Abstract
Three-dimensional ex vivo cell cultures mimic physiological in vivo growth conditions thereby significantly contributing to our understanding of tumor cell growth and survival, therapy resistance and identification of novel potent cancer targets. In the present study, we describe advanced three-dimensional cell culture methodology for investigating cellular survival and proliferation in human carcinoma cells after cancer therapy including molecular therapeutics. Single cells are embedded into laminin-rich extracellular matrix and can be treated with cytotoxic drugs, ionizing or UV radiation or any other substance of interest when consolidated and approximating in vivo morphology. Subsequently, cells are allowed to grow for automated determination of clonogenic survival (colony number) or proliferation (colony size). The entire protocol of 3D cell plating takes ~1 h working time and pursues for ~7 days before evaluation. This newly developed method broadens the spectrum of exploration of malignant tumors and other diseases and enables the obtainment of more reliable data on cancer treatment efficacy.
Keywords: molecular therapeutics, 3D cell culture, extracellular matrix, clonogenicity, proliferation, therapy testing
Introduction
The role of targeted therapies in medical oncology has tremendously increased over the last ten years. A high number of novel molecular substances have already been approved for clinical use and several compounds are in ongoing trials at present (1,2). Despite some very successful therapeutics like the tyrosine kinase inhibitor imatinib, which greatly ameliorated the outcome of patients suffering from chronic myelogenous leukemia (3), several of the molecular drugs have not met the expectations from preclinical data when applied clinically. We reason that one cause for this discrepancy could be that many drugs are tested under non-physiological two-dimensional (2D) cell culture conditions not sufficiently reflecting the microenvironment in vivo.
Three-dimensional (3D) cell culture models are in use for several decades now. Amongst scientists from various fields of biology and medicine, the culturing of cells in three dimensions opened new avenues of experimentation and thinking. Aside from its potential for tissues engineering, our understanding of cell biology has reached a new dimension ranging from gene expression to protein-protein interactions and signal transduction. The groundbreaking work of Bissell and co-workers and many others strikingly exhibited the essence of 3D growth conditions for single cells and higher order multicellular organisms (4–7).
Today, a large body of literature evidently demonstrates that the response of 3D grown cells to external stress and stimuli such as drug treatment or exposure to ionizing radiation more reliably reflects the cell response in vivo than the results obtained under 2D cell monolayer growth conditions (4,8–16). This effect could be due to both, the change in morphology and the activation of integrins and other cell adhesion receptors by binding to the ECM components, which strongly impact on cell behavior, functionality, gene and protein expression, protein-protein interactions, signal transduction and cellular sensitivity to cytotoxic stress (7,15,17–28). For in vitro investigation, cell phenotype and molecular processes can be conserved in 3D ECM-based scaffolds. This understanding gains particular relevance in the field of translational research. An example of even higher clinical relevance is a whole genome gene expression analysis of 3D grown human breast cancer cell lines, which was elegantly used to demonstrate predictive power for the probability of relapse and overall survival of breast cancer patients (12,22).
By keeping in mind the heterogeneous distribution and expression patterns of ECM proteins in the different types of human malignancies, cell phenotypes of normal epithelial cells and cancer cells can be reproducibly maintained or restored by culturing them in laminin-rich basement membrane extracellular matrix (lrECM; Matrigel) (7,29). Either embedded or ‘on top’ with subsequent lrECM overlay, the lrECM isolated from the Engelbreth-Holm-Swarm mouse sarcoma provides a broad spectrum of applications for 3D cell investigations including measurement of apoptosis, cell proliferation, malignant transformation and differentiation. A variety of published protocols explains how cells can be isolated from lrECM gels for protein expression and functional exploration or examined in situ using microscopy on living cells or histology (immunohistochemistry, immunofluorescence) on fixed cells, organotypic cell cultures or tissues (23,29).
Cell survival in vitro is often measured in terms of apoptosis, dye exclusion or proliferation. Although more time consuming, the colony forming assay has been shown to reliably determine tumor cell kill and reflect tumor control, whereas proliferation assays are used to explore tumor growth delay (30,31). Consequently, the colony forming assay is the gold standard for all disciplines for evaluating dose-effect relationships between e.g. drug concentration or radiation dose and cell survival (32).
However, to date, there is no existing assay to determine clonogenic cell survival as well as tumor proliferation under 3D cell culture conditions in a large scale for drug efficacy testing. On this basis, in the present study, we describe a high-throughput 3D lrECM based cell culture technique that greatly broadens the spectrum of already existing 3D cell culture protocols and enables a robust, reliable and reproducible analysis of the cancer cell response to cytotoxic drugs, targeted therapeutics or different kinds of radiation.
Materials and methods
Cell culture
FaDu, A549 and DLD1 carcinoma cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The origin and stability of the cells were routinely monitored by short tandem repeat analysis (microsatellites). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; PAA Laboratories GmbH, Coelbe, Germany) supplemented with 10% fetal bovine serum (FBS; PAA Laboratories) and 1% non-essential amino acids (PAA Laboratories) at 37°C in a humidified atmosphere containing 7% CO2. For all experiments asynchronously growing cell cultures were used.
Radiation exposure
Irradiation (X-rays, 200 kV, 20 mA) was performed at room temperature using a Yxlon Y.TU 320 (Yxlon International CT Development GmbH, Hattingen, Germany) containing a 0.5-mm copper filter. For measurement of the absorbed dose a Duplex dosimeter (PTW Freiburg GmbH, Freiburg, Germany) was used. The dose-rate was ~1.3 Gy/min and applied doses ranged from 0 to 4 Gy.
2D colony formation assay
Asynchronously growing cells were trypsinized, counted using a Neubauer counting chamber (Paul Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany) and plated as single cells in 6-well cell culture plates. After 24 h, cells were irradiated with 4 Gy or treated with cisplatin (25 μM) or cetuximab (5 μg/ml; Merck, Darmstadt, Germany) or left untreated. After 1 h cells were washed with 1X PBS to remove cisplatin from the cell culture medium. For determination of long-term survival cells were cultured for 8 days (A549, DLD1) or 11 days (FaDu) enabling colony growth. After fixation with 80% ethanol cells were stained with Coomassie blue (Merck). Counting of cell colonies with >50 cells was performed using a Stemi 2000 microscope (Carl Zeiss, Jena, Germany). Surviving fractions were calculated as follows: numbers of colonies formed/[numbers of cells plated (irradiated) × plating efficiency (unirradiated)]. Each point on survival curves represents the mean surviving fraction from at least three independent experiments.
3D colony formation assay
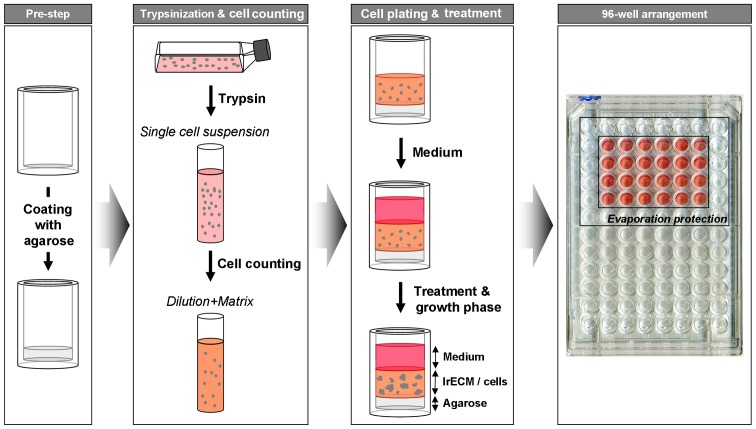
Asynchronously growing cells were trypsinized, counted and mixed with cell culture medium containing 0.5 mg/ml lrECM (cat. no. 354248; BD Biosciences, Heidelberg, Germany). Then, 100 μl of this mixture was placed in 96-well plates precoated with 50 μl of 1% agarose. After 2 h, the cell-lrECM layer was covered with 100 μl of cell culture medium. To prevent evaporation of medium, circumjacent wells were filled with 1X PBS (Fig. 1). After 24 h cells were irradiated with 4 Gy or treated with cisplatin (25 μM) or cetuximab (5 μg/ml) or left untreated similar to 2D cell culture conditions. To withdraw cisplatin from the cell culture, medium was carefully removed without touching the cell-lrECM layer and new cell culture medium was added. This step was repeated five times. Cells were cultured for 8 days (A549, DLD1) or 11 days (FaDu). Cell clusters (with the minimum size of a cell cluster containing 50 cells) were either counted microscopically without staining using a Axiovert 25 with a 2.5× objective (Carl Zeiss) or evaluated automatically as described below.
Figure 1.
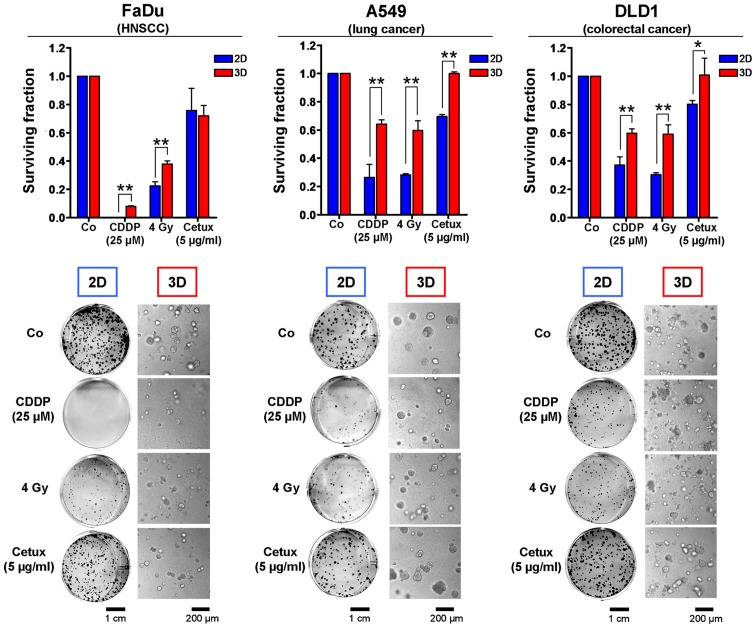
Cell culture conditions affect the tumor cell resistance to irradiation, chemotherapy and molecular therapeutics. Clonogenic cell survival data of a variety of human carcinoma cell lines treated with different clinically applied therapeutics as examples for anticipated results. Two (2D)- or three (3D)-dimensionally grown FaDu squamous cell carcinoma, A549 lung carcinoma and DLD1 colorectal carcinoma cells were treated either with cisplatin (CDDP), X-rays (4 Gy single dose) or the anti-EGFR antibody cetuximab. Images illustrate characteristic growth of cell colonies in 2D and 3D. Data show mean ± SD (n=3; t-test; *P<0.05, **P<0.01).
Automated evaluation of colony number and size
For automated analysis of survival and proliferation, each well was imaged in at least 7 different Z-levels using an Axio Observer microscope with a 2.5× objective ( Carl Zeiss). ImageJ/Fiji (33) was used for image processing and an example ImageJ macro is shown in Table I. Briefly, focus stacking was applied to the Z-level images to yield a single clear image of all 3D colonies. Further processing included background subtraction, median filtering and thresholding steps. A watershed algorithm was used to separate overlapping colonies, and automatic colony counting followed. Tables of object sizes and numbers are written to disk. Overlays of the microscopic images and the segmentation are also saved for quality review. The resulting tables can be further summarized and analyzed using R (34). The example scripts (ImageJ: Table I; R: Table II) automatically process images from multiple wells. The R script generates a histogram of colony size for each well and a summary result table of all wells imaged containing the number of colonies, the average colony area and the total colony area as a measurement of proliferation.
Table I.
Example ImageJ macro to be used on a directory with multiple subdirectories containing images.
| Steps | |
|---|---|
| Step 1 | //ATTENTION: This macro will close all other open images in ImageJ Dialog.create(“ATTENTION”); Dialog.addMessage(“This macro will close all other open images in ImageJ/Fiji!! Please press cancel if there is any unsaved data”); Dialog.show(); |
| Step 2 | //Chose directory containing the image subdirectories dir = getDirectory(“Choose a Directory “); count = 1; list = getFileList (dir); |
| Step 3 | //Chose minimum colony size Dialog.create(“Minimum colony size”); Dialog.addMessage(“Please specify the minimum colony size (area) for counting (in pixels, smaller structures will be ignored)”); Dialog.addNumber(“Minimum colony size”, 600, 0, 5, “area pixels”) Dialog.show(); MinColonySize = Dialog.getNumber(); |
| Step 4 | //Loop through directories for (i=0; i<list.length; i++) { if (endsWith(list[i], “/”)) { //Get files in directory files = getFileList(““+dir+list[i]); |
| Step 5 | //load images and perform focus stacking run(“Image Sequence...”, “open=“ + dir + list[i] + files[0] + “ sort”); run(“Extended Depth of Field (Easy mode)...”, “quality=‘0’ topology=‘0’ show-topology=‘off’ show-view=‘off’”); |
| Step 6 | //Wait for output to open while(!isOpen(“Output”)) { wait(50);} wait(1000); //Just to make sure not too early |
| Step 7 | //Select stacked output selectImage(“Output”); rename(“OStack”); run(“Duplicate...”, “ “); |
| Step 8 | //Substract background, filter and do segmentation run(“8-bit”); run(“Subtract Background...”, “rolling=50 light”); run(“Median...”, “radius=3”); run(“Auto Threshold”, “method=Default white”); run(“Convert to Mask”); run(“Watershed”); rename(“Segmented”); |
| Step 9 | //Count Colonies run(“Analyze Particles...”, “size=MinColonySize -Infinity circularity=0.00–1.00 show=[Overlay Outlines] display clear”); |
| Step 10 | //Save overlay image to disk selectWindow(“OStack”); run(“Select All”); run(“Copy”);selectWindow(“Segmented”); run(“Paste”); run(“Invert”); setFont(“SansSerif”, 32); setColor(120,120,120); setJustification(“left”); drawString(“Min. Colony size: “+ MinColonySize, 10, 50); saveAs(“PNG”, dir + “segmented” + files[0]); |
| Step 11 | //Save results to disk selectWindow(“Results”); saveAs(“Results”, dir+ substring(list[i],0,lengthOf(list[i])-1) + “.csv”); |
| Step 12 | //Close all windows close(“*”);}} |
Each subdirectory should contain multiple images of one well at various levels. Images will be focus stacked and cell colonies will be counted. Output will be a text file for each directory with information on all colonies identified and an image showing the counted colonies for quality check.
Table II.
Example R code to be used to analyze the data.
| Steps | |
|---|---|
| Step 1 | #Set to directory containing the ImageJ output setwd(“C:/Users/xx/Images”) #Adjust this to the directory containing the image subfolders |
| Step 2 | library(“ggplot2”) require(plyr) |
| Step 3 | #load data #get names of csv files, read and add each filename to the dataframe files <- dir(pattern = “*.csv”) data <- read.csv(files[1],header = TRUE) data$file <- files[1] for (i in 2:length(files)){ a <- read.csv(files[i],header = TRUE) a$file <- files[i] data <- rbind(a,data)} |
| Step 4 | #create and output area histograms p <-ggplot(data, aes(x=Area)) + geom_histogram() +scale_x_log10() + facet_wrap(~file) pdf(“histograms.pdf”, , width=8, height=10) print(p) dev.off() |
| Step 5 | #summarize data for each filename (corresponding to each well) and write csv file resultdata <- ddply(data, .(file), summarize, ColonyNumber=length(Area) , MeanColonyArea=mean(Area), TotalColonyArea = sum(Area)) write.csv(resultdata, file = “SummaryCounting.txt”) |
The data are loaded to R and area histograms are created.
Results and Discussion
In the present study, we describe a novel method to measure automatically clonogenic survival and proliferation of cells in a 3D matrix consisting of lrECM which has been reported to mimic physiologic in vivo growth conditions in a better way than conventional 2D cell culture plastic (4–11,14,16). Importantly, this approach can also be used in a high-throughput setting. According to previous data, we found that the response of all three tested human carcinoma cell lines exposed to the chemotherapeutic drug cisplatin (CDDP) or to X-ray irradiation was affected by the growth conditions with cells being significantly more resistant when cultured in 3D (Fig. 1) (9,11,20,24,35). This cell adhesion-mediated radioresistance and therapy resistance might result from a multitude of cellular processes including differences in transcriptional, translational, post-translational processes and signal transduction (8–11,22,36). Not surprising and particularly alerting with regard to molecular drug efficacy is that molecular compounds like the EGFR inhibitor cetuximab are also less effective under 3D growth conditions and that this cellular drug response correlates more closely with in vivo results (Fig. 1) (9–11,37). These data confirm the necessity to test targeted substances and more conventional therapeutics in a 3D matrix-based in vitro assay prior to animal studies to minimize costs, time and effort.
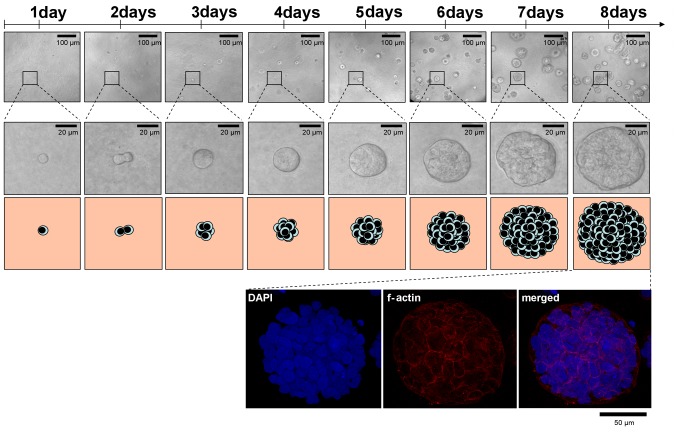
The workflow of plating and treatment of cells for the 3D clonogenic assay is depicted in Figs. 2 and 3. Agarose, cell/lrECM mixture and medium can be applied with a multi-channel pipette allowing time-efficient plating for large-scale analysis. Another advantage over most of the existing matrix-based 3D methods is that the lrECM solution with the concentration of 0.5 mg/ml can be produced with pre-heated medium (37°C) and processed at room temperature for at least 30 min without becoming solid. Therefore, cells do not have to be cooled down which likely provoke a cold stress response and perturb molecular processes (38). To assess the cell number per colony and proliferation of cells embedded into lrECM, we evaluated the number of grown A549 cells over a period of 8 days microscopically (Fig. 4). Phase contrast microscopy and DAPI/f-Actin staining revealed similar proliferation rates of this cell line in a 3D matrix in comparison to 2D monolayer cell cultures (~22 h according to ATCC) with doubling times of about 24 h after a lag phase of 1 day. Importantly, at the time of treatment (1 day after plating), 3D cell cultures are still in the single cell status, a key requirement to measure clonogenic survival (Fig. 4) (30–32).
Figure 2.
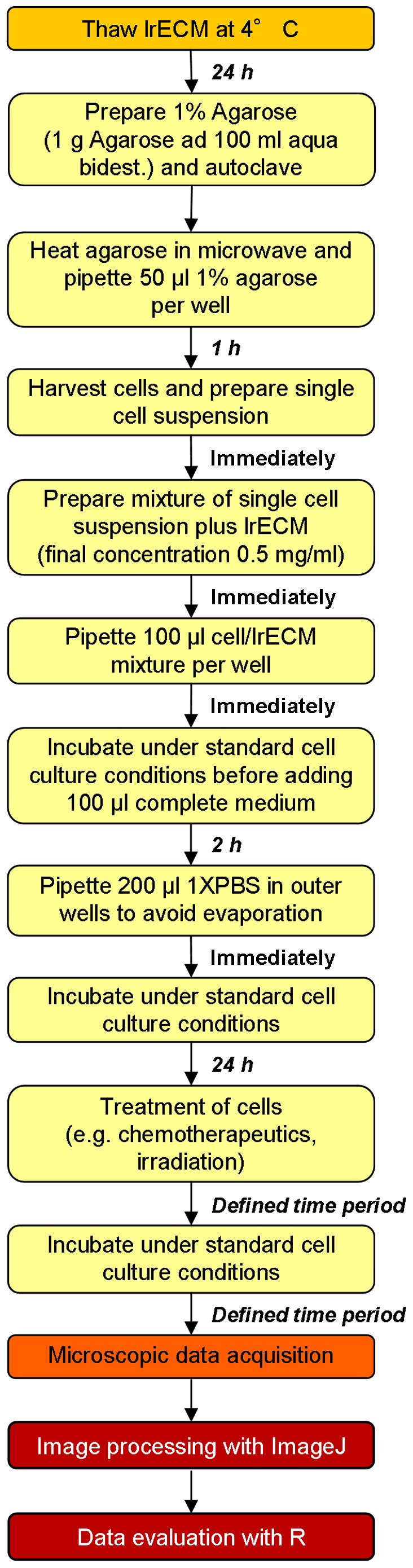
Summary of steps for 3D cell plating and analysis. Detailed workflow of experimental setup including incubation times. See also Figs. 3 and 5 for more information.
Figure 3.
Procedure of 96-well preparation and cell plating into 3D lrECM. To prevent cell adhesion to bottom, wells are coated with agarose. Single cell suspension is prepared from permanent cell culture, diluted with lrECM and pipetted on top of polymerized agarose. Medium is added after polymerization of cell-lrECM mixture. Upon treatment, cell colonies grown from single cells can be counted and/or analyzed microscopically.
Figure 4.
Colonies in 3D lrECM continuously grow out of one single cell. Representative cell colony growth over 8 days illustrated by microscopic pictures and cartoons. Securing a more than 50 cell stage of a single cell colony was achieved by DAPI staining (optional f-actin staining with phalloidin).
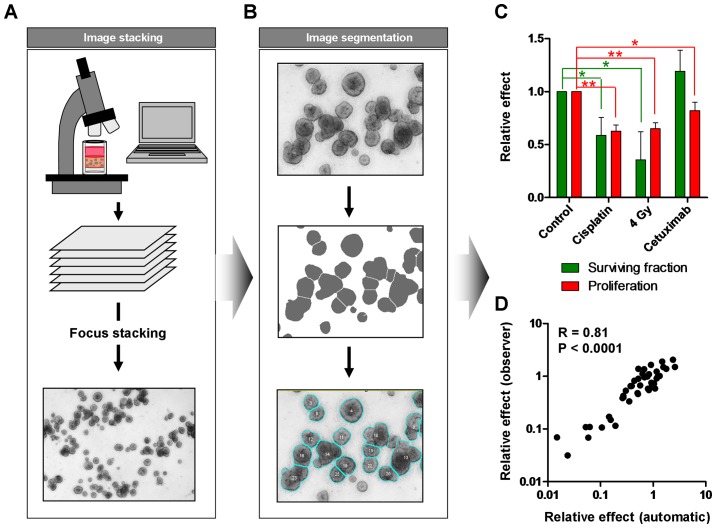
Manually counting of colonies is a time-consuming and error-prone process. Therefore, automated evaluation can reduce the working time and improve the inter-observer reliability and validity of data. As the colonies are in a 3D matrix, we took images of the wells in different Z-levels and performed focus stacking to create a clear image of all colonies (Fig. 5A). After image segmentation, colony number and size were determined (Fig. 5B) enabling the evaluation of the specific treatment effects on tumor cell proliferation as well as on clonogenic survival (Fig. 5C). While irradiation or cisplatin treatment reduced both, the size of the colonies and the colony number, cetuximab mainly affected the tumor growth and had no significant impact on cell survival (Fig. 5C). A comparison with manually counted results showed an excellent correlation (R=0.81) indicating a high reliability of the obtained data (Fig. 5D).
Figure 5.
Automated analysis of 3D colony formation assay can differentiate between effects on survival and proliferation. (A) To account for limited field depth when imaging 3D cell cultures, multiple Z-levels are recorded and merged using an algorithm that preserves the sharpness of each plane (focus stacking). (B) The resulting image is post-processed and segmented to delineate the colonies. (C) Colonies are automatically counted followed by colony size evaluation as surrogate for cell proliferation. Data show mean ± SD (n=3; t-test; *P<0.05, **P<0.01). (D) Comparative analysis of automatically measured colonies and manually measured colonies including correlation calculation (R=0.81; P<0.0001).
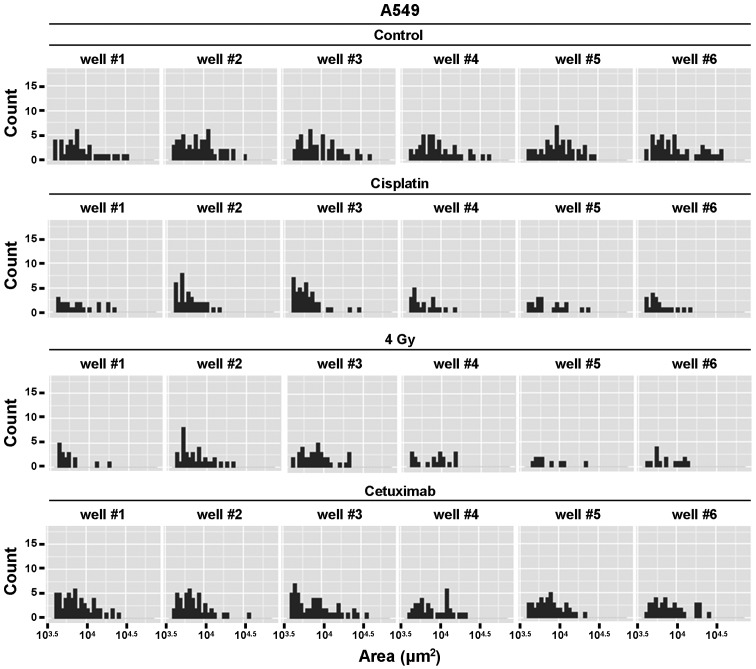
Considering the heterogeneity in human tumors and the role of cancer stem cells for therapy resistance (39), analysis on a single cell base can be crucial to evaluate the potential of targeted therapeutics. With the described technique the size and distribution of every single colony (which grows out of one single cell) could easily be determined and plotted in a histogram (Fig. 6). As shown in Fig. 6, control cell cultures had a wide spectrum of colony sizes with several small and medium-sized but also few larger colonies. In contrast, after exposure to cisplatin, ionizing radiation or cetuximab the distribution shifted to the left resulting in an overall decrease of colony size. These data could give valuable information about the different treatment effects in a tumor cell population.
Figure 6.
Detailed data analysis can reveal effects of cancer treatment on distribution of measured colony sizes on a single-cell-base. With the described method, the size of every single colony under each treatment condition is recorded and the distribution can be analyzed and illustrated with R. Histograms are plotted for each well separately showing the number of colonies (colony count) with a specific colony size (colony area).
In summary, the described protocol is a cost and time-efficient method to analyze the tumor response to cancer therapy in a more physiologic cell culture model. Taking into account that 3D lrECM based assays have been shown to reflect the in vivo conditions more reliably than 2D monolayer cells, it would be beneficial to employ this technique in a large-scale evaluation of molecular compounds prior to in vivo studies.
Acknowledgements
The authors were in part supported by a grant from the Bundesministerium für Bildung und Forschung (BMBF Contracts 03ZIK041 and BMBF-02NUK006B to N.C.), the Deutsche Forschungsgemeinschaft (CO668/4-1 to N.C.), the Deutsche Krebshilfe (108976 to N.C.), the EFRE Europäische Fonds für regionale Entwicklung, Europa fördert Sachsen (100066308) and by the NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research (to I.E.).
References
- 1.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 2.Zhou L, Xu N, Sun Y, Liu XM. Targeted biopharmaceuticals for cancer treatment. Cancer Lett. 2014;352:145–151. doi: 10.1016/j.canlet.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Savage DG, Antman KH. Imatinib mesylate--a new oral targeted therapy. N Engl J Med. 2002;346:683–693. doi: 10.1056/NEJMra013339. [DOI] [PubMed] [Google Scholar]
- 4.Bissell MJ, Weaver VM, Lelièvre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59:1757–1763s. discussion 1763s–1764s. [PubMed] [Google Scholar]
- 5.Weaver VM, Fischer AH, Peterson OW, Bissell MJ. The importance of the microenvironment in breast cancer progression: Recapitulation of mammary tumorigenesis using a unique human mammary epithelial cell model and a three-dimensional culture assay. Biochem Cell Biol. 1996;74:833–851. doi: 10.1139/o96-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, Richards J, Bowman P, Guzman R, Enami J, McCormick K, Hamamoto S, Pitelka D, Nandi S. Sustained growth and three-dimensional organization of primary mammary tumor epithelial cells embedded in collagen gels. Proc Natl Acad Sci USA. 1979;76:3401–3405. doi: 10.1073/pnas.76.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo AT, Mori H, Mott J, Bissell MJ. Constructing three-dimensional models to study mammary gland branching morphogenesis and functional differentiation. J Mammary Gland Biol Neoplasia. 2012;17:103–110. doi: 10.1007/s10911-012-9251-7. [DOI] [PubMed] [Google Scholar]
- 8.Cichon MA, Gainullin VG, Zhang Y, Radisky DC. Growth of lung cancer cells in three-dimensional microenvironments reveals key features of tumor malignancy. Integr Biol Camb. 2012;4:440–448. doi: 10.1039/C1IB00090J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eke I, Schneider L, Förster C, Zips D, Kunz-Schughart LA, Cordes N. EGFR/JIP-4/JNK2 signaling attenuates cetuximab-mediated radiosensitization of squamous cell carcinoma cells. Cancer Res. 2013;73:297–306. doi: 10.1158/0008-5472.CAN-12-2021. [DOI] [PubMed] [Google Scholar]
- 10.Eke I, Storch K, Krause M, Cordes N. Cetuximab attenuates its cytotoxic and radiosensitizing potential by inducing fibro-nectin biosynthesis. Cancer Res. 2013;73:5869–5879. doi: 10.1158/0008-5472.CAN-13-0344. [DOI] [PubMed] [Google Scholar]
- 11.Storch K, Eke I, Borgmann K, Krause M, Richter C, Becker K, Schröck E, Cordes N. Three-dimensional cell growth confers radioresistance by chromatin density modification. Cancer Res. 2010;70:3925–3934. doi: 10.1158/0008-5472.CAN-09-3848. [DOI] [PubMed] [Google Scholar]
- 12.Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Fuente G, Mollinedo P, Grande L, Vazquez-Barquero A, Fernandez-Luna JL. Culture dimensionality influences the resistance of glioblastoma stem-like cells to multi-kinase inhibitors. Mol Cancer Ther. 2014;13:1664–1672. doi: 10.1158/1535-7163.MCT-13-0854. [DOI] [PubMed] [Google Scholar]
- 16.Marushima H, Shibata S, Asakura T, Matsuura T, Maehashi H, Ishii Y, Eda H, Aoki K, Iida Y, Morikawa T, et al. Three-dimensional culture promotes reconstitution of the tumor-specific hypoxic microenvironment under TGFβ stimulation. Int J Oncol. 2011;39:1327–1336. doi: 10.3892/ijo.2011.1142. [DOI] [PubMed] [Google Scholar]
- 17.Park CC, Zhang HJ, Yao ES, Park CJ, Bissell MJ. Beta1 integrin inhibition dramatically enhances radiotherapy efficacy in human breast cancer xenografts. Cancer Res. 2008;68:4398–4405. doi: 10.1158/0008-5472.CAN-07-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damiano JS, Cress AE, Hazlehurst LA, Shtil AA, Dalton WS. Cell adhesion mediated drug resistance (CAM-DR): Role of integrins and resistance to apoptosis in human myeloma cell lines. Blood. 1999;93:1658–1667. [PMC free article] [PubMed] [Google Scholar]
- 19.Eke I, Deuse Y, Hehlgans S, Gurtner K, Krause M, Baumann M, Shevchenko A, Sandfort V, Cordes N. β1 Integrin/FAK/cortactin signaling is essential for human head and neck cancer resistance to radiotherapy. J Clin Invest. 2012;122:1529–1540. doi: 10.1172/JCI61350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed KM, Zhang H, Park CC. NF-κB regulates radioresistance mediated by β1-integrin in three-dimensional culture of breast cancer cells. Cancer Res. 2013;73:3737–3748. doi: 10.1158/0008-5472.CAN-12-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eke I, Zscheppang K, Dickreuter E, Hickmann L, Mazzeo E, Unger K, Krause M, Cordes N. Simultaneous β1 integrin-EGFR targeting and radiosensitization of human head and neck cancer. J Natl Cancer Inst. 2015;107:dju419. doi: 10.1093/jnci/dju419. [DOI] [PubMed] [Google Scholar]
- 22.Fournier MV, Martin KJ. Transcriptome profiling in clinical breast cancer: From 3D culture models to prognostic signatures. J Cell Physiol. 2006;209:625–630. doi: 10.1002/jcp.20787. [DOI] [PubMed] [Google Scholar]
- 23.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eke I, Storch K, Kästner I, Vehlow A, Faethe C, Mueller-Klieser W, Taucher-Scholz G, Temme A, Schackert G, Cordes N. Three-dimensional invasion of human glioblastoma cells remains unchanged by X-ray and carbon ion irradiation in vitro. Int J Radiat Oncol Biol Phys. 2012;84:e515–e523. doi: 10.1016/j.ijrobp.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Liu TJ, LaFortune T, Honda T, Ohmori O, Hatakeyama S, Meyer T, Jackson D, de Groot J, Yung WK. Inhibition of both focal adhesion kinase and insulin-like growth factor-I receptor kinase suppresses glioma proliferation in vitro and in vivo. Mol Cancer Ther. 2007;6:1357–1367. doi: 10.1158/1535-7163.MCT-06-0476. [DOI] [PubMed] [Google Scholar]
- 26.Tancioni I, Uryu S, Sulzmaier FJ, Shah NR, Lawson C, Miller NL, Jean C, Chen XL, Ward KK, Schlaepfer DD. FAK inhibition disrupts a β5 integrin signaling axis controlling anchorage-independent ovarian carcinoma growth. Mol Cancer Ther. 2014;13:2050–2061. doi: 10.1158/1535-7163.MCT-13-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawakami K, Fujita Y, Kato T, Mizutani K, Kameyama K, Tsumoto H, Miura Y, Deguchi T, Ito M. Integrin β4 and vinculin contained in exosomes are potential markers for progression of prostate cancer associated with taxane-resistance. Int J Oncol. 2015;47:384–390. doi: 10.3892/ijo.2015.3011. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda Y, Kawamoto Y, Teduka K, Peng WX, Yamamoto T, Ishiwata T, Naito Z. Morphological and cytoskeletal alterations of nervous system tumor cells with different culturing methods. Int J Oncol. 2011;38:1253–1258. doi: 10.3892/ijo.2011.945. [DOI] [PubMed] [Google Scholar]
- 29.Nelson CM, Inman JL, Bissell MJ. Three-dimensional lithographically defined organotypic tissue arrays for quantitative analysis of morphogenesis and neoplastic progression. Nat Protoc. 2008;3:674–678. doi: 10.1038/nprot.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roper PR, Drewinko B. Comparison of in vitro methods to determine drug-induced cell lethality. Cancer Res. 1976;36:2182–2188. [PubMed] [Google Scholar]
- 31.Salmon SE, Hamburger AW, Soehnlen B, Durie BG, Alberts DS, Moon TE. Quantitation of differential sensitivity of human-tumor stem cells to anticancer drugs. N Engl J Med. 1978;298:1321–1327. doi: 10.1056/NEJM197806152982401. [DOI] [PubMed] [Google Scholar]
- 32.Puck TT, Morkovin D, Marcus PI, Cieciura SJ. Action of x-rays on mammalian cells. II. Survival curves of cells from normal human tissues. J Exp Med. 1957;106:485–500. doi: 10.1084/jem.106.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Team RC. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. Open access available http//cran.r-project.org, 2014. [Google Scholar]
- 35.Eke I, Leonhardt F, Storch K, Hehlgans S, Cordes N. The small molecule inhibitor QLT0267 Radiosensitizes squamous cell carcinoma cells of the head and neck. PLoS One. 2009;4:e6434. doi: 10.1371/journal.pone.0006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, Schäfer KL, Baldus SE, Huckenbeck W, Piekorz RP, Knoefel WT, et al. Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines. PLoS One. 2013;8:e59689. doi: 10.1371/journal.pone.0059689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth T, Paraiso KHT, Hearn K, Rodriguez-Lopez AM, Munck JM, Haarberg HE, Sondak VK, Thompson NT, Azab M, Lyons JF, et al. Inhibition of HSP90 by AT13387 delays the emergence of resistance to BRAF inhibitors and overcomes resistance to dual BRAF and MEK inhibition in melanoma models. Mol Cancer Ther. 2014;13:2793–2804. doi: 10.1158/1535-7163.MCT-14-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roobol A, Carden MJ, Newsam RJ, Smales CM. Biochemical insights into the mechanisms central to the response of mammalian cells to cold stress and subsequent rewarming. FEBS J. 2009;276:286–302. doi: 10.1111/j.1742-4658.2008.06781.x. [DOI] [PubMed] [Google Scholar]
- 39.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]