Abstract
The aim of the present study was to investigate the role of Cyclic-nucleotide Response Element-Binding (CREB) family members and related nuclear transcription factors in the radiation response of human B lymphoma cell lines (Daudi and Ramos). Unlike the more radiosensitive Daudi cells, Ramos cells demonstrated only a moderate increase in early apoptosis after 3–5 Gy irradiation doses, which was detected with Annexin V/PI staining. Moreover, a significant and dose-dependent G2/M phase accumulation was observed in the same cell line at 24 h after both ionizing radiation (IR) doses. Western blot analysis showed an early increase in CREB protein expression that was still present at 3 h and more evident after 3 Gy IR in Ramos cells, along with the dose-dependent upregulation of p53 and NF-κB. These findings were consistent with real-time RT-PCR analysis that showed an early- and dose-dependent upregulation of NFKB1, IKBKB and XIAP gene expression. Unexpectedly, pre-treatment with SN50 did not increase cell death, but cell viability. Taken together, these findings let us hypothesise that the early induction and activation of NF-κB1 in Ramos cells could mediate necrotic cell death and be linked to other molecules belonging to CREB family and involved in the cell cycle regulation.
Keywords: CREB, nuclear transcription factors, RAMOS cell line, human lymphoma cell lines, ionizing radiation
Introduction
Ionizing radiation (IR) is commonly used in cancer therapy although it is known to induce DNA damage, which in turn can lead to gene mutation, chromosome aberrations and cell death (1–3). The processes of apoptosis and necrosis are among the most common types of cell death in response to IR but neoplastic cells could also counteract radiation-induced damage to the membrane and repair DNA damage by activating survival-signalling pathways (4–7). Whether the cell survives or undergoes apoptosis, a number of genes controlling complex pathways are activated (8,9). Among the proteins involved in cell response to IR a role has been assigned to a number of nuclear transcription factors including cyclic-nucleotide response element-binding (CREB) protein and nuclear factor kappa B (NF-κB) (6,10,11). CREB is a member of the CREB/ATF family of transcription factors playing key roles in cell proliferation, differentiation, apoptosis and survival (12–15). Moreover, a number of studies have disclosed the implication of CREB/ATF proteins in regulating lymphoblastic cell proliferation/survival but only a few studies have investigated the CREB/ATF protein response to IR (7,16). Therefore, the aim of the present study was to investigate the effect of IR on CREB/ATF and NF-κB signalling pathway and their possible role in the radiation response of Ramos B cells, an Epstein-Barr virus (EBV)-negative cell line derived from an American Burkitt's lymphoma (17). This specific cell line was chosen due to the important role traditionally played by radiotherapy in the management of several lymphomas, also in combination with chemotherapeutic drugs (18–20). Moreover, Ramos B cells carry a non-functional p53 tumour suppressor protein and constitutively express the transcription factor NF-κB (21), raising the possibility that both factors may contribute to the radiation response of Burkitt's lymphoma cells.
Materials and methods
Cell cultures and irradiation protocol
Ramos B cells, an Epstein-Barr virus (EBV)-negative cell line derived from an American Burkitt's lymphoma (17) and Daudi B cells, an EBV-positive cell line derived from Burkitt's lymphoma (22), were purchased from the American Type Culture Collection (ATCC; LGC Standards S.r.l, Milan, Italy). Both cell lines were grown in suspension at the optimal cell density of 3×105–1×106 cells/ml in HEPES-buffered RPMI-1640 (Gibco Laboratories Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 5 mM glutamine, penicillin/streptomycin (50 IU/ml and 50 μg/ml, respectively). Exponentially growing cells, showing a viability of at least 95%, were administered 3 and 5 Gy at room temperature by a Mevatron M linear accelerator (photonic energy of 6 MV; dose rate 3 Gy/min). Some cellular pools were treated with the NF-κB/IκB pharmacological inhibitor SN50 (Alexis Biochemicals, Lausen, Switzerland) added at the final concentration of 30 μg/ml 45 min before irradiation (23). Cell cytotoxicity was assayed with trypan blue dye exclusion test and by the evaluation of the cell cycle and apoptosis.
Cell cycle analysis
Approximately 0.5×106 cells per experimental condition were harvested, fixed in 70% cold ethanol and kept overnight at 4°C. Cells were then re-suspended in 20 μg/ml propidium iodide (PI) and 100 μg/ml RNase, final concentrations. Cell cycle profiles (10,000 cells) were analysed with a FC500 flow cytometer with the FL3 detector in a linear mode using the CXP software (Beckmann Coulter, Miami, FL, USA). Data were analysed with the MultiCycle software (Phoenix Flow Systems, San Diego, CA, USA).
Annexin V/PI detection of apoptotic and necrotic cells in flow cytometry
To assess apoptosis, a commercial Annexin V-FITC/PI kit (Bender MedSystems GmbH, Vienna, Austria) was used according to the manufacturer's instructions, as previously detailed (24). Briefly, the cells were gently re-suspended in binding buffer and incubated for 10 min at room temperature in the dark with Annexin V-FITC. Samples were then washed and supra-vitally stained with PI (5 μg/ml). Analyses were performed with a FC500 flow cytometer with the FL1 and FL3 detector in a log mode using CXP analysis software (Beckmann Coulter). For each sample, 10,000–20,000 events were collected. Viable cells were Annexin Vneg/PIneg (unlabelled), early apoptotic cells were Annexin Vpos/PIneg, late apoptotic and necrotic cells were Annexin Vpos/PIpos and Annexin Vneg/PIpos, respectively.
Light microscopy analysis of apoptotic and necrotic cells
Daudi and Ramos cells were fixed for 3 h at 4°C in a mixture of 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.6. The cell pellets were dehydrated in alcohol at progressively higher concentrations and embedded in Spurr resin (British Biocell Ltd., Cardiff, UK) according to standard procedures. Semithin sections were counterstained with Toluidine blue to display cell morphology and observed under a Leica DM4000 microscope (Leica microsystems S.p.A., Milan, Italy).
Total RNA extraction
For RNA extraction, 5×106 cells/sample were washed with 1X phosphate-buffered saline (PBS) and centrifuged 10 min at 1,200 rpm at 4°C. Each cell pellet was re-suspended in 1 ml of TRI reagent (Sigma-Aldrich, St. Louis, MO, USA), and centrifuged for 10 min at 10,000 rpm at 4°C. The supernatant was added to 200 μl of chloroform, then shaken vigorously, incubated on ice for 15 min and centrifuged for 20 min at 13,200 rpm at 4°C. RNA in aqueous phase was precipitated with 500 μl of isopropanol, stored for 30 min at −20°C and pelleted by centrifugation at 13,200 rpm for 20 min at 4°C. RNA pellet was washed with 75% ethanol, air dried and re-suspended in RNase-free water. Contaminating DNA was removed using 2 units of RNase-free DNase I (DNA-free kit; Ambion, Austin, TX, USA). RNA concentration was determined by spectrophotometer reading at 260 nm and its purity was assessed by the ratio at 260 and 280 nm readings (Eppendorf BioPhotometer, Hamburg, Germany). To evaluate the quality of extracted RNA, the samples were tested by electrophoresis on agarose gels and visualised by staining with ethidium bromide, under UV light.
Reverse transcription (RT) and real-time RT-polymerase chain reaction (real-time RT-PCR)
High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) was used to reverse transcribe 1 μg of RNA in a reaction volume of 20 μl. Cell samples were incubated in a 2720 Thermal Cycler (Applied Biosystems) initially at 25°C for 10 min, then at 37°C for 2 h and finally at 85°C for 5 sec. Gene expression was determined with real-time PCR using TaqMan probe-based chemistry. Reactions were performed in 96-well plates on an ABI PRISM 7900 HT Fast Real-Time PCR system (Applied Biosystems). TaqMan probes and PCR primers were obtained from Applied Biosystems [TaqMan Gene Expression assays (20X): Hs00765730_m1 for the nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (NFKB1, p105); Hs01042010_m1 for v-rel reticuloendotheliosis viral oncogene homolog A (avian) (RELA, p65); Hs00233287_m1 for inhibitor of kappa light polypeptide gene enhancer B-cells, kinase beta (IKBKB); Hs00234387_m1 for caspase-3 (CASP3); Hs00236913_m1 for X-linked inhibitor of apoptosis (XIAP)]. Glyceraldehyde-3-phosphate dehydrogenase, GAPDH (Applied Biosystems, part no. 4333764F), was used as the housekeeping gene. Each amplification reaction was performed with 10 μl of TaqMan Fast Universal PCR Master Mix (2X), no AmpErase UNG (Applied Biosystems), 1 μl of primer probe mixture, 1 μl of cDNA and 8 μl of nuclease-free water. No-template control was used to check for contamination. Thermal cycling conditions were: 95°C for 20 sec, followed by 40 cycles of amplification at 95°C for 1 sec and 60°C for 20 sec. Real-time RT-PCR analysis was performed in three independent experiments. Amplification was carried out in triplicate for each cDNA sample in relation to each of the investigated genes. Sequence Detection System software version 2.3 (Applied Biosystems) elaborated gene expression data. The comparative 2−ΔΔCt method was used to quantify the relative abundance of mRNA (relative quantification, RQ). This method uses a calibrator sample to enable a comparison of gene expression levels in different samples. The obtained values indicate the changes in gene expression in the sample of interest by comparison with the calibrator sample, after normalisation to the housekeeping gene. Means ± standard error mean (SEM) of data deriving from RQ were determined for each experimental group.
Western blotting and densitometric analysis
Cells lysates (20 μg) were electrophoresed and transferred to nitrocellulose membranes. Nitrocellulose membranes were then blocked in 5% non-fat milk or 5% BSA, 10 mmol/l Tris-HCl pH 7.5, 100 mmol/l NaCl, 0.1% Tween-20, and probed with the following primary antibodies (work dilution 1:1,000): CREB, pCREB, pATF1, pHistone H2A.X (all purchased from Cell Signaling Technology, Beverly, MA, USA); p53, NF-κB, Bcl-2, pcdc2, caspase-3, PARP (all purchased from Santa Cruz Biotechnology, Santa Cruz, CA, USA); β-actin and β-tubulin (purchased from Sigma-Aldrich), and incubated in the presence of specific enzyme conjugated IgG horseradish peroxidase. Immunoreactive bands were identified using the ECL detection system (Amersham International, Buckinghamshire, UK) and analysed with densitometry. Densitometric values, expressed as integrated optical intensity (IOI), were estimated in the ChemiDoc XRS system using Quanti One 1-D analysis software (Bio-Rad Laboratories, Richmond, CA, USA). Values obtained were normalized based on densitometric values of internal β-actin or β-tubulin. Statistical analysis was performed using the analysis of variance (ANOVA). Results are expressed as means ± SD. Values of p<0.05 were considered statistically significant.
Immunofluorescence staining
Cytocentrifuged cells were fixed with 3.7% paraformaldehyde, blocked with 10% normal donkey serum. Samples were then incubated with the following primary antibodies (working diluition, 1:500): NF-κB, pCREB, ATF2, cyclin D1 (Cell Signaling Technology); ATF3 and cyclin A1 (Santa Cruz Biotechnology). Samples were then incubated with IgG-FITC and IgG-TRITC (working dilution, 1:100) (Jackson ImmunoResearch, West Grove, PA, USA) as secondary antibodies. The nuclei were counterstained with DAPI (Vector Laboratories, Inc., Burlingame, CA). All observations were performed using a Zeiss Axioscope light microscope equipped with a Coolsnap Videocamera to acquire images to analyze with MetaMorph 6.1 software (Universal Imaging Corp, Downingtown, PA, USA).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 5.01 for Windows (Graphpad Software Inc., San Diego, CA, USA). Means ± SEM or SD were determined for each experimental group. Data were analysed with one-way analysis of variance (ANOVA), followed by Newman-Keuls multiple comparison test. The level of statistical significance was set as p<0.05.
Results
IR exposure leads to different types of cell death in Burkitt's lymphoma cell lines
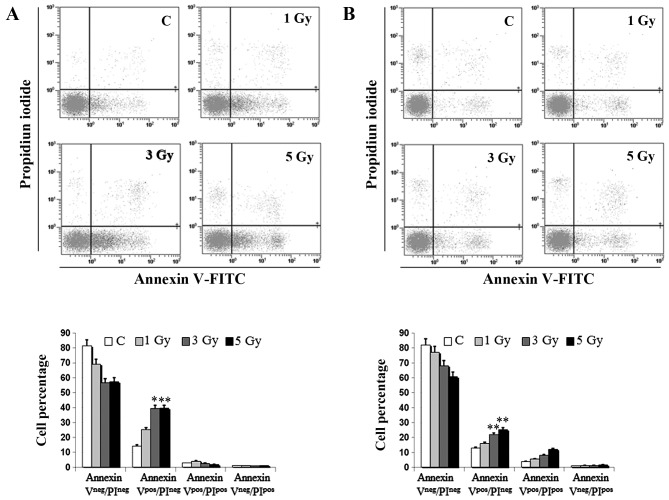
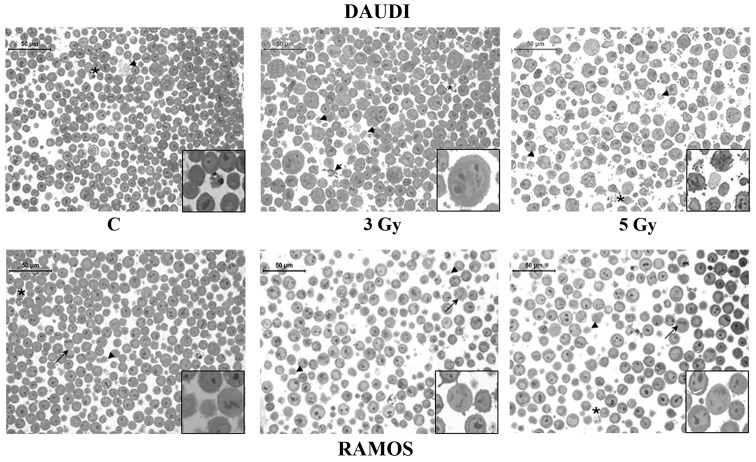
We first investigated IR effects on cell viability of two B lymphoid cell lines cultured in vitro, Daudi cells and Ramos cells. To evaluate the incidence of apoptotic cell death we performed both Annexin V/PI staining in flow cytometry and light microscopy analyses. As a matter of fact, no significant changes in cell viability were induced by any IR dose in cells examined at 1 h after irradiation (data not shown). However, the percentage of early apoptotic Daudi cells (Annexin Vpos/PIneg) was ~4-fold higher than that of unirradiated controls at 24 h, after administration of both the intermediate (3 Gy) and the highest IR dose (5 Gy), and significantly higher than that of irradiated Ramos cells (p<0.05) (Fig. 1). In the latter cell line the percentage of both early apoptotic cells (Annexin Vpos/PIneg) and late apoptotic cells (Annexin Vpos/PIpos) increased in a dose-dependent manner reaching at most a 2-fold increase compared to unirradiated controls (Fig. 1B). These preliminary observations on cell viability indicated a different radio-sensitivity of the two investigated lymphoid cell lines, which was also confirmed by morphological analyses performed in light microscopy (Fig. 2). A significant finding was the presence of huge multi-nucleate cells in Daudi cell line treated with the intermediate dose (Fig. 2).
Figure 1.
Flow cytometric analysis of dead cells in Daudi and Ramos cells. Annexin V/PI in vivo detection with flow cytometry of early apoptotic (Annexin Vpos/PIneg), late apoptotic (Annexin Vpos/PIpos) and necrotic cells (Annexin Vneg/PIpos) at 24 h in (A) Daudi cells and (B) Ramos cells left unirradiated or (C) exposed to different IR doses, as indicated. Bar graph represents the means ± SEM of the cell percentage derived from 4 independent experiments (*p<0.05, **p<0.01 vs. unirradiated control).
Figure 2.
Morphologic analysis of Daudi cells and Ramos cells. Toluidine blue-stained semithin sections showing apoptotic and necrotic features at 24 h in Daudi cells and Ramos cells left unirradiated (C) or exposed to 3 and 5-Gy IR doses, as indicated. Arrowheads (▶) point at necrotic cells, asterisks (*) at apoptotic cells, and arrows (→) at mitotic cells. Representative fields of 1 out of 3 different experiments are shown. Original magnification, ×40. Scale bar, 50 μm. The insets show the different sample features at a higher magnification (×80).
Burkitt's lymphoma cell lines with a different radio-sensitivity respond to irradiation with different cell cycle distributions
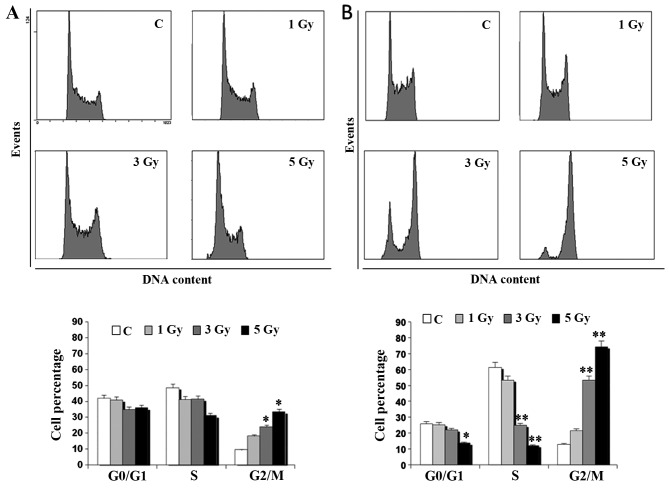
Cell cycle analysis in flow cytometry demonstrated that the different radio-sensitivity of Daudi cells and Ramos cells was related to a different profile of the cell cycle (Fig. 3). Indeed, an evident dose-dependent accumulation of Ramos cells in the G2/M phase of the cell cycle was observed at 24 h whereas no dramatic change in the cell cycle profile of Daudi cells was found. In particular, the mean percentage of Ramos cells in the G2/M phase was significantly higher than that of unirradiated controls (p<0.01), being ~5-fold higher after exposure to 3 Gy and even 7-fold higher after irradiation with 5 Gy (Fig. 3B). The percentage of S phase cells was significantly lower than in unirradiated controls at 24 h after irradiation (p<0.01). On the contrary, the percentage of Ramos cells in G1 phase did not significantly differ from that of unirradiated control cells, except when exposed to the highest IR dose (p<0.05). Notably, more radio-sensitive Daudi cells displayed a lower accumulation in the G2/M phase at 24 h compared to the less radio-sensitive Ramos cells (p<0.01) (Fig. 3).
Figure 3.
Flow cytometric analysis of cell cycle distributions in Daudi cells and Ramos cells. Cell cycle profiles obtained at 24 h in (A) Daudi cells and (B) Ramos cells left unirradiated or exposed to different IR doses, as indicated. Bar graphs represent the means ± SEM of the cell percentage derived from 4 independent experiments (*p<0.05 vs. unirradiated control).
Radio-resistant cell lines respond to irradiation with changes in nuclear transcription factors and cell cycle regulatory molecules
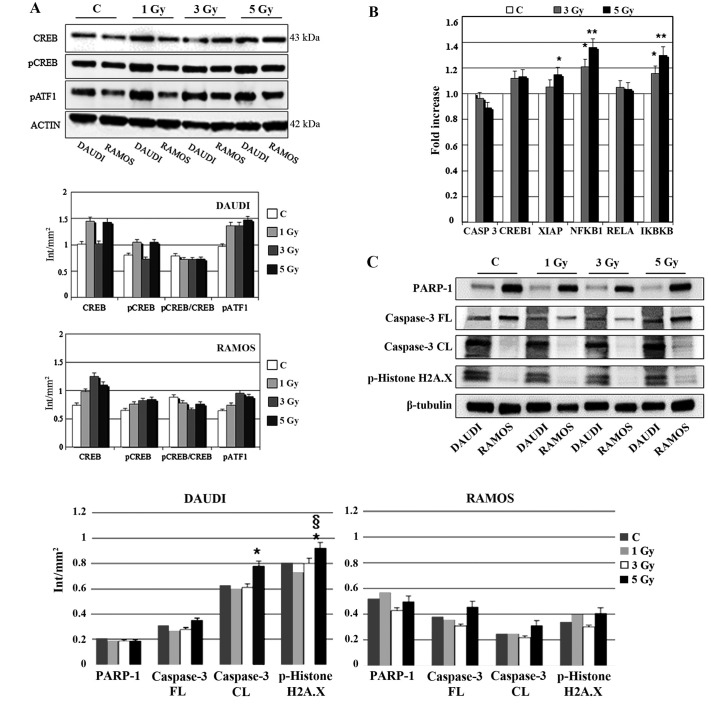
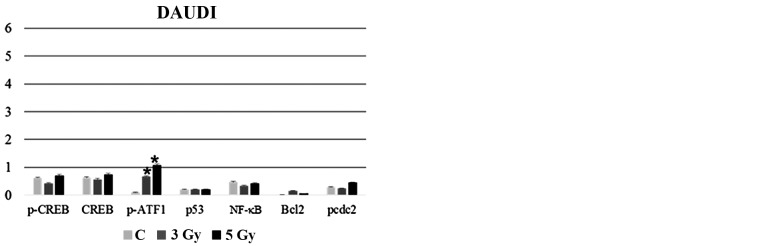
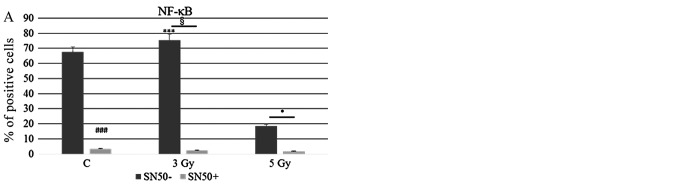
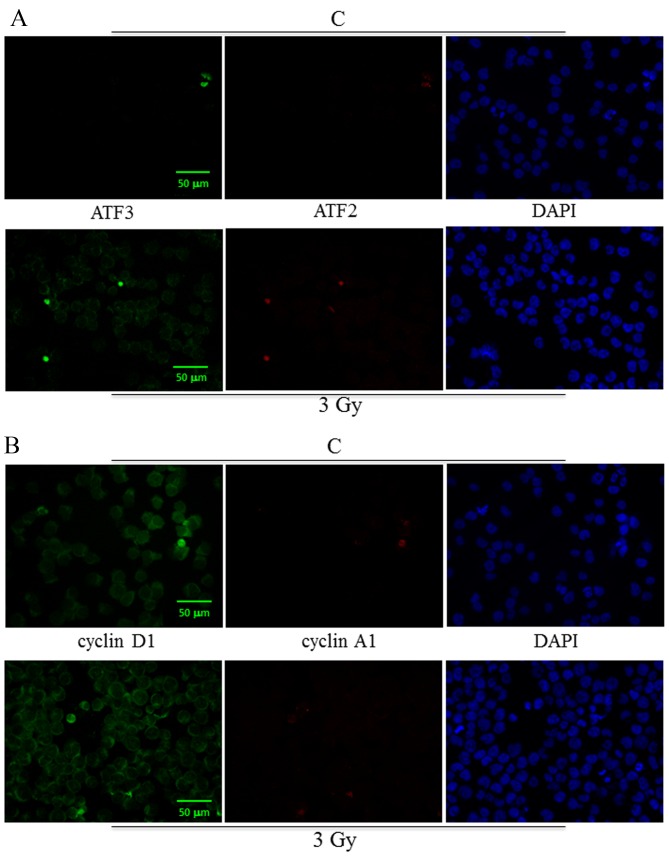
To assess molecular changes underlying biological effects we first performed western blot analysis at 30 min after IR exposure. As shown in Fig. 4A, Ramos cell line displayed an early increase (30 min) in CREB protein expression more evident after 3 Gy and paralleled by CREB gene induction (Fig. 4B). Instead, in the case of XIAP, NFKB1 and IKBKB genes there was a significant and dose-dependent induction in comparison with unirradiated controls (Fig. 4B). Furthermore, 3 or 5-Gy IR doses did not affect caspase-3 (CASP3) mRNA levels compared to unirradiated controls (Fig. 4B). Notably, at the same time interval (30 min) after irradiation, Daudi cell line exhibited a reduced expression of PARP-1 in comparison with Ramos cell lines (p<0.05), along with an increased expression of cleaved caspase-3 after exposure to 5 Gy IR (p<0.05) (Fig. 4C); moreover, this cell line displayed a significantly increased expression of pHistone H2A.X after 5 Gy IR when compared to unirradiated control and 5-Gy irradiated Ramos cells (p<0.05) (Fig. 4C) indicating a higher incidence of DNA damage. To follow-up nuclear transcription factor changes we performed western blot analysis at later time intervals (1, 3 and 24 h) observing the concomitant and partially dose-dependent upregulation of CREB, pCREB, p53, NF-κB at 3 h in Ramos cells (p<0.05), whereas pATF1 was significantly upregulated in Daudi radio-sensitive cells in a dose-dependent manner (p<0.05) (Fig. 5). Immunofluorescence experiments demonstrated the nuclear translocation of NF-κB (Fig. 6A) and the activation of CREB after 3 Gy IR (Fig. 6B) with an evident localization of pCREB even in apoptotic nuclei. As expected, after the administration of SN50, a specific pharmacological inhibitor of p50 nuclear translocation, the expression and nuclear translocation of NF-κB detected in Ramos cells after radiation exposure was downregulated (Fig. 6A). Notably, the treatment with SN50 did not induce significant changes either in cell apoptosis or in cell cycle distribution (data not shown) but in the percentage of viable Ramos cells particularly after the intermediate IR dose (Table I). Similarly to pCREB, other members of the family, namely ATF2 and ATF3, were exclusively localised inside apoptotic nuclei in irradiated samples (Fig. 7A), suggesting the involvement of these nuclear transcription factors in apoptotic cell death. Moreover, besides ATF3, another molecule involved in cell cycle regulation, cyclin D1, was found upregulated in Ramos cell line treated with 3 Gy IR dose (Fig. 7B) whereas the labelling was less intense upon 5-Gy irradiation (data not shown). Instead, no labelling for cyclin A1 was found either in control or in irradiated samples (Fig. 7B).
Figure 4.
Effects of different IR doses on nuclear transcription factors and cell cycle regulatory molecules. (A) Western blot analysis of CREB, pCREB and pATF1 in Daudi cells and Ramos cells at 30 min after exposure to different radiation doses, as indicated. Samples were normalised by incubating membranes in the presence of mouse β-actin monoclonal antibody. The most representative blot of six separate experiments is shown. (B) Relative gene expression of CASP3, CREB1, XIAP, NFKB1, RELA and IKBKB in Ramos cells, 30 min after IR exposure, as determined with real-time RT-PCR. Data was first normalised to GAPDH mRNA levels and then expressed as relative to control (calibrator sample, defined as 1). The graph bars represent the means ± SEM of relative quantification data derived from 3 independent experiments (*p<0.05, **p<0.01 vs. unirradiated control). y-axis, fold change. (C) Western blot analysis of PARP-1, caspase-3 (FL, full length; CL, cleaved forms), pHistone H2A.X in Daudi cells and Ramos cells at 30 min after exposure to different radiation doses, as indicated. Samples were normalised by incubating membranes in the presence of mouse β-tubulin monoclonal antibody. The most representative blot of three separate experiments is shown (*p<0.05 vs. unirradiated control).
Figure 5.
Effects of different IR doses on nuclear transcription factors and cell cycle regulatory molecules. Densitometric analysis of CREB, pCREB, pATF1, p53, NF-κB, Bcl-2 and pcdc2 in (A) Daudi cells and (B) Ramos cells at 3 h after exposure to different radiation doses, as indicated. Samples were normalised by incubating membranes in the presence of mouse β-actin monoclonal antibody. Significant differences (*p<0.05) are seen for Ramos cells vs. Daudi cells and for Daudi cells vs. unirradiated control (*p<0.05).
Figure 6.
Statistical histograms show the MFI (Mean Fluorescence Intensity) obtained through immunofluorescence labelling of (A) NF-κB and (B) pCREB in Ramos cells at 24 h after exposure to different IR doses, as indicated, in the absence or presence of SN50. Significant differences are seen in the mean percentage of NF-κB or CREB positive cells (***p<0.01 vs. unirradiated control; and *p<0.05 −SN50 vs. +SN50).
Table I.
Ramos cell viability at 24 h after irradiation with the indicated doses.
| Sample | Means ± SD |
|---|---|
| C 0 h | 100 |
| C 24 h | 167.3±23.2 |
| 3 Gy 24 h | 70.7±7.1 |
| 5 Gy 24 h | 70.3±10.0 |
| C SN50 24 h | 150.7±22.0 |
| 3 Gy SN50 24 h | 96.0±13.5 |
| 5 Gy SN50 24 h | 70.7±25.2 |
Values are expressed as the mean percentage ± SD of 3 different experiments. p<0.05, C vs. 3 Gy; C vs. 5 Gy; C SN50 vs. 3 Gy SN50; 3 Gy vs. 3 Gy SN50; p<0.01, C SN50 vs. 5 Gy SN50.
Figure 7.
Immunofluorescence labelling of ATF3, ATF2, cyclin D1, cyclin A1 in Ramos cells at 24 h after exposure to 3-Gy IR dose. Green and red fluorescence single emissions are shown in the panels: (A) green (ATF3) and red (ATF2); (B) green (cyclin D1) and red (cyclin A1). Nuclei were counterstained with DAPI staining. Representative fields of 1 out of 3 different experiments are shown. Original magnification, ×20. Scale bar, 50 μm.
Discussion
Radiotherapy is the therapy of choice for a number of lymphomas. Current clinical trial efforts are aimed at optimizing doses of radio/chemotherapy, with the goal of maintaining excellent long-term disease control, while limiting late toxicities (25). In the present study, we show that human Burkitt lymphoma cell lines (Daudi cells and Ramos cells) are mildly sensitive to low radiation doses currently implemented in radiotherapy regimens (1–5 Gy) since they display a slight reduction in cell viability, both due to necrotic and apoptotic cell death, and a concomitant cell cycle block in the G2/M phase aimed at repairing IR-induced cell damage. In particular, Ramos cells, which express a mutated form of p53 and a constitutive activation of the NF-κB pathway, respond to IR with an early increase in CREB protein expression (30 min) more evident after being exposed to a dose of 3 Gy and still present at 3 h when expression of p53 and NF-κB is also upregulated in a dose-dependent manner. Consistently, a significant dose-dependent induction of NFKB1 and of other survival genes, namely XIAP and IKBKB, is observed prior to protein expression. Previous studies have shown that some tumour cells exposed to radiation or certain chemotherapeutic drugs exhibit an enhanced activation of the NF-κB pathway (6,26,27), whereas inhibition of NF-κB improves the apoptotic response to radiation therapy (28). In addition, we previously demonstrated that K562 erythroleukaemia cells, known for their resistance to IR-induced apoptosis (9,29), carry out a survival strategy driven by protein kinase C (PKC) δ/NF-κB pathway activation, that helps the cells resist potentially genotoxic agents (30). Unexpectedly, in the present study we observed that the pharmacologic inhibition of NF-κB activation does not increase radiosensitivity. On the contrary, the cell viability of Ramos cells irradiated with 3 Gy increased, indicating different dose-related effects of NF-κB activation. This is related to the presence of pCREB and other family members in apoptotic nuclei and is consistent with previous findings indicating that NF-κB affects cell sensitivity to radio/chemotherapy in a cell type-specific manner (31). It is worth noting that besides NF-κB and CREB, other nuclear transcription factors, like p53 and, partially, pcdc2, are concomitantly upregulated. In particular, p53 and cdc2 have been involved in the G2 cell cycle block after IR (20,32,33) and a number of studies have shown that tyrosine phosphorylation of the human cdc2 prevents entry into mitosis in the presence of damaged or unreplicated DNA (34). On the other hand, Wuerzberger-Davis et al (34) have demonstrated that T leukemia cells are able to respond to IR injury with the NF-κB pathway activation that results in G2/M cell cycle arrest, whereas, cells accumulating in the G1 phase might undergo cell death, as demonstrated in our previous studies (6,35). The best explored link between NF-κB activation and cell cycle progression involves cyclin D1, a cyclin which is expressed relatively early in the cell cycle and which is crucial to commitment to DNA synthesis. In particular, cyclin D1 expression plays a role, not only in maintaining the integrity of the G1/S checkpoint following IR exposure, but also in the activation of molecular pathways resulting in programmed cell death (36). Moreover, cyclin D, together with cyclin A and p53, are CREB target genes crucially involved in the cell cycle regulation (15,37,38). Notably, both cyclins are genes inducible by NF-κB (39,40) and in particular cyclin A is upregulated in cell lines, transgenic mice and patient bone marrow that show increased CREB levels. ATF2 forms a complex with CRE-binding protein (CREB) and stimulates cyclin D1 promoter activity (41). In the present study, we observed the upregulation of both cyclin D1 and ATF3 when the intermediate dose of IR was applied and the presence of both ATF2 and ATF3 at nuclear level in Ramos cells undergoing apoptotic cell death. This is particularly interesting in the light of the recently published observations showing that oncogenic or tumour suppressor activities of ATF2 are associated with its nuclear/cytosolic subcellular localization (42).
Our findings highlight for the first time the involvement of CREB/ATF family members in the radiation response of lymphoid neoplastic cell lines and suggest a pro-apoptotic role of CREB and a pro-necrotic role of NF-κB in our in vitro system at least after 3 Gy IR dose. This should be taken into account in light of the increasing use of NF-κB inhibitors in the treatment of different types of human cancer (43). Ongoing studies are focused on making further functional correlations in order to better understand the role of these stress responses in radio-sensitivity/resistance mechanisms.
Acknowledgements
The authors wish to thank Professor G. Ausili Cefaro who allowed the realization of the irradiation protocol.
References
- 1.Di Pietro R, Falcieri E, Centurione L, Centurione MA, Mazzotti G, Rana R. Ultrastructural patterns of cell damage and death following gamma radiation exposure of murine erythroleukemia cells. Scanning Microsc. 1994;8:667–673. [PubMed] [Google Scholar]
- 2.Di Pietro R, Centurione L, Santavenere E, Centurione MA, Sanità Di Toppi G, Zamai L, Rana R. Ionizing radiation-induced apoptosis and DNA repair in murine erythroleukemia cells. Scanning Microsc. 1996;10:253–259. [PubMed] [Google Scholar]
- 3.Suzuki K, Yamashita S. Low-dose radiation exposure and carcinogenesis. Jpn J Clin Oncol. 2012;42:563–568. doi: 10.1093/jjco/hys078. [DOI] [PubMed] [Google Scholar]
- 4.Miscia S, Di Baldassarre A, Alba Rana R, Di Pietro R, Cataldi A. Engagement of DNA polymerases during apoptosis. Cell Prolif. 1997;30:325–340. doi: 10.1111/j.1365-2184.1997.tb00945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cataldi A, Zauli G, Di Pietro R, Castorina S, Rana RA. Involvement of the pathway PI-3-kinase/AKT 1 in the establishment of the survival response to ionising radiation. Cell Signal. 2001;13:1–7. doi: 10.1016/s0898-6568(01)00147-4. [DOI] [PubMed] [Google Scholar]
- 6.Cataldi A, di Giacomo V, Rapino M, Genovesi D, Rana RA. Cyclic nucleotide response element binding protein (CREB) activation promotes survival signal in human K562 erythroleukemia cells exposed to ionising radiation/etoposide combined treatment. J Radiat Res (Tokyo) 2006;47:113–120. doi: 10.1269/jrr.47.113. [DOI] [PubMed] [Google Scholar]
- 7.Cataldi A, Di Giacomo V, Rapino M, Zara S, Rana RA. Ionizing radiation induces apoptotic signal through protein kinase Cdelta (delta) and survival signal through Akt and cyclic-nucleotide response element-binding protein (CREB) in Jurkat T cells. Biol Bull. 2009;217:202–212. doi: 10.1086/BBLv217n2p202. [DOI] [PubMed] [Google Scholar]
- 8.Miura Y. Oxidative stress, radiation-adaptive responses, and aging. J Radiat Res (Tokyo) 2004;45:357–372. doi: 10.1269/jrr.45.357. [DOI] [PubMed] [Google Scholar]
- 9.Di Pietro R, Fang H, Fields K, Miller S, Flora M, Petricoin EC, Dveksler G, Rana RA, Grimley PM. Peroxiredoxin genes are not induced in myeloid leukemia cells exposed to ionizing radiation. Int J Immunopathol Pharmacol. 2006;19:517–524. doi: 10.1177/039463200601900307. [DOI] [PubMed] [Google Scholar]
- 10.Shaywitz AJ, Greenberg ME. CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Venkataraman SL, Dodson GE, Mabb AM, LeBlanc S, Tibbetts RS. Direct regulation of CREB transcriptional activity by ATM in response to genotoxic stress. Proc Natl Acad Sci USA. 2004;101:5898–5903. doi: 10.1073/pnas.0307718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Pietro R, di Giacomo V, Caravatta L, Sancilio S, Rana RA, Cataldi A. Cyclic nucleotide response element binding (CREB) protein activation is involved in K562 erythroleukemia cells differentiation. J Cell Biochem. 2007;100:1070–1079. doi: 10.1002/jcb.21106. [DOI] [PubMed] [Google Scholar]
- 14.Cho EC, Mitton B, Sakamoto KM. CREB and leukemogenesis. Crit Rev Oncog. 2011;16:37–46. doi: 10.1615/CritRevOncog.v16.i1-2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Auria F, Di Pietro R. Role of CREB protein family members in human haematological malignancies. In: Gali-Muhtasib H, editor. Cancer Treatment/Book 2. InTech; Rijeka, Croatia: 2013. pp. 201–226. [Google Scholar]
- 16.Deng X, Elzey BD, Poulson JM, Morrison WB, Ko SC, Hahn NM, Ratliff TL, Hu CD. Ionizing radiation induces neuroendocrine differentiation of prostate cancer cells in vitro, in vivo and in prostate cancer patients. Am J Cancer Res. 2011;1:834–844. [PMC free article] [PubMed] [Google Scholar]
- 17.Klein G, Giovanella B, Westman A, Stehlin JS, Mumford D. An EBV-genome-negative cell line established from an American Burkitt lymphoma; receptor characteristics. EBV infectibility and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology. 1975;5:319–334. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- 18.Skvortsova I, Popper BA, Skvortsov S, Saurer M, Auer T, Moser R, Kamleitner H, Zwierzina H, Lukas P. Pretreatment with rituximab enhances radiosensitivity of non-Hodgkin's lymphoma cells. J Radiat Res (Tokyo) 2005;46:241–248. doi: 10.1269/jrr.46.241. [DOI] [PubMed] [Google Scholar]
- 19.Sancilio S, Grill V, Di Pietro R. A combined approach with rituximab plus anti-TRAIL-R agonistic antibodies for the treatment of haematological malignancies. Curr Pharm Des. 2008;14:2085–2099. doi: 10.2174/138161208785294582. [DOI] [PubMed] [Google Scholar]
- 20.Impicciatore G, Sancilio S, Miscia S, Di Pietro R. Nutlins and ionizing radiation in cancer therapy. Curr Pharm Des. 2010;16:1427–1442. doi: 10.2174/138161210791033932. [DOI] [PubMed] [Google Scholar]
- 21.Camus S, Higgins M, Lane DP, Lain S. Differences in the ubiquitination of p53 by Mdm2 and the HPV protein E6. FEBS Lett. 2003;536:220–224. doi: 10.1016/S0014-5793(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Bassat H, Goldblum N, Mitrani S, Klein G, Johansson B. Concanavalin A receptors on the surface membrane of lymphocytes from patients with African Burkitt's lymphoma and lymphoma cell lines. Int J Cancer. 1976;17:448–454. doi: 10.1002/ijc.2910170406. [DOI] [PubMed] [Google Scholar]
- 23.Sancilio S, Di Giacomo V, Quaglietta AM, Iacone A, Angelucci D, Tatasciore U, Rana RA, Cataldi A, Zauli G, Di Pietro R. TRAIL promotes a pro-survival signal in erythropoietin-deprived human erythroblasts through the activation of an NF-kB/IkBalpha pathway. J Biol Regul Homeost Agents. 2011;25:375–386. [PubMed] [Google Scholar]
- 24.Caravatta L, Sancilio S, di Giacomo V, Rana R, Cataldi A, Di Pietro R, Di Pietro R. PI3-K/Akt-dependent activation of cAMP-response element-binding (CREB) protein in Jurkat T leukemia cells treated with TRAIL. J Cell Physiol. 2008;214:192–200. doi: 10.1002/jcp.21186. [DOI] [PubMed] [Google Scholar]
- 25.Campbell BA, Connors JM, Gascoyne RD, Morris WJ, Pickles T, Sehn LH. Limited-stage diffuse large B-cell lymphoma treated with abbreviated systemic therapy and consolidation radiotherapy: Involved-field versus involved-node radiotherapy. Cancer. 2012;118:4156–4165. doi: 10.1002/cncr.26687. [DOI] [PubMed] [Google Scholar]
- 26.Zauli G, Sancilio S, Cataldi A, Sabatini N, Bosco D, Di Pietro R. PI-3K/Akt and NF-kappaB/IkappaBalpha pathways are activated in Jurkat T cells in response to TRAIL treatment. J Cell Physiol. 2005;202:900–911. doi: 10.1002/jcp.20202. [DOI] [PubMed] [Google Scholar]
- 27.Sabatini N, Di Pietro R, Rapino M, Sancilio S, Comani S, Cataldi A. PI-3-kinase/NF-kappaB mediated response of Jurkat T leukemic cells to two different chemotherapeutic drugs, etoposide and TRAIL. J Cell Biochem. 2004;93:301–311. doi: 10.1002/jcb.20166. [DOI] [PubMed] [Google Scholar]
- 28.Dolcet X, Llobet D, Pallares J, Matias-Guiu X. NF-κB in development and progression of human cancer. Virchows Arch. 2005;446:475–482. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- 29.Papazisis KT, Zambouli D, Kimoundri OT, Papadakis ES, Vala V, Geromichalos GD, Voyatzi S, Markala D, Destouni E, Boutis L, et al. Protein tyrosine kinase inhibitor, genistein, enhances apoptosis and cell cycle arrest in K562 cells treated with gamma-irradiation. Cancer Lett. 2000;160:107–113. doi: 10.1016/S0304-3835(00)00569-3. [DOI] [PubMed] [Google Scholar]
- 30.Cataldi A, Rapino M, Centurione L, Sabatini N, Grifone G, Garaci F, Rana R. NF-kappaB activation plays an anti-apoptotic role in human leukemic K562 cells exposed to ionizing radiation. J Cell Biochem. 2003;89:956–963. doi: 10.1002/jcb.10560. [DOI] [PubMed] [Google Scholar]
- 31.Pajonk F, Pajonk K, McBride WH. Inhibition of NF-kappaB, clonogenicity, and radiosensitivity of human cancer cells. J Natl Cancer Inst. 1999;91:1956–1960. doi: 10.1093/jnci/91.22.1956. [DOI] [PubMed] [Google Scholar]
- 32.Winters ZE, Ongkeko WM, Harris AL, Norbury CJ. p53 regulates Cdc2 independently of inhibitory phosphorylation to reinforce radiation-induced G2 arrest in human cells. Oncogene. 1998;17:673–684. doi: 10.1038/sj.onc.1201991. [DOI] [PubMed] [Google Scholar]
- 33.Sabatini N, Di Giacomo V, Rapino M, Rana R, Garaci G, Cataldi A. JNK/p53 mediated cell death response in K562 exposed to etoposide-ionizing radiation combined treatment. J Cell Biochem. 2005;95:611–619. doi: 10.1002/jcb.20392. [DOI] [PubMed] [Google Scholar]
- 34.Wuerzberger-Davis SM, Chang PY, Berchtold C, Miyamoto S. Enhanced G2-M arrest by nuclear factor-{kappa}B-dependent p21waf1/cip1 induction. Mol Cancer Res. 2005;3:345–353. doi: 10.1158/1541-7786.MCR-05-0028. [DOI] [PubMed] [Google Scholar]
- 35.Eleuterio E, Di Giuseppe F, Sulpizio M, di Giacomo V, Rapino M, Cataldi A, Di Ilio C, Angelucci S. Proteome analysis of X-ray irradiated human erythroleukemia cells. Biochim Biophys Acta. 2008;1784:611–620. doi: 10.1016/j.bbapap.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Jin P, Gu Y, Morgan DO. Role of inhibitory CDC2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol. 1996;134:963–970. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pardo FS, Su M, Borek C. Cyclin D1 induced apoptosis maintains the integrity of the G1/S checkpoint following ionizing radiation irradiation. Somat Cell Mol Genet. 1996;22:135–144. doi: 10.1007/BF02369903. [DOI] [PubMed] [Google Scholar]
- 38.Cheng L, Li L, Qiao X, Liu J, Yao X. Functional characterization of the promoter of human kinetochore protein HEC1: Novel link between regulation of the cell cycle protein and CREB family transcription factors. Biochim Biophys Acta. 2007;1769:593–602. doi: 10.1016/j.bbaexp.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 40.Magné N, Toillon RA, Bottero V, Didelot C, Houtte PV, Gérard JP, Peyron JF. NF-kappaB modulation and ionizing radiation: Mechanisms and future directions for cancer treatment. Cancer Lett. 2006;231:158–168. doi: 10.1016/j.canlet.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Beier F, Lee RJ, Taylor AC, Pestell RG, LuValle P. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc Natl Acad Sci USA. 1999;96:1433–1438. doi: 10.1073/pnas.96.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau E, Ronai ZA. ATF2 - at the crossroad of nuclear and cytosolic functions. J Cell Sci. 2012;125:2815–2824. doi: 10.1242/jcs.095000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozakai N, Kikuchi E, Hasegawa M, Suzuki E, Ide H, Miyajima A, Horiguchi Y, Nakashima J, Umezawa K, Shigematsu N, et al. Enhancement of radiosensitivity by a unique novel NF-κB inhibitor, DHMEQ, in prostate cancer. Br J Cancer. 2012;107:652–657. doi: 10.1038/bjc.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]