Vol. 158: 363–375
Cheng M.-C., Hsieh E.-J., Chen J.-H., Chen H.-Y., and Lin T.-P. Arabidopsis RGLG2, Functioning as a RING E3 Ligase, Interacts with AtERF53 and Negatively Regulates the Plant Drought Stress Response.
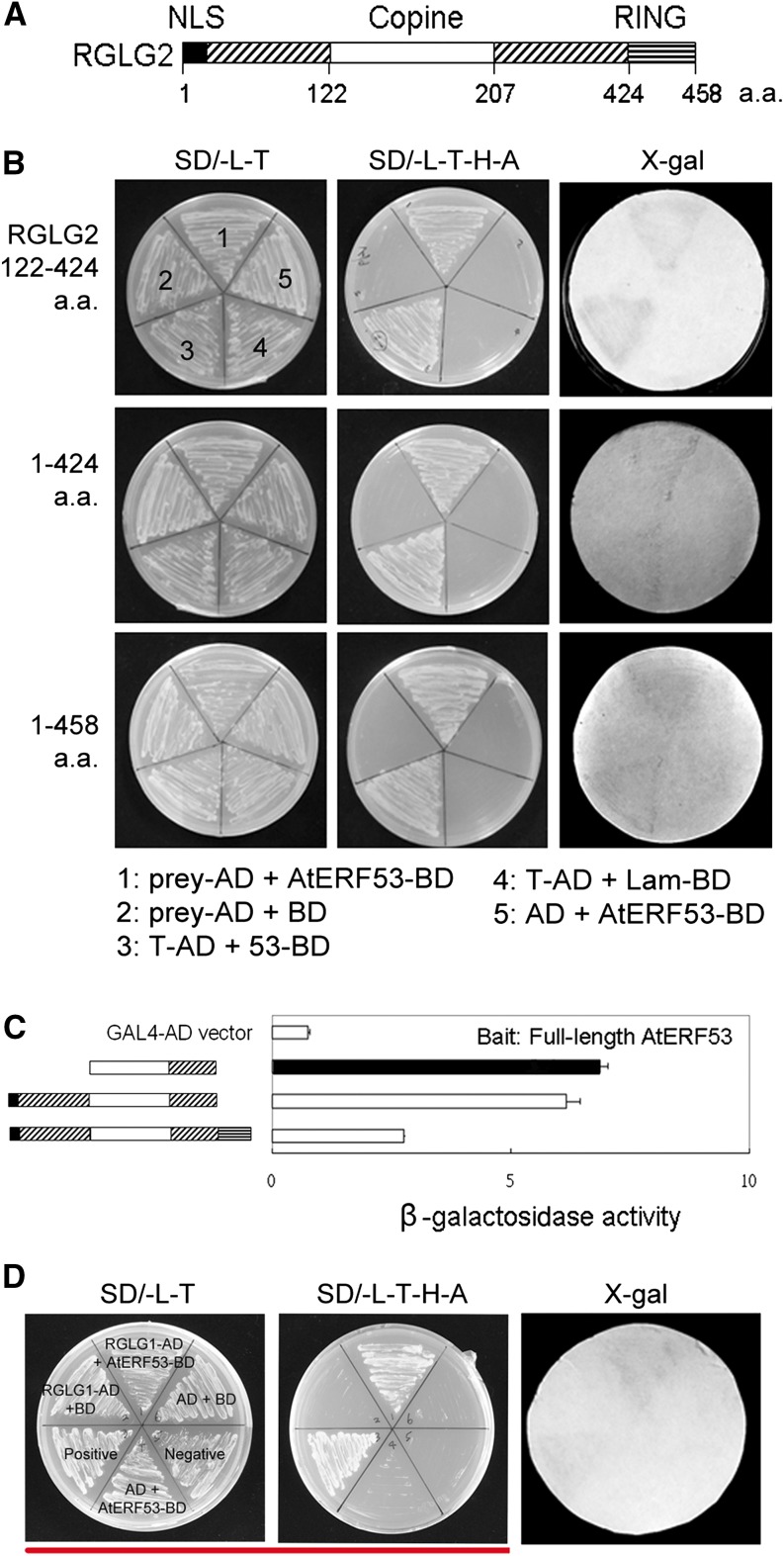
In Figure 3, the wrong insertion was used for 3D. The same image was used for both RGLG21-424/AtERF53 Y2H in Figure 3B and RGLG1/AtERF53 Y2H in Figure 3D. The corrected figure is shown below. The figure legend is unaltered from the original version. Similar results were obtained with each experiment, leading to a substantial number of similar figures. The correction does not affect any of the conclusions of the article.
Figure 3.
Identification of RGLG interaction with AtERF53 via a yeast two-hybrid analysis. A, Diagram of the specific domains of RGLG2. B, The five yeast clones transformed with the bait, AtERF53, and different lengths of RGLG2 growing on the selective medium and exhibiting β-galactosidase activity. a.a., Amino acids; AD, the pGADT7 vector; BD (for the DNA-binding domain), the pGBKT7 vector; Lam/BD, human lamin C fused to the pGBKT7 vector as a negative control. Every plate has the same labeling as the first labeled plate. C, Localization of the RGLG2 protein interaction domain with AtERF53 using yeast two-hybrid assays. β-Galactosidase activity was measured for each transformant. Error bars indicate sd (n = 3). The enzyme activity of the empty vector was defined as 1.0. The diagram for each construct is indicated in the left panel. D, Yeast cells transformed with RGLG1-AD and AtERF53-BD growing on the selective medium and exhibiting β-galactosidase activity.