Abstract
How breast diversity is generated is a fascinating and fundamental question with important clinical implications. It is clear that the diversity of phenotypes displayed by breast cancer cells reflects the array of cell types present in the disease-free breast epithelium, including luminal, basal, and stem cells. Therefore, it is hypothesized that the molecular regulators governing normal development of the breast epithelium may double as engines of breast tumor diversity. In the past few years, a deepened understanding of the mammary epithelial hierarchy has prompted the search for the cellular precursors of breast tumors. At the same time, the use of novel experimental strategies as well as the new technology of massively parallel sequencing have provided insight into the origin and evolution of breast tumors. Here, we review the current understanding of the basis of the intrinsic subtypes and the sources of intertumor heterogeneity.
Keywords: Breast cancer, tumor heterogeneity, cell-of-origin, mammary gland
INTRODUCTION
Breast tumors exhibit striking genetic and phenotypic diversity. Features that vary widely among breast cancers include proliferation rate, invasiveness, metastatic potential, response to anticancer therapy, and the presence or absence of specific oncogenic driver mutations. Indeed, each of the ‘hallmarks of cancer’ originally identified by Weinberg and Hanahan (1,2) represents a potential axis of heterogeneity along which breast tumors may be distributed. Breast cancer diversity exists at several levels, encompassing differences both between tumors from different patients (inter-tumor heterogeneity) and among cancer cells within of a single tumor (intra-tumor heterogeneity).
For many years, breast cancers have been studied and classified based on their histologic appearance and the presence or absence of select biomarkers, including hormone receptors and cytokeratins. Since the early 2000s, the revelation that breast cancers can be robustly classified into discrete molecular subtypes based on their global gene expression profiles has fundamentally shaped the current understanding of inter-tumor heterogeneity (3–5). At least six distinct molecular subtypes of breast cancer have been identified on the basis of gene expression profiling, which include luminal A, luminal B, HER2-enriched, basal-like, and claudin-low tumors, as well as a normal breast-like group (for an excellent overview of molecular subtypes, see reference 6). Since these subtypes were originally derived from unsupervised hierarchical clustering of global gene expression data, they are not defined by arbitrary selection of specific histopathologic features, and are therefore referred to as the “intrinsic” subtypes. Though the intrinsic subtypes fall quite short of encompassing the full extent of cancer diversity, in general tumors that fall within the same subtype behave in similar ways and exhibit similar sensitivity to therapy (7–10). In addition, the fact that breast cancers show long-term consistency with regard to molecular subtype and exhibit a high concordance rate between primary tumors and recurrences/metastases (11) suggests that each subtype may represent a stable biological state. Dissecting the biology of these very distinct disease entities has become a major focus of contemporary breast cancer research.
THE MAMMARY EPITHELIAL HIERARCHY
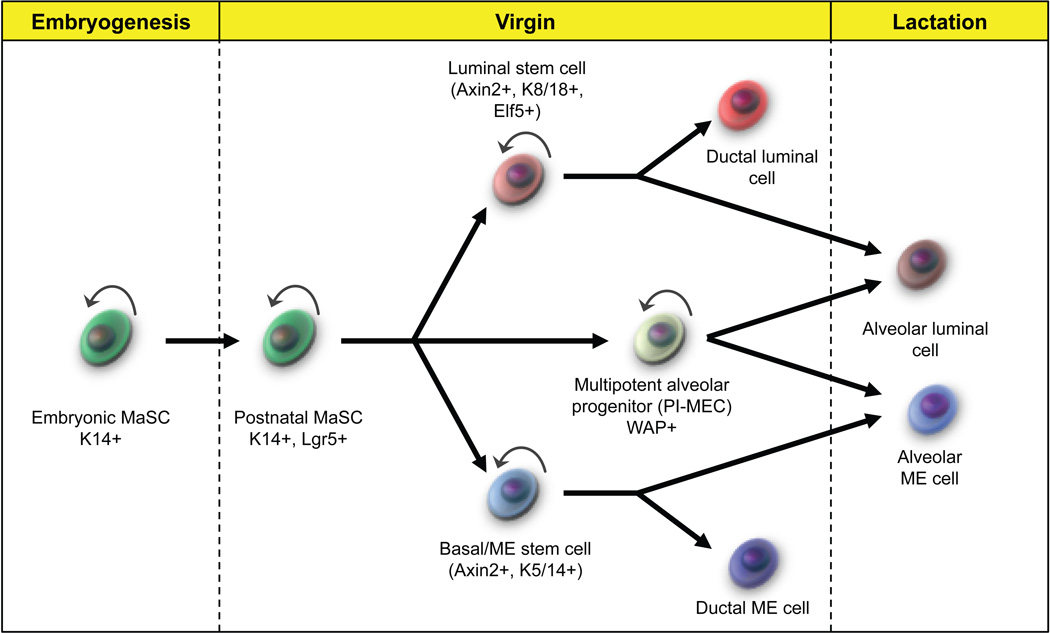
The roots of breast tumor heterogeneity lie in the developmental hierarchy of the normal mammary gland (Figure 1). The mammary epithelium is a bilayered structure, containing an inner layer of luminal cells and an outer layer of basal or myoepithelial cells (basal/ME). Anatomically, the gland is a tree-like structure consisting of a network of ducts and lobules. Milk is produced by secretory luminal cells in the lobules, while smooth muscle contraction of the ductal and lobular myoepithelial cells is responsible for milk release (12,13). The epithelium undergoes cycles of regeneration and regression with successive pregnancies. In light of this observation, the existence of a long-lived mammary tissue stem cell (MaSC) has been postulated for almost 60 years. Using the cleared fat pad assay pioneered by DeOme, several pioneering groups demonstrated that transplanting fragments of mammary tissue or bulk suspensions of MECs could serially regenerate entire epithelial trees in the recipient mice, suggesting the existence of long-lived progenitor cells within the tissue (14–16).
FIGURE 1. The mammary epithelial hierarchy.
A simplified schematic depicting the known relationships between stem, progenitor, and mature cell populations with the mammary epithelium. Bipotent MaSCs have been identified in lineage tracing in both embryonic and adult glands. More recently, unipotent stem cells have also been identified in both the luminal and basal lineages. There is also evidence for a long-lived, multipotent alveolar progenitor cell population (PI-MECs) which expands during pregnancy and survives involution. However, recent evidence suggests this population may only contribute to the luminal alveolar lineage.
In 2006, it was shown that these mammary stem cells, or MaSCs, were nearly exclusively contained within the CD24+/CD29high/CD49fhigh population of basal/myoepithelial cells. Singly transplanted cells from this fraction occasionally generated entire outgrowths in recipient mice, a bona fide demonstration of MaSC function and bipotency (16,17). However, a majority the basal/ME cells were unable to generate outgrowths. This may be an indication that the basal/ME epithelial compartment is heterogeneous, containing both stem and non-stem cells. On the other hand, it may reflect incomplete or sporadic activation of MaSCs in the transplant assay and/or technical limitations of the assay itself.
Historically, studying MaSCs in the human gland has been difficult because of a lack of a comparable assay to the cleared fat pad transplant. However, even before the definitive identification and isolation of mouse MECs, the existence of a human analog was suspected based on contiguous regions of identical X-chromosome inactivation in human mammary lobules (19). Pioneering work in this area involved in vitro colony-forming assays, where sorted subsets of human MECs are grown in tissue culture and their progeny are characterized. Using this approach, several groups reported the existence of “bipotent” human mammary stem cells that are able to generate both luminal and basal progeny (Stingl et al. 2001, another references). Recently, with the development of a model allowing the growth of human mammary epithelial cells (HMECs) in humanized mouse stroma (termed human-in-mouse or HIM), it has become possible to evaluate the behavior of these putative MaSCs in vivo as well (22). In particular, a CD49f+/EpCAM− population of human basal/ME cells, analagous to the CD24+/CD29high population in mice, is also enriched for repopulating ability in HIM xenografts (23). Alternatively, one group identified an aldehyde dehydrogenase (ALDH)-positive subpopulation in the human epithelium with enhanced ability to generate epithelial structures in the HIM model (24). In general, engraftment potential is much lower in HIM transplants than in orthotopic mouse transplantation, likely because of inadequate supporting stromal milieu or residual immune activity in recipient mice. Hence, unlike in mice, the existence of a bona fide human MaSC capable of generating a fully functional tissue from a single cell has yet to be definitively demonstrated.
Evidence also exists for unipotent stem or progenitor cells that maintain the luminal or basal cell population. Currently, the dominant paradigm is a hierarchical model of mammary development, with bipotent MaSCs residing at the apex of the hierarchy, and dedicated luminal and myoepithelial progenitors giving rise to terminally differentiated progeny. Human luminal progenitor cells display a CD49f+/EpCAM+ immunophenotype, while their mouse counterparts are CD24high/CD49flow/CD61+/Sca1−/CD133low/− (20,25,26). No specific combination of markers has yet been identified to purify myoepithelial progenitors, but these cells can be derived from serial passage of the MaSC-enriched fraction, implying that they indeed lie downstream in the hierarchy (20).
Until recently the evidence for the bipotentiality of MaSCs was limited to the observation that these cells could clonally generate both lineages in fat pad transplantation assays, which may or may not be an accurate reflection of the behavior of these cells in situ. Recently, several groups have taken advantage of sophisticated in vivo lineage tracing approaches to study the behavior of MaSCs in their native tissues. These studies have made use of lineage-specific inducible Cre alleles to allow for labeling of specific sets of cells at different developmental stages. Using this approach, multiple groups have identified a population of K14-expressing basal cells that makes long-term contributions to the luminal lineage (25–27). A small subset of basal cells marked by Procr expression seem to be highly enriched for stem cells, since clonal lineage tracing analysis demonstrated that 93% of labeled Procr+ clones contained both luminal and basal cells after a 6-week chase. However, using a very similar lineage-tracing approach that also employed an inducible K14−Cre allele, van Keymeulen et al. (28) recently reported the existence of self-renewing, unipotent luminal and basal stem cells within the mammary gland. These cells did not appear to contribute to the opposite lineage to any significant extent, suggesting that MaSCs were not essential for maintenance of the mammary gland in situ under normal physiologic conditions. Likewise, Prater et al. (29) recently described a set of cells labeled by smooth muscle actin (SMA) that behave similarly and remain restricted to the myoepithelial lineage. Therefore, the role of MaSCs in vivo and the relevance of the fat pad transplantation assay is currently a topic of some debate.
Determinants of breast cancer heterogeneity
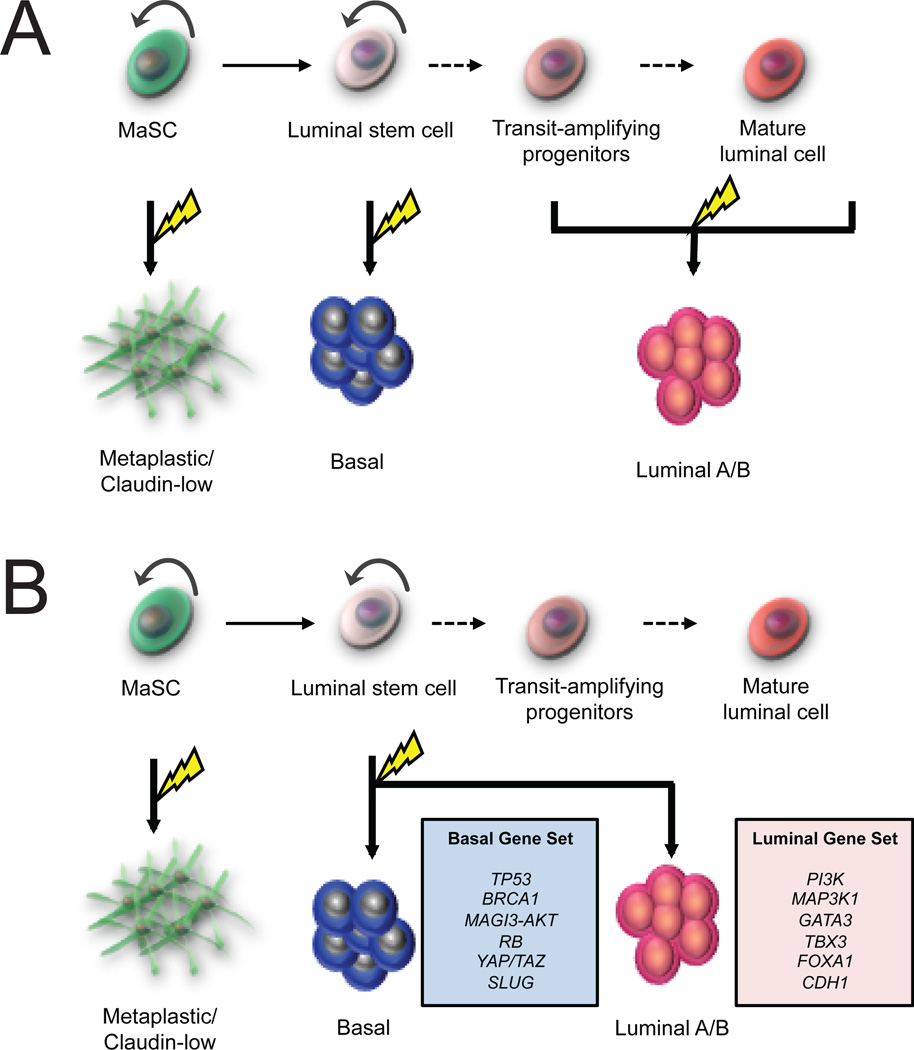
How does the mammary hierarchy relate to breast tumor heterogeneity? All breast tumors arise from the accumulation of oncogenic hits in a genetically normal precursor. Breast tumors may differ in either the nature of those genetic mutations, or in the identity of the initiating cell. This precursor, or “cell-of-origin” undergoes clonal expansion during the earliest stage of tumor progression, and therefore has a critical impact on the behavior and progression of the resulting tumor. Presumably, the characteristics of the normal precursor cell are epigenetically passed on to the tumor cells (and their progeny) following transformation (Figure 2A). As such, the identity of the cell-of-origin for various tumor types has been a topic of great interest.
FIGURE 2. Cellular origins of luminal and basal tumors.
A, Cell-of-origin model of breast tumor heterogeneity. In this model, luminal tumors arise from transformation of more committed progenitors, and maintain their differentiation during tumor progression. On the other hand, basal-like tumors arise from earlier progenitors or from unipotent luminal stem cells. B, Genetic mutation model. In this scenario, basal and luminal tumors can both arise in similar precursors but the nature of the oncogenic signal determines the eventual phenotype of the tumor.
Notably, the cell-of-origin concept is closely related to, but quite distinct from, the cancer stem cell concept (30). The cell of origin is the normal cell which receives the initial genetic insult that eventually results in a full-blown tumor, while the cancer stem cell is operationally defined as the cell that can maintain tumorigenicity and seed metastases. All tumors have a cell-of-origin, but they may not necessarily contain cancer stem cells. Also, since the CSC phenotype could emerge at some later stage of tumor progression, these two cells need not be identical or even similar. This has been a source of some confusion in the literature, since cancer stem cells are also frequently referred to as tumor-initiating cells.
As stated earlier, a second and equally important determinant of tumor phenotype is the particular collection of mutations that lead to oncogenesis, different from tumor to tumor. In recent years, massively parallel DNA sequencing technologies has enabled whole-exome analysis of a large number of human tumors. Comprehensive molecular profiles of breast and other cancer types have revealed associations between the panel of mutated genes and particular tumor subtypes (31–33). Recent data suggests that many frequently mutated genes may act as determinants of tumor differentiation in addition to exerting oncogenic effects. Therefore, through a combination of genetic and cell-of-origin effects, both epigenetic and genetic influences can serve as engines of tumor diversity.
MaSCs as precursors of breast tumors
From where in this complex cellular hierarchy do breast tumors originate? Is there a single tumor precursor for all breast tumors, or can multiple cell types initiate a tumor? If so, which cells in the mammary gland are likely tumor precursors for which types of breast cancer?
Historically, MaSCs were theorized to play a key role in breast tumor initiation. In theory, these cells should have a lower threshold for oncogenic transformation than progenitor cells (which lack self-renewal) or differentiated cells (which have lost both self-renewal and proliferative capacity). Additionally, MaSCs are long-lived within the epithelium and therefore are at increased risk for acquiring the stepwise genetic alterations required for transformation, whereas transit-amplifying progenitors and terminally differentiated cells presumably have a more limited lifespan.
There is convincing evidence that MaSCs may be the targets of transformation in certain mouse models of breast cancer (34–36). Several lines of evidence suggest that MaSCs are the precursors of the spontaneous mammary tumors which arise in transgenic MMTV-Wnt1 mice. These tumors are notable for considerable intra-tumor heterogeneity, containing luminal-like and basal-like cells. Originally, the presence of a shared secondary deletion of Pten in both types of cells suggested a common bipotent precursor, though a recent deep-sequencing study demonstrated that only half of MMTV-Wnt1 tumors follow this type of hierarchy (37). In any event, mammary glands from all MMTV-Wnt1 mice have a massively expanded basal/MaSC subpopulation prior to tumorigenesis (36,38–40). In addition, these tumors have a gene expression profile that is similar to the MaSC-enriched basal epithelial subpopulation (41). Other mammary tumor models driven by deletion of Apc or mutant B-catenin also show preneoplastic changes consistent with stem cell overexpansion, suggesting that tumors in these mice likely arise from MaSC transformation (42,43). Indeed, conditional deletion of Apc produces tumors only when targeted to Krt14-expressing basal cells/MaSCs, although the tumors were molecularly distinct from MMTV-Wnt1 tumors (44).
Collectively, these findings are reflective of the role of the Wnt signaling pathway in regulating the expansion and proliferation of MaSCs and of stem cells in general (25,45). To what extent, however, do Wnt-driven mouse mammary tumor models accurately recapitulate human tumor development? While there is substantial evidence that aberrant Wnt signaling may contribute to tumor progression and metastasis, recurrent Wnt-activating mutations in B-catenin or APC appear to be relatively rare in breast cancer. As such, most human tumors seem to follow a distinct oncogenic pathway compared with MMTV-Wnt1 mouse tumors, and the generalizability of the MMTV-Wnt1 model to human breast cancer is an open question. Certainly, aberrant Wnt signaling is a critical component of breast tumor biology, and it is possible that human tumors may indeed be initiated from the transformation of MaSCs, but this has not yet been definitively demonstrated.
Origin of BRCA1-associated breast cancer
The origin of BRCA1-associated breast cancer has been a topic of recent interest. Tumors arising in BRCA1 carriers are unusual in that they are usually of the basal subtype, which is relatively uncommon in sporadic cases (~15%). These tumors also tend to be quite heterogenous, and express markers associated with mammary stem cells including KRT5/6, KRT14 and TP63, while lacking expression of hormone receptors (46). In the past, perceived similarities between basal-like breast cancer cells and MaSCs fueled speculation that these tumors are initiated by the transformation of MaSCs (47–49). This notion was supported by direct evidence from mouse models that indicated that selective loss of Brca1 in basal cells led to the generation of basal-like tumors (50). Recently, however, the initiating cell for these murine tumors has been investigated in more detail. Molyneux et al. (51) employed a model where deletion of the BRCA1 tumor suppressor was targeted to the basal/MaSC population with a Krt14-Cre allele (on a p53-heterozygous background) or to luminal ER- cells with a BRG1-Cre allele. Interestingly, while deletion BRCA1 in basal cells/MaSCs did indeed lead to basal-like tumor formation, the tumors were malignant adenomyoepitheliomas that did not resemble the invasive adenocarcinomas most commonly in human BRCA1-associated cancer. However, when deletion of BRCA1 was specifically targeted to to β-lactoglobulin-expressing luminal cells using a Blg-Cre allele, they generated high-grade invasive ductal carcinomas (IDC) similar to those seen in BRCA1 mutation carriers. Moreover, the majority of these tumors showed a basal-like phenotype, expressing basal-specific markers such as Krt14 and Tp63. This result suggested that luminal cells, rather than MaSCs, may be the more likely precursors for basal-like disease, at least in the setting of BRCA1 loss.
Interestingly, in humans, BRCA1 haploinsufficiency is associated with lineage commitment defects prior to tumorigenesis. BRCA1 mutation carriers harbor altered luminal progenitor cell phenotypes (23,52). These luminal progenitors cells are characterized by decreased expression of mature luminal differentiation markers and abnormal expression of markers of basal epithelial cells (52). This observation has been interpreted as a defect in luminal lineage commitment and maturation, the cause of which appears to be aberrant accumulation of the Slug transcriptional repressor in luminal progenitors. In normal tissues, Slug represses luminal differentiation in basal cells and is rapidly turned over via protesomal degradation (52–54). In BRCA1-mutant tissues, however, Slug protein is aberrantly stabilized in luminal cells. Therefore, BRCA1 loss leads to abnormal maturation and differentiation of the luminal lineage, and subsequently the formation of basal-like tumors.
Origin of sporadic breast cancer
The revelation that BRCA1-associated tumors likely arise from luminal progenitors, rather than MaSCs, raises the question of whether sporadic breast tumors also arise from luminal precursors. Recent work suggests that this indeed may be the case. Apart from the Brca1/p53 example, luminal cells have been implicated as targets of transformation in other mouse models as well. Tao et al. (26) used an innovative approach where oncogenes could be delivered to the mammary epithelium in a cell type-specific manner. By introducing the Etv6-NTRK3 fusion oncogene into Krt8-expressing luminal cells, they observed a range of tumor phenotypes with variable expression of luminal and basal markers. A caveat is that this fusion protein is typically present at high frequency only in secretory breast cancer, a rare subtype.
Aberrations in the PI3K pathway occur in a large fraction of sporadic breast cancers, including luminal and basal tumors (32,33,55). Meyer et al. (56) generated a mouse model in which a constituitively active mutant form of PI3K was conditionally expressed in the luminal population driven either by WAP (which targets alveolar progenitors) or MMTV (which targets a heterogeneous group of progenitor cells). Interestingly, tumors initiated from either population were heterogeneous, and displayed evidence of both luminal and basal differentiation. A recent study by Melchor et al. (57) expanded on these findings by using a conditional knockout approach to examine the phenotype of tumors driven by either Brca2 or Pten deletion (either with or without concommitant p53 loss). In this study, variation of the tumor-initiating mutation as well as the cell of origin allowed for untangling of the effects of these two factors on tumor phenotype. In line with the previous findings, the cell-of-origin determined tumor phenotype in Brca2-deleted mice. However, interestingly, deletion of Pten and/or p53 in an identical subset of BLG-expressing, ER-negative luminal cells led to distinct phenotypes depending on the initiating lesion. While deletion of Brca2 in this population always resulted in a basal-like tumor, deletion of Pten or p53 resulted in tumors that were variably luminal A/B, basal-like, or the so-called “normal breast-like” subtype. Thus, much of the spectrum of heterogeneity seen in human tumors can be recapitulated in mouse tumors that are initiated in a luminal cell of origin.
Even so, mouse models are inherently limited in their ability to fully represent all incarnations of breast cancer in human patients. To investigate the cell of origin for human tumors Keller et al. (58) isolated luminal cells from human breast reduction tissues on the basis of cell surface marker expression, induced tumorigenesis by lentiviral transduction of the cells with multiple combinations of oncogenes, and studied the resulting tumors that formed in immunodeficient mice. Interestingly, the tumors that formed were variably luminal-like or basal-like, encompassing much of the heterogeneity seen in sporadic human tumors. On the other hand, paralleling the findings of Molyneux et al., transformation of CD10-expressing basal/ME cells generated tumors that did not resemble common forms of human cancer. Instead, these tumors were poorly differentiated carcinomas with metaplastic features, and were molecularly similar to claudin-low breast cancer. Similar tumor xenografts generated from the transformation of cultured mammary epithelial cells (HMECs), which are derived from the CD10+ fraction and display a predominately basal phenotype. When transduced with telomerase, oncogenic KRAS and SV40 large T antigen (termed HMLER), these cells form poorly differentiated or metaplastic carcinomas in mice that show heterogenous areas of squamous, papillary, giant cell, and glandular histologies (58,59). Claudin-low breast tumors often display similar features (7). Therefore, luminal progenitors likely serve as the origin of luminal and basal-like human breast cancers, while MaSCs or basal/ME progenitor cells likely initiate claudin-low tumors.
Genetic determinants of luminal and basal subtype
If transformation of luminal progenitors with the same oncogenes can variably lead to either a basal-like or luminal-like tumor, as in the examples cited previously, what ultimately determines tumor phenotype? One possibility is that the choice of promoters used in the previously discussed studies (i.e. Wap, Krt18, Blg) target a heterogeneous population of cells, and that cell-of-origin effects remain dominant in determining whether the tumor displays a luminal or a basal-like phenotype (Figure 2B). Indeed, recent evidence suggests that the luminal progenitor population is heterogeneous. In mice, most luminal progenitors appear to lack expression of estrogen receptor, but at least a portion of ER+ cells also show clonogenicity (60). In humans, several distinct subsets of luminal progenitors have been identified, including a cryptic ERBB3-expressing class that does not appear to be present in all individuals (61). However, whether these varying cell types contribute to different subtypes of breast cancer remains an open question.
An alternate possibility is that distinct initiating and/or secondary mutations occurring in each precursor cell are critical in determining the eventual tumor phenotype. In recent years, the development of massively-parallel sequencing technologies has made genome- or exome-wide sequencing of breast tumors a reality. This technology has been used to compare large numbers of individual tumors in attempt to identify recurrently mutated genes. Alternatively, deep or even single-cell sequencing has been used to reconstruct the life history of individual cancers. These studies have revealed significant differences in the mutational profiles of luminal-like and basal-like tumors. The most intuitive explanation for these associations is, of course, that particular oncogenic mutations may lead a developing tumor to adopt a particular differentiation state early in tumorigenesis. This notion is supported by the previously discussed findings where cells derived from BRCA1-mutation carriers show aberrant differentiation (52).
Luminal A/B cancers exhibit distinct mutational profiles, including mutations in PI3K, MAP3K1, GATA3, FOXA1 and TBX3, key regulators of luminal differentiation in the normal mammary gland (33). A number of frequently mutated genes in luminal A/B tumors play a key role in luminal epithelial differentiation and/or hormone receptor signaling, including GATA3, FOXA1, and TBX3. In particular GATA3 is known to play an essential role in the terminal differentiation of luminal progenitor cells (24,62). Meanwhile, FOXA1 plays a critical role in ER-dependent transcription (62). Though it remains to be proven, it seems highly likely that aberrant functioning of these genes play some role in the determination of luminal tumor phenotype.
The mutational profile of basal-like tumors is notable in that there is a higher number of somatic mutations per tumor, but a lower number of frequently mutated genes and a higher degree of genomic instability (33,63). Clearly, this unique mutational profile indicates that basal-like breast cancer is a fundamentally different disease than luminal cancer, and is more reminiscent of non-mammary tumor types such as serous ovarian carcinoma, which has a similar mutational and phenotypic profile (33,64). A striking finding is the extremely high rate of p53 mutations in basal-like disease (over 80% in the TCGA dataset), which was the only gene mutated at high frequency in this subtype. Is, then, the loss of p53 a defining event in the genesis of basal-like breast cancer? Recently, there is direct evidence from mouse models that p53 loss can bias tumors toward a more basal-like or mesenchymal fate. Liu et al. () reported that double deletion of both Pten and p53 led to the generation of triple-negative tumors that shared many features with the claudin-low subset, regardless of whether the initial deletion of Pten and p53 was driven by the MMTV or WAP promoters.
In light of these findings, it is likely that early loss of p53 may lead to secondary mutations that alter MEC phenotype and lead to basal-like tumor formation. Based on their mutational profile, basal-like tumors are hypothesized to undergo rapid clonal evolution, and recent deep sequencing studies suggest that this may indeed be the case (32,66). The loss of the DNA repair mechanisms orchestrated by p53 may allow basal-like tumors to acquire secondary mutations which impart characteristics that would provide a competitive advantage over neighboring clones - such as rapid growth, invasiveness, and loss of cell-cell contacts – all of which are also features of basal-like tumors. There is some evidence that recurrent secondary mutations, amplifications or deletions also appear to contribute to the basal-like phenotype. Amplification of TAZ, a key component in the Hippo signaling pathway, is a frequent occurrence in basal-like tumors and has been shown to regulate the basal/MaSC phenotype during mammary gland development (67,68).
Role of cellular plasticity in basal-like breast tumor development
Does the luminal origin of basal-like breast cancer imply that de-differentiation of luminal cells is an essential component of basal tumor initiation?
On the one hand, as has been pointed out previously, it may simply be the case that the tumor subtypes are inaccurately named. Lim et al. (23) directly compared the gene expression profiles of normal mammary epithelial subsets (i.e. basal/MaSC, luminal progenitor, and mature luminal cells) to those of breast tumors falling into each of the intrinsic subtypes, including basal, luminal A/B, and claudin-low. They found that the luminal A and B subtypes were most similar to the EpCAM+/CD49f− mature luminal cells, while unexpectedly, the luminal progenitor gene expression signature was most highly associated with the basal-like subtype. Meanwhile, the MaSC-signature was closely in line with the claudin-low subtype. This observation has since been noted by other groups as well (6). In fact, many of the immunohistochemical and molecular markers that define the basal-like subtype, such as cytokeratins 14 and cytokeratin 5/6, are also expressed by luminal cells in human tissues (69). Therefore, the so-called “basal-like” gene expression signature may actually be a luminal progenitor signature. Likewise, the claudin-low tumors show similarities with the basal/MaSC subpopulation of epithelial cells, a finding that is in line with the generation of metaplastic, poorly-differentiated tumors from basal/ME precursors.
On the other hand, it is possible that under the influence of certain oncogenic mutations, luminal cells become developmentally plastic and dedifferentiate to re-acquire a basal or stem-like phenotype, thereby giving rise to a basal-like tumor. Some have suggested that such a differentiation may involve a complete or partial epithelial-to-mesenchymal transition (EMT), where luminal cells lose apicobasal polarity, become motile and express markers characteristic of basal or mesenchymal cells. An EMT signature is enriched in basal-like tumors and to a greater degree in claudin-low tumors (7). Moreover, several groups have shown that EMT is intimately linked with an undifferentiated or embryonic stem-like state (70,71). However, many breast tumor specimens fail to show evidence of EMT, and the in vivo relevance of this process has been questioned (72,73). Nonetheless, amplification or overexpression of several EMT inducers have been identified in basal-like tumors, including alterations in SNAI2, SNAI1, TWIST1, and YAP/TAZ (74,67).
Cellular plasticity has also been shown to occur by stochastic mechanisms in cancerous cells as well as in normal breast epithelial cells. Recently, Chaffer et al. (75) reported that a subpopulation of mammary epithelial cells retained the capacity to spontaneously generate stem-like cells in vitro. When transformed, these cells were enriched for stem cell markers and exhibited enhanced tumorigenicity in xenotransplantation assays. Similar cell-state transitions have been observed in cultured breast cancer cell lines, where normal or malignant non-stem cells purified by FACS and cultured separately were observed to regenerate the stem cell population at a rate too rapid to be explained by cell sorting impurities (53,76). Since the in vitro tissue culture microenvironment is presumably more or less homogenous, these transitions were hypothesized likely to occur randomly rather than in a directed manner. The transitions were modelled by Gupta et al. as a Markov process, where the cells stochastically transition between luminal-like, basal-like and stem-like states at characteristic frequencies.
Recently, Phillips et al. (53) extended these findings to normal epithelial cells as well, and showed that expression of the transcription factor Slug was critical for interconversion of more differentiated luminal and basal cells to stem cells. Interestingly, mice lacking Slug were completely protected against tumor development when crossed with the MMTV-myc strain, suggesting that these cell-state transitions governed by Slug were critical for tumorigenesis. As a caveat, the in vivo prevalence of stochastic transitions between stem and non-stem cells in breast cancer has not yet been explored. Recently, however, several groups have reported in vivo evidence of stochastic interconversion between stem-like and non-stem cells in other cancer types, including Wnt-driven intestinal tumors (77).
Origin of HER2-enriched breast cancer
Regarding the pathogenesis, cell-of-origin and genetic determinants of HER2-enriched breast tumors, considerably less is known. This subtype is more heterogeneous with respect to hormone receptor expression than the other subtypes, with the majority being negative for hormone receptors but a sizeable minority showing expression of ER or PR. Furthermore, only 70% of these tumors are clinically HER2-positive (7). At a molecular level, however, all HER2-enriched tumors are defined by high expression of a common set of HER2-regulated genes. Therefore, it is tempting to speculate that HER2-enriched tumors driven by a common oncogenic signaling pathway (i.e. dysregulation of HER2/EGFR signaling) and may not necessarily share a common cell of origin. Efforts to model HER2+ disease have made extensive use of the MMTV-neu mouse (78).
To date, the most likely candidates for transformation in these mice are the so-called parity induced mammary epithelial cells (PI-MECs). These cells are identified as lacZ-expressing cells that arise in nulliparous WAP-Cre/Rosa26-lacZ mice, undergo significant expansion at pregnancy, and survive involution (79–81). PI-MECs were originally described as multipotent alveolar progenitors that contribute to the basal and luminal layers of alveoli in subsequent cycles of pregnancy and lactation. Recent work, however, suggests that PI-MECs reside in the luminal layer and exclusively give rise to ER- secretory alveolar luminal cells (82).
Several lines of evidence support the notion that PI-MECs are the targets of MMTV-neu tumorigenesis. First, mice deficient in cyclin D1 activity cannot maintain a functional PI-MEC population, and additionally are resistant to tumorigenesis when crossed with HER2-neu mice (83). Second, conditional ablation of PI-MECs was shown to reduce the onset of tumor development in MMTV-neu mice (84). Could HER2-enriched breast tumors develop from a similar population present in human tissues? To date, this has not been definitively demonstrated for several reasons. First, the existence of PI-MECs in human tissues has not been proven. Currently, no specific markers are available to prospectively isolate PI-MECs, so there is no way to identify or study these cells in human epithelia. Second, the extent to which the MMTV-neu model represents HER2-enriched breast cancer is unclear, as these tumors have a global gene expression profile that is more similar to the luminal A and B subtypes (41,85,86). Thus, the definitive origin of HER2-positive tumors remains to be elucidated.
SUMMARY AND CONCLUDING REMARKS
Recent years have witnessed impressive advances in the understanding of how breast tumors are initiated. The emerging consensus is that the most common subtypes, including luminal A/B and basal-like tumors likely arise as a result of transformation of a luminal progenitor cell of origin. In contrast, rare metaplastic and claudin-low breast tumors may have a different origin, either from a unipotent myoepithelial stem cell or a MaSC. While a few of the genetic determinants that drive basal or luminal tumor phenotype have been elucidated, such as BRCA1 loss, many more remain to be discovered. Importantly, a majority of basal-like tumors do not show loss of BRCA1 expression or BRCA1 mutation; in these tumors, a different genetic driver may allow the luminal progenitor cell of origin to transdifferentiate or adopt basal-like features during tumor development. Lastly, the origin of HER2-enriched breast cancer remains somewhat of a mystery. Better models of HER2-mediated tumorigenesis are needed to resolve the life history of this subtye.
In this review, we have focused our discussion mainly on inter-tumor heterogeneity, but also acknowledge that it is only one facet of the tremendous diversity exhibited by breast tumors. Individual clones and even single cells within each breast tumor may differ markedly in their oncogenic makeup and in the range of phenotypes manifest by the tumor cells, a feature that is usually not captured by ‘omics’ approaches such gene expression profiling. Nonetheless, the intrinsic subtypes represent fundamentally distinct disease processes with very different origins and patterns of evolution. In the future, a better understanding of the basic biology of each subtype should lead to an enhanced ability to diagnose and treat women with different forms of breast cancer.
REFERENCES
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sørile T, Eisen MB, Van De Rijn M, Jeffrey SS, Ress CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5) doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Molecular Oncology. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Research. 2010;12(5) doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clinical Cancer Research. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 9.Cheang MCU, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, et al. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65(20):9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 12.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes and Development. 2014;28(11):1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macias H, Hinck L. Mammary gland development. Wiley Interdisciplinary Reviews: Developmental Biology. 2012;1(4):533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshino K, Gardner WU. Transplantability and life span of mammary gland during serial transplantation in mice [30] Nature. 1967;213(5072):193–194. doi: 10.1038/213193a0. [DOI] [PubMed] [Google Scholar]
- 15.Daniel CW, De Ome KB, Young JT, Blair PB, Faulkin LJ., Jr The in vivo life span of normal and preneoplastic mouse mammary glands: a serial transplantation study. Proc Natl Acad Sci U S A. 1968;61(1):53–60. doi: 10.1073/pnas.61.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeOme KB, Faulklin LJ, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules. Cancer Res. 1959;19(5):515–520. [PubMed] [Google Scholar]
- 17.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 18.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 19.Tsai YC, Lu Y, Nichols PW, Zlotnikov G, Jones PS, Smith HS. Contiguous patches of normal human mammary epithelium derived from a single stem cell: Implications for breast carcinogenesis. Cancer Res. 1996;56(2):402–404. [PubMed] [Google Scholar]
- 20.Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67(2):93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- 21.Stingl J, Raouf A, Emerman JT, Eaves CJ. Epithelial progenitors in the normal human mammary gland. J Mammary Gland Biol Neoplasia. 2005;10(1):49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- 22.Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101(14):4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 24.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176(1):19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asselin-Labat M, Vaillant F, Shackleton M, Bouras T, Lindeman GJ, Visvader JE. Delineating the epithelial hierarchy in the mouse mammary gland. Cold Spring Harbor Symposia on Quantitative Biology. 2008;73:469–478. doi: 10.1101/sqb.2008.73.020. [DOI] [PubMed] [Google Scholar]
- 25.Van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/β-catenin- responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11(3):387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 26.Tao L, Van Bragt MPA, Laudadio E, Li Z. Lineage tracing of mammary epithelial cells using cell-type-specific cre-expressing adenoviruses. Stem Cell Rep. 2014;2(6):770–779. doi: 10.1016/j.stemcr.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506(7488):322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 28.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 29.Prater MD, Petit V, Alasdair RI, Giraddi RR, Shehata M, Menon S, et al. Mammary stem cells have myoepithelial cell properties. Nature Cell Bio. 2014;16(10):942–950. doi: 10.1038/ncb3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 31.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smalley M, Ashworth A. Stem cells and breast cancer: A field in transit. Nature Reviews Cancer. 2003;3(11):832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- 35.Polyak K. Breast cancer: Origins and evolution. J Clin Invest. 2007;117(11):3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100(26):15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508(1):113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci U S A. 2004;101(12):4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu BY, Kim YC, Leatherberry V, Cowin P, Alexander CM. Mammary gland development requires syndecan-1 to create a β-catenin/TCF-responsive mammary epithelial subpopulation. Oncogene. 2003;22(58):9243–9253. doi: 10.1038/sj.onc.1207217. [DOI] [PubMed] [Google Scholar]
- 40.Vaillant F, Asselin-Labat M, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. The mammary progenitor marker CD61/β3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68(19):7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- 41.Lim E, Wu D, Pal B, Bouras T, Asselin-Labat M, Vaillant F, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Research. 2010;12(2) doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imbert A, Eelkema R, Jordan S, Feiner H, Cowin P. ΔN89β-catenin induces precocious development, differentiation, neoplasia in mammary gland. J Cell Biol. 2001;152(3):555–568. doi: 10.1083/jcb.153.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moser AR, Mattes EM, Dove WF, Lindstrom MJ, Haag JD, Gould MN. Apc(Min), a mutation in the murine APC gene, predisposes to mammary carcinomas and focal alveolar hyperplasias. Proc Natl Acad Sci U S A. 1993;90(19):8977–8981. doi: 10.1073/pnas.90.19.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuraguchi M, Ohene-Baah NY, Sonkin D, Bronson RT, Kucherlapati R. Genetic mechanisms in Apc-mediated mammary tumorigenesis. PLoS Genetics. 2009;5(2) doi: 10.1371/journal.pgen.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25(2):254–264. doi: 10.1016/j.ceb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19(2):264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 47.Foulkes WD. BRCA1 functions as a breast stem cell regulator. J Med Genet. 2004;41(1):1–5. doi: 10.1136/jmg.2003.013805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A. 2008;105(5):1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foulkes WD, Stefansson IM, Chappuis PO, Bégin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95(19):1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Holstege H, Van Der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104(29):12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molyneux G, Geyer FC, Magnay F, McCarthy A, Kendrick H, Natrajan R, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7(3):403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 52.Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8(2):149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, et al. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Reports. 2014;2(5):633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer DS, Brinkhaus H, Müller U, Müller M, Cardiff RD, Bentires-Alj M. Luminal expression of PIK3CA mutant H1047R in the mammary gland induces heterogeneous tumors. Cancer Res. 2011;71(13):4344–4351. doi: 10.1158/0008-5472.CAN-10-3827. [DOI] [PubMed] [Google Scholar]
- 57.Melchor L, Molyneux G, Mackay A, Magnay F, Atienza M, Kendrick H, et al. Identification of cellular and genetic drivers of breast cancer heterogeneity in genetically engineered mouse tumour models. J Pathol. 2014;233(2):124–137. doi: 10.1002/path.4345. [DOI] [PubMed] [Google Scholar]
- 58.Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, et al. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci U S A. 2012;109(8):2772–2777. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes and Development. 2001;15(1):50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Booth BW, Smith GH. Estrogen receptor-α and progesterone receptor are expressed in label-retaining mammary epithelial cells that divide asymmetrically and retain their template DNA strands. Breast Cancer Research. 2006;8(4) doi: 10.1186/bcr1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shehata M, Teschendorff A, Sharp G, Novcic N, Russell IA, Avril S, et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Research. 2012;14(5) doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 Maintains the Differentiation of the Luminal Cell Fate in the Mammary Gland. Cell. 2006;127(5):1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Ruijter TC, Veeck J, De Hoon JPJ, Van Engeland M, Tjan-Heijnen VC. Characteristics of triple-negative breast cancer. J Cancer Res Clin Oncol. 2011;137(2):183–192. doi: 10.1007/s00432-010-0957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellis MJ, Perou CM. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discovery. 2013;3(1):27–34. doi: 10.1158/2159-8290.CD-12-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu JC, Voisin V, Wang S, Wang DY, Jones RA, Datti A, Uehling D, et al. Combined deletion of Pten and p53 in mammary epithelium accelerates triple-negative breast cancer with dependency on eEF2K. EMBO Mol Med. 2014;6(12):1542–1560. doi: 10.15252/emmm.201404402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skibinski A, Breindel JL, Prat A, Galván P, Smith E, Rolfs A, et al. The Hippo Transducer TAZ Interacts with the SWI/SNF Complex to Regulate Breast Epithelial Lineage Commitment. Cell Reports. 2014;6(6):1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Q, Zhang N, Gray RS, Li H, Ewald AJ, Zahnow CA, et al. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes and Development. 2014;28(5):432–437. doi: 10.1101/gad.233676.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gusterson BA, Ross DT, Heath VJ, Stein T. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Research. 2005;7(4):143–148. doi: 10.1186/bcr1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mani SA, Guo W, Liao M, Eaton EN, Ayyanan A, Zhou AY, et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morel A, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3(8) doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tarin D. The fallacy of epithelial mesenchymal transition in neoplasia. Cancer Res. 2005;65(14):5996–6000. doi: 10.1158/0008-5472.CAN-05-0699. [DOI] [PubMed] [Google Scholar]
- 73.Chui MH. Insights into cancer metastasis from a clinicopathologic perspective: Epithelial-Mesenchymal Transition is not a necessary step. International Journal of Cancer. 2013;132(7):1487–1495. doi: 10.1002/ijc.27745. [DOI] [PubMed] [Google Scholar]
- 74.Sarrió D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68(4):989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 75.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 77.Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1–2):25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 78.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89(22):10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner K, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: Its role in functional adaptation and tissue renewal. Development. 2002;129(6):1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 80.Boulanger CA, Wagner K, Smith GH. Parity-induced mouse mammary epithelial cells are pluripotent, self-renewing and sensitive to TGF-β1 expression. Oncogene. 2005;24(4):552–560. doi: 10.1038/sj.onc.1208185. [DOI] [PubMed] [Google Scholar]
- 81.Matulka LA, Triplett AA, Wagner K. Parity-induced mammary epithelial cells are multipotent and express cell surface markers associated with stem cells. Dev Biol. 2007;303(1):29–44. doi: 10.1016/j.ydbio.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 82.Chang TH, Kunasegaran K, Tarulli GA, De Silva D, Voorhoeve PM, Pietersen AM. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Research. 2014;16(1) doi: 10.1186/bcr3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, et al. Cyclin D1 Kinase Activity Is Required for the Self-Renewal of Mammary Stem and Progenitor Cells that Are Targets of MMTV-ErbB2 Tumorigenesis. Cancer Cell. 2010;17(1):65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Henry MD, Triplett AA, Oh KB, Smith GH, Wagner K. Parity-induced mammary epithelial cells facilitate tumorigenesis in MMTV-neu transgenic mice. Oncogene. 2004;23(41):6980–6985. doi: 10.1038/sj.onc.1207827. [DOI] [PubMed] [Google Scholar]
- 85.Hollern DP, Andrechek ER. A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to human breast cancer. Breast Cancer Research. 2014;16(3) doi: 10.1186/bcr3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfefferle AD, Herschkowitz JI, Usary J, Harrell JC, Spike BT, Adams JR, et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 2013;14(11) doi: 10.1186/gb-2013-14-11-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]