Abstract
Eukaryotes rely on efficient distribution of energy and carbon skeletons between organs in the form of sugars. Glucose in animals and sucrose in plants serve as dominant distribution forms. Cellular sugar uptake and release require vesicular and/or plasma membrane transport proteins. Humans and plants use related proteins from three superfamilies for sugar translocation: the major facilitator superfamily (MFS), the sodium solute symporter Family (SSF; only animal kingdom), and SWEETs1-5. SWEETs carry mono- and disaccharides6 across vacuolar or plasma membranes. Plant SWEETs play key roles in sugar translocation between compartments, cells, and organs, notably in nectar secretion7, phloem loading for long distance translocation8, pollen nutrition9, and seed filling10. Plant SWEETs cause pathogen susceptibility by sugar leakage from infected cells3,11,12. The vacuolar AtSWEET2 sequesters sugars in root vacuoles; loss-of-function increases susceptibility to Pythium infection13. Here we show that its orthologue, the vacuolar glucose transporter OsSWEET2b from rice, consists of an asymmetrical pair of triple-helix-bundles (THBs), connected by an inversion linker helix (TM4) to create the translocation pathway. Structural and biochemical analyses show OsSWEET2b in an apparent inward (cytosolic) open state forming homomeric trimers. TM4 tightly interacts with the first THB within a protomer and mediates key contacts among protomers. Structure-guided mutagenesis of the close paralogue SWEET1 from Arabidopsis identified key residues in substrate translocation and protomer crosstalk. Insights into the structure-function relationship of SWEETs is valuable for understanding the transport mechanism of eukaryotic SWEETs and may be useful for engineering sugar flux.
SWEET transporters are unique in that they have seven predicted transmembrane (TM) domains with two internal THBs. Prokaryotic SemiSWEETs, which also transport mono- or disaccharides4,14-16, contain only a single THB4,14. SWEETs likely arose by gene duplication of the prokaryotic unit in concert with the insertion of an inversion linker helix4. Structural analyses demonstrated that two SemiSWEETs dimerize in a parallel configuration to create a translocation pore14-16. Based on the SemiSWEET structure, a single SWEET unit with its two THBs was proposed to form the translocation path14. However, single SWEETs may not function autonomously, because they can form functionally important oligomers4. Low sequence conservation and the unpredictable effect of the inversion linker TM domain on the structure of eukaryotic SWEETs limit our mechanistic understanding of the transport process.
To elucidate the molecular transport mechanism of eukaryotic SWEETs and determine the relevance of oligomerization for sugar transport, we carried out a systematic analysis of eukaryotic SWEETs to identify candidates suitable for structural and biochemical studies. OsSWEET2b from rice Oryza sativa featured desirable biochemical properties. Phylogenetically, OsSWEET2b belongs to Clade I3,6,7, which includes the founding family member, Arabidopsis thaliana SWEET1 (AtSWEET1), a plasma membrane hexose transporter3 (Extended Data Fig. 1), and the closest relative of OsSWEET2b, AtSWEET2, a transporter involved in vacuolar retention of sugars and pathogen resistance13. Similar to AtSWEET2, OsSWEET2b-EGFP localized to vacuolar membranes when expressed in yeast (Fig. 1a). Consistent with the intracellular localization, OsSWEET2b did not complement the glucose uptake deficiency of a yeast plasma membrane hexose transporter mutant (Extended Data Fig. 2).
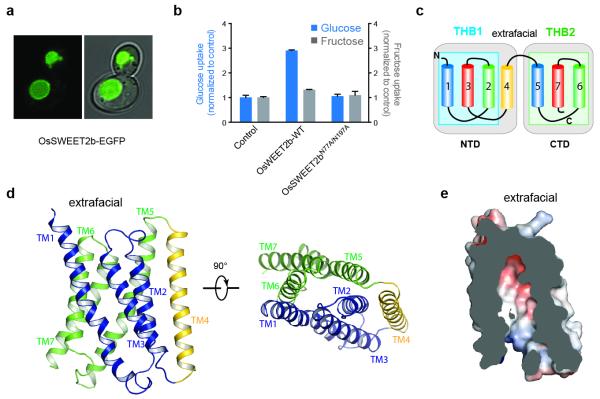
Figure 1. Localization and structure of OsSWEET2b.
a, OsSWEET2b-EGFP localizes to the vacuolar membrane in yeast. b, Transport activity of OsSWEET2b was examined by a liposome uptake assay. Glucose uptake is shown as bars in blue. Liposomes with OsSWEET2b showed elevated glucose uptake compared with control liposomes without protein. Mutation of two putative pore-lining asparagine residues abolished transport activities (±SEM, n=4). Fructose uptake is shown as bars in grey (±SEM, n=3). c, Membrane topology diagram of OsSWEET2b. TM4 and THB1 constitute the N-terminal domain (NTD) while THB2 forms the C-terminal domain (CTD). d, Ribbon representation of the OsSWEET2b protomer. THB1 is coloured in blue, THB2 in green, and TM4 in yellow. e, Slab view of OsSWEET2b in an inward (cytoplasmic) open conformation. Surface is coloured according to electrostatic potential.
To directly test activity, OsSWEET2b was purified and reconstituted into lipid vesicles. Vesicles containing OsSWEET2b showed elevated glucose transport activity compared to control vesicles; activity was abolished using OsSWEET2b mutations known to block SWEET transport activity14 (Fig. 1b). By contrast, fructose uptake was marginal and close to the detection limit (Fig. 1b). A chimera between the N-terminal (first four TMs) domain of OsSWEET2b and the C-terminal (last three TMs) domain of OsSWEET1a, a close Clade I homolog of OsSWEET2b, resulted in robust glucose transport activity (Extended Data Fig. 3). Together, these results implicate OsSWEET2b in glucose transport, similar to other Clade I and II SWEET transporters. Since AtSWEET1 is closely related to OsSWEET2b, can be measured in a variety of heterologous transport assays, and has the greatest quantity of available functional data, it was used for structure-function studies directed by the OsSWEET2b structure, below.
We crystalized and solved OsSWEET2b structures in two forms at 3.1 Å (P21) and 3.7 Å (P212121) resolution (Table 1). Experimental phases were determined by single-wavelength anomalous dispersion (SAD) on selenomethionine-containing crystals (Extended Data Fig. 5a). For one asymmetric unit of either crystal form, three OsSWEET2b molecules are related by a non-crystallographic three-fold axis perpendicular to the membrane plane, forming a trimeric assembly (Fig. 3a & 3b; Extended Data Fig. 5b & 5c). Individual OsSWEET2b copies superimposed with a root-mean-square-deviation (RMSD) of less than 0.8 Å, indicating nearly identical overall structures.
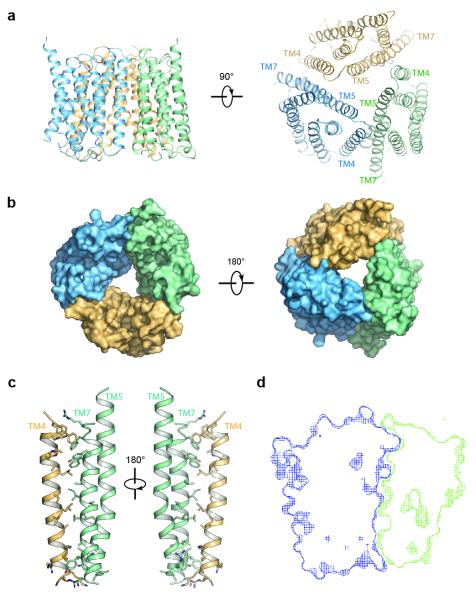
Figure 3. Structure of the OsSWEET2b trimer.
a, Two orthogonal views of the OsSWEET2b trimer in ribbon representation. b, Surface representations of OsSWEET2b viewed from the intrafacial (cytosolic; left) and extrafacial (luminal; right) side. c, Close-up view of the trimer interface. TM4 from one protomer and TM5 and TM7 from the neighbouring protomer are shown as ribbon representation. Side chains of residues participating in the interaction are shown as sticks. d, Cross-section view of the trimer interface. The two interacting protomers are presented as blue and green surface meshes.
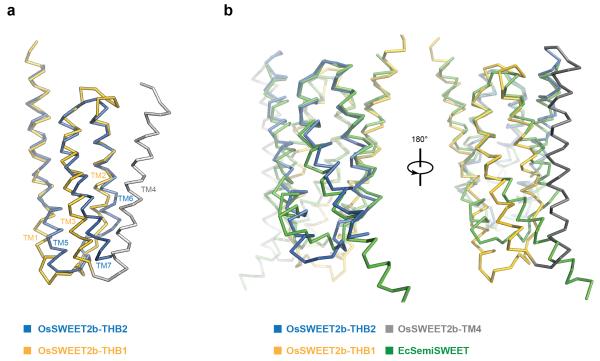
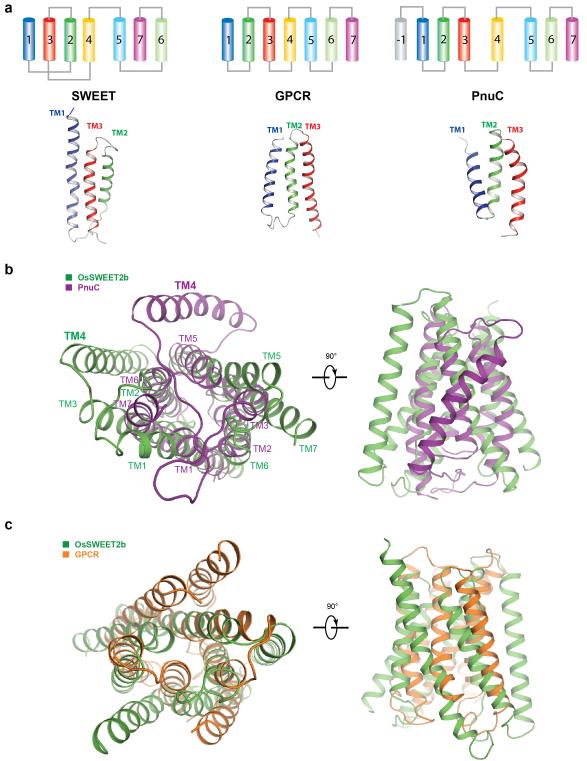
Each OsSWEET2b protomer is composed of seven TM helices. The N- and C-terminal three TM helices form THBs with a characteristic 1-3-2 helix arrangement and a similar fold as in SemiSWEET THBs14-16 (Fig. 1c & 1d). Comparison of OsSWEET2b with SemiSWEET revealed that six TMs in an OsSWEET2b protomer resemble the functional dimer of SemiSWEET where the N-terminal THB1 and C-terminal THB2 enclose a putative transport route (Fig. 2a). This indicates that a SWEET protomer is sufficient to form the pore. TM4, designated as an inversion linker, spans the membrane and orients THB2 parallel to THB1, creating a similar configuration as in homodimeric SemiSWEETs14. TM4 packs intimately against THB1 to constitute the N-terminal domain of SWEET, but barely contacts THB2 within the same protomer (Fig. 1c & 1d). This feature can explain previously puzzling observations that co-expression of AtSWEET1 split as THB1+TM4 and THB2 reconstituted transport activity, while THB1 and TM4+THB2 did not4. To accommodate TM4, the intrafacial (cytosolic) half of THB1 bends away from where TM4 binds, causing significant structural divergence of THB1 and THB2 (Extended Data Fig. 6a). Together, the membrane topology of SWEET shows drastically different connectivity and spatial TM arrangement when compared to other known heptahelical membrane proteins, in particular G-protein coupled receptors or PnuC vitamin transporters17-19 (Extended Data Fig. 7).
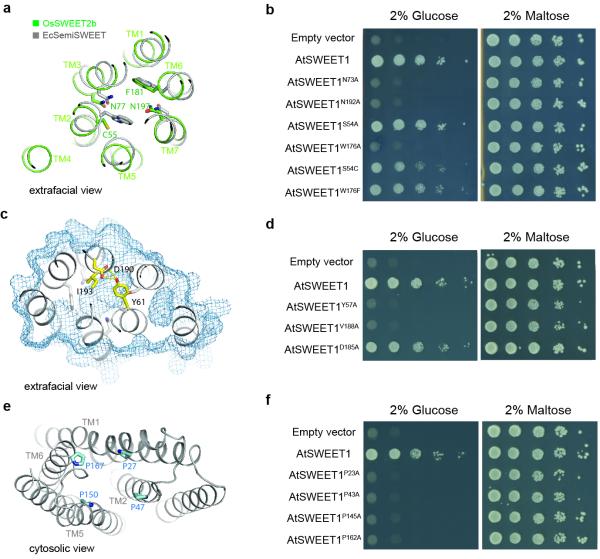
Figure 2. Key elements of the transport pathway.
a, Comparison of the substrate binding pocket of OsSWEET2b (green) to that of EcSemiSWEET (gray). Key residues in the binding pocket are shown as sticks. b, AtSWEET1 complements the defective growth in glucose of EBY4000, a hexose transport-deficient yeast strain. W176A, N73A, and N192A mutations in AtSWEET1 abolished glucose-dependent growth. Growth on maltose was not affected by the different mutants. c, Extrafacial (extracellular) gate. Residues that cap the aqueous cavity from the extrafacial side form the extrafacial gate with side chains shown as sticks in yellow. Residues in the substrate-binding pocket are coloured in grey. The surface of the protein is shown as pale blue mesh. d, Effect of mutations in the extrafacial gate of AtSWEET1. Alanine substitution of V188, but not the conserved D185, in the extrafacial gate of AtSWEET1 abolished growth in glucose. Alanine substitution of Y57 leads to a growth defect on glucose, possibly due to mistargeting4. e, Intrafacial hinge points. The four proline residues (shown as sticks) lining the transport pathway are near the same level in the membrane. f, The intrafacial (cytosolic) gate is required for sugar transport. Mutations in the conserved prolines that form the intrafacial gate of AtSWEET1 (P23A, P43A, P145A and P162A) eliminate the growth of the EBY4000 yeast strain in synthetic medium supplemented with glucose.
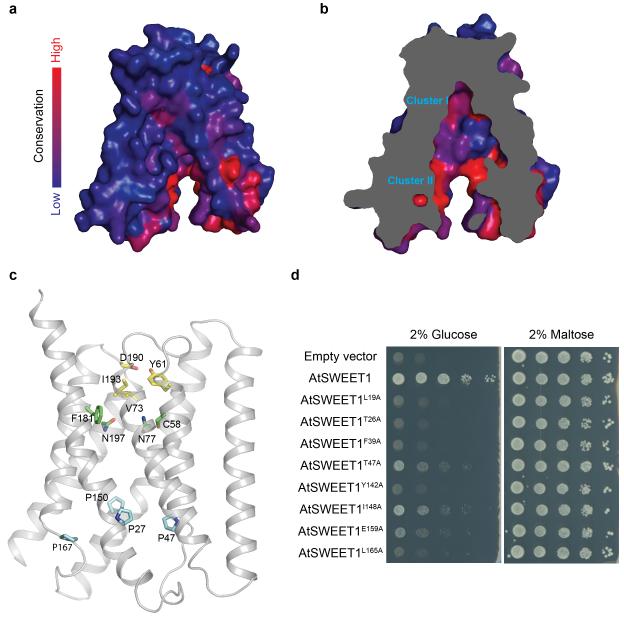
Phosphorylation of the C-terminus of a plasma membrane SWEET support that it faces the cytosol20. By analogy, we predict that the C-termini of the vacuolar OsSWEET2b face the cytosol (intrafacial side), while the extrafacial side locates in the vacuolar lumen (Fig. 1c). Between the N- and C-terminal domains of each protomer, a large solvent-accessible cavity extends from the cytosolic side to the top of the molecule where it is sealed from the vacuolar side by an extrafacial gate (Fig. 1e). The structure thus appears to represent an inward (cytosolic) open conformation. A major constituent of the extrafacial gate is Tyr61 on THB1 (Fig. 2c & Extended Data Fig. 8c), hydrogen-bonding with Asp190 and interacting with Gln132 (Fig. 2c). This Tyr-Asp pair is highly conserved (Extended Data Fig. 4). The Ile193 side chain also forms part of the luminal gate (Fig. 2c). Replacing residues equivalent to Tyr61 and Ile193 in AtSWEET1 (Tyr57 and Val188) abolished transport activity, confirming their importance (Fig. 2d). Consistent with roles in transport, the majority of conserved residues of plant SWEETs line the cavity, with two highly conserved clusters near the predicted substrate-binding pocket and the putative intrafacial gate (Extended Data Fig. 8a & 8b). A series of hydrophobic residues line the transport pathway, potentially lowering resistance for sugar passage (Fig. 1e).
Structural comparison of OsSWEET2b with EcSemiSWEET in an inward-open conformation16 reveals common themes and distinct features. THB1 of OsSWEET2b differs significantly from the protomer of EcSemiSWEET (RMSD = 4.1 Å), while THB2 superimposes better (RMSD = 2.4 Å; Extended Data Fig. 6b). OsSWEET2b residues involved in possible substrate binding or extrafacial gate formation are positioned close to those in EcSemiSWEET16 (Fig. 2a). This indicates that heptahelical eukaryotic SWEETs and tri-helical prokaryotic SemiSWEETs share common sugar binding pockets and transport routes. Consistent with that, the extrafacial half of OsSWEET2b overlays better with EcSemiSWEET than the intrafacial half.
The transport pathway and putative substrate-binding pocket in OsSWEET2b are highly asymmetrical, contrasting the symmetry of the ancestral SemiSWEET homodimers. In SemiSWEETs, pairs of tryptophans and asparagines are invariant, forming the binding pocket and facilitating transport14-16. In OsSWEET2b, two highly conserved asparagine residues (Asn77 and Asn197) surround the putative substrate-binding pocket at equivalent positions (Fig 2a & Extended Data Fig. 8c). Both are essential for AtSWEET1 activity14 (Fig. 2b). In contrast, no tryptophan pairs were found along the cavity. Instead, Cys58 on THB1 and Phe181 on THB2 occupy equivalent positions in OsSWEET2b, while a Ser54 and Trp176 are the equivalents in AtSWEET1 (Fig. 2a & Extended Data Fig. 4). Replacing Trp176 of AtSWEET1 with alanine abolished activity, whereas alanine- or cysteine-substitution of Ser54 had a minimal effect on activity (Fig. 2b). Furthermore, substituting Trp176 in AtSWEET1 with phenylalanine had little effect on transport activity (Fig. 2b). Together these results indicate that one aromatic residue in THB2, but not THB1, is essential for transport, highlighting the asymmetrical configuration of SWEET. Thus, the two THBs diverged to create an asymmetric binding pocket in eukaryotic SWEETs.
Near the intrafacial side, TM1, TM2, TM5, and TM6 are the main constituents of the transport route (Fig. 2e). Notably, in each of these four TMs, a proline that either terminates the helix or induces a kink was found facing the transport route and located at a similar level in the membrane (Fig. 2e). Substituting any of the four prolines caused loss of AtSWEET1 activity (Fig. 2f, Extended Data Fig. 9). These observations, together with the strict conservation across all Arabidopsis SWEETs, support important roles of the proline tetrad in transport. The proline ring might enable concerted structural rearrangements associated with transitions between conformational states, possibly as hinges for gating the transport pathway. Interestingly, a highly conserved PQ motif is found at the equivalent position on TM1 in SemiSWEETs14-16, with glutamine forming a key component of the cytosolic gate through cross-protomer hydrogen bonds. The absence of a glutamine next to the proline in eukaryotic SWEETs suggests that cross-protomer hydrogen bonds are not required to form its cytosolic gate, which is likely made possible by covalent linkage through TM4. Residues immediately above and below the proline ring in the pore are highly conserved, and substitution with alanine reduced activity (Extended Data Fig. 8d), indicating that neighbouring residues may also contribute to the intrafacial gate.
Our structural and functional observations delineate the transport route to within a single SWEET protomer. Crystal structures reveal three OsSWEET2b units forming a trimer (Fig. 3a & 3b). Interactions between the protomers are primarily mediated by TM4 of one protomer with TM7 as well as the C-terminal half of TM5 of the neighbouring protomer (Fig. 3a & 3c). Several lines of evidence support the physiological relevance of trimerization. First, the relative position of the three protomers is compatible with a trimer in the membrane plane (Fig. 3a). Second, the interface between protomers is extensive, burying ~1700 Å2 between two protomers with a shape complementarity index score of 0.69, similar as in typical antibody-antigen pairs21 (Fig. 3d). Third, identical trimeric assembly was observed in two crystal forms with different packing modes, indicating that trimer formation is independent of specific crystal packing (Extended Data Fig. 5b & 5c).
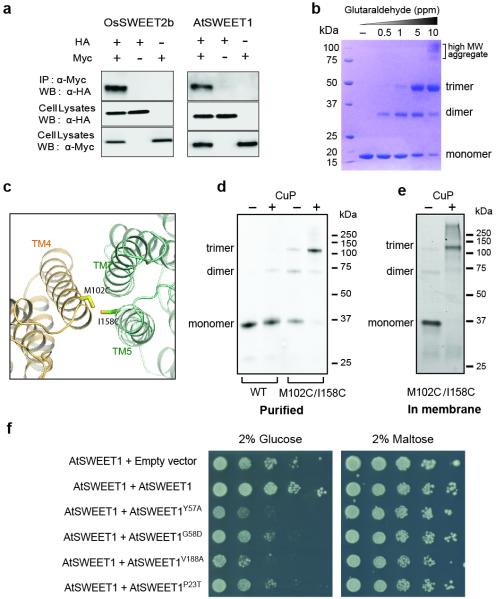
To validate the trimeric assembly, we performed biochemical characterizations. Co-immunoprecipitation experiments suggest that both OsSWEET2b and AtSWEET1 form homo-oligomers (Fig. 4a). Furthermore, glutaraldehyde cross-linked OsSWEET2b into trimers in a concentration-dependent manner (Fig. 4b), supporting trimer formation in solution. To dissect whether the observed trimer of OsSWEET2b represents its quaternary structure in the membrane, we carried out site-directed disulfide-bridge cross-linking studies. The double cysteine mutant with a pair of cysteines at the trimer interface (Fig. 4c) cross-linked via disulfide bridges under oxidizing conditions in both detergent micelles and in the isolated membrane, as shown by the newly formed crosslink product at the expected trimer position on SDS-PAGE (Fig. 4d & 4e). Together, these data support trimeric assembly as observed in crystal structures to represent the in vivo quaternary structure.
Figure 4. Trimer formation by OsSWEET2b.
a, Co-immunoprecipitation (Co-IP) of OsSWEET2b and AtSWEET1. Left, c-Myc-tagged OsSWEET2b pulls down HA-tagged OsSWEET2b; right, c-Myc-tagged AtSWEET1 pulls down HA-tagged AtSWEET1. b, Cross-link of purified OsSWEET2b in detergent micelle. Increasing amounts of glutaraldehyde (0, 0.5, 1, 5, 10 ppm) were incubated with the purified OsSWEET2b. The samples were analysed by SDS-PAGE. c, The design of cysteine pair mutations at the interface of the trimer. M102 and I158 were mutated to cysteines (side chains shown as sticks). d, Cysteine-directed cross-linking of OsSWEET2b in detergent solution. GFP-tagged OsSWEET2b with M102C/I158C mutations was purified and oxidized in air or with copper phenanthroline (CuP). Proteins separated on SDS-PAGE were imaged by in-gel fluorescence of GFP. e, Cysteine-directed cross-linking of OsSWEET2b in membrane. Cell membranes containing M102C/I158C mutant were untreated or treated with CuP. OsSWEET2b-GFP was visualized by in-gel fluorescence. f, Mutations in the residues at the gates of AtSWEET1 have a dominant negative effect on transport. EBY4000 yeast co-expressing wild-type AtSWEET1 and either an empty vector control or wild-type AtSWEET1 were able to grow on medium supplemented with glucose as the sole carbon source. Co-expression with an AtSWEET1-Y57A or G58D mutant prevents growth on glucose. Similarly, co-expression with mutants that abolish transport and that are localized in the intrafacial (P23T) or extrafacial (V188A) gates prevent growth on glucose. As a control, growth on maltose was not affected.
The trimeric assembly of OsSWEET2b provides a structural basis for explaining observations from split-GFP and split-ubiquitin experiments that SWEETs homo- or hetero-oligomerize4. Mutagenesis identified dominant-negative mutations of AtSWEET1 that inhibit the activity of co-expressed wild-type protein; finding originally thought to indicate transport route formation from multiple subunits4. The trimeric structure of OsSWEET2b offers a new explanation: each protomer encompasses a transport route, though protomers might be coupled allosterically. Two previously identified dominant-negative mutations, Y57A and its neighbouring G58D, were expected to disrupt the extrafacial gate or interfere with its conformational transitions given that Y57 is a major component of this gate. Though mistargeting of the Y57 mutant complicates the interpretation, the G58 mutant is properly localized to the plasma membrane, and has a similar dominant-negative effect. This suggests that interference with the proper function of the extrafacial gate in one protomer may hinder activity of other protomers, consistent with allosteric coupling. To further assess the role of the extrafacial gate, we examined V188, the other main contributor to this gate. As shown above, the V188A mutation abolishes transport activity in AtSWEET1 (Fig. 2d). When co-expressed with wild-type AtSWEET1, the V188A mutant almost completely inhibited activity of the wild type form (Fig. 4f). In addition, we confirmed that the P23T mutation causes dominant negative inhibition of a co-expressed wild-type transporter4, possibly due to its effect on the cytosolic gate. These gate mutations might impair conformational state transitions, as shown in other transporters22,23. Together, these results show that gate mutations cause dominant-negative effects and support that structure and function are coupled among SWEET protomers. As a variety of SWEETs form homo- or hetero- oligomers4, this coupling may be more generally applicable, expanding the properties and regulatory states of SWEET sugar transporters.
The proposed structural coupling of protomers may affect conformational states and cause functional coupling, which bears similarities to cooperativity found in other classes of transporters. For instance, human GLUT1 forms tetramers in which each protomer transports glucose, however protomers interact cooperatively such that two subunits exist in outward-open and two as inward-open states24. Similarly, protomers of triple-barrelled ammonium transporters (AMTs) are allosterically coupled in a way that transport activity is feedback-inhibited by its own substrate via phosphorylation25,26.
Membrane transporters with internal symmetry are widespread; notable examples include MFS, LeuT-fold, AMT, CLC, and ABC transporters27,28. These transport proteins are thought to have evolved from simple “half-transporters” containing a single repeat via gene duplication and subsequent fusion28. The availability of the structures of OsSWEET2b and several SemiSWEET transporters provides a structural framework to understand the link between half-transporters and full-transporters with internal repeats. In eukaryotic SWEETs, covalent fusion of two THBs through an inversion TM creates an asymmetrical translocation pathway, greatly enriching their structural features compared to the simpler symmetrical SemiSWEETs. Covalent fusion in eukaryotic SWEETs thus expands their functional capacity to accommodate diverse substrates, enhance substrate specificity, or fine-tune activity.
METHODS
Protein expression and purification
The Oryza sativa OsSWEET2b coding sequence was cloned into a modified pPICZ-C vector (Invitrogen Life Technologies) with C-terminal 3C protease cleavage site, EGFP and His-tag. To obtain diffracting crystals, the intrinsically disordered and poorly conserved 15 residues at the C-terminal after TM7 were removed (Extended Data Fig. 4). Transformed Pichia pastoris were grown until OD600 10 and induced at 27°C. Membrane proteins of lysed cells were extracted by 3% dodecyl-β -D-maltoside (DDM, Anatrace) in the presence of protease inhibitor cocktail at 4°C for 2 hours. The insoluble fractions were removed by centrifugation at 16,000 rpm for 45 min. The supernatant was incubated with cobalt resin at 4°C for 2 hours. The resin was washed with buffer containing 0.3% n-decyl-β-D-maltoside (DM) and 20 mM imidazole. The target protein was cleaved off from the tag by incubating with 3C protease. For crystallization experiments, the protein surface was modified by reductive methylation29. The protein was further purified by gel filtration (Superdex 200, GE Healthcare) in buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl and 0.5% n-nonyl-β-D-glucopyranoside (NG, Anatrace). The peak fraction was collected and concentrated to 10 mg/ml for crystallization trials. For nonspecific cross-linking, the protein was extracted and purified in DDM. After the cobalt affinity purification and 3C protease cleavage, the protein was directly subject to further purification by gel filtration in 10 mM HEPES-NaOH (pH 8.0), 150 mM NaCl and 0.05% DDM.
To produce selenomethionine-labeled OsSWEET2b protein, sf9 insect cells (Spodoptera frugiperda) in methionine-free medium were infected by baculovirus (Bac-to-Bac system, Invitrogen) containing OsSWEET2b construct and were supplemented with 100 mg/L selenomethionine after 12 hours. Cells were harvested after 48 hours before the membrane fractions were isolated by ultracentrifugation. The protein was purified in the same manner as the unlabelled protein.
Crystallization and structural determination
The crystals were grown using the hanging drop method by mixing protein with an equal volume of crystallization solution at 22°C. Plate-like crystals (Form I and SeMet crystal) were grown in 0.1 M MES (pH 6.5), 30% PEG400, 10mM MnCl2, 5% ethanol. Cubic crystals (Form II crystal) were grown in 0.1 M MES (pH 6.0), 27% PEG400. The Form I crystals were directly flash-frozen in liquid nitrogen. The Form II crystals were cryo-protected and dehydrated by gradually increasing PEG400 concentration to 44% and then flash-frozen in liquid nitrogen.
The diffraction data were collected at 24ID-C/E and 23ID-B/D of the Advanced Photon Source and 5.0.2 of Advanced Light Source. Native data set was processed using HKL200030. The selenomethionine anomalous data were collected from multiple crystals and integrated using XDS31 as implemented in RAPD (https://github.com/RAPD/RAPD). The eight most correlated partial datasets from three crystals, as determined by the Pearson’s coefficient between common Bragg reflections, were combined with POINTLESS and scaled with AIMLESS32. The Se sites and initial phases were determined by SHELX C/D/E33 and improved by Phaser-EP34. The map was improved by Resolve35 and was of sufficient quality to identify and trace transmembrane helices with the aid of Se positions. The model built into the map was directly refined against the native data set in P21 space group (Form I). The resulting model was subject to iterative refinement with NCS restrains in Phenix and manual rebuilding in Coot36,37. The final model has 98.8% residues in the favored region with the rest in the allowed region from the Ramachandran plot. For P212121 data set (Form II), the molecular replacement solution was found using the refined model from Form I by Phaser38. The model was first refined by DEN and then iteratively by refine.phenix37,39. In the final model, 95.9% residues are in the favored region and the rest are in the allowed region. Structure quality was assessed by MolProbity and figures were prepared based on higher resolution P21 form using PyMOL40,41.
Cross-linking
For nonspecific cross-linking, purified protein in DDM (around 1 mg/ml in 15 μl) was incubated with various concentrations of glutaraldehyde (0, 0.00005%, 0.0001%, 0.0005%, 0.001%) at room temperature for 8 hours. The reaction was quenched by addition of 50 mM Tris-Cl, pH 8.0. The sample was mixed with equal volume of SDS loading buffer, separated on 10% SDS-PAGE gels and stained with Coomassie blue. For site-directed cysteine cross-linking of protein in detergent micelles, the GFP-tagged protein was purified by cobalt affinity column in 0.05% DDM and incubated with 0.3 mM CuSO4 and 0.9 mM o-phenanthroline for 2 hours at 4°C. The reaction was quenched by 50 mM EDTA. For site-directed cysteine cross-linking in membrane, the membrane fraction was prepared and incubated with 0.3 mM CuSO4 and 0.9 mM o-phenanthroline for 2 h at 4°C. The reaction was stopped by the addition of 50 mM EDTA. All samples were mixed with equal volume of SDS loading buffer and separated on 10% SDS-PAGE gels. The gels were imaged using a BioRad imaging system with excitation and emission wavelengths of 488 nm and 522 nm, respectively.
Liposome-based uptake assays
Liposomes were prepared according to a published protocol42. Purified OsSWEET2b was reconstituted into E. coli polar lipid (Avanti) with a protein to lipid ratio of 1:80 in KPM6.5 buffer (50 mM potassium phosphate, 2 mM MgSO4, pH 6.5). The proteoliposomes were extruded through 400 nm membrane and harvested by ultracentrifugation at 100,000g for 1 hr. The proteoliposomes were washed twice with ice-cold KPM6.5 buffer and resuspended in the same buffer to a final concentration of 100 mg/ml immediately before the assay. For each uptake assay, 2 μl of proteoliposomes were diluted into 100 μl of assay buffer containing 0.2 μCi of D-[14C]glucose in the presence of 5 mM cold glucose. The reaction mixtures were incubated for 2 hr or 3 hr at RT, and then rapidly filtered through 0.22 μm cellulose acetate filter membranes (Millipore). The filter membranes were immediately washed with 2 ml ice-cold reaction buffer without any substrate and taken for liquid scintillation counting. The uptake of fructose was performed similarly. All the experiments were repeated at least three times independently.
Yeast complementation assay and fluorescence imaging
The transport activity of AtSWEET1 and its mutants was tested by complementing the growth defect of the hexose uptake mutant yeast strain EBY4000 [hxt1-17Δ::loxP gal2Δ::loxP stl1Δ::loxP agt1Δ::loxP ydl247wΔ::loxP yjr160cΔ::loxP]43. The coding sequence for AtSWEET1 and mutants were cloned in the vectors pDRf1-GW, p112A1NE-GW and pDRf1-GFP GW vectors for yeast expression. Expression of the transgene in p112A1NE is under the control of the ADH1 promoter, while overexpression in the pDRf1 vector is regulated by the strong PMA promoter. Yeast was transformed using the lithium acetate/SS carrier DNA/PEG method44. Transformants were selected on synthetic medium (0.17% YNB and 0.5% (NH4)2SO4) supplemented with 2% maltose and auxotrophic requirements. Cells were grown in liquid synthetic medium overnight and then diluted to an OD600 of ~0.2 in water before plating. Ten-fold serial dilutions were spotted on medium containing auxotrophic requirements and 2% glucose as the sole carbon source or 2% maltose as a control. Plates were incubated at 30°C for 4 days, and imaged using a CanoScan scanner (Canon). Representative samples from three separate transformations are shown.
Confocal imaging of EGFP tagged AtSWEET1 and mutants was performed on a Leica TCS SP5 microscope with a HCX PL APO lambda blue 63X 1.40 OIL UV objective, 488 nm excitation and 492–600 nm emission. Images were autocontrasted and overlaid with Fiji (http://fiji.sc). Cells were grown overnight in liquid synthetic medium supplemented with 2% maltose and auxotrophic requirements, and resuspended in water before imaging.
Co-Immunoprecipitation
OsSWEET2b and AtSWEET1 coding sequences without stop codon were Gateway-cloned (Life Technologies) into the yeast expression vectors pDR-GW- Trp1-4xMyc and pDR-GW- Leu2-3xHA carrying either the TRP1 or the LEU2 auxotrophic markers. These vectors were derived from pDRf1GW 45 by replacing the URA3 marker by a TRP1 (pDR-GW-TRP1) or a LEU2 marker (pDR-GW-Leu2) to allow for coexpression and by adding 4 copies of the c-Myc tag or 3 copies of the HA-tag. Tagged proteins were expressed in the protease-deficient Saccharomyces cerevisiae strain BJ5465 [MATa ura3-52 trp1 leu2-delta1 his3-delta200 pep4::HIS3 prb1-delta1.6R can1 GAL] (ATCC). Yeast coexpressing either cMyc- and HA-tagged OsSWEET2b, or cMyc- and HA-tagged ATSWEET1, were selected on synthetic medium (0.17% YNB and 0.5% (NH4)2SO4) supplemented with 2% glucose without leucine and tryptophan. Cells were grown in liquid synthetic medium until OD600 0.4-0.6, and harvested by centrifugation at 4,000×g for 10min. Cell lysates were extracted in buffer containing 50mM Tris pH 7.5, 150mM NaCl, 5mM EDTA, 10% glycerol, 0.1% NP-40, 1% Triton X-100, 0.1% SDS and protease inhibitor cocktail (Roche). Cell lysates were incubated for 4 hrs with 1ug anti-Myc (Santa Cruz Biotechnology; Cat. # sc-789) antibody in the presence of Protein A/G agarose beads (Santa Cruz Biotechnology). The resulting complexes were washed four times with a washing buffer containing 50mM Tris pH 7.5, 150mM NaCl, 5mM EDTA, 0.1% NP-40, 1% Triton X-100 and protease inhibitor cocktail, followed by acid elution using 0.2M glycine pH 2.6 on ice for 10min. Eluates were neutralized by adding 1M Tris pH 7.5. Anti-HA antibody (1:1000, Roche; Cat. # 11 867 423 001) and secondary anti-rat-HRP (1:10000, Pierce Cat #. 31470) and anti-rabbit-HRP (1:10000, Bio-Rad Cat #. 1706515) were used for immunoblotting.
Transporter activity assay using FRET glucose sensor in HEK293T cells
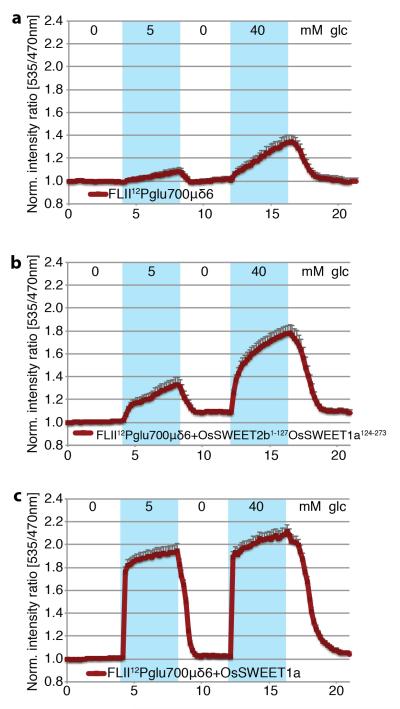
Procedures for cell culture, transfection, image acquisition and FRET sensor analysis have been described previously46. Briefly, HEK293T cells (ATCC® CRL-11268™, without further authentication or test of mycoplasma contamination) were co-transfected with a plasmid carrying the glucose sensor FLII12Pglu700μδ6 only, and either OsSWEET1a or a chimeric plasmid OsSWEET2b1-127OsSWEET1a124-273 carrying OsSWEET2b (1-127aa) fused with OsSWEET1a (124-273aa), which was obtained by overlapping PCR. One day after transfection, imaging was carried out at the following settings: exposure time 300ms, gain 3, binning 2, and time interval 10s. Data were normalized to the initial ratio.
Extended Data
Extended Data Figure 1. Phylogenetic tree for Clade I and II SWEETs from Arabidopsis and rice.
a, Molecular phylogenetic analysis was performed by the Maximum Likelihood method. The evolutionary history was inferred using the Maximum Likelihood method based on the JTT matrix-based model47. The tree with the highest log likelihood (−6353.2408) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbour-joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with the superior log likelihood value. The analysis involved 24 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 208 positions in the final dataset. Evolutionary analyses were conducted in MEGA648. ‘n/t’ represents not-tested. b, Percentage identity and similarity between Arabidopsis and rice SWEETs in Clade I were performed using NCBI BLASTP.
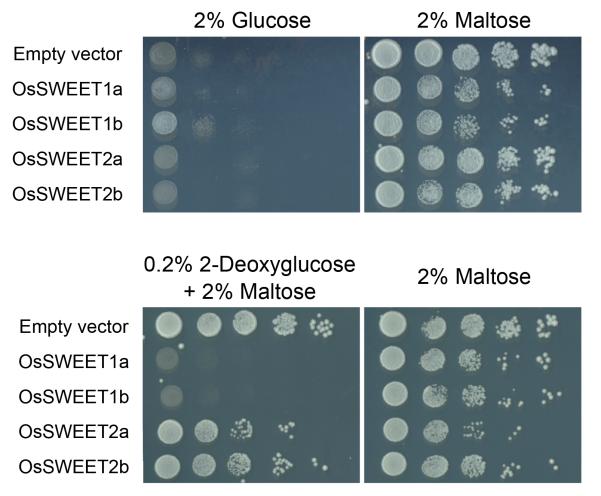
Extended Data Figure 2. Functional analysis of SWEET activities by a yeast growth assay.
When tested for complementation of the growth defect of the EBY4000 mutant strain, OsSWEET1a and OsSWEET1b showed limited growth on glucose, while OsSWEET2a and OsSWEET2b did not show growth. When tested on the toxic glucose analogue 2-deoxyglucose, only OsSWEET1a and OsSWEET1b failed to grow, suggesting that they may be glucose transporters. In contrast, OsSWEET2a and OsSWEET2b were able to grow in the presence of the 2-deoxyglucose, possibly because they are localized to the vacuole membrane and are not able to mediate uptake of the sugar analogue.
Extended Data Figure 3. Functional characterization of OsSWEET2b.
a, HEK293T cells expressing the FRET glucose sensor FLII12Pglu700μδ6 by itself served as negative controls. Glucose uptake activity of the OsSWEET2b/OsSWEET1a chimera (b) and OsSWEET1a (c) were reported by the co-expressed sensor FLII12Pglu700μδ6 (±SEM, n=12). The experiments were repeated four times. Representative results from one experiment are shown.
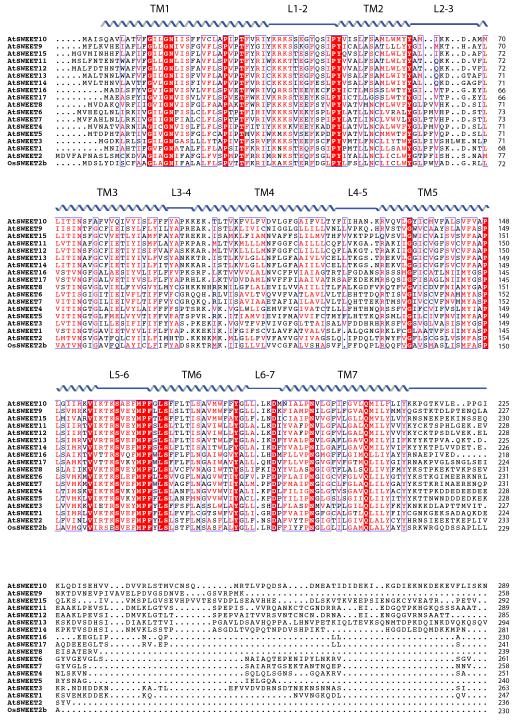
Extended Data Figure 4. Sequence alignment of AtSWEETs and OsSWEET2b.
Sequences of SWEETs were aligned using Clustal Omega. Secondary structure assignment based on OsSWEET2b structure is indicated above the alignment.
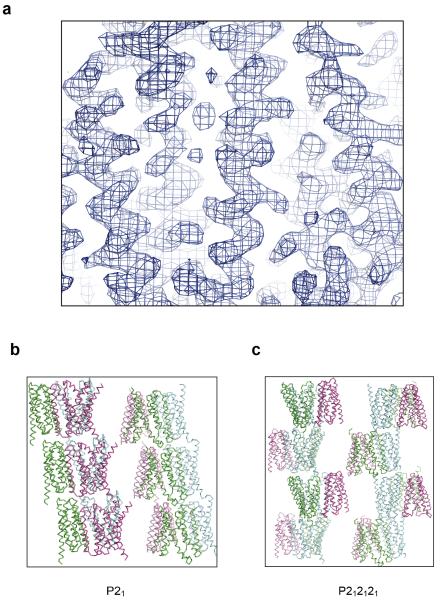
Extended Data Figure 5. Experimental electron density map and crystal packing of two crystal forms.
a, The electron density map is contoured at σ1.5 and coloured in blue. b, Crystal lattice structure of OsSWEET2b in the P21 space group (Form I). c, Crystal lattice structure of OsSWEET2b in the P212121 space group (Form II). Each protomer within a trimer is shown in blue, purple, or cyan.
Extended Data Figure 6. Comparison of THBs of OsSWEET2b and EcSemiSWEET.
a, Comparison of THB1 and THB2 of OsSWEET2b. THB1 of OsSWEET2b (yellow) was superimposed onto THB2 of OsSWEET2b (blue). The inversion linker TM4 is coloured in grey. b, Superposition of OsSWEET2b (THB1 in yellow, THB2 in blue, and TM4 in grey) to EcSemiSWEET (green).
Extended Data Figure 7. Comparison of SWEET, PnuC, and GPCR.
a, Membrane topology diagram of SWEET (left), GPCR (middle), and PnuC (right) is shown with ribbon representations of their respective three TM unit (bottom). The same color scheme is used for TMs. b, Structural comparison of OsSWEET2b and PnuC. OsSWEET2b is shown in green, and PnuC is in purple. c, Structural comparison of OsSWEET2b and a GPCR (metabotropic glutamate receptor). OsSWEET2b is shown in green, and GPCR in orange.
Extended Data Figure 8. Conservation of SWEET and key residues in the transport pathway of OsSWEET2b.
a, Conservation surface mapping of OsSWEET2b, which is coloured according to the degree of conservation of the surface residues of 527 analysed SWEET sequences. b, The cut-away view of OsSWEET2b shows the degree of the conservation of residues lining the transport route. Two clusters with higher conservation are labelled and correspond with the presumptive sugar binding site (I) and the intrafacial gate (II). c, Ribbon representation of OsSWEET2b with selected residues in the transport pathway are shown as sticks. Extrafacial gate residues are coloured in yellow, substrate binding pocket residues in green, and intrafacial hinge residues in cyan. d, Amino acids flanking the critical prolines in the intrafacial gate are also essential for AtSWEET1 activity. Alanine substitution of residues immediately above and below the conserved prolines that form the intrafacial gate in AtSWEET1 reduce the transport of glucose. Growth of the EBY4000 strain is unaffected in maltose. These results suggest that mutations in residues flanking the intrafacial gate have a similar effect as mutations of the conserved prolines.
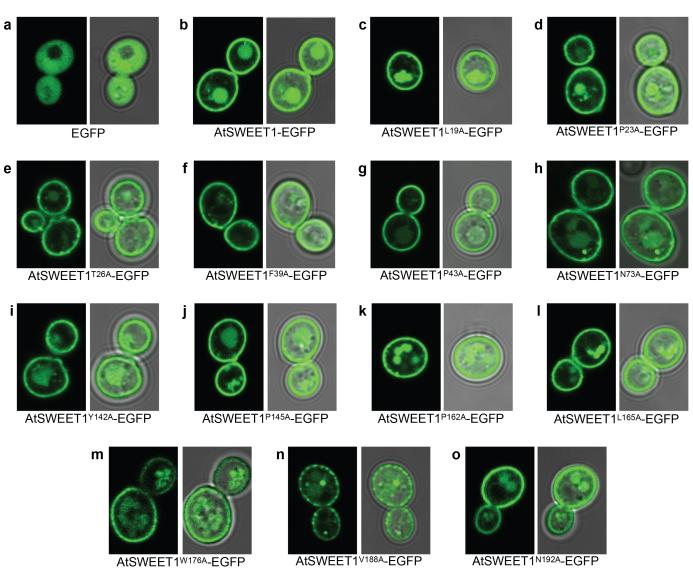
Extended Data Figure 9. Membrane localization of residues that abolish glucose transport in AtSWEET1.
a-o, Fluorescence and overlaid transmitted light images of yeast expressing an AtSWEET1-EGFP fusion (b) and its mutants that did not grow on glucose (c-o). The mutants failed to complement the growth defect of the EBY4000 strain despite proper targeting to the plasma membrane.
Extended Data Table 1.
Data collection and refinement statistics.
| Native-I | Native-II | SeMet | |
|---|---|---|---|
| Data Collection | |||
| Space group | P21 | P212121 | P21 |
| Cell dimensions (Å) | |||
| a, b, c (Å) | 51.83,142.24,81.57 | 89.98,95.49,150.51 | 51.75, 142.50,80.95 |
| α, β, γ(°) | 90.00,93.36,90.00 | 90.00,90.00,90.00 | 90.00, 94.10,90.00 |
| Wavelength (Å) | 1.07170 | 1.00000 | 0.97916 |
| Resolution (Å) | 50.00-3.10 | 50.00-3.66 | 142.51-3.50 |
| (3.15-3.10) | (3.72-3.66) | (3.69-3.50) | |
| Rmerge (%) | 9.2 (65.4) | 8.7 (>1) | 25.9 (>1) |
| I/σ(I) | 16.2 (1.6) | 12.6 (1.0) | 13.3 (2.4) |
| Completeness (%) | 98.9 (96.2) | 97.8 (99.7) | 100.0 (100.0) |
| Redundancy | 6.6 (5.4) | 5.5 (4.4) | 33.3 (34.1) |
| Refinement | |||
| Resolution | 30.00-3.10 | 30.00-3.69 | |
| (3.27-3.10) | (3.82-3.69) | ||
| Unique reflections | 19435 (1489) | 13252 (522) | |
| Rwork/Rfree | 25.4/29.1 | 25.6/30.7 | |
| Number of atoms | |||
| Protein | 4924 | 4860 | |
| Ligands/PEG | 93 | 153 | |
| B-factors (Å2) | |||
| Protein | 97.27 | 136.26 | |
| Ligands/PEG | 103.65 | 140.18 | |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.009 | 0.008 | |
| Bond angles (°) | 1.466 | 1.236 |
High-resolution shell is shown in parentheses.
Acknowledgements
We thank the staff at Beamline 23ID-B and 23ID-D (APS, Argonne National Laboratory) and at Beamline 5.0.2 (Advanced Light Source) for assistance at the synchrotron. This work was made possible by support from Stanford University, the Harold and Leila Y. Mathers Charitable Foundation and the Alfred P. Sloan Foundation to LF and the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences at the US Department of Energy (DOE DE-FG02-04ER15542) to WBF. The functional characterization of OsSWEET2b was supported by grants from the National Science Foundation (IOS-1258018) to WBF. LSC was supported by the National Science Foundation Postdoctoral Research Fellowship in Biology (1401855). SL was supported by the National Natural Science Foundation of China (31300618). Part of this work was conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team, supported by a grant from the National Institute of General Medical Sciences (NIH, P41 GM103403). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) and the Office of Science by Argonne National Laboratory, was supported by U.S. DOE (DE-AC02-06CH11357).
Footnotes
Author Contributions. WBF and LF conceived and designed experiments. YT performed expression, purification, crystallization, data collection, crystallography and biochemical experiments, SL performed liposome uptake experiments, YX contributed to initial characterization and biochemical characterizations, LSC, L-QC, J-SE performed functional experiments, KP assisted crystallography, LF contributed to initial characterization, data collection and crystallography. YT, LC, L-QC, J-SE, SL, WBF and LF analysed the data. LF and WBF wrote the manuscript.
The crystallographic coordinates and structural factors are deposited into Protein Data Bank with accession numbers 5CTG and 5CTH.
Authors declare no competing financial interests.
REFERENCES
- 1.Wright EM. Glucose transport families SLC5 and SLC50. Mol. Aspects Med. 2013;34:183–196. doi: 10.1016/j.mam.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Cura AJ, Carruthers A. Role of monosaccharide transport proteins in carbohydrate assimilation, distribution, metabolism, and homeostasis. Compr. Physiol. 2012;2:863–914. doi: 10.1002/cphy.c110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen LQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xuan YH, et al. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA. 2013;110:E3685–3694. doi: 10.1073/pnas.1311244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of sugars. Annu. Rev. Biochem. 2015;84:865–894. doi: 10.1146/annurev-biochem-060614-033904. [DOI] [PubMed] [Google Scholar]
- 6.Eom JS, et al. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015;25:53–62. doi: 10.1016/j.pbi.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Lin IW, et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature. 2014;508:546–549. doi: 10.1038/nature13082. [DOI] [PubMed] [Google Scholar]
- 8.Chen LQ, et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335:207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- 9.Sun MX, Huang XY, Yang J, Guan YF, Yang ZN. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013;26:83–91. doi: 10.1007/s00497-012-0208-1. [DOI] [PubMed] [Google Scholar]
- 10.Chen LQ, et al. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell. 2015;27:607–619. doi: 10.1105/tpc.114.134585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015;82:632–643. doi: 10.1111/tpj.12838. [DOI] [PubMed] [Google Scholar]
- 12.Cohn M, et al. Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector-mediated induction of a SWEET sugar transporter in cassava. Mol. Plant Microbe. Interact. 2014;27:1186–1198. doi: 10.1094/MPMI-06-14-0161-R. [DOI] [PubMed] [Google Scholar]
- 13.Chen H-Y, et al. Vacuolar SWEET2 transporter reduces glucose efflux to limit pathogen propagation in roots. Plant J. 2015 [Google Scholar]
- 14.Xu Y, et al. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature. 2014;515:448–452. doi: 10.1038/nature13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Crystal structure of a bacterial homologue of SWEET transporters. Cell Res. 2014;24:1486–1489. doi: 10.1038/cr.2014.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Nishizawa T, Yamashita K, Ishitani R, Nureki O. Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter. Nat. Commun. 2015;6:6112. doi: 10.1038/ncomms7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yee DC, et al. The transporter-opsin-G protein-coupled receptor (TOG) superfamily. FEBS J. 2013;280:5780–5800. doi: 10.1111/febs.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaehme M, Guskov A, Slotboom DJ. Crystal structure of the vitamin B3 transporter PnuC, a full-length SWEET homolog. Nat. Struct. Mol. Biol. 2014;21:1013–1015. doi: 10.1038/nsmb.2909. [DOI] [PubMed] [Google Scholar]
- 19.Jaehme M, Guskov A, Slotboom DJ. The twisted relation between Pnu and SWEET transporters. Trends Biochem. Sci. 2015;40:183–188. doi: 10.1016/j.tibs.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Niittyla T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX. Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics. 2007;6:1711–1726. doi: 10.1074/mcp.M700164-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 22.Kazmier K, et al. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat. Struct. Mol. Biol. 2014;21:472–479. doi: 10.1038/nsmb.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481:469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Zutter JK, Levine KB, Deng D, Carruthers A. Sequence determinants of GLUT1 oligomerization: analysis by homology-scanning mutagenesis. J. Biol. Chem. 2013;288:20734–20744. doi: 10.1074/jbc.M113.469023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan L, et al. Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell. 2013;25:974–984. doi: 10.1105/tpc.112.108027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanquar V, et al. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell. 2009;21:3610–3622. doi: 10.1105/tpc.109.068593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forrest LR, Kramer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta. 2011;1807:167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Keller R, Ziegler C, Schneider D. When two turn into one: evolution of membrane transporters from half modules. Biol. Chem. 2014;395:1379–1388. doi: 10.1515/hsz-2014-0224. [DOI] [PubMed] [Google Scholar]
- 29.Rypniewski WR, Holden HM, Rayment I. Structural consequences of reductive methylation of lysine residues in hen egg white lysozyme: an X-ray analysis at 1.8-A resolution. Biochemistry. 1993;32:9851–9858. doi: 10.1021/bi00088a041. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. In: Methods Enzymol. Carter Charles W., Jr., editor. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA, USA: 2008. http://www.pymol.org. [Google Scholar]
- 42.Veenhoff LM, Poolman B. Substrate recognition at the cytoplasmic and extracellular binding site of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem. 1999;274:33244–33250. doi: 10.1074/jbc.274.47.33244. [DOI] [PubMed] [Google Scholar]
- 43.Wieczorke R, et al. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464:123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 44.Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 45.Loque D, Lalonde S, Looger LL, von Wiren N, Frommer WB. A cytosolic trans-activation domain essential for ammonium uptake. Nature. 2007;446:195–198. doi: 10.1038/nature05579. [DOI] [PubMed] [Google Scholar]
- 46.Hou BH, et al. Optical sensors for monitoring dynamic changes of intracellular metabolite levels in mammalian cells. Nat. Protoc. 2011;6:1818–1833. doi: 10.1038/nprot.2011.392. [DOI] [PubMed] [Google Scholar]
- 47.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]