Abstract
Clear-cell renal cell carcinoma (ccRCC) is the most common subtype of RCC (70–80%) and is associated with poor prognosis in 40% of cases mainly due to metastasis in the course of the disease. RASSF1, with its isoforms RASSF1A and RASSF1C, is a tumor suppressor gene which has not been fully analyzed in ccRCC yet. The epigenetic downregulation of RASSF1A is commonly associated with promoter hyper-methylation. The aim of the present study was to compare the ccRCC outcomes with the expression of RASSF1A and RASSF1C. Tissues were obtained from 86 ccRCC patients. RASSF1A and RASSF1C mRNA levels were assessed in tumor and matched normal kidney tissue, and in 12 samples of local metastases by quantitative PCR (qPCR). RASSF1A and RASSF1C proteins levels were semi-quantified in 58 samples by western blot analysis and their tissue localization was assessed by immunohistochemistry. Hypermethylation of RASSF1A promoter was measured by high-resolution-melting methylation-specific qPCR. RASSF1A mRNA levels were 4 and 5 times lower in 66% of tumor and 75% metastasized samples. RASSF1A hypermethylation was found in 40% of analyzed T cases. RASSF1A protein expression was 5 or 20 times decreased in 70% tumor and 75% metastatic samples, respectively. RASSF1A hypermethylation, mRNA and protein levels were associated with TNM progression and higher Fuhrman's grading. Decreased RASSF1A expression, hyper-methylation, TNM and Fuhrman's grading were associated with poorer overall survival (OS). Cox hazard ratio (HR) analysis revealed predictor role of RASSF1A mRNA levels on OS and progression-free survival (PFS) in relation to Fuhrman's grading (OS HR=2.25, PFS HR=2.93). RASSF1C levels were increased in ccRCC; no correlations with clinicopathological variables were found. We conclude that RASSF1C gene is not involved in ccRCC progression and we propose that the measurements of RASSF1A mRNA levels in paired tumor-normal kidney tissue could serve as a new prognostic factor in ccRCC.
Keywords: cancer progression, clear cell renal cell carcinoma, gene expression, metastatic disease, outcome, prognostic factor, qPCR, RASSF1A
Introduction
Clear cell renal carcinoma (ccRCC) is the most frequent RCC subtype and is characterized by high mortality of 40 within 5 years, due to late diagnosis and distant metastases found in 30 (1) to 80 (2) of RCC patients at a time of examination or within the course of the disease. Among patients who undergo radical resection for clinically localized disease, future meta-static disease will develop in 20–40 of the ccRCC cases (3). The search for new molecular targets is continuing due to high mortality rate of advanced RCC (4).
The 3p chromosomal region contains tumor suppressor genes (TSG) whose downregulation is involved in cancer progression: VHL (5), FHIT (6) and RASSF1 (7). RASSF1 [Ras association (RalGDS/AF-6) domain family member 1] gene encodes RASSF1A and RASSF1C proteins which function as intracellular signal transducers (8). RASSF1A mRNA levels were decreased in at least 37 types of tumors (8) with promoter hypermethylation as the common mechanism of its underexpession (9).
Since no quantitative analysis of RASSF1A gene expression has yet been performed in ccRCC we decided to assess RASSF1A mRNA and protein levels in tumor, normal kidney tissue and metastases. We also analyzed the methylation status of RASSF1A promoter by a novel quantitative technique. Moreover, we checked mRNA and protein levels of RASSF1C gene in matched tumor-normal kidney and metastasized samples of ccRCC patients.
Materials and methods
Patients and samples
Tissue samples were collected from 86 ccRCC patients who underwent radical nephrectomy at the Department of Urology, Medical University of Gdansk, Poland, between January 2011 and September 2013. The clinical data of patients is presented in Table I. The study was approved by the local ethics committee; written consent was obtained before the surgery from each patient.
Table I.
Details of qPCR assays.
| Assay | Primer sequences | Amplicon size/CpGs product (bp) | qPCR efficiency (%) | qPCR reaction conditions | qPCR reaction content |
|---|---|---|---|---|---|
| RASSF1A promoter methylation | 5′-TTTTTTTAGGATTTAGATTGGG-3′ 5′-CCTACACCCAAATTTCCATTAC-3′ |
71/4 | 101 | 95°C, 5 min; 42× (95°C, 5 sec; 55°C, 10 sec; 72°C, 10 sec; 75°C, 10 sec, sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.1°C | 5 μl SensiFast HRM (BioLine, London, UK), 300 nM each primer, ∑ 10 μl |
| RASSF1A mRNA level measurment | 5′-CTCGTCTGCCTGGACTGTTGC-3′ 5′-TCAGGTGTCTCCCACTCCACAG-3′ |
101 | 98.5 | 95°C, 3 min; 37× (95°C, 5 sec; 58°C, 10 sec; 72°C, 10 sec; 75°C, 10 sec, sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | 5 μl SensiFast NoRox SYBR-Green (BioLine), 200 nM each primer, ∑ 10 μl |
| RASSF1C mRNA level measurment | 5′-TACTGCAGCCAAGAGGACTCGG-3′ 5′-TCAGGTGTCTCCCACTCCACAG-3′ |
116 | 92.3 | 95°C, 3 min; 37× (95°C, 5 sec; 58°C, 10 sec; 72°C, 10 sec; 75°C, 10 sec, sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | 5 μl SensiFast NoRox SYBR-Green (BioLine), 200 nM each primer, ∑ 10 μl |
| GUSB mRNA level measurment for qPCR normalization | 5′-ATGCAGGTGATGGAAGAAGTGGTG-3′ 5′-AGAGTTGCTCACAAAGGTCACAGG-3′ |
177 | 99.6 | 95°C, 3 min; 35× (95°C, 5 sec; 57°C, 10 sec; 72°C, 10 sec; 75°C, 10 sec, sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | 5 μl SensiFast NoRox SYBR-Green (BioLine), 200 nM each primer, ∑ 10 μl |
Sample acquisition
Samples were obtained according to our previous report (10) with some modifications. Briefly, dissected tissue samples of primary ccRCC tumor (n=86, named T), normal kidney (n=86, named C as controls) and adrenal gland or the whole lymph node (n=12, named M), were collected in the operating room and placed immediately in approximately five volumes of RNAlater (Ambion Inc., Austin, TX, USA).
RNA isolation and cDNA synthesis
We utilized the same methodology as previously described (10) with some modifications. In short, ExtractMe Total RNA kit (DNAGdansk, Gdansk, Poland) was used for RNA extraction, followed by spectro-photometric (NanoDrop ND-1000; Thermo Fisher Scientific, Fitchburg, WI, USA) and electrophoretic (Bioanalyzer 2100 apparatus; Agilent Technologies, Santa Clara, CA, USA) analysis of RNA. After DNA removal (Turbo DNA-free kit; Ambion, Austin, TX, USA), 2 μg total RNA was reversibly transcribed with the use of RevertAid reverse transcriptase (Fermentas-Thermo Fischer Scientific) and 0.5 μg dT18 primers (Sigma-Aldrich, Munich, Germany) in a total volume of 20 μl.
Assessment of RASSF1A and RASSF1C mRNA expression
Quantitative real-time PCR (qPCR) technique was applied to measure mRNA level of RASSF1A and RASSF1C genes in all samples after normalization to the GUSB reference gene level in each sample (10). List of primers and details regarding time-temperature protocol are described in Table II. All reactions were run in duplicate using 1 μl of 4X diluted cDNA and SensiFast Sybr™ No-ROX kit (BioLine, London, UK) chemistry in a total volume of 10 μl. Each gene assay was run on a separate plate (StepOnePlus apparatus; Life Technologies-Applied Biosystems, Grand Island, NY, USA) with non-template control (water instead of cDNA) and 10X diluted pooled cDNA as a run-to-run precision control. Data was acquired by StepOne Software ver. 2.2 and geometric mean of Ct (threshold cycle) values were used for comparable expression analysis. The Livak's method was utilized for quantification: Ratio = 2−ΔΔCt to obtain raw expression data for each sample, followed by calibration to average expression data of control samples (fold change; control sample = 1).
Table II.
Clinicopathological features of ccRCC patients and association between RASSF1A and RASSF1C mRNA levels and clinical data.
| RASSF1A qPCR results (%) | RASSF1C qPCR results (%) | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Patients (n=86) | Subgroups | Low (≤0.266) | High (>0.266) | P-value (low vs. high)a | Low (≤0.191) | High (>0.191) | P-value (low vs. high)a |
| Age (years) | ≤62 | 31 (36) | 14 (16) | 0.65 | 14 (16) | 31 (36) | 1.00 |
| 62.16±11.24 | >62 | 26 (30) | 15 (18) | 12 (14) | 29 (34) | ||
| Range, 33–83 | |||||||
| Gender | Female (n=38) | 23 (27) | 15 (17) | 0.36 | 13 (15) | 25 (29) | 0.49 |
| Male (n=48) | 34 (40) | 14 (16) | 13 (15) | 35 (41) | |||
| Tumor size (cm) | ≤7 (n=50) | 33 (38) | 17 (20) | 1.00 | 15 (18) | 35 (41) | 1.00 |
| >7 (n=36) | 24 (28) | 12 (18) | 11 (13) | 24 (28) | |||
| Fuhrman's | |||||||
| histological grade | 1 + 2 (n=37) | 16 (19) | 21 (24) | 0.0002 | 11 (13) | 26 (30) | 1.00 |
| 3 + 4 (n=49) | 41 (42) | 8 (9) | 14 (16) | 35 (41) | |||
| TNM stage | |||||||
| Non-metastatic | T1-2N-0M0 | 24 (28) | 21 (24) | 0.011 | 14 (16) | 31 (36) | 1.00 |
| Metastatic | T1-2N1M0 | ||||||
| T3-N0-1M0 | |||||||
| T4-N0-2M0 | |||||||
| T1-4N2M0 | |||||||
| T1-4N0-2M1 | 33 (39) | 8 (9) | 12 (14) | 29 (34) | |||
P-values were calculated by Fisher's 2×2 test.
DNA extraction, bisulfite modification and acquisition of control DNA
DNA was isolated from each specimen (~2 mg) to a total volume of 20 μl followed by bisulfide modification according to manufacturer's protocol (DNA Methylation-Direct™ kit; Zymo Research, Irvine, CA, USA). Final elution was performed with the use of 50 μl of ddH2O, and the DNA concentration was assessed with NanoDrop ND-1000. For the generation of a dilution series of control DNA standards, fully methylated (named MD) and unmethylated (UMD) human genomic DNA (HCT116 cell line, DKO strain, Human Methylated & Non-methylated DNA Set; Zymo Research) were utilized.
Methylation sensitive high resolution melting quantitative PCR (MS-HRM-qPCR) assessment of RASSF1A promoter methylation status
Methylation was assessed in samples with the use of methylation specific-high resolution melting (MS-HRM) (9). Primers sequences were designed using MethPrimer software (http://www.urogene.org/methprimer/); primers, reaction mixtures and time-temperature conditions are listed for each promoter region interrogated in Table I. MS-qPCR reactions were set on StepOnePlus (Life-Technologies) apparatus and after conventional 42-cycle amplification, post-PCR products were checked using HRM application (details in Table I) with the use of HRM software ver. 3.1 (Life-Technologies). For each run, matched DNA from T, C and M samples were set; standard dilutions of MD and UMD were made to 100, 50, 25, 10 and 0% of MD in UMD and used in the same PCR plate as well as no template control. The final methylation results divided samples into two groups: homogenous and heterogenous methylated DNA; homogenous DNA was further semi-quantitatively divided into intervals, according to MD standards (0–10%; 10–25%; 25–50% and 50–100%). Heterogenously methylated DNA samples were excluded from the analysis, due to limitation of the method (9,11).
Western blot analysis
Protein lysates were prepared with Mammalian Cell Extraction kit (BioVision, Milpitas, CA, USA). The lysates (10 μg) were loaded to a 10% Mini-Protean TGX gel (Bio-Rad Laboratories, Hercules, CA, USA), resolved by SDS-PAGE and transferred to a PVDF membrane using the Trans-Blot Turbo system (Bio-Rad Laboratories). Membranes were stained with 0.1% Ponceau S to ensure equal loading after transfer, and subsequently blocked with 5% albumin fraction V in TBS buffer with 0.1% Tween-20 (TBST) for 1 h at room temperature (RT). After washing with TBST, membranes were incubated (overnight, 4°C) with specific primary antibodies in 2% albumin/TBS: 1:2,000 rabbit anti-RASSF1A (#bs-1234R; Bioss, Inc., Woburn, MA, USA); mouse polyclonal anti-RASSF1C (1:1,000) (#ab24419; Abcam, Cambridge, UK) and anti-GAPDH peroxidise-conjugated IgM 1:50,000 (#G9295; Sigma-Aldrich, St. Louis, MO, USA). After triple washing with TBST, blots were incubated (2 h, RT) with horseradish peroxidase-conjugated secondary antibodies: 1:15,000 anti-rabbit IgG or anti-mouse IgG (Sigma-Aldrich). Following triple washing with TBST, immunoreactive bands were detected on medical X-ray film (Agfa HealthCare, Mortsel, Belgium) using chemiluminescent peroxidase substrate (Sigma-Aldrich). Densitometric analyses of immunoreactive protein bands was performed with Quantity One software (Bio-Rad Laboratories) and calculated as units = Intensity/mm2. After normalization to GAPDH protein units for each sample, the semi-quantitate results for either tumor or metastasized samples were obtained as a ratio = mean unitsT/M/mean unitsC for RASSF1A or RASSF1C proteins.
Immunohistochemistry for RASSF1A and RASSF1C proteins
Formalin-fixed paraffin-embedded tissue sections (6 μm) from side tissues were deparaffinized and hydrated through xylenes and graded alcohol series. After antigen retrieval using hot acidic citrate buffer (Epitope Retrieval Solution pH 6.0; Leica Biosystems Newcastle Ltd., Newcastle upon Tyne, UK) samples were blocked for endogenous peroxidase activity by using 3% hydrogen peroxide for 10 min. Sections were then incubated with 2.5% normal horse serum [ImmPRESS anti-rabbit Ig (peroxidase) polymer detection kit; Vector Laboratories, Inc., Burlingame, CA, USA] to block non-specific binding of immunoglobulin. Immunohistochemical (IHC) staining was performed using anti-RASSF1A rabbit anti-human polyclonal antibody (1:100) (#bs-1234R; Bioss, Woburn, MA, USA) or mouse polyclonal anti-RASSF1C (1:100) (#ab24419; Abcam). After 2-h incubation with primary antibodies at room temperature, slides were washed in PBS and incubated with an appropriate secondary antibody [ImmPRESS anti-rabbit Ig (peroxidase) polymer detection kit or ImmPRESS anti-mouse Ig (peroxidase) polymer detection kit] for 30 min. Slides were rinsed in PBS and immunoreactive cells were visualized by addition of 3,3′-diaminobenzidine solution (DAB peroxidase substrate kit; Vector Laboratories) and counterstained with hematoxylin. Sections were then dehydrated, mounted in DPX mounting medium and viewed under a Nikon Eclipse E800 light microscope with Lucia G software. The specificity of the IHC staining was determined by a negative control, which was prepared under the same conditions as mentioned above, replacing primary antibodies with 2.5% normal horse serum [ImmPRESS anti-rabbit Ig (peroxidase) polymer detection kit].
Statistical analysis
Statistics was performed with the use of GraphPad Prism ver. 6.05 (GraphPad Software, Inc., San Diego, CA, USA) and Statistica ver. 10c (Statsoft Inc., Tulsa, OK, USA). Non-parametric Mann-Whitney U and Kruskal-Wallis ANOVA tests were used to compare clinical and molecular data since most data did not pass D'Agostino and Pearson omnibus test. Fisher's 2×2 exact test was used to analyse relationships between the subgroups. Spearman's correlation or multivariate regression were utilized for testing the associations between two or three variables. The Cox-Mantel proportional hazard regression model was used to evaluate the effect of explorative variables on survival of ccRCC patients. First, univariate Cox regression analysis for every single variable was performed. Secondly, variables with a P-value <0.05 were included into multivariate Cox regression analysis with a variable selection via backward elimination. All associations were presented as hazard ratios (HR) with their 95% confidence interval (CI) and P-values (12). Variables for overall survival (OS) and progression-free survival (PFS) rates were calculated separately. Kaplan-Meier estimations were performed to describe survival rates.
Results
Clinicopathological characteristics of patients
Of 86 ccRCC patients (mean age, 62.1±11.2 years) (Table II), 37 were diagnosed as stage I (T1-2N0M0), 8 as stage II (T2N0M0), 12 as stage III (T1-2N1M0 or T3N0-1M0) and 29 as stage IV (T4N0-2M0 or T1-4N2M0 or T1-4N0-2M1). At the time of surgery 47.7% ccRCC patients were diagnosed with local or distant metastases. According to Fuhrman's nuclear grading 4 patients were grade 1, 32 grade 2, 23 grade 3 and 26 were grade 4. None of the patients had undergone chemotherapy or radiotherapy before the nephrectomy. The mean follow-up period was 21 months (range, 3–48), to date 45 patients were alive (52); all deaths (except for 1 patient) were related to ccRCC progression. Median overall survival (OS) rate was 12 months. During follow-up metastases occurred in 38 (44) patients while the median progression free-survival (PFS) rate was 6 months.
Expression of RASSF1A and RASSF1C genes at the mRNA level
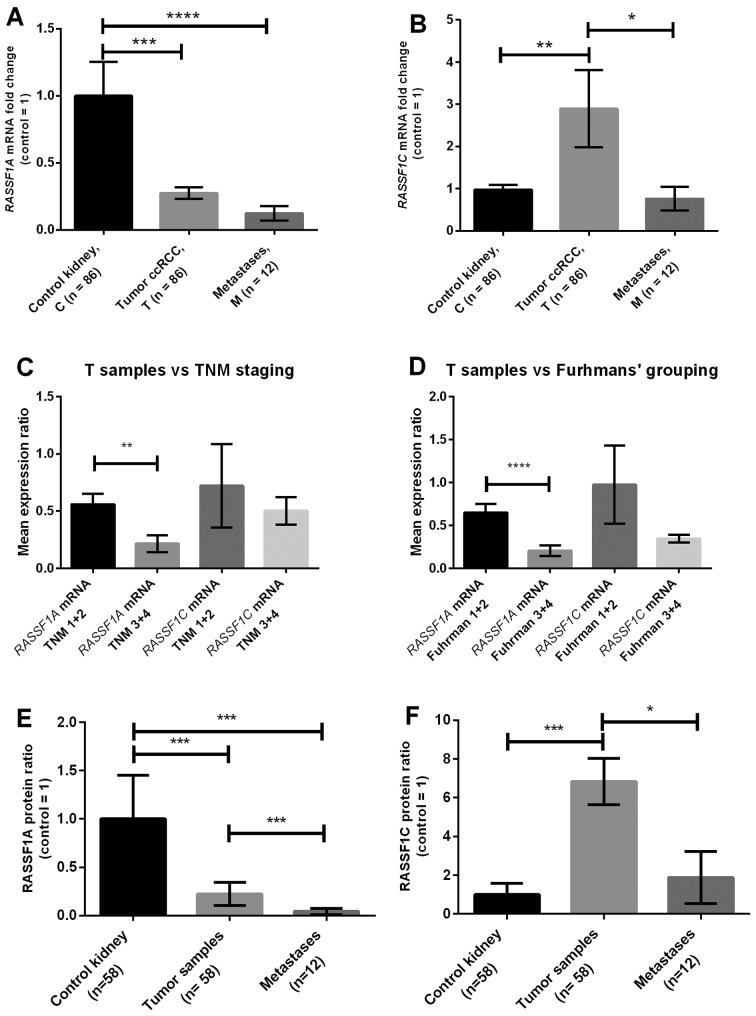
As shown in Fig. 1A, RASSF1A mRNA level in T and M samples were ~4 and 5 times lower vs. C samples, respectively. The mRNA levels of RASSF1C were ~3 times higher in tumor in comparison to either C or M samples, respectively (Fig. 1B).
Figure 1.
RASSF1A and RASSF1C gene expression in ccRCC. (A) RASSF1A and (B) RASSF1C mRNA levels in tissue samples of ccRCC patients were assessed by qPCR. (C) Plots and (D) show gene expression in tumor samples related to TNM and Fuhrman's grading. (E and F) RASSF1A and RASSF1C protein levels assessed by western blot analysis. Bars and whiskers represent mean ± SEM normalized to control kidney samples. *P<0.05; **P<0.01; ***P<0.001; ****P< 0.0001 between groups (Mann-Whitney U test).
After setting-up the threshold value based on median values of RASSF1A or RASSF1C expression levels in control samples decreased level of RASSF1A mRNA was noted in 66 of T and 75 of M samples, whereas RASSF1C expression was upregulated in 76 T and 33 M samples.
Patients with higher nuclear grades (Fuhrman's 3+4) and metastatic ccRCC (TNM 3+4) showed downregulation of RASSF1A (Table II). The mRNA levels of RASSF1A were ~3 and 4 times lower in TNM 3+4 and Fuhrman's 3+4 than in TNM 1+2 or Fuhrman's 1+2 groups, respectively (Fig. 1C and D). No correlations between clinical data and RASSF1C gene expression were found (Table II and Fig. 1C and D).
RASSF1A promoter methylation status
According to the analysis of MD/UMD standards the results of HRM-MS-qPCR were qualified into four grades: 1, 0–10% methylation; 2, 10–25%; 3, 25–50%; 4, 50–100% (data not shown). We assessed >25% methylation as hypermethylation status finding 35 (60%) unmethylated and 23 (40%) hypermethylated T samples. A significant negative correlation between increasing grades of methylation and RASSF1A mRNA levels was found (Fig. 3). In hypermethylated T samples ~3.5 times lower RASSF1A mRNA level than in non-methylated T cases was observed. Hypermethylation of RASSF1 promoter was associated only with high TNM status (Table III).
Figure 3.
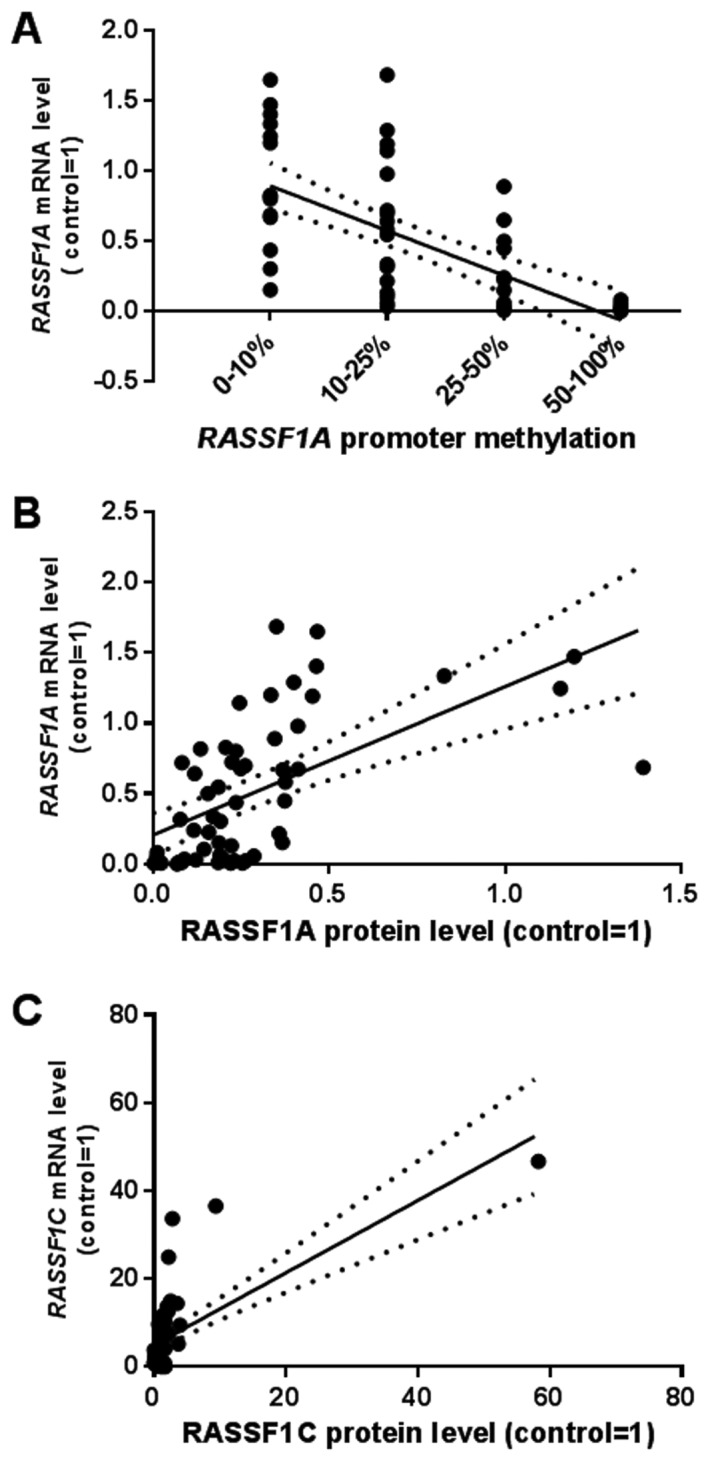
Correlation plots between either RASSF1A promother methylation, methylation and mRNA (A) or mRNA and protein levels of (B) RASSF1A and (C) RASSF1C. Details in the text.
Table III.
Association between RASSF1A promoter methylation, protein expression of RASSF1A and RASSF1C and data of patients.
| Patients/specimens/qPCR results Variables, n | RASSF1A promoter methylation and protein assessment | RASSF1C protein assessment | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Unmethylation vs. hypermethylation (%) | Protein level relative to control samples (%) | Protein level relative to control samples (%) | |||||||
|
|
|
|
|||||||
| Unmethylated (%) | Hypermethylated (%) | P-valuea | Low (≤10.09b) | High (>10.09b) | P-valuea | Low (>3.03b) | High (≤3.03b) | P-valuea | |
| Age (years) | |||||||||
| ≤62 (n=29) | 17 (29) | 12 (21) | 1.00 | 24 (41) | 5 (9) | 0.35 | 24 (41) | 5 (9) | 1.00 |
| >62 (n=29) | 18 (31) | 11 (19) | 20 (34) | 9 (16) | 24 (41) | 5 (9) | |||
| Gender | |||||||||
| Female (n=280) | 14 (24) | 14 (24) | 0.18 | 22 (38) | 6 (10) | 0.76 | 24 (41) | 4 (8) | 0.73 |
| Male (n=30) | 21 (36) | 9 (16) | 22 (38) | 8 (14) | 24 (41) | 6 (10) | |||
| Tumor size (cm) | |||||||||
| ≤7 (n=34) | 19 (33) | 15 (26) | 0.43 | 25 (43) | 9 (16) | 0.76 | 28 (47) | 7 (12) | 0.51 |
| >7 (n=24) | 16 (28) | 8 (13) | 19 (33) | 5 (8) | 21 (36) | 3 (5) | |||
| Fuhrman's histological grade | |||||||||
| 1 + 2 (n=25) | 17 (29) | 8 (14) | 0.41 | 15 (26) | 10 (17) | 0.028 | 19 (33) | 6 (10) | 0.3 |
| 3 + 4 (n=33) | 18 (31) | 15 (26) | 29 (50) | 4 (7) | 29 (50) | 4 (7) | |||
| TNM stage groupingc | |||||||||
| Non-metastatic (n=32) | 24 (41) | 8 (14) | 0.01 | 20 (33) | 12 (20) | 0.006 | 27 (44) | 5 (8) | 1.00 |
| Metastatic n=26 | 11 (19) | 15 (26) | 27 (41) | 2 (6) | 24 (40) | 5 (8) | |||
| RASSF1A/RASSF1C qPCR expression level | |||||||||
| Low (≤0.266 for RASSF1A) or (≥0.19 for RASSF1C) | 17 (29) | 16 (28) | 0.17 | 31 (53) | 2 (3) | 0.0004 | 20 (34) | 0 | 0.01 |
| High (>0.266 for RASSF1A) or (<0.19 for RASSF1C) | 18 (31) | 7 (12) | 13 (22) | 12 (21) | 28 (48) | 10 (18) | |||
P-values were calculated by Fisher's 2×2 test;
protein content was expressed in densitometric units;
TNM division as described in Table I.
RASSF1A promoter hypermethylation was found in 5/8 of metastasized samples in which RASSF1A mRNA levels were ~15 times lower than in non-methylated metastasized samples (Fig. 2). No correlations between methylation status of metastases and clinical data were found.
Figure 2.
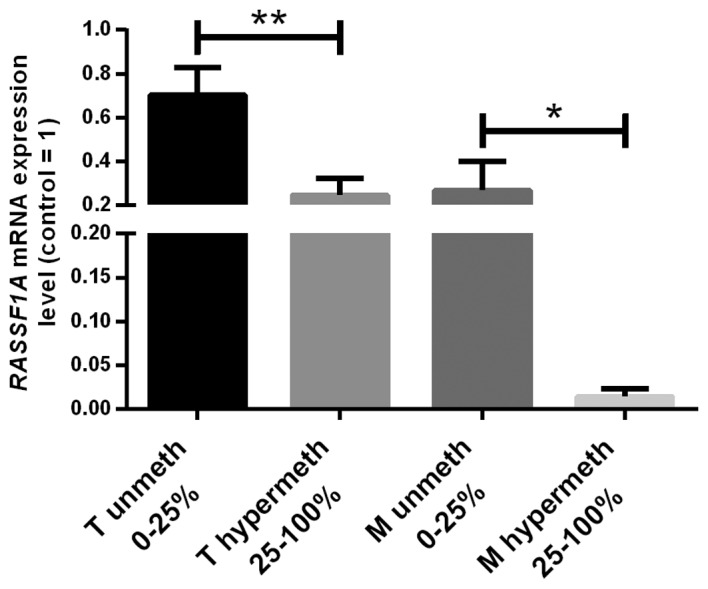
Quantitative comparison between RASSF1A mRNA levels in tumor and metastasized samples divided by methylation of the RASSF1A gene promoter. Impact of RASSF1A promoter methylation on the gene expression. mRNA results of kidney tumor and metastasized samples (T, M) were divided according to DNA methylation; 25% DNA methylation was treated as threshold. Mann-Whitney U test was applied: *P<0.05; **P<0.01 between groups.
RASSF1A and RASSF1C protein analysis
RASSF1A and RASSF1C protein levels were assessed by western blot analysis in paired 58 ccRCC and control samples, and 12 metastases analyzed for RASSF1A hypermethylation. The average RASSF1A protein levels in T and M samples were ~5 and 20 times lower than in control tissue, respectively (Figs. 4 and 1E). After setting-up threshold level 44/58 T (76) and 10/12 M (83) samples showed decreased RASSF1A protein level (Fig. 1E and Table III). Decreased RASSF1A level in tumor samples was associated with higher Fuhrman's grade and high TNM grades (Table III).
Figure 4.
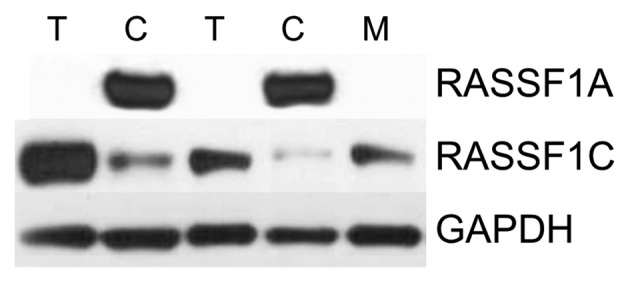
Analysis of RASSF1A and RASSF1C proteins in ccRCC by western blot analysis. Semi-quantitative analysis of RASSF1A and RASSF1C proteins in tumor (T), control kidney (C ) and metastasized (M) samples normalized to GAPDH protein level. Lines 1 and 2 represent biopsies from patient characterized by TNM 3 and Fuhrman's 2 grade, whereas lines 3–5 represent biopsies from patient with TNM 4, Fuhrman's 3 grade.
RASSF1C protein level was on average ~6 times higher in T vs. C samples with similar levels in M and C (Figs. 4 and 1F). However, high RASSF1C level was found only in 10/58 (20) T and 3/12 (25) M samples (Fig. 1F and Table III).
Positive correlations were observed between mRNA and protein levels (rs=0.66 for RASSF1A and rs=0.69 for RASSF1C; P<0.001, Spearman's test).
The analysis of possible indirect association between RASSF1A promoter methylation → decreased mRNA level → decreased protein level revealed significant relationship between three measured variables (multivariable regression; P<0.001, b=−0.63; Fig. 5).
Figure 5.
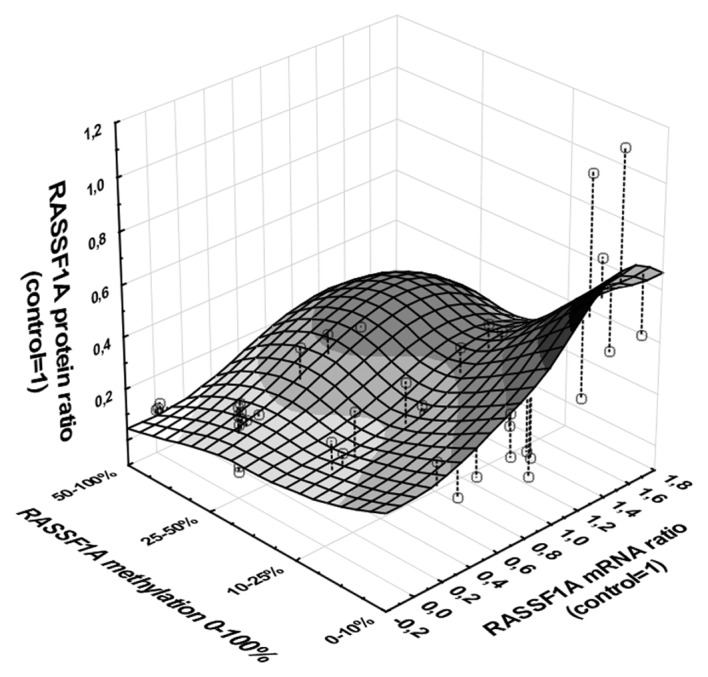
Association between RASSF1A promoter methylation and expression at mRNA and protein levels in ccRCC tumor samples. Graphic presentation of multivariate regression analysis of RASSF1A expression pattern; promoter methylation, mRNA level and protein level. XYZ plot represent results of 58 T, C and 12 M biopsies; single results are shown by empty dots. Darkening area represents increasing association between variables (white area for <0.1 association to black area for >0.7 association). Regression analysis with methylation as an independent variable: b=−0.63, P<0.001.
Tissue localization of RASSF1A and RASSF1C proteins
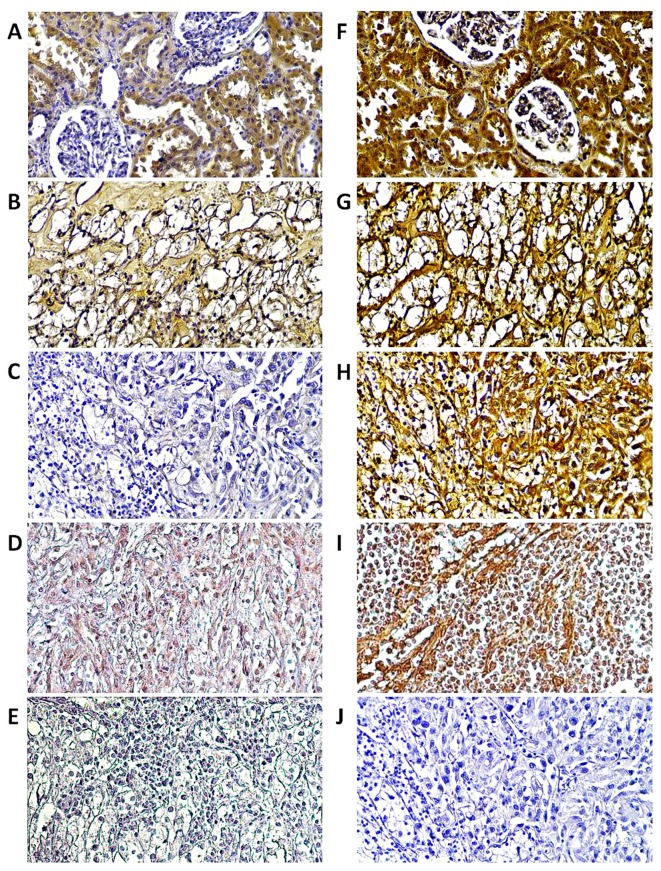
Immunohistochemical staining for RASSF1A and RASSF1C proteins was performed on twelve paired T and C samples and six M samples. As presented in Fig. 6, both proteins showed intense immunoreactivity in epithelial cells of healthy kidney whereas in tumor and metastasized samples RASSF1A was barely noticeable (Fig. 6B–D). Strong immunoreactivity of the RASSF1C protein was found in all studied samples (Fig. 6F–I).
Figure 6.
Immuhistochemical localization of RASSF1A and RASSF1C proteins in ccRCC. Immuhistochemical localization of (A–D) RASSF1A and RASSF1C (F–I) proteins in ccRCC. Normal kidney (A and F), tumor kidney of TNM 3 and Fuhrman's grade 2 (B and G) or TNM 4, Fuhrman's grade 4 (C and H) or metastasized lymph node (D and I) of two ccRCC patients (according to Fig. 5) are shown. Strong reaction was observed for RASSF1C in all samples, RASSF1A is characterized by strong presence in epithelial cells of control kidney; weak expression was observed in tumor and mestastized samples as compared to negative control (primary antibody was omitted) of (J) either tumor or (E) metastasized lymph node samples. Magnification, ×20.
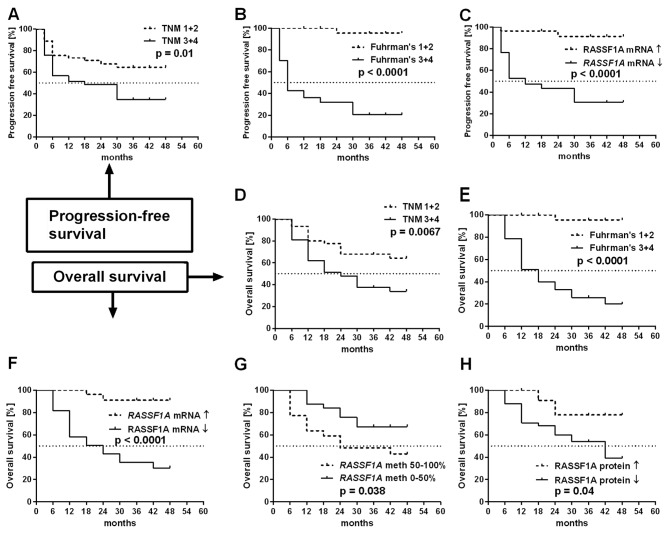
Survival analysis
We found that overall and progression-free survival were strongly associated with higher TNM and Fuhrman's grades of ccRCC patients (Fig. 7A, B, D and E). Patients with decreased RASSF1A mRNA levels showed significantly shorter OS and PFS rates than patients with high RASSF1A mRNA levels (Fig. 7C and F). Furthermore, higher RASSF1A promoter methylation status and lower RASSF1A protein levels were associated with shorter OS (Fig. 7G and H).
Figure 7.
Kaplan-Meier's survival analysis for ccRCC patients related to clinicopathological and molecular data. Progression-free survival plots for (A and B) 86 or 58 (C) ccRCC patients. Overall survival plots in 86 (D–F) or 58 (G and H) ccRCC patients.
It was noted, that the levels of RASSF1C were not associated with either OS or PFS rates of ccRCC patients (plots not shown).
Multivariate analysis using the Cox proportional hazard model indicated that the classification based on RASSF1A mRNA level was the independent predictor of OS and PFS in ccRCC patients when assessed by Fuhrman's histological grade (Tables IV and V).
Table IV.
Univariable and multivariable Cox regression analysis of overall survival rate of ccRCC patients.
| Univariable analysis | Multivariable analysis | |||
|---|---|---|---|---|
|
|
|
|||
| Parameters | P-value | HR (95 CI) | P-value | HR (95 CI) |
| Gender | ||||
| Female vs. male | 0.069 | 2.38 (0.93–6.11) | ||
| Age (years) | ||||
| >62 vs.≤62 | 0.42 | 0.69 (0.27–1.71) | ||
| Tumor size (cm) | ||||
| >7 vs.≤7 | 0.46 | 0.71 (0.29–1.75) | ||
| Tumor grade | ||||
| T3+4 vs. T1+2 | 0.0001 | 8.37 (2.76–25.35) | 0.28 | 1.95 (0.57–6.64) |
| Histological grade | ||||
| F3+4 vs. F1+2 | <0.0001 | 34.08 (4.50–258.08) | 0.007 | 18.28 (2.19–152.39) |
| RASSF1A mRNA levels | ||||
| (≤0.266) vs. (>0.266) | 0.004 | 6.06 (1.76–20.91) | 0.02 | 2.25 (0.62–8.12) |
| RASSF1A methylation | ||||
| (>25) vs. (≤25%) | 0.02 | 3.00 (1.18–7.65) | 0.22 | 1.88 (0.68–5.18) |
| RASSF1A protein levels | ||||
| (≤10.09) vs. (>10.09) | 0.32 | 0.54 (0.15–1.85) | ||
| RASSF1C mRNA levels | ||||
| (>0.191) vs. (≤0.191) | 0.83 | 0.91 (0.34–2.37) | ||
| RASSF1C protein levels | ||||
| (>3.03) vs. (≤0.191) | 0.35 | 2.01 (0.46–8.72) | ||
Table V.
Univariable and multivariable Cox regression analysis of progression free-survival rate of ccRCC patients.
| Univariable analysis | Multivariable analysis | |||
|---|---|---|---|---|
|
|
|
|||
| Parameters | P-value | HR (95 CI) | P-value | HR (95 CI) |
| Gender | ||||
| Female vs. male | 0.049 | 2.37 (1.01–5.64) | 0.55 | 1.33 (0.52–3.39) |
| Age (years) | ||||
| >62 vs.≤62 | 0.19 | 0.57 (0.25–1.32) | ||
| Tumor size (cm) | ||||
| >7 vs.≤7 | 0.61 | 0.81 (0.35–1.84) | ||
| Tumor grade | ||||
| T3+4 vs. T1+2 | 0.003 | 3.57 (1.51–8.47) | 0.73 | 1.67 (0.47–2.88) |
| Histological grade | ||||
| F3+4 vs. F1+2 | 0.005 | 33.86 (4.54–252.35) | 0.006 | 18.81 (2.26–156.32) |
| RASSF1A mRNA levels | ||||
| (≤0.266) vs. (>0.266) | 0.001 | 10.40 (2.43–44.51) | 0.019 | 2.93 (0.59–14.43) |
| RASSF1A methylation | ||||
| (>25%) vs. (≤25%) | 0.28 | 1.56 (0.69–3.55) | ||
| RASSF1A protein levels | ||||
| (≤10.09) vs. (>10.09) | 0.43 | 0.65 (0.22–1.9) | ||
| RASSF1C mRNA levels | ||||
| (>0.191) vs. (≤0.191) | 0.67 | 0.82 (0.34–2.01) | ||
| RASSF1C protein levels | ||||
| (>3.03) vs. (≤0.191) | 0.12 | 4.83 (0.64–35.85) | ||
Discussion
RASSF1A has been suggested to function as a tumor suppressor gene: its decreased expression at mRNA or protein levels was observed in almost all studied human cancers (8,13,14), however, only a few groups were analyzed for its role in ccRCC.
To the best of our knowledge, the present study is the first report of decreased RASSF1A mRNA levels in ccRCC. To date, the decreased RASSF1A expression assessed by the QPCR method in ovarian cancer (15), non-small cell lung cancer (NSCLC) (16), breast and lung cancers (17) and esophageal squamous cell (ESCC) (18) was associated with progression of cancer and poor patient outcome (15,16,18). Thus, lower transcription of RASSF1A seems to be a wider phenomenon.
Epigenetic alterations occur frequently in various cancer types with hypermethylation of the CpG islands being a frequent cause of gene silencing. Unexpectedly, when we assessed hypermethylation of RASSF1A promoter region in ccRCC, we found a relatively low number of hypermethylated tumor samples, probably due to high DNA heterogeneity in ccRCC tissue as observed by other groups (19,20). Although other authors reported high association of RASSF1A methylation with increased risk of RCC, it has to be noted that this was attributed only to serum DNA but not cancer tissue (21). In the present study probably the more homogeneous histology of twelve ccRCC metastasized samples resulted in the increased OR similarly to data on serum DNA (22–24). In other malignancies the hypermethylation of RASSF1A was related to the progression of breast (25), NSCLC (26), prostate (7), pancreatic (27), ESCC (18) and colorectal (28) cancers.
The reliable measurement of the gene expression should involve at least two levels of quantification, since the DNA → mRNA → protein sequence often becomes deregulated in cancer cells (29). The analysis of protein level is necessary for the realistic evaluation of alterations of the gene expression both in normal and cancer tissues. The decreased levels of RASSF1A in ccRCC described in the study present a novel observation. The finding of the relation between low levels of RASSF1A in ccRCC samples and progression of cancer (TNM and Fuhrman's grading) support the role of RASSF1A as a tumor suppressor gene also in ccRCC, similarly to other types of cancer (30,31).
The immunohistochemical evaluation of protein expression in tissue sections is a standard method in cancer studies. Our qualitative observations of decreased RASSF1A immunoreactivity in ccRCC tissue are in line with the results of the tissue microarrays (TMA) study which showed that low RASSF1A tissue expression was associated with poorer outcome (32). Other authors found that downregulation of RASSF1A immunoexpression was associated with early RCC formation (19).
We present novel data on the RASSF1C expression at mRNA and protein level in ccRCC. Their increased expression in ccRCC did not correlate with cancer progression on the contrary to breast cancer and ESCC (18,33). Thus, RASSF1C expression probably does not play oncogenic role in ccRCC. The epigenetic methylation does not play any role in RASSF1C gene expression since its promoter region does not contain CpG islands (27).
In conclusion, the results of the present study suggest that measurement of RASSF1A mRNA levels in paired tumor-normal kidney tissue could be used as a new prognostic factor in ccRCC, whereas the involvement of RASSF1C gene in ccRCC progression was not confirmed.
Acknowledgements
The present study was supported by the National Science Centre (grant no: 2012/05/B/NZ4/02735); the Ministry of Science and Higher Education (grant IP2010 045970 and grant no: GUMed: 02-017/07); the system project ‘InnoDoktorant-Scholarships for Ph.D. students, VIth edition’. The project is co-financed by the European Union in the frame of the European Social Fund. Authors wish to thank Michal Czarniak, for the laboratory support.
References
- 1.Novara G, Martignoni G, Artibani W, Ficarra V. Grading systems in renal cell carcinoma. J Urol. 2007;177:430–436. doi: 10.1016/j.juro.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 2.Massari F, Bria E, Maines F, Milella M, Giannarelli D, Cognetti F, Pappagallo G, Tortora G, Porta C. Adjuvant treatment for resected renal cell carcinoma: Are all strategies equally negative? Potential implications for trial design with targeted agents. Clin Genitourin Cancer. 2013;11:471–476. doi: 10.1016/j.clgc.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Keizman D, Rouvinov K, Sella A, Gottfried M, Maimon N, Kim JJ, Eisenberger MA, Sinibaldi V, Peer A, Carducci MA, et al. Is there a ‘trial effect’ on outcome of patients with meta-static renal cell carcinoma treated with sunitinib? Cancer Res Treat. 2015 Mar 5; doi: 10.4143/crt.2014.289. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Mijn JC, Mier JW, Broxterman HJ, Verheul HM. Predictive biomarkers in renal cell cancer: Insights in drug resistance mechanisms. Drug Resist Updat. 2014;17:77–88. doi: 10.1016/j.drup.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Nyhan MJ, O'Sullivan GC, McKenna SL. Role of the VHL (von Hippel-Lindau) gene in renal cancer: A multifunctional tumour suppressor. Biochem Soc Trans. 2008;36:472–478. doi: 10.1042/BST0360472. [DOI] [PubMed] [Google Scholar]
- 6.Haroun RA, Zakhary NI, Mohamed MR, Abdelrahman AM, Kandil EI, Shalaby KA. Assessment of the prognostic value of methylation status and expression levels of FHIT, GSTP1 and p16 in non-small cell lung cancer in Egyptian patients. Asian Pac J Cancer Prev. 2014;15:4281–4287. doi: 10.7314/APJCP.2014.15.10.4281. [DOI] [PubMed] [Google Scholar]
- 7.Daniunaite K, Jarmalaite S, Kalinauskaite N, Petroska D, Laurinavicius A, Lazutka JR, Jankevicius F. Prognostic value of RASSF1 promoter methylation in prostate cancer. J Urol. 2014;192:1849–1855. doi: 10.1016/j.juro.2014.06.075. [DOI] [PubMed] [Google Scholar]
- 8.Chan JJ, Katan M. PLCɛ and the RASSF family in tumour suppression and other functions. Adv Biol Regul. 2013;53:258–279. doi: 10.1016/j.jbior.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): A new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41. doi: 10.1093/nar/gkm013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wierzbicki PM, Klacz J, Rybarczyk A, Slebioda T, Stanislawowski M, Wronska A, Kowalczyk A, Matuszewski M, Kmiec Z. Identification of a suitable qPCR reference gene in metastatic clear cell renal cell carcinoma. Tumour Biol. 2014;35:12473–12487. doi: 10.1007/s13277-014-2566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikeska T, Candiloro IL, Dobrovic A. The implications of heterogeneous DNA methylation for the accurate quantification of methylation. Epigenomics. 2010;2:561–573. doi: 10.2217/epi.10.32. [DOI] [PubMed] [Google Scholar]
- 12.Avădănei ER, Wierzbicki PM, Giuşcă SE, Grigoraş A, Amălinei C, Căruntu ID. Macrophage profile in primary versus secondary liver tumors. Folia Histochem Cytobiol. 2014;52:112–123. doi: 10.5603/FHC.2014.0014. [DOI] [PubMed] [Google Scholar]
- 13.van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wierzbicki PM, Rybarczyk A. The Hippo pathway in colorectal cancer. Folia Histochem Cytobiol. 2015;53:105–119. doi: 10.5603/FHC.a2015.0015. [DOI] [PubMed] [Google Scholar]
- 15.Fu LJ, Zhang SL. Expression of RASSF1A in epithelial ovarian cancers. Eur Rev Med Pharmacol Sci. 2015;19:813–817. [PubMed] [Google Scholar]
- 16.Pastuszak-Lewandoska D, Kordiak J, Migdalska-Sęk M, Czarnecka KH, Antczak A, Górski P, Nawrot E, Kiszałkiewicz JM, Domańska D, Brzeziańska-Lasota E. Quantitative analysis of mRNA expression levels and DNA methylation profiles of three neighboring genes: FUS1, NPRL2/G21 and RASSF1A in non-small cell lung cancer patients. Respir Res. 2015;16:76. doi: 10.1186/s12931-015-0230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeves ME, Firek M, Chen ST, Amaar Y. The RASSF1 gene and the opposing effects of the RASSF1A and RASSF1C isoforms on cell proliferation and apoptosis. Mol Biol Int. 2013;2013:145096. doi: 10.1155/2013/145096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo W, Cui L, Wang C, Guo Y, Shen S, Kuang G, Dong Z. Decreased expression of RASSF1A and up-regulation of RASSF1C is associated with esophageal squamous cell carcinoma. Clin Exp Metastasis. 2014;31:521–533. doi: 10.1007/s10585-014-9646-5. [DOI] [PubMed] [Google Scholar]
- 19.Peters I, Rehmet K, Wilke N, Kuczyk MA, Hennenlotter J, Eilers T, Machtens S, Jonas U, Serth J. RASSF1A promoter methylation and expression analysis in normal and neoplastic kidney indicates a role in early tumorigenesis. Mol Cancer. 2007;6:49. doi: 10.1186/1476-4598-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu GS, Lai CY, Xu Y, Bu CF, Su ZX. Aberrant methylation of RASSF1A gene contribute to the risk of renal cell carcinoma: A meta-analysis. Asian Pac J Cancer Prev. 2015;16:4665–4669. doi: 10.7314/apjcp.2015.16.11.4665. [DOI] [PubMed] [Google Scholar]
- 22.de Martino M, Klatte T, Haitel A, Marberger M. Serum cell-free DNA in renal cell carcinoma: A diagnostic and prognostic marker. Cancer. 2012;118:82–90. doi: 10.1002/cncr.26254. [DOI] [PubMed] [Google Scholar]
- 23.Onay H, Pehlivan S, Koyuncuoglu M, Kirkali Z, Ozkinay F. Multigene methylation analysis of conventional renal cell carcinoma. Urol Int. 2009;83:107–112. doi: 10.1159/000224878. [DOI] [PubMed] [Google Scholar]
- 24.Kawai Y, Sakano S, Suehiro Y, Okada T, Korenaga Y, Hara T, Naito K, Matsuyama H, Hinoda Y. Methylation level of the RASSF1A promoter is an independent prognostic factor for clear-cell renal cell carcinoma. Ann Oncol. 2010;21:1612–1617. doi: 10.1093/annonc/mdp577. [DOI] [PubMed] [Google Scholar]
- 25.Hagrass HA, Pasha HF, Shaheen MA, Abdel Bary EH, Kassem R. Methylation status and protein expression of RASSF1A in breast cancer patients. Mol Biol Rep. 2014;41:57–65. doi: 10.1007/s11033-013-2837-3. [DOI] [PubMed] [Google Scholar]
- 26.Nawaz I, Qiu X, Wu H, Li Y, Fan Y, Hu LF, Zhou Q, Ernberg I. Development of a multiplex methylation specific PCR suitable for (early) detection of non-small cell lung cancer. Epigenetics. 2014;9:1138–1148. doi: 10.4161/epi.29499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malpeli G, Amato E, Dandrea M, Fumagalli C, Debattisti V, Boninsegna L, Pelosi G, Falconi M, Scarpa A. Methylation-associated down-regulation of RASSF1A and up-regulation of RASSF1C in pancreatic endocrine tumors. BMC Cancer. 2011;11:351. doi: 10.1186/1471-2407-11-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang HL, Zhang Y, Liu P, Zhou PY. Aberrant promoter methylation of RASSF1A gene may be correlated with colorectal carcinogenesis: A meta-analysis. Mol Biol Rep. 2014;41:3991–3999. doi: 10.1007/s11033-014-3267-6. [DOI] [PubMed] [Google Scholar]
- 29.Kristensen VN, Lingjærde OC, Russnes HG, Vollan HK, Frigessi A, Børresen-Dale AL. Principles and methods of integrative genomic analyses in cancer. Nat Rev Cancer. 2014;14:299–313. doi: 10.1038/nrc3721. [DOI] [PubMed] [Google Scholar]
- 30.Joo MK, Kim KH, Park JJ, Yoo HS, Choe J, Kim HJ, Lee BJ, Kim JS, Bak YT. CpG island promoter hypermethylation of Ras association domain family 1A gene contributes to gastric carcinogenesis. Mol Med Rep. 2015;11:3039–3046. doi: 10.3892/mmr.2014.3055. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, Li H, Shi T, Ma X, Wang B, Xu H, Ai X, Ju Z, Wang C, Zhang G, et al. Relationship between the expression of RASSF1A protein and promoter hypermethylation of RASSF1A gene in bladder tumor. J Huazhong Univ Sci Technolog Med Sci. 2008;28:182–184. doi: 10.1007/s11596-008-0217-3. [DOI] [PubMed] [Google Scholar]
- 32.Tezval H, Merseburger AS, Matuschek I, Machtens S, Kuczyk MA, Serth J. RASSF1A protein expression and correlation with clinicopathological parameters in renal cell carcinoma. BMC Urol. 2008;8:12. doi: 10.1186/1471-2490-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeves ME, Baldwin SW, Baldwin ML, Chen ST, Moretz JM, Aragon RJ, Li X, Strong DD, Mohan S, Amaar YG. Ras-association domain family 1C protein promotes breast cancer cell migration and attenuates apoptosis. BMC Cancer. 2010;10:562. doi: 10.1186/1471-2407-10-562. [DOI] [PMC free article] [PubMed] [Google Scholar]