpi. At 48 h, expression of ZmTPS1 was repressed, consistent with previously reported microarray analysis (Huffaker et al., 2011).
Figure 6.
Inducible gene expression pattern of ZmTPS1, ZmKSL3, and ZmKSL5. A to D, Quantitative RT-PCR analysis for gene expression of three KSs with treatment of: A, F. graminearum spores inoculation; B, MeJA + EP; C, ABA; and D, MeJA. An2 was used as positive control. Leaves were collected at 0 (blue), 12 (red), 24 (green), and 48 (purple) h with treatment for all analysis except ABA treatment, in which root was treated and collected for quantitative RT-PCR analysis.
Treatment with abscisic acid (ABA) or a combination of jasmonic acid (JA) and ethephon (EP) has been reported to induce kauralexin accumulation, as well as expression of An2/ZmCPS2 (Schmelz et al., 2011), which produces the ent-CPP substrate of KSs (Harris et al., 2005). Thus, the expression of ZmTPS1, ZmKS(L3)/D5, and ZmKSL5 in response to these defense hormones was investigated. Upon combined treatment with MeJA and EP, ZmKSL5 transcripts accumulated at 12 h and 24 h after treatment. ZmTPS1 also showed increased transcript levels after 12 h, while An2/ZmCPS2 was found to exhibit increased transcript levels after 24 h (Fig. 6B). By contrast, ZmKS(L3)/D5 transcript levels were slightly repressed. ABA strongly induced expression of An2/ZmCPS2 in the roots of maize seedlings, along with that of ZmKSL5 and ZmKS(L3)/D5, although not ZmTPS1 (Fig. 6C). Treatment with JA has been reported to not induce kauralexin accumulation (Schmelz et al., 2011). Nevertheless, it does induce expression of An2/ZmCPS2, as well as ZmTPS1, ZmKS(L3)/D5, and ZmKSL5 (Fig. 6D). Consistent with the induction observed here, the promoters for all three of these KS(L)/TPS-e genes contain various defense hormone response elements (Supplemental Table S2). In any case, the inducible transcription of ZmTPS1 and ZmKSL5, as well as ZmKS(L3)/D5, is correlated to some extent with that of An2/ZmCPS2 (Fig. 6), which indicates that these may be involved in more specialized labdane-related diterpenoid biosynthesis, such as the production of the kauralexin A series.
DISCUSSION
Here it was found that maize contains a tandem array of genes, ZmTPS1, ZmKSL3, and ZmKSL5 (Fig. 3), which all exhibit KS activity (Fig. 4). Although ZmTPS1 was originally suggested to be responsible for maize sesquiterpene emissions (Schnee et al., 2002), this role has since been assigned to the herbivore-induced ZmTPS10 instead (Schnee et al., 2006). Moreover, despite loss of the usual N-terminal γ-domain associated with the KS(L)/TPS-e subfamily, both ZmTPS1 and ZmKSL5 not only phylogenetically cluster with this subfamily (Fig. 2), but also are targeted to the plastids (Supplemental Fig. S2), consistent with a role in di- rather than sesquiterpenoids biosynthesis.
While this tandem array falls within the region that has been mapped for the GA-deficient and KS-associated d5 mutation (Beavis et al., 1991), only the expression of ZmKSL3 seems to be affected in d5 plants (Fig. 5), and reduced expression of ZmTPS1 or ZmKSL5 does not affect maize plant growth (Supplemental Fig. S4). Accordingly, ZmKSL3 corresponds to D5, and only this KS appears to be involved in production of GA, clarifying its metabolism. Notably, it has been previously reported that d5 plants exhibit some residual production of ent-kaurene (Hedden and Phinney, 1979), which is consistent with the KS activity of the unaffected ZmTPS1 and ZmKSL5 reported here. Although ZmTPS1 and ZmKSL5 exhibited constitutive expression in seedlings, they cannot compensate for the lack of ZmKS(L3)/D5 absence in d5 because they are not expressed in most actively growing tissues (Supplemental Fig. S6), similar to what has been described for the expression of the ent-CPP producing OsCPS1 and OsCPS2 from rice (Toyomasu et al., 2015), where only OsCPS1 is involved in GA metabolism (Sakamoto et al., 2004).
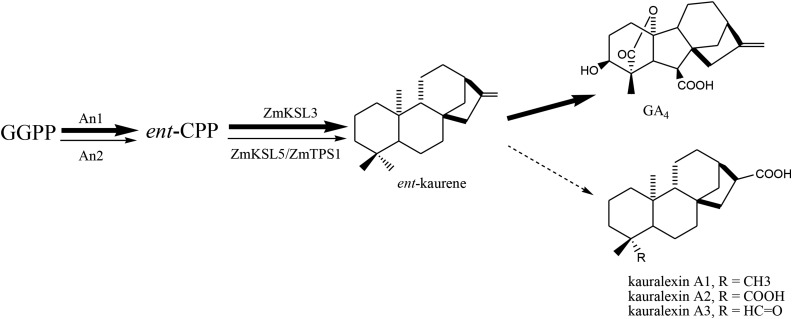
In addition to being a precursor to GAs, ent-kaurene is an intermediate in the production of many other labdane-related diterpenoid natural products such as the maize kauralexins. A role for ZmTPS1 and ZmKSL5 in such more specialized metabolism is implied by the inducible transcription observed here, with a role in production of kauralexins suggested by the closely related expression pattern of ZmKSL5 with that for An2/ZmCPS2, which has a known role in such biosynthesis (Fig. 6). Although it has been suggested that kauralexins are derived only from ent-isokaurene (Schmelz et al., 2014), from these results it seems likely that kauralexins A1 to A3 are derived from ent-kaurene instead, with loss of the exocyclic C = C during transformation of C17 to the characteristic carboxylic acid (Fig. 7), offering some insight into maize phytoalexin biosynthesis.
Figure 7.
GA and kauralexin A biosynthesis in maize. An1 and An2 were identified as specific ent-CPP producing CPSs for GA and kauralexin biosynthesis, respectively. Bold arrows indicate the GA biosynthetic steps.
Nevertheless, maize must contain an ent-isokaurene synthase for at least production of kauralexins B1 to B3. Rice has already been shown to encode such an enzyme, OsKSL5 and/or OsKSL6 (Kanno et al., 2006; Xu et al., 2007), which fall into the early diverging KSL clade of the TPS-e subfamily in cereals that is associated with more specialized diterpenoid metabolism (Fig. 2). This clade also contains several KSLs from maize, and it seems likely that one of these (ZmKSL1, ZmKSL2, and/or ZmKSL4) serves as the ent-isokaurene synthase required for kauralexin biosynthesis, although further investigations are required to verify this.
In any case, from the results reported here, it is possible to speculate about the origins of this maize array of KSs. This seems to arise from two tandem gene duplication events (Supplemental Fig. S8). The original KS, with a role in GA biosynthesis, likely resembled ZmKS(L3)/D5 and, given the presence of MeJA response elements in the promoters of all three genes, this ancestral gene may have already had a role in more specialized metabolism. Initial gene duplication would have then led to some subfunctionalization, with loss of the N-terminal γ-domain in one copy, although with retention of the plastid targeting sequence. This seems to have occurred with the copy that assumed an increased role in more specialized metabolism, and this gene underwent an additional tandem gene duplication event leading to ZmTPS1 and ZmKSL5. The role of this secondary ancestral gene in more specialized metabolism is suggested by the presence of stress-responsive ABA response elements and W-box sequences in the promoters of both ZmTPS1 and ZmKSL5, which are not present in that of ZmKS(L3)/D5 (Supplemental Table S2). Interestingly, the presence of ABA response elements and inducible gene expression with ABA treatment is consistent with the recently suggested role for kauralexins in drought tolerance (Vaughan et al., 2014). Nevertheless, the somewhat different expression patterns observed for ZmTPS1 and ZmKSL5 suggests that these may play varied roles in maize diterpenoid metabolism.
Regardless of the exact roles of ZmTPS1 and ZmKSL5, the results reported here clarify maize diterpenoid metabolism. In particular, despite the presence of a tripartite tandem array of KSs, GA biosynthesis primarily relies on just ZmKS(L3)/D5. On the other hand, the inducible expression of these KSs indicates a role in more specialized metabolism, with the closely matching coexpression between ZmKSL5 and An2/ZmCPS2 suggesting a role for this more specifically in kauralexin (i.e. phytoalexin) biosynthesis. In turn, the use of ent-kaurene as an intermediate provides insight into the production of these phytoalexins.
MATERIALS AND METHODS
Plant and Fungal Materials
Maize (Zea mays) cultivars Mo17 and B73 were grown on sterile water-agar medium for 10 d, and these seedlings used for fungal infection and RNA extraction. Fusarium graminearum was grown on potato dextrose agar medium at 28°C for 3 d, and spores collected by washing the mycelium with sterile water, the concentration of which was adjusted to 1 × 106 mL−1 with 0.1% Tween 20 carrier solution for plant infection. Spores were sprayed on the maize leaves carefully scratched on the surface with a scalpel, then kept at 100% humidity under a transparent cover for 24 h. N. benthamiana was germinated and grown in soil at 22°C with 16 h day/8 h night photoperiod for 4 to 5 weeks, ready for agroinfiltration.
Gene Cloning and Recombinant Constructs
Plant seedlings inoculated with F. graminearum spores for 24 h were used for RNA extraction, which was reversely transcribed to cDNA using M-MLV reverse transcriptase (Takara). The coding sequence for ZmTPS1, ZmKSL3, and ZmKSL5 was amplified from this cDNA and ligated into pGM-T vector (Tiangen). After sequencing and confirmation by comparison to the genomic data on-line (NCBI, MaizeGDB, Phytozome 10.0), all three genes were subcloned into pET28a, with removal of the N-terminal transit peptide sequence (67 amino acids for ZmTPS1, 47 amino acids for ZmKSL5, and 53 amino acids for ZmKSL3, respectively), for recombinant expression. The full-length open reading frames of all three genes also were subcloned into pCAMBIA 2300-eGFP with or without stop codon (ZmTPS1 and ZmKSL5) for transient expression under control of the cauliflower mosaic virus 35S promoter in N. benthamiana.
Recombinant Biochemical Analysis
Each TPS was characterized by expression in a modular metabolic engineering system (Cyr et al., 2007). Accordingly, ZmTPS1, ZmKSL3, or ZmKSL5 in pET28a was cotransformed with pGGeC or pMBIS into BL21 Escherichia coli. As previously described, upon induction pGGeC leads to the production of ent-CPP (Cyr et al., 2007), while pMBIS leads to the production of (E,E)-farnesyl diphosphate (Martin et al., 2003), enabling analysis of KS or sesquiterpene synthase activity, respectively. Cultures were inoculated from three to five cotransformed colonies, with 5 mL (liquid LB media) starter cultures grown at 37°C and 200 rpm overnight, which were then transferred into 50 mL LB medium for analysis. Recombinant expression was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside when OD600 reached 0.6 to 0.8, and the culture fermented at 16°C overnight. The culture was extracted twice with an equal volume of hexanes, and this organic extract separated and dried down by rotary evaporation. The residue was resuspended with 100 μL hexanes, and 1 μL was injected for GC-MS analysis, which was performed using a model Number 6890-5973 (Agilent Technologies) with detection by a quadruple mass spectrometer in EI mode using the following temperature program: 70°C for 2 min, then increase to 250°C at 10°C/min, which was held for 2 min. To verify the production of ent-kaurene, OsKS was incorporated into the same system to make an authentic standard for direct comparison of retention time and mass spectra.
Transient Expression in N. benthamiana
Transient overexpression of ZmTPS1, ZmKSL3, and ZmKSL5 in N. benthamiana was carried out as described by Brückner and Tissier (2013). Briefly, Agrobacterium tumefaciens strain GV3101 was transformed with pCAMBIA 2300-eGFP constructs, with or without stop codons, and the resulting recombinant strains infiltrated into 5-week-old N. benthamiana, either with or without An2, as well as P19 to suppress posttranscriptional silencing (Voinnet et al., 2003). Leaves were collected 5 d after infiltration for extraction, which was carried out by grinding in liquid N2, followed by swirling with hexanes. The hexane extracts were filtered, concentrated by rotary evaporation, and fractionated by the silica gel column chromatography, with elution by a 20:1 mixture of hexanes and ethyl acetate. The eluted fractions were dried under N2 gas, and the residues were dissolved in hexanes for GC-MS analysis as above. Subcellular localization was analyzed using a model Number A1R/A1 confocal laser scanning microscope (Nikon), with leaves collected 4 d after infiltration, as previously described by Vaughan et al. (2013).
Gene Expression Analysis
Ten-day-old Mo17 and B73 seedlings were separated into root and shoot tissues, which were used for constitutive gene expression analysis. Mo17 seedlings also were used for pathogen inoculation and phytohormone treatment. Before all treatments, the seedlings were wounded, using a scalpel to scratch three 2- to 3-cm incisions on the leaves. F. graminearum spores (1 × 106 mL−1) were sprayed on the seedlings in 0.1% Tween 20 carrier solution. For phytohormone treatment, each seedling was sprayed with 2 mL of 50 μm MeJA solely or combined with 16.5 μm EP in 0.1% Tween 20. A quantity of 100 μm ABA was used to treat roots of the Mo17 seedling. All treated seedlings were covered with a transparent plastic dome to keep the humidity at roughly 100%. Leaf and root samples were collected at 0, 12, 24, and 48 h after inoculation or treatment. Total RNA was isolated with TRNzol (Tiangen) followed the manufacture protocol, with cDNA synthesis carried out as recommended by the M-MLV reverse transcriptase kit from Takara. Semiquantitative reverse transcription (RT)-PCR was performed to analyze constitutive gene expression of the three TPS genes in Mo17 and B73 root and aboveground tissues. Quantitative RT-PCR was performed to check the inducible expression of the three TPS genes following fungal pathogen infection and phytohormone treatment, which was carried out on a CFX96 (Bio-Rad) using the SsoFast Eva Green Supermix (Bio-Rad). Ef1α was used as endogenous control. Semiquantitative RT-PCR for ZmTPS1, ZmKSL5, An2, and Ef1α was carried out using the same primers used for quantitative PCR (Supplemental Table S3). All amplicons were sequenced to verify primer specificity.
Gene Expression and Sequencing in d5 Mutant
Seeds for the d5 mutant and its inbred parental line (A188) were obtained from MaizeGDB (stock nos. 214C-d5 and Ames 22443, respectively). The parental (wild type) and d5 mutant were grown in a potting soil mix under the same conditions as Mo17 and B73. After 10 d, leaves were collected for gDNA and RNA extraction for gene expression and sequencing analysis by semiquantitative RT-PCR as above. To verify that the observed dwarfish was due to GA deficiency, 40 mg/L GA3 was applied to a 10-d-old d5 seedling, and phenotype was observed daily for 2 weeks. The genomic sequence of ZmKSL3 was amplified from d5, as well as B73 and the parental line A188, and sequenced.
Identification of ZmKSL5 and ZmTPS1 Mutants
Uniform Mu mutants were acquired for ZmKSL5 (UFMu-08573) and ZmTPS1 (UFMu-06794) from MaizeGDB. The parental line (W22) and mutants were grown in a potting soil mix under the same conditions as above. After 10 d, RNA was extracted from leaves for cDNA synthesis. Mutant plants were identified through quantitative RT-PCR, and allowed to grow further to verify the lack of an obvious growth defect.
Bioinformatics Analysis
All maize putative diterpene synthases and other cereal KS(L)s were analyzed using the CLC Sequence Viewer 7.0 (CLC bio) to produce a rooted phylogenetic tree from their amino-acid sequences, using the default settings, with CmKS and AtKS as the outgroup. The gene accession numbers are as follows: maize KS(L)s (ZmKSL1, AFW61735; ZmKSL2, DAA54948; ZmKSL3, DAA36069; ZmKSL4, DAA49845; ZmKSL5, NP_001141888; ZmTPS1, AAO18435); rice KS(L)s (OsKS, NP_001053841; OsKSL2, LC033788; OsKSL4, AY616862; OsKSL5, DQ823352; OsKSL6, ABH10733; OsKSL7, DQ823354; OsKSL8, AB118056; OsKSL10, DQ823355; OsKSL11, DQ100373); wheat KS(L)s (TaKSL1, AB597957; TaKSL2, AB597958; TaKSL3, AB597959; TaKSL4, AB597960; TaKSL5, AB597961; TaKS(L6), AB597962); barley (Hordeum vulgare) KS (HvKS, AY551436); AtKS (AAC39443); and Cucurbita maxima KS (CmKS, U43904). Subcellular location was predicted using ChloroP (www.cbs.dtu.dk/services/ChloroP/), Predotar (urgi.versailles.inra.fr/predotar/predotar.html), PCLA (www.andrewschein.com/cgi-bin/pclr/pclr.cgi), and iPSORT (ipsort.hgc.jp/), with transit peptide sequence cutoff setting using amino-acid sequence of tested genes. Promoter analysis was carried out using 1.5-kb upstream sequences from the initiating codons of ZmTPS1, ZmKSL3, and ZmKSL5, which were downloaded from phytozome 10.0, with cis-regulatory element prediction performed using PlantCARE (bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE (www.dna.affrc.go.jp/PLACE/). Heat map of ZmKSL and An2 gene expression in B73 tissues was plotted with the gplots package of the R program.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AFW61735, DAA54948, DAA36069, DAA49845, NP_001141888, AAO18435, NP_001053841, LC033788, AY616862, DQ823352 ABH10733, DQ823354, AB118056, DQ823355, DQ100373, AB597957, AB597958, AB597959, AB597960, AB597961, AB597962, AY551436, AAC39443, and U43904.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GC-MS analysis of ZmTPS1 sesquiterpene products.
Supplemental Figure S2. Plastidic localization of ZmTPS1, ZmKSL5, and ZmKSL3.
Supplemental Figure S3. Maize d5 mutant seedlings and height restoration by GA treatment.
Supplemental Figure S4. ZmTPS1 and ZmKSL5 mutants did not show dwarf phenotype.
Supplemental Figure S5. GC-MS analysis of KS activity for ZmTPS1 and ZmKSL5 in d5.
Supplemental Figure S6. Heat map of KSL gene expression in B73 with An2.
Supplemental Figure S7. Constitutive gene expression pattern of ZmTPS1, ZmKSL3, and ZmKSL5.
Supplemental Figure S8. Proposed evolution of maize tandem KS gene array.
Supplemental Table S1. Plastid localization prediction results.
Supplemental Table S2. Predicted cis-regulatory elements in 1.5-kb promoter region of three KS genes.
Supplemental Table S3. Primers used for semiquantitative (S) and quantitative RT-PCR.
Sequence Alignment S1. Alignment of ZmKSL5 CDS in B73 and d5.
Sequence Alignment S2. Alignment of ZmTPS1 CDS in B73 and d5.
Sequence Alignment S3. Alignment of genomic sequence of ZmKSL3 in B73 and d5.
Supplementary Material
Acknowledgments
We thank Bin Wei for heat map plotting.
Glossary
- ABA
abscisic acid
- CPP
copalyl diphosphate
- CPS
CPP synthase
- ent-CPP
ent-copalyl diphosphate
- EP
ethephon
- hpi
hours postinfection
- JA
jasmonic acid
- KS
ent-kaurene synthase
- KSL
KS-like
- MeJA
methyl jasmonate
- RT
reverse transcription
- TPS
terpene synthase
Footnotes
Articles can be viewed without a subscription.
References
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Beavis WD, Grant D, Albertsen M, Fincher R (1991) Quantitative trait loci for plant height in four maize populations and their associations with qualitative genetic loci. Theor Appl Genet 83: 141–145 [DOI] [PubMed] [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandle JE, Telmer PG (2007) Steviol glycoside biosynthesis. Phytochemistry 68: 1855–1863 [DOI] [PubMed] [Google Scholar]
- Brückner K, Tissier A (2013) High-level diterpene production by transient expression in Nicotiana benthamiana. Plant Methods 9: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ (2007) A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc 129: 6684–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Köllner TG, Gershenzon J (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637 [DOI] [PubMed] [Google Scholar]
- Fleet CM, Yamaguchi S, Hanada A, Kawaide H, David CJ, Kamiya Y, Sun T-P (2003) Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol 132: 830–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol Biol 59: 881–894 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H (2006) Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett 580: 6175–6181 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phinney BO (1979) Comparison of ent-kaurene and ent-isokaurene synthesis in cell free systems from etiolated shoots of normal and dwarf-5 maize seedlings. Phytochemistry 18: 1475–1479 [Google Scholar]
- Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Hillwig ML, Xu M, Toyomasu T, Tiernan MS, Wei G, Cui G, Huang L, Peters RJ (2011) Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J 68: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Kaplan F, Vaughan MM, Dafoe NJ, Ni X, Rocca JR, Alborn HT, Teal PE, Schmelz EA (2011) Novel acidic sesquiterpenoids constitute a dominant class of pathogen-induced phytoalexins in maize. Plant Physiol 156: 2082–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Otomo K, Kenmoku H, Mitsuhashi W, Yamane H, Oikawa H, Toshima H, Matsuoka M, Sassa T, Toyomasu T (2006) Characterization of a rice gene family encoding type-A diterpene cyclases. Biosci Biotechnol Biochem 70: 1702–1710 [DOI] [PubMed] [Google Scholar]
- Katsumi M, Phinney BO, Jefferies PR, Henrick CA (1964) Growth response of the d-5 and an-1 mutants of maize to some kaurene derivatives. Science 144: 849–850 [DOI] [PubMed] [Google Scholar]
- Margis-Pinheiro M, Zhou X-R, Zhu Q-H, Dennis ES, Upadhyaya NM (2005) Isolation and characterization of a Ds-tagged rice (Oryza sativa L.) GA-responsive dwarf mutant defective in an early step of the gibberellin biosynthesis pathway. Plant Cell Rep 23: 819–833 [DOI] [PubMed] [Google Scholar]
- Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21: 796–802 [DOI] [PubMed] [Google Scholar]
- Mellon JE, West CA (1979) Diterpene biosynthesis in maize seedlings in response to fungal infection. Plant Physiol 64: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJ. (2013) Gibberellin phytohormone metabolism. In Bach T, Rohmer M, eds, Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches. Springer, New York, pp 233––249. [Google Scholar]
- Peters RJ. (2010) Two rings in them all: the labdane-related diterpenoids. Nat Prod Rep 27: 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney BO. (1956) Growth response of single-gene dwarf mutants in maize to gibberellic acid. Proc Natl Acad Sci USA 42: 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE (1999) Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. Plant J 19: 411–421 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ (2014) Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J 79: 659–678 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Kaplan F, Huffaker A, Dafoe NJ, Vaughan MM, Ni X, Rocca JR, Alborn HT, Teal PE (2011) Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc Natl Acad Sci USA 108: 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schnee C, Köllner TG, Gershenzon J, Degenhardt J (2002) The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-β-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol 130: 2049–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Köllner TG, Held M, Turlings TC, Gershenzon J, Degenhardt J (2006) The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc Natl Acad Sci USA 103: 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Sun TP. (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–R345 [DOI] [PubMed] [Google Scholar]
- Tezuka D, Ito A, Mitsuhashi W, Toyomasu T, Imai R (2015) The rice ent-KAURENE SYNTHASE LIKE 2 encodes a functional ent-beyerene synthase. Biochem Biophys Res Commun 460: 766–771 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Usui M, Sugawara C, Kanno Y, Sakai A, Takahashi H, Nakazono M, Kuroda M, Miyamoto K, Morimoto Y, et al. (2015) Transcripts of two ent-copalyl diphosphate synthase genes differentially localize in rice plants according to their distinct biological roles. J Exp Bot 66: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MM, Christensen S, Schmelz EA, Huffaker A, McAuslane HJ, Alborn HT, Romero M, Allen LH, Teal PE (2014) Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ 38: 2195–2207 [DOI] [PubMed] [Google Scholar]
- Vaughan MM, Wang Q, Webster FX, Kiemle D, Hong YJ, Tantillo DJ, Coates RM, Wray AT, Askew W, O’Donnell C, et al. (2013) Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 25: 1108–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya NM, Coates RM, Peters RJ (2007) Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68: 312–326 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Zhou K, Xu M, Tiernan M, Xie Q, Toyomasu T, Sugawara C, Oku M, Usui M, Mitsuhashi W, Chono M, et al. (2012) Functional characterization of wheat ent-kaurene(-like) synthases indicates continuing evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ (2014) To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu Rev Plant Biol 65: 259–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.