Two soybean protein thiol disulfide oxidoreductases achieve cooperative and efficient oxidative protein folding by relaying an oxidizing equivalent supplied from endoplasmic reticulum oxidoreductin-1a.
Abstract
Most proteins produced in the endoplasmic reticulum (ER) of eukaryotic cells fold via disulfide formation (oxidative folding). Oxidative folding is catalyzed by protein disulfide isomerase (PDI) and PDI-related ER protein thiol disulfide oxidoreductases (ER oxidoreductases). In yeast and mammals, ER oxidoreductin-1s (Ero1s) supply oxidizing equivalent to the active centers of PDI. In this study, we expressed recombinant soybean Ero1 (GmERO1a) and found that GmERO1a oxidized multiple soybean ER oxidoreductases, in contrast to mammalian Ero1s having a high specificity for PDI. One of these ER oxidoreductases, GmPDIM, associated in vivo and in vitro with GmPDIL-2, was unable to be oxidized by GmERO1a. We therefore pursued the possible cooperative oxidative folding by GmPDIM, GmERO1a, and GmPDIL-2 in vitro and found that GmPDIL-2 synergistically accelerated oxidative refolding. In this process, GmERO1a preferentially oxidized the active center in the a′ domain among the a, a′, and b domains of GmPDIM. A disulfide bond introduced into the active center of the a′ domain of GmPDIM was shown to be transferred to the active center of the a domain of GmPDIM and the a domain of GmPDIM directly oxidized the active centers of both the a or a′ domain of GmPDIL-2. Therefore, we propose that the relay of an oxidizing equivalent from one ER oxidoreductase to another may play an essential role in cooperative oxidative folding by multiple ER oxidoreductases in plants.
In eukaryotes, many secretory and membrane proteins fold via disulfide bond formation in the endoplasmic reticulum (ER). Seed storage proteins of major crops, such as wheat, corn, rice, and beans, which are important protein sources for humans and domestic animals, are synthesized in the ER of the endosperm or cotyledon. A number of seed storage proteins fold by the formation of intramolecular disulfide bonds (oxidative folding) and are transported to and accumulate in protein bodies (Kermode and Bewley, 1999; Jolliffe et al., 2005). In contrast to normally folded proteins, misfolded and unfolded proteins are retained in the ER and degraded by an ER-associated degradation or vacuolar system (Smith et al., 2011; Pu and Bassham, 2013). Therefore, quick and efficient oxidative folding of nascent seed storage proteins is needed for their accumulation in protein bodies.
During this process, protein disulfide isomerase (PDI; EC 5.3.4.1) and other ER protein thiol disulfide oxidoreductases (ER oxidoreductases) are thought to catalyze the formation and isomerization of disulfide bonds in nascent proteins (Hatahet and Ruddock, 2009; Feige and Hendershot, 2011; Lu and Holmgren, 2014). After phylogenetic analysis of the Arabidopsis genome, 10 classes of ER oxidoreductases (classes I–X) were identified (Houston et al., 2005). Among them, class I ER oxidoreductase, a plant PDI ortholog, has been studied in a wide variety of plants. Class I ER oxidoreductases have two catalytically active domains a and a′, containing active centers composed of Cys-Gly-His-Cys and two catalytically inactive domains b and b′. An Arabidopsis ortholog of class I ER oxidoreductases is required for proper seed development and regulates the timing of programmed cell death by chaperoning and inhibiting Cys proteases (Andème Ondzighi et al., 2008). OaPDI, a PDI from Oldenlandia affinis, a coffee family (Rubiaceae) plant, is involved in the folding of knotted circular proteins (Gruber et al., 2007). The rice ortholog (PDIL1-1) was suggested to be involved in the maturation of the major seed storage protein glutelin (Takemoto et al., 2002). Furthermore, rice PDIL1-1 plays a role in regulatory activities for various proteins that are essential for the synthesis of grain components as determined by analysis of a T-DNA insertion mutant (Satoh-Cruz et al., 2010).
The oxidative refolding ability of class I ER oxidoreductases was confirmed in recombinant soybean (GmPDIL-1) and wheat proteins produced by an Escherichia coli expression system established from cDNAs (Kamauchi et al., 2008; Kimura et al., 2015).
Class II and III ER oxidoreductases have an a–b–b′–a′ domain structure. Class II ER oxidoreductases have an acidic amino acid-rich sequence in the N-terminal region ahead of the a domain. Recombinant soybean (GmPDIL-2) and wheat class II ER oxidoreductases have oxidative refolding activities similar to that of class I (Kamauchi et al., 2008; Kimura et al., 2015). Class III ER oxidoreductases contain the nonclassical redox-center Cys-X-X-Ser/Cys motifs, as opposed to the more traditional CGHC sequence, in the a and a′ domains. Recombinant soybean (GmPDIL-3) and wheat proteins lack oxidative refolding activity in vitro (Iwasaki et al., 2009; Kimura et al., 2015). Class IV ER oxidoreductases are unique to plants and have an a–a′–ERp29 domain structure, which is homologous to the C-terminal domain of mammalian ERp29 (Demmer et al., 1997).
Recombinant soybean class IV ER oxidoreductases (GmPDIS-1 and GmPDIS-2) and wheat class IV ER oxidoreductase possess an oxidative refolding activity that is weaker than that of classes I and II (Wadahama et al., 2007; Kimura et al., 2015). Class V ER oxidoreductases are plant orthologs of mammalian P5 and have an a–a′–b domain structure. A rice class V ER oxidoreductase, consisting of PDIL2 and PDIL3, plays an important role in the accumulation of the seed storage protein Cys-rich 10-kD prolamin (crP10; Onda et al., 2011). Recombinant soybean class V ER oxidoreductase, GmPDIM and wheat class V ER oxidoreductase possess an oxidative refolding activity similar to that of class IV (Wadahama et al., 2008; Kimura et al., 2015). In the soybean, GmPDIL-1, GmPDIL-2, GmPDIM, GmPDIS-1, and GmPDIS-2 were found to associate transiently with a seed storage precursor protein, proglycinin, in the ER of the cotyledon by coimmunoprecipitation experiments, suggesting that multiple ER oxidoreductases are involved in the folding of the nascent proglycinin.
The disulfide bond in the active center of ER oxidoreductases is reduced as a result of catalyzing disulfide bond formation in an unfolded protein. The reduced active center of PDI was discovered to be oxidized again by ER oxidoreductin-1 (Ero1p) in yeast (Frand and Kaiser, 1998; Pollard et al., 1998). Ero1p orthologs are present universally in eukaryotes. Yeast and flies have a single copy of the ERO1 gene, which is essential for survival (Frand and Kaiser, 1998; Pollard et al., 1998; Tien et al., 2008). Mammals have two genes encoding Ero1-α (Cabibbo et al., 2000) and Ero1-β (Pagani et al., 2000) that function as major disulfide donors to nascent proteins in the ER, but are not critical for survival (Zito et al., 2010). Domain a of yeast PDI is the most favored substrate of yeast Ero1p (Vitu et al., 2010), whereas a′ of human PDI is specifically oxidized by human Ero1-α (Chambers et al., 2010) and Ero1-β (Wang et al., 2011). Electrons from Cys residues of the active centers of PDI are transferred to oxygen by Ero1 (Tu and Weissman, 2004; Sevier and Kaiser, 2008). The reaction mechanisms of yeast Ero1p and human Ero1s have been intensively investigated; their regulation by PDI has been extensively studied as well (Tavender and Bulleid, 2010; Araki and Inaba, 2012; Benham et al., 2013; Ramming et al., 2015). Only rice Ero1 (OsERO1) has been identified as a plant ortholog of Ero1p (Onda et al., 2009). OsERO1 is necessary for disulfide bond formation in rice endosperm. The formation of native disulfide bonds in the major seed storage protein proglutelin was demonstrated to depend upon OsERO1 by RNAi knockdown experiments. However, no plant protein thiol disulfide oxidoreductases that are oxidized by a plant Ero1 ortholog have been identified to date.
In this study, we show that multiple soybean ER oxidoreductases can be activated by a soybean Ero1 ortholog (GmERO1a). In addition, we propose a synergistic mechanism by which GmPDIM and GmPDIL-2 cooperatively fold unfolded proteins using oxidizing equivalents provided by GmERO1 in vitro.
RESULTS
Identification and Characterization of Soybean Ero1
The GmERO1a cDNA and GmERO1b cDNA encoding the soybean Ero1 orthologs were cloned. GmERO1a and GmERO1b encoded proteins composed of 465 amino acids containing 19 Cys residues (Fig. 1A and Supplemental Fig. S1). GmERO1 proteins were expressed as a single 53-kD band in soybean root, stem, leaf, and cotyledon (Fig. 1B). The band shifted to 46 kD under nonreducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), suggesting that GmERO1 proteins have intramolecular disulfide bonds. We confirmed that GmERO1 proteins, which have a putative transmembrane region (Trp-16–Ser-34) near the N termini and two putative N-glycosylated Asn residues, are type I membrane-bound glycoproteins targeted to the ER. When the extract from the cotyledon was treated with endoglycosidase H or F, the size of GmERO1 proteins decreased from 53 kD to 50 kD (Fig. 1C), suggesting that one or more high-Man type N-glycan(s) were attached to GmERO1 proteins. GmERO1 proteins were recovered in the precipitate of the cell homogenate, but not the supernatant, after ultra-centrifugation at 100,000g, and pretreatment with Triton X-100 caused solubilization of GmERO1 proteins in the supernatant along with soybean calnexin (Fig. 1D). In confocal microscopic images, GmERO1 was observed upon immunostaining to colocalize with GmPDIS-1, which is localized in the ER (Wadahama et al., 2007; Fig. 1E).
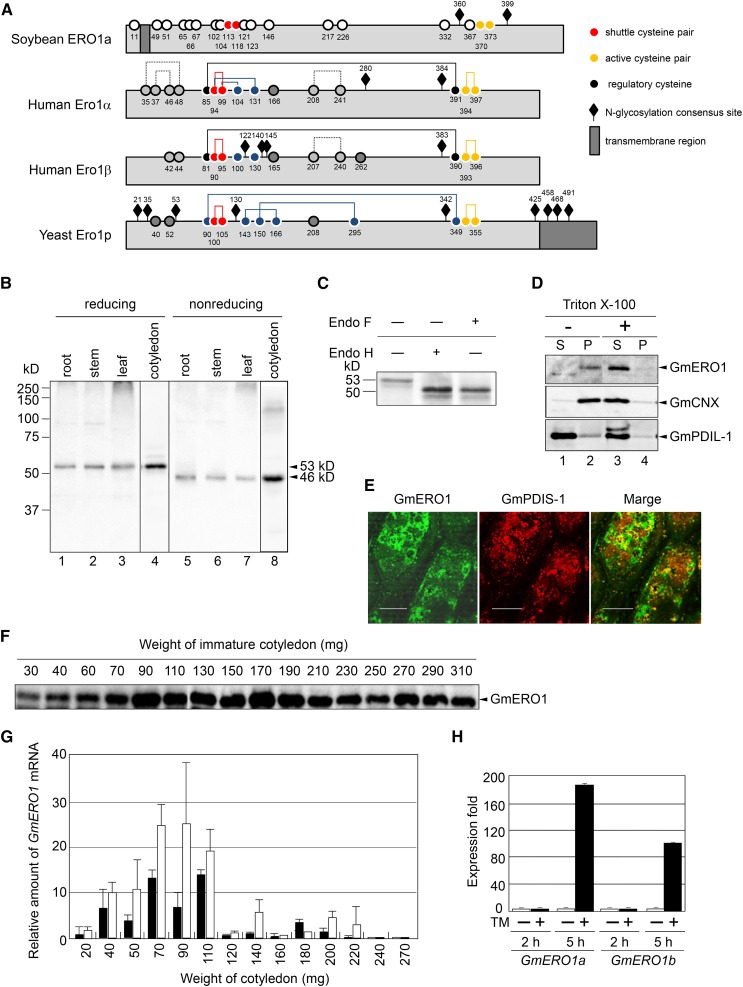
Figure 1.
Identification of soybean Ero1. A, Schematic representation of Ero1 orthologs. Numbered circles represent the positions of the Cys residues. Lines indicate disulfide bonds. B, GmERO1 is ubiquitously expressed in tissues. The proteins extracted from root (4.5 μg protein; lanes 1 and 5), stem (10 μg protein; lanes 2 and 6), leaf (30 μg protein; lanes 3 and 7), and immature cotyledon (100 mg, 30 μg protein; lanes 4 and 8) were separated under reducing (lanes 1–4) and nonreducing SDS-PAGE (lanes 5–8) and subjected to western-blot analysis with an antiserum prepared against recombinant GmERO1a. C, GmERO1 is N-glycosylated. GmERO1 in the immature cotyledon (30 mg) was detected by western-blot analysis with the anti-GmERO1 serum after treatment with (+) or without (−) endoglycosidase H (Endo H) or endoglycosidase F (Endo F). The 53-kD band of GmERO1 was shifted to 50 kD after Endo H and Endo F treatment. D, GmERO1 is a membrane-bound protein. An immature cotyledon (100 mg) was homogenized by sonication. The homogenate was centrifuged at 100,000g for 2 h at 4°C in the absence (−; lanes 1 and 2) or presence (+) of 1% Triton X-100 (lanes 3 and 4). GmERO1 protein in the supernatant (S; lanes 1 and 3) and pellet (P; lanes 2 and 4) was detected by western-blot analysis with anti-GmERO1 serum, GmCNX serum, or anti-GmPDIL-1 serum. E, GmERO1 localizes in the ER. The immature cotyledon (195 mg) was fixed and embedded in resin. The sections were cut with a microtome and immunostained with anti-GmERO1 guinea pig serum and anti-GmPDIS-1 rabbit serum, and observed under a confocal microscope. Bars = 20 μm. F, Expression of GmERO1 in cotyledons during development. The proteins (25 μg) extracted from cotyledons were analyzed by western blot with anti-GmERO1a serum. G, Relative amounts of GmERO1a (black bars) and GmERO1b (white bars) mRNA in cotyledons during development were quantified by real-time PCR. Data are represented as mean ± se of n = 3. H, Expression levels of GmERO1a and GmERO1b mRNA are up-regulated under ER stress. Young soybean leaves were treated with (+, black bars) or without (−, white bars) 5 μg/mL TM for 2 or 5 h. After treatment, GmERO1a and GmERO1b mRNA were quantified by real-time PCR. Data are represented as mean ± se of n = 3. TM, tunicamycin.
The expression levels of GmERO1 proteins and their mRNAs were high when seed storage proteins β-conglycinin and glycinin were synthesized (70–100 mg and 150–200 mg cotyledon weight, respectively; Fig. 1, F and G). GmERO1a and GmERO1b mRNAs were up-regulated following ER stress induced by tunicamycin (Fig. 1H). The ER stress response elements (Oh et al., 2003; Iwata and Koizumi, 2005; Iwata et al., 2008; Hayashi et al., 2013; Sun et al., 2013) were not found within the 2500-bp upstream and downstream sequences of the transcriptional region of either GmERO1a or GmERO1b.
Recombinant GmERO1a Oxidizes Various ER Oxidoreductases
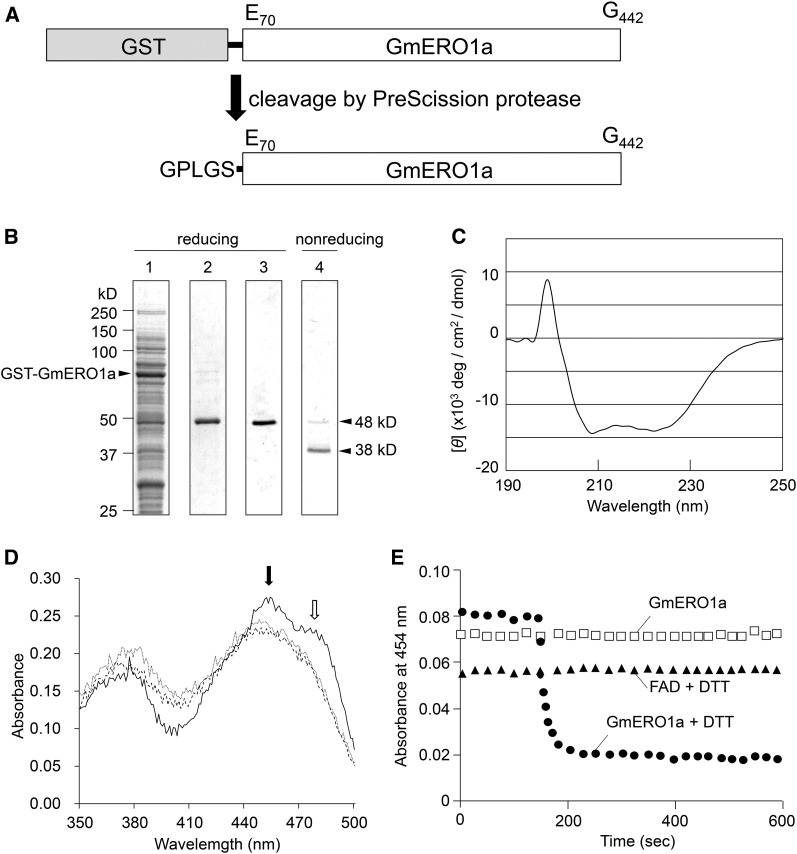
The soluble and folded 48-kD recombinant GmERO1a, which is missing 69 N-terminal residues including the putative transmembrane region and the C-terminal 23 residues (Fig. 2A), was expressed in E. coli. Its mobility as visualized by SDS-PAGE shifted to 38 kD under nonreducing conditions, suggesting that the recombinant protein has intramolecular disulfide bonds (Fig. 2B). The recombinant protein had circular dichroism spectra typical of a well-folded protein (Fig. 2C). The UV-visible absorption spectrum of the recombinant GmERO1a showed a typical unresolved flavin envelope with a maximum A454 and a shoulder at 485 nm (Fig. 2D). The spectrum, identical to that of authentic flavin adenine dinucleotide (FAD), was obtained after denaturation with guanidine hydrochloride, indicating that FAD is noncovalently bound to recombinant GmERO1a. The molar ratio of GmERO1a to bound FAD was calculated to be 1:1 with a molecular extinction coefficient of 12.9 mm−1 cm−1. The FAD moiety bound to GmERO1a was reduced by dithiothreitol (DTT; Fig. 2E), suggesting that FAD bound to GmERO1a acts as a cofactor to transfer electrons from a substrate.
Figure 2.
Expression of recombinant GmERO1a. A, Cleavage of recombinant GST-fused GmERO1a by PreScission protease. The gray box indicates GST, and the black line denotes the PreScission cleavage sequence. The N-terminal GST was cleaved from the fusion protein after protease treatment. B, Expression and purification of recombinant GmERO1a. Recombinant GST-fused GmERO1a expressed in E. coli (lane 1) was purified by a glutathione Sepharose 4B affinity column chromatography. GmERO1a was cleaved by PreScission protease and eluted from the column (lane 2), followed by gel filtration chromatography (lanes 3 and 4). Proteins in each eluate were separated by reducing SDS-PAGE (lanes 1–3) and nonreducing SDS-PAGE (lane 4), and stained with Coomassie Brilliant Blue. C, Far-UV circular dichroism spectrum of recombinant GmERO1a in 20 mm Tris-HCl buffer (pH 7.4) containing 150 mm NaCl and 10% glycerol was obtained using a J-720 spectropolarimeter in a 1-mm path-length cell with a scan speed of 20 nm/min at 14°C. D, Absorbance spectrum of recombinant GmERO1a was measured in the absence (black line) or presence (gray line) of 6 m guanidine hydrochloride. Free FAD spectrum (dashed line) was also measured. Black and white arrows show the maximum A454 and the shoulder at 485 nm of GmERO1a, respectively. E, GmERO1a-bound FAD is reduced by DTT. Absorbance changes at 454 nm of 10 μm GmERO1a (circles) and 10 μm free FAD (triangles) were monitored in the presence of 2 mm DTT. DTT was added at zero time. GmERO1a in the absence of DTT was also measured (rectangles).
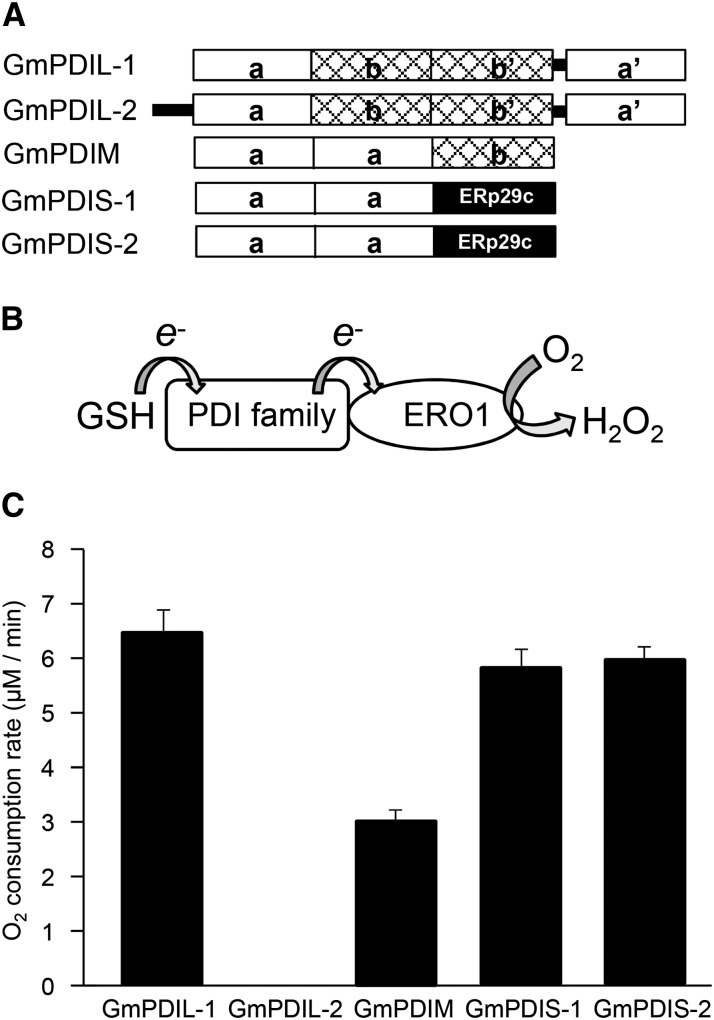
We next determined the ability of recombinant GmERO1a to oxidize five soybean ER oxidoreductases (Fig. 3A). Oxidation of ER oxidoreductases by GmERO1a was monitored by oxygen consumption (Fig. 3B). The reaction was performed in the presence of glutathione (GSH), as a substrate for oxidation by ER oxidoreductases oxidized by GmERO1a. In contrast to human Ero1α and Ero1β, which predominantly oxidize PDI, GmEro1a oxidized GmPDIL-1, GmPDIS-1, GmPDIS-2, and GmPDIM to comparable levels (Fig. 3C and Supplemental Fig. S2). GmPDIL-2 was negligibly oxidized by GmERO1a.
Figure 3.
GmERO1a shows broad specificity for ER oxidoreductases. A, Schematic representation of soybean ER oxidoreductases. Boxes represent domains. B, Schematic representation of the continuous oxidation of PDI family members by Ero1 in the presence of GSH and the reduction of O2. C, Oxygen consumption rate of GmERO1a (1 μm) in the presence of 3 μm each ER oxidoreductase and 10 mm GSH at 25°C. Data are represented as mean ± se of n = 3.
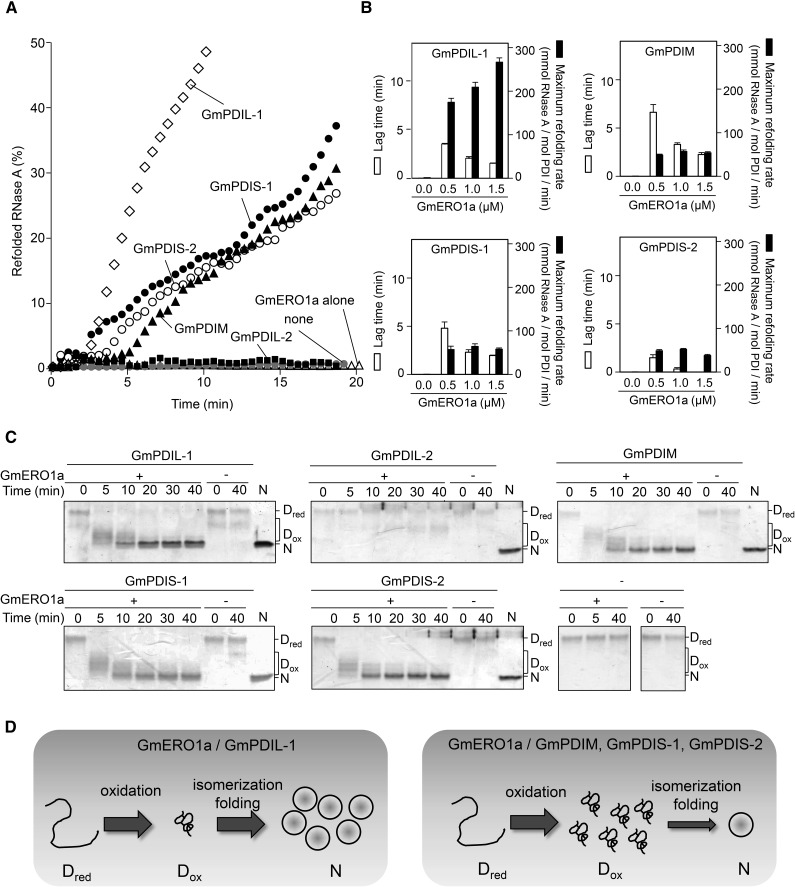
Oxidative refolding by these ER oxidoreductases was determined in the presence of GmERO1a. ER oxidoreductases other than GmPDIL-2 were able to refold reduced and denatured RNase A (Fig. 4A). Lag times prior to the initiation of refolding (Fig. 4B, white bars) and refolding rates (black bars) differed between the ER oxidoreductases. The highest refolding rate was observed in the presence of GmPDIL-1. The rate of refolding by GmPDIL-1 increased with increasing concentrations of GmERO1a. The rates of refolding by GmPDIM, GmPDIS-1, and GmPDIS-2 were slower than that by GmPDIL-1, and underwent very little change at different concentrations of GmERO1a. In contrast to the refolding rates, the lag times decreased with increasing concentrations of GmERO1a. These results suggest that dithiol oxidation is rate limiting in the reaction by GmPDIL-1, whereas disulfide isomerization was rate limiting in the reactions by GmPDIM, GmPDIS-1, and GmPDIS-2.
Figure 4.
Reconstitution of oxidative protein folding with ER oxidoreductases and GmERO1a in vitro. A, GmERO1a (1 μm) without (GmERO1a alone), or with 3 μm of each ER oxidoreductase, was incubated with reduced and denatured RNase A (8 μm), and the recovered RNase A activity was assayed. “None” shows refolding of RNase A alone. B, Lag time (white bars) and maximum refolding rate (black bars) of reduced and denatured RNase A (8 μm) by 3 μm each ER oxidoreductase in the presence of GmERO1a. Data are represented as mean ± se of n = 3. C, Refolding of reduced and denatured RNase A in the absence (−) or presence (+) of GmERO1a and each ER oxidoreductase, quenched with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid, and was analyzed by nonreducing SDS-PAGE. Dred, reduced and denatured RNase A; Dox, denatured RNase A with nonnative disulfides; N, native RNase A. D, Model of refolding of RNase A by ER oxidoreductases in the presence of GmERO1a. Left, GmPDIL-1 transfers nonnative disulfide bonds to Dred. Dox is promptly folded into the native form accompanied by the isomerization of disulfide bonds. Right, GmPDIM, GmPDIS-1, or GmPDIS-2 transfers nonnative disulfide bonds to Dred. Dox accumulates due to rate-limiting isomerization activities of GmPDIM, GmPDIS-1, and GmPDIS-2.
This observation was confirmed by analysis of intermediates during refolding. RNase A molecules during refolding were separated by nonreducing SDS-PAGE after modification of the free thiol groups with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (Fig. 4C). GmPDIL-1 converted most of the reduced and denatured RNase A molecules (Dred) into an intermediate with nonnative disulfide bonds (Dox) within the first 5 min. During the next 15 min, Dox was converted into a native form with four disulfide bonds (N; Fig. 4D, left). GmPDIM, GmPDIS-1, or GmPDIS-2 converted most of the Dred into Dox within the first 5 min, and conversion of Dox into N was slower than by GmPDIL-1 (Fig. 4, C and D, right). Neither Dox nor N was generated by GmPDIL-2 (Fig. 4C).
GmPDIL-2 and GmPDIM Cooperatively Refold Denatured RNase A
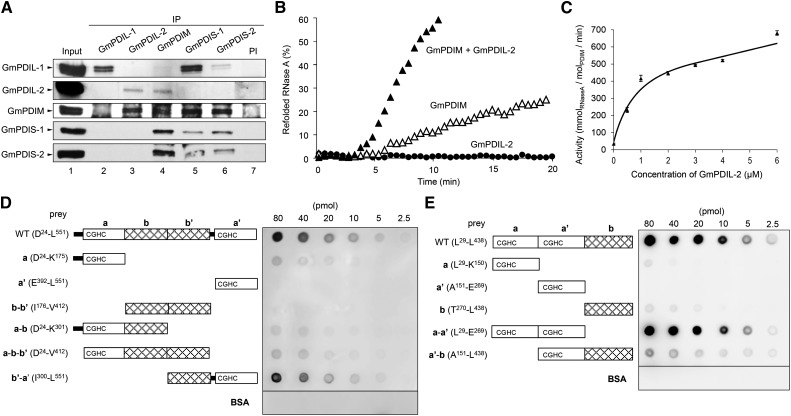
It was previously found that GmPDIS-1, GmPDIM, GmPDIL-1, and GmPDIL-2 associate with nascent proglycinin in the ER, suggesting that these ER oxidoreductases cooperatively fold glycinin in vivo (Wadahama et al., 2007, 2008; Kamauchi et al., 2008). Associations between these ER oxidoreductases in the ER were detected by coimmunoprecipitation. GmPDIM and GmPDIS-1, GmPDIM and GmPDIS-2, GmPDIS-1 and GmPDIS-2, and GmPDIM and GmPDIL-2 were coimmunoprecipitated (Fig. 5A). Therefore, we examined the refolding of RNase A when these ER oxidoreductases were present simultaneously. No combination of GmPDIM, GmPDIS-1, and GmPDIS-2 showed an additive effect on the refolding rate (Supplemental Fig. S3). In contrast, the refolding rate of denatured RNase A when GmPDIM and GmPDIL-2 were present simultaneously was almost five times greater than that for GmPDIM alone, although GmPDIL-2 alone had no refolding activity (Fig. 5B). The refolding rate drastically increased until a 1:1 m ratio of GmPDIL-2 to GmPDIM was obtained (Fig. 5C). The association of GmPDIL-2 and GmPDIM was confirmed by far-western-blot analysis (Fig. 5, D and E).
Figure 5.
GmPDIL-2 and GmPDIM form a complex, and synergistically refold RNase A using disulfide bonds produced by GmERO1a. A, Detection of ER oxidoreductase complexes. IP from the cotyledon extract was carried out with each anti-ER oxidoreductase serum or PI. Immunoprecipitates and the cotyledon extract (lane 1) were analyzed by western blot with each anti-ER oxidoreductase serum. B, Effects of the addition of GmPDIL-2 to GmPDIM on the refolding of RNase A. Reduced and denatured RNase A (8 μm) was incubated with 3 μm ER oxidoreductases in the presence of 1 μm GmERO1a. C, GmPDIL-2 concentration dependence of the refolding activity in the presence of 1 μm GmERO1a and 1 μm GmPDIM. Data are represented as mean ± se of n = 3. D, Far-western blot analysis of the association of the GmPDIL-2 (WT) or GmPDIL-2 domain fragment with GmPDIM. Indicated amounts of GmPDIL-2, each GmPDIL-2 domain fragment, or BSA (prey) were dot-blotted and incubated with GmPDIM. Bound GmPDIM was immunostained. E, For GmPDIM (WT), each GmPDIM domain fragment or BSA (prey) was dot-blotted and incubated with GmPDIL-2. Bound GmPDIL-2 was immunostained. BSA, bovine serum albumin; IP, immunoprecipitation; PI, preimmune serum; WT, wild type.
To identify the relevant domains for the binding of GmPDIM and GmPDIL-2, various domain fragments of GmPDIL-2 (Supplemental Fig. S4) and GmPDIM (Supplemental Fig. S5) were subjected to far-western-blot analysis. Only the b′–a′ fragment of GmPDIL-2 bound to GmPDIM with affinity comparable to full GmPDIL-2 (Fig. 5D). The a–a′ fragment of GmPDIM bound with high affinity to GmPDIL-2 (Fig. 5E). Taken together, the binding sites of GmPDIL-2 and GmPDIM are composed of the b′ and a′ domains of GmPDIL-2 and the a and a′ domains of GmPDIM.
The Active Centers of GmPDIL-2 Are Important to Their Cooperative Activity with GmPDIM
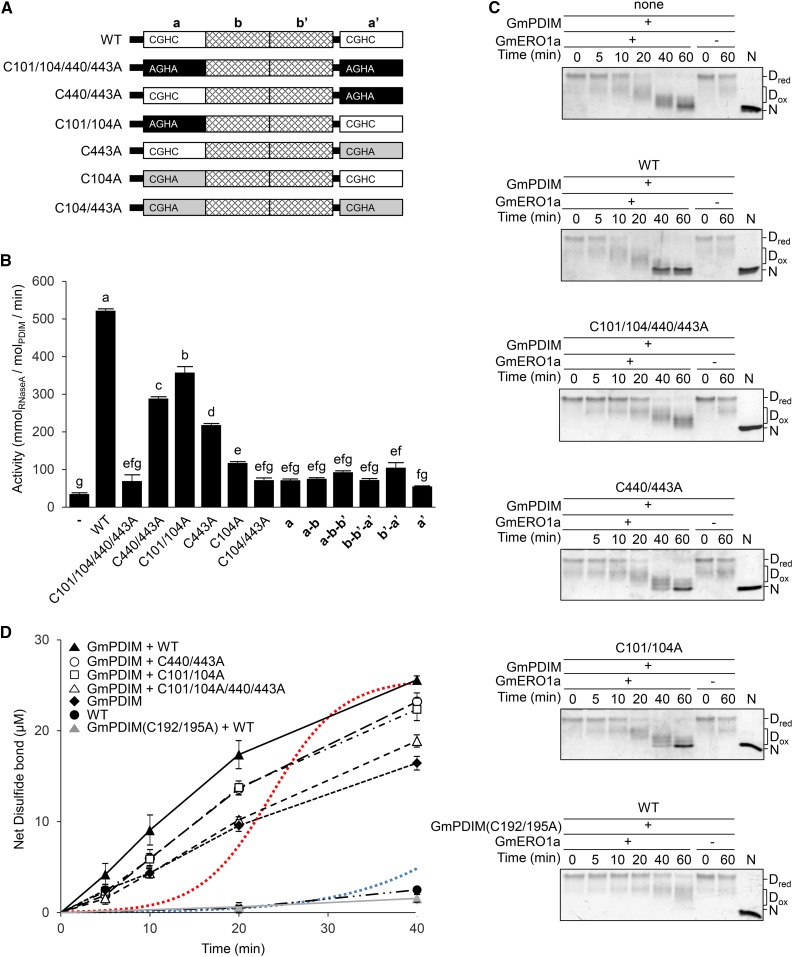
The acceleration of oxidative refolding of RNase A by the cooperation of GmPDIL-2 and GmPDIM depends on the catalytic Cys residues in the a and a′ domains of GmPDIL-2. When four catalytic Cys residues were replaced with Ala [GmPDIL-2(C101/104/440/443A)], the acceleration of oxidative refolding was eliminated (Fig. 6, A and B). Replacing two catalytic Cys residues in either the a or a′ domain with Ala [GmPDIL-2(C440/443A) or GmPDIL-2(C101/104A)] caused a decrease in oxidative refolding activity to 55% or 69% of wild-type levels, respectively. In mutants in which the C-terminal catalytic Cys residue in the a′ and/or a domain was replaced by Ala [GmPDIL-2(C104/443A), GmPDIL-2(C443A), and GmPDIL-2(C104A)], oxidative refolding was slower than that of GmPDIL-2(C440/443A) or GmPDIL-2(C101/104A). The effects of a, a′, a–b, a–b–b′, b–b′–a′, and b′–a′ fragments on the acceleration of oxidative refolding were all very low (Fig. 6B). One reason for the small effects of the domain fragments on the acceleration of oxidative refolding, other than the a–b–b′ fragment, may be their low oxidative refolding activities and disulfide reductase activities seen in the presence of glutathione redox buffer without GmERO1a (Supplemental Fig. S4E).
Figure 6.
Active centers of GmPDIL-2 play vital roles in the acceleration of both refolding and disulfide bond formation. A, Schematic representation of wild-type GmPDIL-2 (WT) and its mutants. “CGHC”, “AGHA”, and “CGHA” represent the amino acid sequences of the active sites of the a and a′ domains. Numbers in the name of the mutants represent Cys residues mutated to Ala. B, Acceleration of refolding by GmPDIL-2 mutants and GmPDIL-2 domain fragments in the GmERO1a/GmPDIM system. Reduced and denatured RNase A (8 μm) was incubated with 1 μm GmPDIM and 4 μm wild-type GmPDIL-2 (WT), its mutants, or domain fragments in the presence of 1 μm GmERO1a. Data are represented as mean ± se of n = 3–5. Bars with the same letters are not significantly different at P < 0.05 by Tukey-Kramer test. C, Disulfide bond formation in reduced and denatured RNase A (8 μm) during cooperative refolding by GmPDIM or GmPDIM(C192/195A) (1 μm) and GmPDIL-2 or GmPDIL-2 mutants (3 μm) in the presence of GmERO1a (1 μm) was analyzed by nonreducing SDS-PAGE. Dred, reduced and denatured RNase A; Dox, denatured RNase A with nonnative disulfides; N, native RNase A. D, Disulfide bond formation in RNase A over time. Reduced and denatured RNase A was incubated with indicated combinations of proteins as in C. The amounts of free thiol groups in the reaction mixtures were measured and net disulfide bonds were calculated by subtracting the free disulfide bonds from total Cys residues. Red and blue dotted lines show the amounts of native disulfide bonds formed in RNase A, which were calculated from the amounts of refolded RNase A. Data are represented as mean ± se of n = 3. WT, wild type.
Oxidative refolding by ER oxidoreductases is achieved through the formation of nonnative disulfide bonds in substrate proteins and their subsequent rearrangement to native bonds as seen in Fig. 4C. Therefore, it was predicted that GmPDIL-2 would stimulate the formation and/or rearrangement of substrate disulfide bonds. The addition of GmPDIL-2 to GmPDIM induced more rapid conversion of Dred into Dox and N than did GmPDIM alone (Fig. 6C). Mutations in the active centers of GmPDIL-2 caused retardation in the conversion into both Dox and N. The rate of disulfide bond formation during the first 10 min of the addition of GmPDIL-2 to GmPDIM was twice that of GmPDIM alone (Fig. 6D), suggesting that GmPDIL-2 stimulates the oxidation of GmPDIM by GmERO1. Mutant GmPDIL-2(C101/104/440/443A) was devoid of the acceleration effect on refolding. Since GmPDIL-2(C440/443A) and GmPDIL-2(C101/104A) showed about one-half the acceleration effect of the wild type, the active centers of the a and a′ domain appeared to act independently. The amount of native disulfide bonds formed during the reaction was calculated from the amount of refolded RNase A. The formation rate of native disulfide bonds during the cooperation of GmPDIL-2 and GmPDIM (Fig. 6D, red curve) was 4-fold higher than GmPDIM alone (Fig. 6D, blue curve), suggesting that GmPDIL-2 accelerates both disulfide bond formation and the rearrangement accompanied with folding.
GmPDIL-2 Accelerates the Oxidation of GmPDIM by GmERO1a
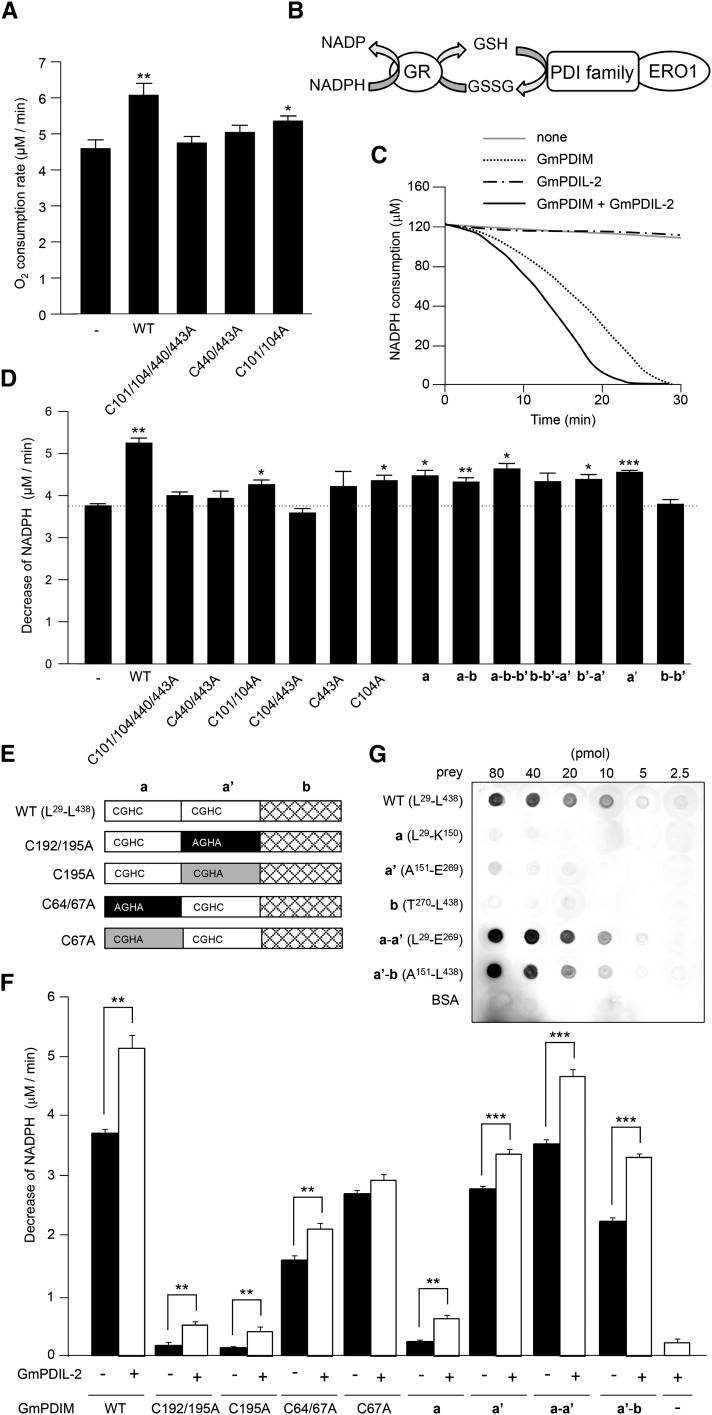
From the results shown in Fig. 6, it was suggested that GmPDIL-2 accelerates the oxidation of GmPDIM by GmERO1a. To confirm this possibility, oxidation of GmPDIM by GmERO1a was measured in the presence of GmPDIL-2. In this experiment, GSH was used as a substrate. Addition of GmPDIL-2 caused an increase in O2 consumption rate to 130% of that without GmPDIL-2 (Fig. 7A). The acceleration effect was lost upon mutation of the active center of the a′; GmPDIL-2(C440/443A) and GmPDIL-2(C101/104/440/443A) showed no acceleration effects. The acceleration effect of GmPDIL-2(C101/104A) was lower than that of wild-type GmPDIL-2. Similar effects by GmPDIL-2 and its active-center mutations were observed in experiments coupled with glutathione disulfide reductase and NADPH (Fig. 7, B–D). Fragments a and a′ showed an acceleration effect of one-half that of the wild type. Fragment b–b′ showed no effect. The attachment of the b, b′, or b–b′ domain to the a or a′ domain had no positive effect.
Figure 7.
GmPDIL-2 accelerates the oxidation of GmPDIM by GmERO1a. A, Oxygen consumption was monitored during incubation of 2 μm GmERO1a and 3 μm GmPDIM without or with 3 μm GmPDIL-2 or its mutants in the presence of 10 mm GSH. Data are represented as mean ± se of n = 6. *, P < 0.05; **, P < 0.01 compared with the reaction without GmPDIL-2 (Welch’s t test). B, Schematic representation of the coupling reaction of oxidation by ERO1 and the reduction of GSSG by GR in the presence of GSH and NADPH. C, NADPH consumption over time in the presence of 1 μm GmERO1a and 3 μm GmPDIM, 3 μm GmPDIL-2, or 3 μm GmPDIM and 3 μm GmPDIL-2 in the presence of 3 mm GSH, 120 μm NADPH, and 1 U/mL GR. D, Decrease in NADPH in the presence of indicated combinations of proteins as in C. Data are represented as mean ± se of n = 10 (without GmPDIL-2); n = 3–4 (with wild-type GmPDIL-2 and its mutants). *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with the reaction without GmPDIL-2 (−; Welch’s t test). E, Schematic representation of wild-type GmPDIM (WT) and its mutants. Numbers in the name of mutants represent Cys residues mutated to Ala. F, Oxidation of GmPDIM (WT), its mutants, or its domain fragments by GmERO1a without or with GmPDIL-2 was measured as in C. Data are represented as mean ± se of n = 3–6. **, P < 0.01; ***, P < 0.001 (Welch’s t test). G, Far-western-blot analysis of the association of GmPDIM or its domain fragment with GmERO1a. Indicated amounts of GmPDIM (WT), each domain fragment or BSA (prey) was dot-blotted and incubated with GmERO1a. Bound GmERO1a was immunostained. BSA, bovine serum albumin; GR, glutathione-disulfide reductase; WT, wild type.
GmERO1a Preferentially Oxidizes the Active Center in the a′ Domain of GmPDIM; GmPDIM Oxidizes Both Active Centers in GmPDIL-2
From the results shown in Fig. 7, B–D, we envisioned a pathway by which GmPDIM, oxidized by GmERO1, transfers disulfide bonds to the active centers of GmPDIL-2. To test this hypothesis, we determined which active center of GmPDIM is oxidized by GmERO1a using active-center mutants of GmPDIM (Fig. 7, E and F, black bars). Both GmPDIM(C192/195A) and GmPDIM(C195A) were negligibly oxidized by GmERO1a. Therefore, no disulfide bond formation in reduced and denatured RNase A by GmPDIM(C192/195A) and GmPDIL-2 was detected (Fig. 6, C and D). In contrast to GmPDIM(C192/195A) and GmPDIM(C195A), GmPDIM(C64/67A) and GmPDIM(C67A) were oxidized by GmERO1a, although at a slower rate than wild-type GmPDIM. These findings suggest that GmERO1a specifically oxidizes the active center in the a′ domain of GmPDIM. This finding was corroborated by the fact that the a′ fragment, but not the a fragment, was oxidized at 74% of the oxidation rate of wild-type GmPDIM by GmERO1a. The oxidation rate of the a′–b fragment was slightly slower than that of the a′ fragment, and the a–a′ fragment was oxidized at almost same rate as wild-type GmPDIM, suggesting that the b domain does not contribute to oxidation by GmERO1a. Oxidation of every GmPDIM mutant by GmERO1 was accelerated by GmPDIL-2 (Fig. 7F, white bar). Association of GmPDIM and GmERO1a was detected by far-western-blot analysis (Fig. 7G). The a, a′, or b fragment of GmPDIM hardly associated with GmERO1a. However, either the a–a′ or a′–b fragment associated with GmERO1a at an affinity comparable to wild-type GmPDIM.
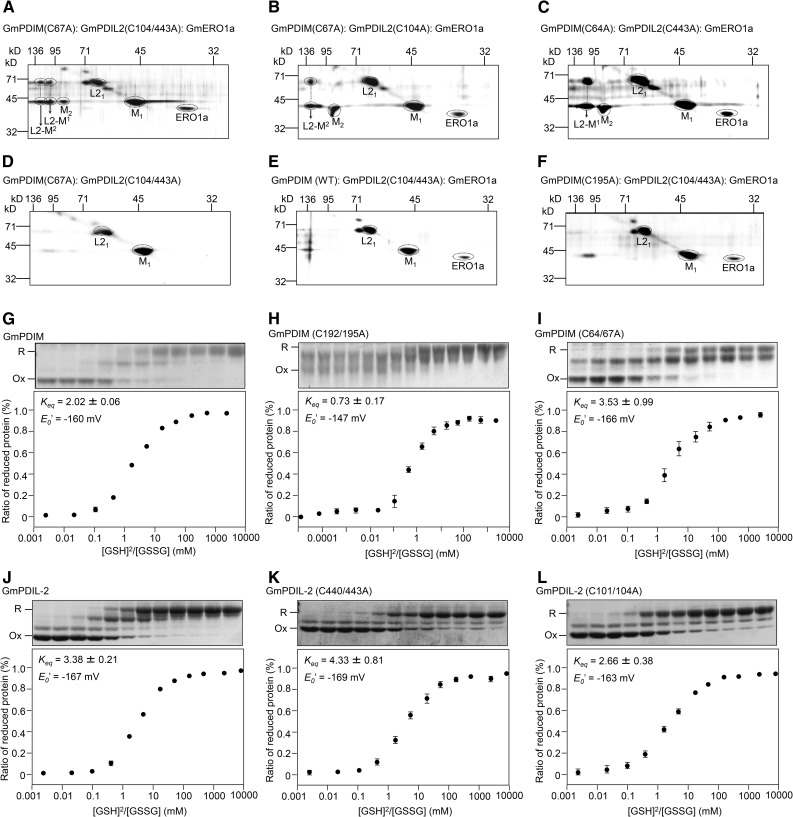
To confirm that the disulfide bond in the active center of the a′ domain of GmPDIM formed by GmERO1a is transferred to the active center of GmPDIL-2 via the active center of the a domain of GmPDIM, we analyzed mixed disulfide complexes of the active-center mutants of GmPDIM and GmPDIL-2. When GmPDIM(C67A) and GmERO1a were incubated with GmPDIL-2(C104/443A), two types of mixed disulfide complexes (L2-M1 and L2-M2) of GmPDIM(C67A) and GmPDIL-2(C104/443A), and the GmPDIM(C67A) dimer were detected (Fig. 8A). When GmPDIM(C67A) and GmERO1a were incubated with GmPDIL-2(C104A) or GmPDIL-2(C443A), only one mixed disulfide complex, L2-M2 (Fig. 8B) or L2-M1, (Fig. 8C) was formed. Neither the mixed disulfide complex nor the GmPDIM(C67A) dimer was generated in the absence of GmERO1a (Fig. 8D). When wild-type GmPDIM and GmERO1a were incubated with GmPDIL-2(C104/443A), only a few mixed disulfide complexes were detected (Fig. 8E). In addition, no mixed disulfide complexes were formed from GmPDIM(C195A) (Fig. 8F). From these results, L2-M1 and L2-M2 were deduced to be mixed disulfide complexes, which were linked by disulfide bonds between Cys-67 of GmPDIM and either Cys-443 or Cys-104 of GmPDIL-2.
Figure 8.
GmPDIM oxidizes the active centers of GmPDIL-2. A, The mixed disulfide of the GmPDIM and GmPDIL-2 mutants. GmPDIM(C67A) (6 μm), 3 μm GmPDIL-2(C104/443A), and 1 μm GmERO1a were incubated at 25°C for 60 min and separated by two-dimensional electrophoresis. M1, GmPDIM monomer; L21 GmPDIL-2 monomer; M2, GmPDIM dimer; L2M1 and L2M2, mixed disulfide complex of GmPDIL-2 mutants and GmPDIM(C64A). B, C, D, E, F, Indicated combinations of proteins were incubated and analyzed as described in A. G, H, I , J, K, L, Assay of redox equilibrium constants of GmPDIM, GmPDIL-2, and their mutants. Coomassie Brilliant Blue stainings of the SDS-polyacrylamide gels are shown above the redox graphs. Data are represented as mean ± se of n = 3.
The results shown in Fig. 7F suggest that a disulfide bond introduced into the active center of the a′ domain of GmPDIM is transferred to the active center of the a domain of GmPDIM. Then the oxidized GmPDIM can oxidize the active center of either the a or a′ domain of GmPDIL-2 (Fig. 8, A–F). The direction of a redox reaction between functional groups depends on their redox potentials, i.e. a functional group with higher redox potential oxidizes a functional group with lower redox potential. To obtain the redox potentials of GmPDIM and GmPDIL-2, their redox equilibrium constants were determined. Redox equilibrium constants of GmPDIM and GmPDIL-2 were 2.0 mm and 3.4 mm, respectively (Fig. 8, G and J). Since these are the mean values of two active sites, redox equilibrium constants of the redox-inactive mutants of GmPDIM and GmPDIL-2 were determined (Fig. 8, H, I, K, and L). GmPDIM(C192/195A) showed the lowest redox equilibrium constant (Keq = 0.7) among the mutant proteins. The reduction potential E0′ values calculated from each redox equilibrium constant of GmPDIM(C192/195A), GmPDIM(C64/67A), GmPDIL-2(C440/443A), and GmPDIL-2(C101/104A) were −147 mV, −166 mV, −169 mV, and −163 mV, respectively. These values suggest that the a domain of GmPDIM has a tendency to oxidize both the active centers of the a and a′ domains of GmPDIL-2.
DISCUSSION
In this study, we found that the plant Ero1 ortholog has a broad substrate specificity. GmERO1a oxidizes, not only the conserved typical ER oxidoreductase GmPDIL-1, but also GmPDIM, GmPDIS-1, and GmPDIS-2. Human Ero1α and Ero1β preferentially oxidize PDI. Therefore, another pathway that oxidizes other ER oxidoreductases likely exists in human. Peroxiredoxin-4 primarily oxidizes two PDI family members, ERp46 and P5, using H2O2 generated during the oxidation of PDI by human Ero1α (Sato et al., 2013). Conversely, in the plant ER, a peroxiredoxin-4 ortholog has not been found. In rice, a lack of OsEro1 causes the aggregation of glutelin because of deficient disulfide bond formation (Onda et al., 2009), suggesting that disulfide bond formation depends on the Ero1 system for protein folding in the plant ER. Therefore, plant Ero1 may induce efficient oxidative folding in the ER by supplying disulfide bonds to multiple ER oxidoreductases.
Among the soybean ER oxidoreductases examined in this study, only GmPDIL-2 was hardly oxidized by GmERO1a. The Keq values of the active centers in the a and a′ domains of GmPDIL-2 were 4.33 and 2.66, respectively. Since these values are similar to that of the a′ domain of GmPDIM (Keq = 3.53), which was preferentially oxidized by GmERO1a, the lack of oxidation by GmERO1 is not due to the reduction potential of the active centers of GmPDIL-2. One reason may be the low affinity between GmERO1a and GmPDIL-2 (data not shown).
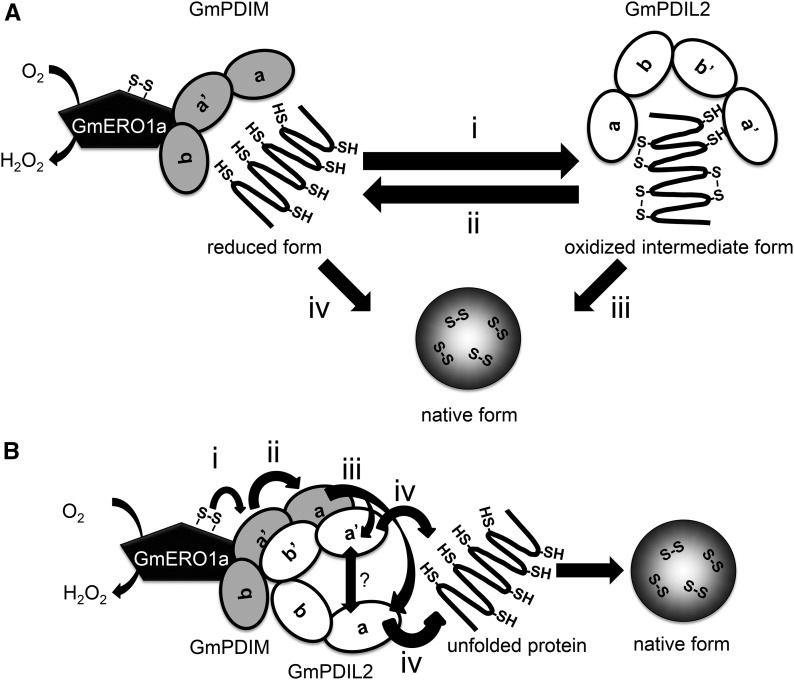
We found GmPDIL-2 associate with GmPDIM in vivo. GmPDIL-2 can associate with GmPDIM to cooperatively fold denatured RNase A in vitro. Oxidative folding proceeds via two steps: (Step 1) introduction of transient, nonnative disulfide bonds and (Step 2) their isomerization into native disulfide bonds. The oxidative folding activity of GmPDIM was lower than that of GmPDIL-2 in the presence of glutathione buffer without GmERO1a (Supplemental Fig. S4E and Supplemental Fig. S5D). Therefore, there may be a pathway in which GmPDIM forms nonnative disulfide bonds in the substrate and then GmPDIL-2 folds and rearranges these disulfide bonds into native bonds (Fig. 9A).
Figure 9.
Models of cooperation of GmPDIM and GmPDIL-2. A, Role-sharing model of GmPDIM and GmPDIL-2. GmPDIM oxidized by GmERO1a introduces transient disulfide bonds in an unfolded substrate protein (i). Transient disulfide bonds in the substrate protein are reduced by the reduced form of GmPDIL-2 (ii). Reduced thiols are rearranged into native disulfide bonds, mainly by GmPDIL-2 (iii), but partially by GmPDIM (iv). B, model of oxidative relays from GmERO1a to GmPDIM, GmPDIL-2, and substrate. GmERO1a oxidizes the active center in the a′ domain of GmPDIM (i). The formed disulfide bond is transferred sequentially from the a′ domain of GmPDIM to the a domain of GmPDIM (ii), either the a or a′ domain of GmPDIL-2 (iii), and substrate (iv). Transferring of a disulfide bond between the a and a′ domains of GmPDIL-2 is unclear. Both a and a′ domains of GmPDIM and both b′ and a′ domains of GmPDIL-2 are essential for the association of GmPDIM and GmPDIL-2.
In addition to synergistic effects on folding, the addition of GmPDIL-2 to GmPDIM accelerated the formation of disulfide bonds in RNase A. GmPDIL-2 may increase the net substrates for GmPDIM by reducing nonnative disulfide bonds and stimulating folding. However, even when GSH was used as a substrate, GmPDIL-2 increased the rate of oxidation of GmPDIM by GmERO1, suggesting that increasing the rate of oxidation does not depend on substrate folding only. We found that the active center of the a domain of GmPDIM oxidized the active center of either the a or a′ domain of GmPDIL-2 (Fig. 8, A–F). The increase in the oxidation rate of GmPDIM by GmERO1a may be explained by the transfer of disulfide bonds from GmPDIM to the associated GmPDIL-2, in addition to substrate proteins. The absolute concentration of glutathione in the ER is unknown; however, a consensus exists that the total glutathione concentration in the ER is around 9 to 10 mm and that the ratio of [GSH]2/[GSSG] in the ER is around 3:1 (Hatahet and Ruddock, 2009). From these values, the reduction potential for ER glutathione is estimated as −191 mV. Under these redox conditions in the ER, the active centers of GmPDIM and GmPDIL-2 would be largely reduced. This means that GmPDIM, once oxidized by GmERO1a, would oxidize GmPDIL-2.
Using the active-center mutants, a model of the pathway of disulfide bond transfer from GmPDIM to GmPDIL-2 can be constructed (Fig. 9B). First, GmERO1a entirely oxidizes the a′ domain of GmPDIM (1). The preference of GmERO1a for the a′ domain of GmPDIM is probably due to the reduction potential and the affinity between GmERO1a and the a′ domain of GmPDIM. Since the reduction potential of the a′ domain was lower than that of the a domain, the a′ domain will be more easily oxidized by GmERO1a. Then, the oxidized GmPDIM transfers a disulfide bond to GSH or GmPDIL-2 mainly from the a domain (2), because the oxidation of GmPDIM(C64/67A) and GmPDIM(C67A) was significantly lower than that of wild-type GmPDIM (Fig. 7F). This finding suggests that the disulfide bond transferred to the a′ domain of GmPDIM from GmERO1a is intramolecularly transferred to the a domain. The oxidation of the a domain by the a′ domain is thermodynamically contradictory, as the apparent redox equilibrium constant of the a domain of GmPDIM(C192/195A) (0.73) was lower than that of the a′ domain of GmPDIM(C64/67A) (3.53). If the a′ domain of GmPDIM is oxidized, the redox equilibrium constant of the a and/or a′ domain may be altered by conformational changes such that the a domain becomes susceptible to oxidation by the a′ domain. In human PDI, the a′ domain was first oxidized by Ero1-α, which then oxidizes the a domain, and the reduced a′ domain is oxidized by Ero1-α. During the sequential transfer of disulfide bonds, PDI dynamically changes its structure from a closed form to an open form (Nakasako et al., 2010; Serve et al., 2010; Wang et al., 2013). Subsequently, disulfide bonds are transferred from the GmPDIM to either the a or a′ domain of GmPDIL-2 (3) and from the oxidized GmPDIL-2 to a protein substrate (4).
Human PDI has been shown to oxidize other ER oxidoreductases, such as ERp47, ERp57, and P5 (Araki et al., 2013, Oka et al., 2015). Therefore, Ero1-α and PDI are thought to constitute a regulatory hub for the regulation of the redox states of ER oxidoreductases. In contrast, multiple soybean ER oxidoreductases are able to be oxidized by GmERO1a. Therefore, in plants, it is thought that Ero1 and ER oxidoreductases constitute a robust network for disulfide bond transfer and the regulation of redox homeostasis in the ER. The relay of oxidizing equivalents from one ER oxidoreductase to another may play an essential role for cooperative oxidative folding by multiple ER oxidoreductases.
MATERIALS AND METHODS
Plants
Soybean (Glycine max L. Merrill cv Jack) seeds were planted in 5-l pots and grown in a controlled environmental chamber at 25°C under 16-h day/8-h night cycles. All samples taken were immediately frozen and stored in liquid nitrogen until use.
cDNA Cloning of GmERO1a and GmERO1b
The cDNAs transcribed from GmERO1a and GmERO1b were amplified from total RNA extracted from 100 mg immature soybean cotyledon by real-time PCR using forward primer 5′-CCTCAGTGTTCTTCAGCGATCTTCTTCTGGGTCTG-3′ and reverse primer 5′-CCAAACCACGACAGTGGTTTTCCTATGTCTAGTC-3′, and forward primer 5′-ATGGTGAAAGCGGAGATTGAGAAAAAGGGTTGCAGC-3′ and reverse primer 5′-CCACGACGGTGGTTTTCCTATGTCTAGTCTCTCTAG-3′, respectively. The amplified DNA fragments were subcloned into the pUC19 vector. The inserts in the plasmid vectors were sequenced.
Preparation of Recombinant GmERO1a
An expression plasmid encoding glutathione-S-transferase (GST-fused GmERO1a E70-G422 was constructed as follows. The DNA fragment was amplified from GmERO1a cDNA by PCR using primers 5′-GCGATGGATCCTATGAAACTGTGGATCGTCTTAATG-3′ and 5′-GCGATGTCGACTATCCTCCTTCCATGATTCTTTCAGC-3′. The amplified DNA fragment was subcloned into the pGEX6p-2 vector (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The recombinant protein has GST linked to the amino terminus. Rosseta-gami cells (Takara Bio, Kusatsu, Shiga, Japan) were transformed with the GST-fusion vector described above. Expression of recombinant GST-GmERO1a was induced in LB containing 0.5 mm isopropyl thiogalactoside, 20 μm FAD, 100 μg/mL ampicillin, 15 μg/mL kanamycin, and 12.5 μg/mL tetracycline at 15°C for 120 h. The cells were collected by centrifugation and disrupted by sonication in phosphate-buffered saline. Recombinant GST-GmERO1a was adsorbed to a glutathione Sepharose 4B column and digested by PreScission protease (GE Healthcare). The eluted recombinant GmERO1a was purified by gel filtration chromatography on a TSK gel G3000SW column (Tosoh, Tokyo, Japan) equilibrated with 20 mm Tris-HCl buffer (pH 7.4) containing 150 mm NaCl and 10% glycerol as described previously (Wadahama et al., 2007). The concentration of purified recombinant GmERO1a was determined by amino acid analysis with nor-Leu as an internal standard.
Western-Blot Analysis
Roots, leaves, and stems were collected from 10-d-old seedlings. Seeds were collected from plants, and cotyledons were isolated. Tissues were frozen in liquid nitrogen and then ground into a fine powder with a micropestle SK-100 (Tokken, Chiba, Japan). Proteins were extracted by boiling for 5 min in SDS-PAGE buffer (Laemmli, 1970) containing a 1% cocktail of protease inhibitors (Sigma-Aldrich, St. Louis, MO). To cleave N-glycans, proteins were extracted from the cotyledons in 0.1% SDS/50 mm phosphate buffer (pH 5.5). Protein concentration in the sample was measured with a RC DC protein assay kit (Bio-Rad Laboratories, Hercules, CA). Proteins (0.4 mg) were treated with 10 mu endoglycosidase F or endoglycosidase H (Sigma-Aldrich) at 37°C for 16 h. To separate the supernatant and membrane fractions, the frozen cotyledons from three 100-mg seeds in liquid nitrogen were crushed with a micropestle SK-100 and sonicated with a Sonifier II (Branson Ultrasonics, Danbury, CT) in 200 mm Tris-HCl, pH 7.8, five times for 30 s on ice. The homogenate was placed into a cell strainer (BD Biosciences, San Jose, CA) and centrifuged at 824g for 40 min at 4°C. The filtrated suspension was divided two fractions, diluted with 200 mm Tris-HCl, pH 7.8, with or without 1% Triton X-100, and centrifuged at 100,000g for 1 h at 4°C. Proteins were subjected to SDS-PAGE (Laemmli, 1970) and blotted onto a polyvinylidene difluoride membrane. Blots were immunostained first with antiserum and then with horseradish peroxidase-conjugated IgG antiserum (Promega, Fitchburg, WI) as the secondary antibody. Anti-GmERO1 serum and anti-soybean calnexin (GmCNX) serum were prepared using recombinant GmERO1a and recombinant C-terminal-truncated GmCNX (Supplemental Fig. S6), respectively, by Anti-GmPDIL-1 serum (Operon Biotechnologies, Tokyo, Japan) that was prepared previously (Kamauchi et al., 2008). Blots were developed with the Western Lightning Chemiluminescence Reagent (Perkin Elmer Life Sciences, Waltham, MA).
Confocal Microscopy
Cotyledons from developing soybean seeds (195 mg) were cut into 3 × 3 × 1 mm cubes. The pieces of tissue were fixed with 4% formaldehyde for 2 h at room temperature. The fixed cotyledon pieces were dehydrated with a series of ethanol dilutions, embedded in Historesin (Leica Microsystems, Wetzlar, Germany), and sliced into sections. The sections were stained with anti-GmPDIS-1 rabbit serum (Wadahama et al., 2007) and then biotin-anti-rabbit IgG goat serum (Cortex Biochem, San Leandro, CA), followed by incubation with Cy5-streptavidin (GE Healthcare). For detection of GmERO1, specimens were stained with guinea pig antiserum against recombinant GmERO1a, followed by staining with Cy3-conjugated anti-guinea pig IgG goat serum (Chemicon International, Temecula, CA). The specimens were examined on a FV1000-d laser scanning confocal imaging system (Olympus, Tokyo, Japan).
Measurement of mRNA
Total RNA was isolated from young leaves treated with or without 5 μg/mL tunicamycin at 25°C for 2 or 5 h using an RNeasy Plant Mini kit (Qiagen, Venlo, The Netherlands). Quantification of mRNA was performed by real-time PCR with a Thermal Cycler Dice Real Time System with SYBR Premix Ex Taq (Takara Bio). Forward primers 5′-CATGCTCTCTTCTGAGTATTTGTCA-3′ and 5′-TCATCCAAGAAATGGGACCT-3′ and reverse primers 5′-CAGTGGTTTTGGTATGTCTAGTCTCTC-3′ and 5′-CGACGGTGGTTTTCCTATGT-3′ were used for detection of GmERO1a mRNA and GmERO1b, respectively.
Far-UV Circular Dichroism Analysis
Circular dichroism spectra of recombinant proteins in 20 mm Tris-HCl buffer (pH 7.4) containing 150 mm NaCl and 10% glycerol were obtained using a J-720 spectropolarimeter (JASCO, Tokyo, Japan) in a 1-mm path-length cell with a scan speed of 20 nm/min at 14°C.
Endoplasmic Reticulum Oxidoreductase Oxidation Assays
Oxygen consumption was measured using a Clark-type oxygen electrode system (OXYT-1; Hansatech Instruments, King’s Lynn, Norfolk, UK). All experiments were performed at 25°C in 100 mm [4-(2-hydroxyethyl)-1-piperazinyl]ethanesulfonic acid (HEPES; pH 7.5), 2 mm CaCl2, and 150 mm NaCl. Catalytic oxygen consumption was initiated by the addition of GmERO1a in a reaction mixture containing recombinant ER oxidoreductases or its variants, and 10 mm GSH.
The activity was measured using a coupled assay following the decrease in A340 due to the consumption of NADPH by glutathione reductase (Sigma-Aldrich) with the reaction being started by the addition of GmERO1a to initiate the reaction (Nguyen et al., 2011; Sato et al., 2013). A molar extinction coefficient of 6200 m−1 cm−1 for NADPH was used for calculations. All experiments were performed in 100 mm HEPES (pH 7.5) containing 150 mm NaCl and 2 mm CaCl2.
RNase A Refolding Assays
Thiol oxidative refolding activity was assayed as previously described by measuring RNase activity following regeneration of the active form of the enzyme from its reduced and denatured form in the presence of the recombinant proteins (Creighton, 1977; Lyles and Gilbert, 1991). Reduced and denatured RNase A was prepared as described previously (Creighton, 1977). Each reaction mixture contained 100 mm HEPES buffer (pH 7.5), 150 mm NaCl, 2 mm CaCl2, 0.5 mm glutathione disulfide (GSSG), 2 mm GSH, reduced RNase A, and recombinant oxidoreductases and their variants, and were incubated at 25°C. The formation of active RNase A was measured spectrophotometrically by monitoring the hydrolysis of the RNase A substrate cCMP at 284 nm. The isomerase activities of oxidoreductases were calculated as described previously (Kulp et al., 2006). Briefly, isomerase activity was determined from the linear increase in the amount of enzymatically active RNase A with time after a lag. Oxidation of free thiol residues to disulfides on reduced RNase A were also determined by measuring the amounts of free thiol groups in the reaction mixtures (Ellman, 1959).
Gel-Based RNase A Refolding Experiments
RNase A oxidation analyses were performed by the addition of 1 μm GmERO1a in 100 mm HEPES buffer (pH 7.5), 150 mm NaCl, 2 mm CaCl2, and 3 μm recombinant ER oxidoreductases and their variants containing 8 μm denatured and reduced RNase A. At the indicated time points, free thiols were blocked by the addition of SDS buffer containing 8 mm 4-acetamido-4-maleimidylstilbene-2,2 disulfonic acid and separated on a 15% polyacrylamide gel by SDS-PAGE without reducing reagent. Proteins were detected by Coomassie Brilliant Blue staining after electrophoresis.
Immunoprecipitation Experiments
Cotyledons (each 100 mg) were homogenized with a Dounce homogenizer at 4°C in 20 mm HEPES buffer (pH 7.2) containing 150 mm NaCl, 1% digitonin, and 1% cocktail of protease inhibitors (Sigma-Aldrich). The homogenate was placed on ice for 1 h and centrifuged for 30 min at 10,000g at 4°C. Immunoprecipitation was performed at 4°C for 1 h with preimmune serum or anti-GmPDIL-1, anti-GmPDIL-2, anti-GmPDIM, anti-GmPDIS-1, or anti-GmPDIS-2 serum. The immunoprecipitate was collected with protein A-conjugated Sepharose beads (Sigma-Aldrich), washed with 20 mm HEPES buffer (pH 7.2) containing 150 mm NaCl, and subjected to western-blot analysis using a specific antiserum against PDI family proteins as primary antibodies and peroxidase-conjugated ImmunoPure Recombinant Protein A (Pierce Biotechnology, Rockford, IL).
Dot Far-Western-Blot Analysis
Purified recombinant ER oxidoreductases and their variants as prey proteins were spotted onto a nitrocellulose membrane (GenScript, Piscataway, NJ) in a volume of 4 μL. The membrane was dried, rinsed twice, and blocked with 20 mm Tris-HCl (pH 8) containing 150 mm NaCl, 0.05% Tween 20, and 5% nonfat dry milk (blocking solution) at 4°C for 16 h. After blocking, the membrane was incubated in 0.2 μm bait protein in blocking solution without nonfat dry milk (TBS plus Tween 20) for 3 h at 4°C. The membrane was incubated with antibait antiserum and further with horseradish peroxidase-conjugated IgG antiserum (Promega) as the secondary antibody diluted with blocking solution. Blots were washed four times for 20 min with TBS plus Tween 20 and developed using the Western Lightning Chemiluminescence Reagent (Perkin Elmer Life Sciences).
Assay of Mixed Disulfide Complexes
A 4-μL aliquot of purified GmPDIL-2 or GmPDIL-2 variants, and GmPDIM or GmPDIM variants, was incubated at 25°C for 60 min and quenched with N-ethyl maleimide. Proteins were subjected to 7.5% SDS-PAGE under nonreducing conditions. Lanes cut from the first SDS-polyacrylamide gel were incubated in SDS-sample buffer with 5% 2-mercaptoethanol for 120 min at 37°C. The gel slices were then subjected to 12.5% SDS-PAGE under reducing conditions. The separated proteins were stained with a Silver Stain II kit (Wako Pure Chemical Industries, Madrid, Spain).
Measurement of Redox Equilibrium Constants of GmPDIM and GmPDIL-2
GmPDIM, GmPDIL-2, and their variants (1 μm) were incubated with 0.1 mm GSSG and 0.015–28 mm GSH at 25°C for 1 h in 0.1 m sodium phosphate buffer, pH 7.0, containing 1 mm EDTA and 150 mm NaCl. After incubation under N2 at 25°C for 1 h, further thiol-disulfide exchange was prevented by the addition of 10% trichloroacetic acid. The aggregated proteins were precipitated by centrifugation and washed with 100% acetone. The protein pellet was solubilized and incubated in 0.1 m sodium phosphate buffer, pH 7.0, containing 2% SDS and 3 mm methoxypolyethylene glycol-maleimide (mPEG5000-mal; Fluka/Sigma-Aldrich) at 25°C for 30 min. Proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Values for the reduced fraction, oxidized fraction, and intermediate fraction (in the case of wild-type GmPDIM and GmPDIL-2, which have two active sites) were quantified using ImageJ software (National Institutes of Health, Bethesda, MD). The values for the completely oxidized or reduced states were regarded as 0 or 100, respectively, and all intermediate states were recalibrated. In the case of wild-type GmPDIM and GmPDIL-2, the intensity of the band in which one of the two active centers was oxidized was calibrated as one-half of the reduced form. The sum of the completely reduced form and one-half of the half-reduced form was plotted. The Keq was calculated by fitting the recalibrated fraction of the apparent reduced form to the following equation: R = ([GSH]2/[GSSG])/{Keq + ([GSH]2/[GSSG])}, in which R is the relative ratio of the reduced forms. The equilibrium redox potential of proteins was calculated using the Nernst equation [E′0 = E′0(GSH/GSSG) – (RT/nF) * ln Keq] using the glutathione standard potential E′0(GSH/GSSG) of −0.240 V at pH 7.0 and 25°C.
Statistical Analysis
All data are shown as the mean ± se from at least three replicates. Welch’s t tests (two tailed, unpaired) or a Tukey-Kramer test were used for comparisons among more than three experiments.
Accession Numbers
Sequence data from this article can be found in the DDBJ/EMBL/GenBank databases under accession numbers GmERO1a (AB622257), GmERO1b (AB622258), and GmCNX (AB196933).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Amino acid sequences of GmERO1a, GmERO1b, and HsEro1α.
Supplemental Figure S2. Oxidation of ER oxidoreductases by recombinant GmERO1a.
Supplemental Figure S3. Refolding of RNase A by multiple soybean ER oxidoreductases.
Supplemental Figure S4. Expression, purification, and activity of wild-type GmPDIL-2 and its variants.
Supplemental Figure S5. Expression and purification of wild-type GmPDIM and its variants.
Supplemental Figure S6. Expression and purification of recombinant C-terminal truncated soybean calnexin.
Supplemental Table S1. List of primers and template plasmids for PCR of variant preparation.
Glossary
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- BSA
bovine serum albumin
- crP10
Cys-rich 10-kD prolamin
- DTT
dithiothreitol
- Ero
ER oxidoreductin
- GmCNX
anti-soybean calnexin
- GmERO1
recombinant soybean Ero1
- GmPDIL
recombinant soybean
- GmPDIM
ER oxidoreductase
- GmPDIS
recombinant soybean class IV ER oxidoreductase
- GSH
glutathione
- GST
glutathione-s-transferase
- IP
immunoprecipitation
- OsERO1
rice Ero1
- PDI
protein disulfide isomerase
- PDIL
rice ortholog
- PSI
preimmune serum
Footnotes
This work was supported by Grants in Aid for Scientific Research (no. 18658055 and no. 26660111) from the Japan Society for the Promotion of Science, a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences, and a grant from the Takano Life Science Research Foundation.
Articles can be viewed without a subscription.
References
- Andème Ondzighi C, Christopher DA, Cho EJ, Chang S-C, Staehelin LA (2008) Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 20: 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Iemura S, Kamiya Y, Ron D, Kato K, Natsume T, Nagata K (2013) Ero1-α and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J Cell Biol 202: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Inaba K (2012) Structure, mechanism, and evolution of Ero1 family enzymes. Antioxid Redox Signal 16: 790–799 [DOI] [PubMed] [Google Scholar]
- Benham AM, van Lith M, Sitia R, Braakman I (2013) Ero1-PDI interactions, the response to redox flux and the implications for disulfide bond formation in the mammalian endoplasmic reticulum. Philos Trans R Soc Lond B Biol Sci 368: 20110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R (2000) ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem 275: 4827–4833 [DOI] [PubMed] [Google Scholar]
- Chambers JE, Tavender TJ, Oka OBV, Warwood S, Knight D, Bulleid NJ (2010) The reduction potential of the active site disulfides of human protein disulfide isomerase limits oxidation of the enzyme by Ero1α. J Biol Chem 285: 29200–29207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton TE. (1977) Kinetics of refolding of reduced ribonuclease. J Mol Biol 113: 329–341 [DOI] [PubMed] [Google Scholar]
- Demmer J, Zhou C, Hubbard MJ (1997) Molecular cloning of ERp29, a novel and widely expressed resident of the endoplasmic reticulum. FEBS Lett 402: 145–150 [DOI] [PubMed] [Google Scholar]
- Ellman GL. (1959) Tissue sulfhydryl groups. Arch Biochem Biophys 82: 70–77 [DOI] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM (2011) Disulfide bonds in ER protein folding and homeostasis. Curr Opin Cell Biol 23: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA (1998) The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell 1: 161–170 [DOI] [PubMed] [Google Scholar]
- Gruber CW, Čemažar M, Clark RJ, Horibe T, Renda RF, Anderson MA, Craik DJ (2007) A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J Biol Chem 282: 20435–20446 [DOI] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW (2009) Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal 11: 2807–2850 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Takahashi H, Wakasa Y, Kawakatsu T, Takaiwa F (2013) Identification of a cis-element that mediates multiple pathways of the endoplasmic reticulum stress response in rice. Plant J 74: 248–257 [DOI] [PubMed] [Google Scholar]
- Houston NL, Fan C, Xiang JQ, Schulze JM, Jung R, Boston RS (2005) Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol 137: 762–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K, Kamauchi S, Wadahama H, Ishimoto M, Kawada T, Urade R (2009) Molecular cloning and characterization of soybean protein disulfide isomerase family proteins with nonclassic active center motifs. FEBS J 276: 4130–4141 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe NA, Craddock CP, Frigerio L (2005) Pathways for protein transport to seed storage vacuoles. Biochem Soc Trans 33: 1016–1018 [DOI] [PubMed] [Google Scholar]
- Kamauchi S, Wadahama H, Iwasaki K, Nakamoto Y, Nishizawa K, Ishimoto M, Kawada T, Urade R (2008) Molecular cloning and characterization of two soybean protein disulfide isomerases as molecular chaperones for seed storage proteins. FEBS J 275: 2644–2658 [DOI] [PubMed] [Google Scholar]
- Kermode A, Bewley JD (1999) Synthesis, processing and deposition of seed proteins: the pathway of protein synthesis and deposition in the cell. In Shewry P, Casey R, eds, Seed Proteins. Springer, Dordrecht, The Netherlands, pp 807–841 [Google Scholar]
- Kimura S, Higashino Y, Kitao Y, Masuda T, Urade R (2015) Expression and characterization of protein disulfide isomerase family proteins in bread wheat. BMC Plant Biol 15: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp MS, Frickel E-M, Ellgaard L, Weissman JS (2006) Domain architecture of protein-disulfide isomerase facilitates its dual role as an oxidase and an isomerase in Ero1p-mediated disulfide formation. J Biol Chem 281: 876–884 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lu J, Holmgren A (2014) The thioredoxin superfamily in oxidative protein folding. Antioxid Redox Signal 21: 457–470 [DOI] [PubMed] [Google Scholar]
- Lyles MM, Gilbert HF (1991) Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: pre-steady-state kinetics and the utilization of the oxidizing equivalents of the isomerase. Biochemistry 30: 619–625 [DOI] [PubMed] [Google Scholar]
- Nakasako M, Maeno A, Kurimoto E, Harada T, Yamaguchi Y, Oka T, Takayama Y, Iwata A, Kato K (2010) Redox-dependent domain rearrangement of protein disulfide isomerase from a thermophilic fungus. Biochemistry 49: 6953–6962 [DOI] [PubMed] [Google Scholar]
- Nguyen VD, Saaranen MJ, Karala AR, Lappi AK, Wang L, Raykhel IB, Alanen HI, Salo KEH, Wang CC, Ruddock LW (2011) Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J Mol Biol 406: 503–515 [DOI] [PubMed] [Google Scholar]
- Oh D-H, Kwon C-S, Sano H, Chung W-I, Koizumi N (2003) Conservation between animals and plants of the cis-acting element involved in the unfolded protein response. Biochem Biophys Res Commun 301: 225–230 [DOI] [PubMed] [Google Scholar]
- Oka OB, Yeoh HY, Bulleid NJ (2015) Thiol-disulfide exchange between the PDI family of oxidoreductases negates the requirement for an oxidase or reductase for each enzyme. Biochem J 469: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda Y, Kumamaru T, Kawagoe Y (2009) ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc Natl Acad Sci USA 106: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda Y, Nagamine A, Sakurai M, Kumamaru T, Ogawa M, Kawagoe Y (2011) Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell 23: 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R (2000) Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J Biol Chem 275: 23685–23692 [DOI] [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS (1998) Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell 1: 171–182 [DOI] [PubMed] [Google Scholar]
- Pu Y, Bassham DC (2013) Links between ER stress and autophagy in plants. Plant Signal Behav 8: e24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramming T, Okumura M, Kanemura S, Baday S, Birk J, Moes S, Spiess M, Jenö P, Bernèche S, Inaba K, Appenzeller-Herzog C (2015) A PDI-catalyzed thiol-disulfide switch regulates the production of hydrogen peroxide by human Ero1. Free Radic Biol Med 83: 361–372 [DOI] [PubMed] [Google Scholar]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N, Okita TW, Ogawa M, Satoh H, Kumamaru T (2010) Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol 51: 1581–1593 [DOI] [PubMed] [Google Scholar]
- Sato Y, Kojima R, Okumura M, Hagiwara M, Masui S, Maegawa K, Saiki M, Horibe T, Suzuki M, Inaba K (2013) Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci Rep 3: 2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serve O, Kamiya Y, Maeno A, Nakano M, Murakami C, Sasakawa H, Yamaguchi Y, Harada T, Kurimoto E, Yagi-Utsumi M, Iguchi T, Inaba K, et al. (2010) Redox-dependent domain rearrangement of protein disulfide isomerase coupled with exposure of its substrate-binding hydrophobic surface. J Mol Biol 396: 361–374 [DOI] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA (2008) Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta 1783: 549–556 [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yang ZT, Song ZT, Wang MJ, Sun L, Lu SJ, Liu JX (2013) The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J 76: 274–286 [DOI] [PubMed] [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol 128: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavender TJ, Bulleid NJ (2010) Molecular mechanisms regulating oxidative activity of the Ero1 family in the endoplasmic reticulum. Antioxid Redox Signal 13: 1177–1187 [DOI] [PubMed] [Google Scholar]
- Tien AC, Rajan A, Schulze KL, Ryoo HD, Acar M, Steller H, Bellen HJ (2008) Ero1L, a thiol oxidase, is required for Notch signaling through cysteine bridge formation of the Lin12-Notch repeats in Drosophila melanogaster. J Cell Biol 182: 1113–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Weissman JS (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164: 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitu E, Kim S, Sevier CS, Lutzky O, Heldman N, Bentzur M, Unger T, Yona M, Kaiser CA, Fass D (2010) Oxidative activity of yeast Ero1p on protein disulfide isomerase and related oxidoreductases of the endoplasmic reticulum. J Biol Chem 285: 18155–18165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadahama H, Kamauchi S, Ishimoto M, Kawada T, Urade R (2007) Protein disulfide isomerase family proteins involved in soybean protein biogenesis. FEBS J 274: 687–703 [DOI] [PubMed] [Google Scholar]
- Wadahama H, Kamauchi S, Nakamoto Y, Nishizawa K, Ishimoto M, Kawada T, Urade R (2008) A novel plant protein disulfide isomerase family homologous to animal P5 - molecular cloning and characterization as a functional protein for folding of soybean seed-storage proteins. FEBS J 275: 399–410 [DOI] [PubMed] [Google Scholar]
- Wang C, Li W, Ren J, Fang J, Ke H, Gong W, Feng W, Wang CC (2013) Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase. Antioxid Redox Signal 19: 36–45 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhu L, Wang CC (2011) The endoplasmic reticulum sulfhydryl oxidase Ero1β drives efficient oxidative protein folding with loose regulation. Biochem J 434: 113–121 [DOI] [PubMed] [Google Scholar]
- Zito E, Chin K-T, Blais J, Harding HP, Ron D (2010) ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol 188: 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]