Abstract
High-throughput sequencing studies of small RNAs reveal a complex milieu of noncoding RNAs in biological samples. Early data analysis was often limited to microRNAs due to their regulatory nature and potential as biomarkers; however, many more classes of noncoding RNAs are now being recognized. A class of fragments initially excluded from analysis were those derived from transfer RNAs (tRNAs) because they were thought to be degradation products. More recently, critical cellular function has been attributed to tRNA fragments (tRFs), and their conservation across all domains of life has propelled them into an emerging area of scientific study. The biogenesis of tRFs is currently being elucidated, and initial studies show that a diverse array of tRFs are generated from all parts of a tRNA molecule. The goal of this review was to present what is currently known about tRFs and their potential as biomarkers for the earlier detection of disease.
Keywords: tRNA fragments, tRFs, angiogenin, cancer, tRNA biomarkers
What are tRNA Fragments?
High-throughput sequencing studies continue to identify an expanding array of new small noncoding RNAs (ncRNAs).1 These newly identified small ncRNAs are derived from many types of primary RNA transcripts that ultimately give rise to the small, biologically functional molecules of microRNAs (miRNAs), ribosomal RNA (rRNA) fragments, small nucleolar RNA fragments, tRNA fragments (tRFs), and others.2 Early high-throughput sequencing studies of small ncRNAs generally regarded RNA fragments as nothing more than contaminating degradation products and sequences of little interest for biological study.1 It was assumed that these fragments were naturally produced, inert RNAs derived from the functional parental rRNA and tRNA molecules. However, with improving bioinformatic methodologies, these once dismissed RNA fragments are now being recognized for their importance in many biological pathways, including the regulation of translation, stress responses, proliferation, and other aspects influencing human disease.1–5 tRFs are now known to be purposefully processed from both mature and pre-tRNAs. tRFs represent one group of small ncRNAs currently undergoing a new wave of investigation centered primarily on the control mechanisms resulting in their biogenesis and determining their biological function within various cell types and cell growth conditions.1–14
What is Known About the Biogenesis of tRFs?
Biogenesis of tRNAs and tRFs
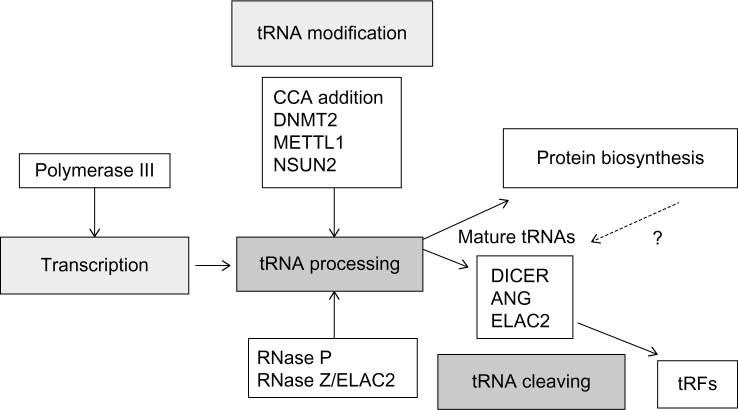
tRNAs are among the most abundant RNA molecules in a given cell, constituting as much as 4%–10% of total cellular RNA.3 The fundamental role of tRNAs, where a charged tRNA delivers an amino acid to a growing peptide chain within the ribosome, is well studied.15 The biogenesis of tRNAs begins with their transcription by RNA polymerase III from a type 2 RNA polymerase III promoter. After transcription, several modifications are made to the tRNA transcript in the form of endonucleitic cleavages, exonucleitic trimming, nontemplated nucleotide additions, and multiple single-base modification events. These events occur prior to the addition of a single amino acid to the 3′ end of the mature tRNA (summarized in Fig. 1). The highly modified and aminoacylated tRNA may then be used within translation.
Figure 1.
An overview of the process leading to tRNA and tRF biogenesis. tRNAs are initially processed from pre-tRNAs into mature tRNA by removing nucleotides that constitute a 5′ leader by RNase P cleavage, and then, the 3′ tail is removed through cleavage of the tRNA primary transcript by RNase Z. A select group of tRNAs harbor introns and must be further processed by a complex known as the tRNA splicing endonuclease, a complex including CLP1 and other proteins (not included in this figure).15 In this figure, representative enzymes are listed as tRNA modifying or cleaving enzymes and are not meant to be inclusive of all such enzymes. It is highly likely that additional modifying and cleaving enzymes for tRNAs will be identified in the future. There is also a yet unanswered question as to whether tRFs are generated after delivering an amino acid or if a subset of the many tRNAs is specifically processed to tRFs without going through the process of amino acylation. Representative tRFs have been identified for all 20 amino acids and for mitochondrial tRNAs as well.
The biogenesis process used by cells to make mature aminoacylated tRNAs consists of many well-regulated steps that result in the interaction of tRNA molecules with several tRNA modification enzymes.5 Paramount to understanding the tRF biological function is the need to understand tRF biogenesis. Recent research has centered on deducing how a tRF is generated from a molecule essential for protein translation and which pathways are used to generate mature tRNAs versus those used to generate various tRFs.
Nomenclature of tRFs
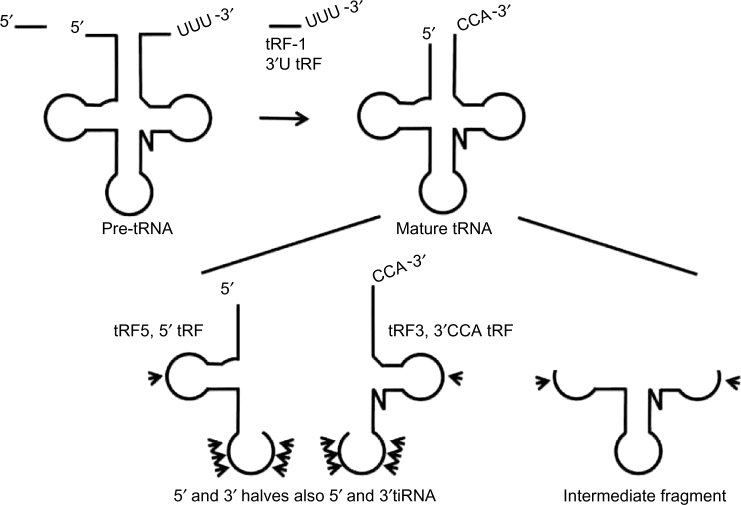
Highlighting the emergence of a new field of scientific study, the nomenclature of tRFs has had a varied beginning.16 In earlier studies of tRFs, the fragments were simply called tRNA halves and included 5′-half-tRNAs as well as 3′-half-tRNAs.7,13,16 In another related study, the tRFs were referred to as tRNA-derived small RNAs.9 Additional labels have been used for tRFs based on their size, expression, or function. In one such study, tRFs expressed in response to stress were called stress-induced tRNAs (tiRNA).13 In another study, tRFs were named for the location of the cleavage site; thus, those cleaved from the 3′ end of the tRNA were called tRF-3.10 Meanwhile, tRFs expressed in response to hormone stimulation have been referred to as sex hormone-dependent tRNA derived RNAs.14 Figure 2 describes frequently identified tRFs and the pathways leading to their biogenesis. As more information becomes available about the function of this group of ncRNAs, a more clear method of naming them will also evolve.
Figure 2.
Nomenclature of tRFs. The nomenclature for tRFs is inconsistent due to their recent identification most often as part of deep-sequencing data sets. Early terminology included tRF-1 or 3′U tRF for the trailing sequence cleaved by RNase Z during maturation.10,70 The tRFs generated following CCA addition and cleaved in the T loop were designated tRF3 or 3′CCA tRF, while the 5′-tRFs generated after cleavage by RNase P were designated tRF-5, 5′-tRFs, or 5′leader-exon tRFs.10,12,70 These fragments are smaller than half-tRNAs and range in size from 13 to 20 base pairs.70 The tRNA halves are larger and the expected size is 30–40 nucleotides; however, this appears to be quite variable.16,37,70 Megel et al proposed a universal naming, where the letter corresponds to the extremity of the tRNA and the number (5 or 3) to the cleavage site, such as tRF3T for a 3′-tRF generated by cleavage in the T loop.16 Utilizing the proposed general naming scheme, the newly identified intermediate tRFs would be tRF53DT. Multiple arrow heads are used to designate regions cleaved by angiogenin (ANG) that are often variable and may occur at any point in the anticodon loop.37 While a single arrow head is used to designate potential, Dicer, RNaseP sites, or possibly novel RNase cleavage.70
The biogenesis of tRFs is highly conserved
Initially regarded as degradation products, tRFs were often excluded from published studies and, as such, were not considered as potential biomarkers in earlier studies.1 However, the expression of tRFs in response to stress is evolutionarily conserved and includes both prokaryotes and eukaryotes, suggesting a functional role of biological importance.3,4,17,18 Furthermore, the processing of tRFs from mature tRNAs has been shown to occur by an evolutionarily conserved group of proteins, the RNases. RNases that have been shown to generate specific types of tRFs, including PrrC, Rny1, Dicer, RNase Z, RNase P, and angiogenin, and it is expected that this list may grow as more fragments are studied.4,6,10,12,17 While these studies have provided the groundwork for understanding that the biogenesis of tRFs is a conserved and regulated process, further investigation is needed to determine the ultimate molecular function of specific tRFs. Additionally, improved detection strategies are needed for the identification of tRFs within biological samples. Once characterized, these tRFs may prove to be robust biomarkers that reveal critical information about a patient’s current state or future risk of the disease.19,20
Interestingly, tRFs observed in many high-throughput studies were assumed to be derived from mature tRNAs because the expected cleavage, nontemplated nucleotide additions, and base modifications required to generate mature tRNAs had taken place prior to the generation of the tRF.1,5 tRNAs are initially processed into mature tRNAs by removing nucleotides that constitute a 5′ leader or 3′ tail through cleavage of the tRNA primary transcript by RNase P and RNase Z, respectively. It has recently been shown that the 3′ trailer sequence may form a subset of the functional tRFs and this subset has been designated as tRF-1, indicating their biogenesis is neither 3′ nor 5′ from the mature tRNA.10 The series of tRF-1 include tRF-1001, which was shown to impair cellular proliferation, when knocked down with siRNA. Thus, biogenesis of pre-tRNAs does result in the formation of a functional class of tRFs.10
Other subsets of tRFs are generated from the splicing of intron sequences and the cleaving of mature tRNAs.15,16 After these cleavage events, the tRNAs gain three additional nucleotides (CCA) at their 3′ ends, which are required for their subsequent aminoacylation.21 Mature tRNAs are also modified at as many as eight or more base pairs during maturation into a biologically functional molecule.5,21,22 These modifications of tRNAs may include, but are not limited to, methylation events such as tRNA-specific modified nucleoside 1-methyladenosine, 1-methylguanosine, N2,N2-dimethylguanosine.5 Interestingly, some nucleotide modifications by methyltransferases have been reported to block cleavage into tRFs, while other modifications of tRNA, specifically a 2′-O-methylation event in the anticodon loop, have been shown to enhance cleavage into tRFs in yeast.5,23–25 However, many of the known mechanisms of processing tRNAs into tRFs have been defined in organisms other than mammals, and these mechanisms will need to be further confirmed for their conservation in higher eukaryotes as the field of tRF biogenesis advances.
Which RNases Cleave tRNAs into Functional tRFs?
Multiple RNases have been identified that cleave tRNAs
The RNases, including Dicer, RNase Z, and angiogenin, have all been studied for their role in cancer cells and as potential biomarkers for predicting cancer risk.26–29 All three enzymes have also been studied using siRNA knockdowns to confirm that they have a role in the generation of tRFs in cancer cells.6–14 In contrast to tRF biogenesis, miRNA biogenesis is a well-characterized process and frequently requires initial processing by Drosha and Dicer to produce mature miRNAs. However, the generation of tRFs is thought to be independent of the Drosha cleavage, while a subset of tRFs are thought to be generated in a Dicer-dependent manner.6,26,30
One example of a tRF generated by Dicer cleavage is the primer-binding small noncoding RNA (PBSncRNA), which was determined to be a tRF derived from tRNA-Lys3.31 Knockdown of PBSncRNA allows replication of human immunodeficiency virus (HIV) and PBSncRNA has been identified in Argonaute 2 (AGO2) complexes, suggesting that the expression of this tRF may target HIV replication. It has been proposed that tRFs may be part of a host response to defend against viral infection; thus, it is hopeful that the function of PBSncRNA will be confirmed to block HIV replication in future studies.
Evidence that RNase Z functions in the biogenesis of tRFs
RNase Z, also known as ELAC2, has been well studied for its role in the production of mature tRNAs by trimming off excess nucleotides at the 3′ end of tRNA primary transcripts. RNase Z has recently been shown to be important in generating a subset of tRFs, particularly tRFs generated from the cleavage of long 3′ end tails from primary tRNAs.7,10 In addition to its role in generating tRFs, the nuclease activity of RNase Z can also be guided by small tRFs to other target RNA molecules inside cells in trans to regulate expression.8 Thus, RNase Z is currently being used to develop a targeted therapeutic approach for cancer treatment based on its ability to recognize and cleave any pre-tRNA-like complex, a method known as tRNase ZL-utilizing efficacious (TRUE) gene silencing.32
The discovery that RNase Z could be engineered to target RNA molecules led Elbarbary et al, to question if RNase Z would bind to endogenous tRFs.7 Immunoprecipitation of RNase Z and isolation of bound RNA showed that a number of ncRNAs were bound to RNase Z. One of the ncRNAs identified in this study was 5′-half-tRNAGlu. The authors went on to show that 5′-half-tRNAGlu was acting as a small guide RNA and directly regulated the expression of the PPM1F mRNA, a protein that has been shown to induce apoptosis in HeLa cells when overexpressed.7
Angiogenin cleaves tRNAs into tRFs during stress
Of the numerous RNases identified in the human genome, one of the best characterized nucleases shown to be used in the processing of tRNAs to tRFs is angiogenin. Angiogenin is a member of the RNase A superfamily and is also known as RNase 5. Angiogenin is well studied for its activity in cleaving mature tRNAs.4 Several studies have shown that angiogenin activity increases its cleavage of tRNAs in response to specific stimuli, such as nutritional deficiency, hypoxia, heat shock, and oxidative stress.4,33–35 In addition, human respiratory syncytial virus (RSV) infection has been shown to stimulate tRF formation through angiogenin cleavage using lung cancer cells as host for the RSV infection.36 Within this study, the infection of lung cancer cells with RSV was shown to specifically increase the formation of tRF–Glu, tRF–Gly and tRF–Lys. Interestingly, infection with a virus from the same family, human metapneumovirus, did not increase the expression of these tRFs, showing that there are both substrate specificity in the generation of tRFs and a complex regulatory mechanism governing the activation of angiogenin to act on tRNAs in order to generate tRFs.36 The specificity of angiogenin in producing tRFs in response to RSV infection was determined by knocking down the expression of the RNA cleaving enzymes such as angiogenin, Dicer, Drosha, RNase Z, and RNase L. Only the knock down of angiogenin significantly reduced tRF formation in response to RSV.36
Mounting evidence from high-throughput sequencing studies suggests that many additional cleavage sites in tRNAs may exist and that one part of the molecule is generally found at higher levels suggesting that these ncRNAs are functionally active.37 Interestingly, the total RNA cleavage of a given tRNA is only about a tenth of the total available tRNA, suggesting that tRFs are generated from a subpopulation of the total mature tRNA for a given tRNA transcript.35,38 Further confirmation that angiogenin and tRNA availability are important in the generation of tRFs came from a comprehensive study by Saikia et al.35 In this study, several conditions, including oxidative stress, hypertonic stress, and increased angiogenin expression, were tested and a microarray-based assay was used to detect tRFs.35 One of the central findings in this study is that different types and lengths of stress result in differing patterns of tRF generation. As this new area of research moves forward, it will be important to validate each testing condition in various cell types rather than to assume that what was found in one set of conditions will be the same in all other cell types and cell growth conditions.
In a study of the protozoan, Tetrahymena thermophila, half-tRNAs were generated during early amino acid starvation.39 In this study, starvation for all amino acids or for any one of the essential amino acids resulted in tRF formation. T. thermophila is an excellent model for studies of amino acid starvation-induced tRNA fragmentation due to the need of this organism to rapidly respond to changing environmental cues. In contrast, studies of tRNA fragmentation in response to starvation in HeLa cells was not corrected by the addition of essential amino acids as it was in Tetrahymena.12,39 However, tRNA fragmentation was blocked by siRNA shutdown of angiongenin.12
The known role of angiogenin in the generation of tRFs may collide with the role of this same protein in tumor immunology. Angiogenin was known to be elevated in the process of angiogenesis as early as 1987 and continues to be a potential biomarker for tumors in numerous tissue sites and a potential target for therapeutic intervention.40–43 Furthermore, circulating antibodies against angiogenin have been found to be a sensitive biomarker for osteosarcoma and are thought to play an important role in tumor immunology for this cancer type.44
What is Known About the Function of tRFs?
tRFs are functionally diverse
Although tRFs have been linked to immune function by their location in exosomes, the specific role of individual tRFs is currently not well characterized.45–47 Early studies examined the collective extracellular tRF function, with one study finding that tRFs in the conditioned media from bladder carcinoma inhibited the growth of endothelial cells.48 However, at that time, it was not possible to characterize the exact tRF responsible for the inhibitory action because methods did not exist to sequence the tRFs in media. Studies confirming the cellular function of tRFs are summarized in Table 1.
Table 1.
Representative studies of tRF function.
| tRF | FUNCTION | CELL TYPE | STUDY |
|---|---|---|---|
| Alteration of cell phenotype in response to tRF expression | |||
| • *5′tRFs | Isolated from the media of a urinary bladder carcinoma cell line and used to inhibit endothelial cell growth | Bovine endothelial cells | 48 |
| • 3′tRFSer | Regulates cell proliferation | HCT116, DU145, LNCaP | 10 |
| • 5′tRFVal | Cleavage of tRNAs during stress | HepG2 | 12 |
| • 5′tRF and 3′tRFHis | Ribosomal bound tRFs change with differing growth conditions | Yeast ribosomes | 56 |
| • 5′tRFGlu, Gly, Lys | Increased tRF expression in response to infection by RSV | A549, primary small alveolar epithelial cells | 36 |
| • *5′ leader | Neurodegeneration motor neuron loss, over expression of 5′ leader tRFs | CLP1 kinase dead mice | 67 |
| • 5′tRFAsp, His, Lys | siRNA to Estrogen and Androgen Receptor reduce expression of 5′ fragments. Androgen receptor dependent cleavage, increases proliferative response | MCF7, BT-474, LNCap LNCap-FGC | 14 |
| • *tRFs from all 20 amino acids | Predicted in drosophila to bind conserved Seed sequences. Showed significant gene ontology enrichment brain activity and aging | Bioinformatic approach not confirmed in cell lines | 68 |
| • 5′tRFGlu,Asp, Gly, Tyr | Displaces YBX1 allowing stabilization of 3′ UTRs of oncogenic transcripts | MDA-LM2, four tRFs transfected in mice | 37 |
| Targeting a specific mRNA through Seed binding in the 3′ UTR | |||
| • 3′tRFLys3 | Loaded into AGO2 complexes and targets HIHIV | HeLa cells | 31 |
| • 3′tRFGly | A DICERER substrate, represses RPA1 | HEK293, normal B cells, lost in subset of lymphomas | 64 |
| Targeting a specific mRNA by acting as a small guide RNA | |||
| • 5′tRFGlu | Down regulates PPM1F transcript | HEK293 | 7 |
| tRFs Regulating Translation | |||
| • *5′-tiRNAs | 5′-tiRNAs but not 3′-tiRNAs have a distinct inhibitory effect on translation | U2OS | 13 |
| • 5′tRFAla | Induces stress granule formation | U2OS | 53 |
| • 5′-tiRNAAla,Cy s | YB1 is the only tiRNA binding protein needed for tiRNA induced stress granule formation | U2OS | 49 |
| • 5′tRFVal | Binds small ribosomal subunit and blocks translation | Haloferax volcanii | 18 |
| • 5′tRFGln | Inhibits protein translation | HeLa | 69 |
Note:
tRFs multiple tRFs.
tRFs contribute to the inhibition of translation
A general function of tRFs was shown to be the inhibition of translation by disrupting the cap-binding complex eIF4F.49 More recently, the function of a subset of tRFs generated by RNase cleavage has been shown to block YB-1 binding of target transcripts.37 The YB-1 protein (YBX1 gene) is an RNA binding protein that is frequently overexpressed in cancer cells and plays a significant role in RNA translation and stability.37,50 YB-1 binds to tRF–Ala and specifically inhibits translation.37 Interestingly, tRF–Ala is one of the tRFs capable of forming a G-quadruplex structure due to the oligo-G nucleotides it contains, and this structure is required for translational repression.51 Earlier studies of G-quadruplex structures showed that they cause resistance to nuclease cleavage and exhibit antiproliferative activity in cancer cells.52
Recent studies have shown that cells treated with mimics of 5′-tRFs form stress granules. The synthetic mimics of 5′-tRFs tRF–Ala, Gly, and Val were all tested on a human osteosarcoma cell line, U2OS cells and stress granules were visualized.53 Stress granules are formed in response to cellular stress, and they play a role in reprogramming the cell to stop transcription and translation in response to cellular stressors.49 The specific set of 5′tRFs have been called tiR-NAs for stress-induced tRFs. Translational inhibition has been linked to 5′-tRFs (5′tiRNAs) by a mechanism, which is independent of phospho-eIF2alpha translational repression.54 Synthetic mimics of 3′tRFs (3′tiRNAs) did not cause stress granule formation. Taken together, these studies confirm that one role of a subset of 5′-tRFs is a rapid response to downregulate RNA translation during stress.54
tRNA modification may alter function of tRFs
Modifications of tRNAs are ubiquitous, with as many as 100 or more different modifications and eight or more modifications per tRNA.22,55 Therefore, it has been hypothesized that modifications may alter the cleavage into tRFs. This is an area that will require a great deal of study in the future and will be enhanced by new techniques, such as AlkB-facilitated RNA methylation sequencing (ARM-seq) described by Cozen et al.5 ARM-seq allows the removal of modifications, so that high-throughput sequencing is not blocked by hard stop RNA modifications.
The loss of one such modifying enzyme, NSUN2, resulted in widespread neurological abnormalities in mice and specifically altered the expression of 5′-tRFs.24 NSUN2 is a tRNA methyltransferase known to be mutated in neurological disorders as well as cancer. This is one of the first confirmations of an enzyme functioning in the regulation of tRNA fragmentation resulting in human disease. With >100 potential modifications, it is expected that tRF expression and function will be altered in many ways as yet undefined. This is an area of research awaiting much study in tumor biology and elsewhere.
tRFs as Potential Biomarkers
Identification and characterization of tRF expression
The variable expression and function of tRFs are just now being elucidated. It will take some time to identify clinical applicability and determine if these small RNAs have potential as biomarkers of disease. Bioinformatic methods to identify tRFs in deep-sequencing studies are rapidly evolving and, in some cases, will allow the mining of historic data sets, where tRFs were initially discarded from analysis.1,56,57 Revisiting data sets, where expression of tRFs may be correlated with clinical characteristics, will allow the identification of potential tRF biomarkers in well-characterized samples. Retrospective studies must be conducted with care and understanding of the complication of tRNA and tRNA-like abundance in the human genome.57 Telonis et al outlined the difficulties in interpreting tRF data in high-throughput sequencing studies.57 Such difficulties include, but are not limited to, the repetitive nature of tRNA sequences in genomes, the complexity of multiple isoacceptors for each amino acid, and the existence of base changes thought to be resulting from modifications. Bioinformatic analysis must be accompanied by a clear description of the criteria used for data set analysis and then be followed by experimental confirmation.
tRFs have been well documented to exist in patient serum and other convenient biological samples at levels similar to miRNAs.46,48,58,59 In prospective studies, consistent methods for patient sample collection and analysis of extracellular RNAs will be critical as biomarker studies of tRFs move forward.60 The methods utilized in each study must be carefully controlled and reported if data are to be comparable across samples and between studies.61 For example, the sample preparation, sample storage, and RNA collection methods must be fully described and consistent for all patients. The method of RNA isolation and amplification, if different between studies, may impact the resulting repertoire of fragments obtained and characterized. The development of biomarkers is a complicated process, and as tRFs make their way into the biomarker pipeline, it will be necessary to apply past experience to develop robust biomarkers with clinical value.62,63
Expression and function of tRFs in cancer
tRF expression has been detected in cancer patient samples from multiple tissue sites and accessible samples.46,48,64 A study of B cell lymphoma found that tRFs were downregulated in lymphoma cell lines and primary biopsies when compared with control B cells.64 In contrast, a study of tRF expression in prostate cancer patients found that tRFs were increased in metastatic samples.59 Studies of tRF expression in cancer cell lines have revealed a diverse array of tRF expression and have been useful as model systems to study the function of tRFs. Mechanistic studies of tRF expression in response to hormones in prostate and breast cancer cells suggest that tRFs enhance cell proliferation and that their expression is tissue dependent.10,14,46,57
tRFs may act like miRNAs, and if confirmed, this could mean that one tRF may regulate multiple mRNA targets.31,64 Defining the biological expression and function of tRFs is reminiscent of the early studies of miRNAs, where much controversy occurred due to the need for novel method development. Given that their level of expression is similar to miRNAs, and as their functions become known, tRFs are expected to provide a new frontier in cancer biomarker development.
Host tRFs regulate viral replication
tRFs have been characterized in RSV, HIV, and Human T-cell lymphotropic virus type 1 (HTLV-1) infection.31,36,65 A pyrosequencing study showed that a tRF derived from tRNA–Lys is increased in cells infected with HIV and that loss of this expression allows HIV replication.31 A different tRF derived from tRNA–Pro was shown to be incorporated into HTLV-1 viral particles and shown to act as a primer for reverse transcriptase during replication.65 The specificity of particular tRFs to viral regulation provides hope that these tRFs may eventually provide novel therapeutic targets to block viral infection.
Pathophysiology of tissue damage is characterized by tRF expression
Oxidative stress results in tissue damage and is associated with many disease states. A recent study has shown that even before DNA damage is detectable, tRFs are detectable. A specific RNA signature, tRNA-specific modified nucleoside 1-methyladenosine, is detected in patients who have kidney damage and correlates with mortality.66 Early detection of tissue damage would be helpful in the detection of many diseases; thus, the emerging field of tRFs provides potential for the discovery of novel biomarkers aimed at earlier detection. More studies are required to determine if tRFs will provide promising new candidates for future biomarkers of health and disease.
Acknowledgments
We would like to acknowledge the staff from the Biomedical Sciences Department for their continued support to our research and publishing efforts, especially Wendy Schwartz and Jeannette Lang.
Footnotes
ACADEMIC EDITOR: Karen Pulford, Editor in Chief
PEER REVIEW: Eight peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1410 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the first draft of the manuscript: LTB. Contributed to the writing of the manuscript: KZ, ABC, KWD. Agreed with manuscript results and conclusions: LTB, KZ, ABC, KWD. Jointly developed the structure and arguments for the paper: KWD, LTB, KZ. Made critical revisions and approved the final version: LTB, KZ, ABC, KWD. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40(14):6787–99. doi: 10.1093/nar/gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawaji H, Nakamura M, Takahashi Y, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16(2):98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 4.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14(10):2095–103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12(9):879–84. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104(46):18097–102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbarbary RA, Takaku H, Uchiumi N, et al. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PLoS One. 2009;4(6):e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbarbary RA, Takaku H, Uchiumi N, et al. Human cytosolic tRNase ZL can downregulate gene expression through miRNA. FEBS Lett. 2009;583:3241–6. doi: 10.1016/j.febslet.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–95. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole C, Sobala A, Lu C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–60. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–42. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 13.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112(29):E3816–25. doi: 10.1073/pnas.1510077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karaca E, Weitzer S, Pehlivan D, et al. Baylor Hopkins Center for Mendelian Genomics Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157(3):636–50. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Megel C, Morelle G, Lalande S, Duchêne AM, Small I, Maréchal-Drouard L. Surveillance and cleavage of eukaryotic tRNAs. Int J Mol Sci. 2015;16(1):1873–93. doi: 10.3390/ijms16011873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–9. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grewal SS. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim Biophys Acta. 2015;1849(7):898–907. doi: 10.1016/j.bbagrm.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Prensner JR, Chinnaiyan AM, Srivastava S. Systematic, evidence-based discovery of biomarkers at the NCI. Clin Exp Metastasis. 2012;29(7):645–52. doi: 10.1007/s10585-012-9507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endres L, Dedon PC, Begley TJ. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015;12(6):603–14. doi: 10.1080/15476286.2015.1031947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durdevic Z, Schaefer M. tRNA modifications: necessary for correct tRNA-derived fragments during the recovery from stress? Bioessays. 2013;35(4):323–7. doi: 10.1002/bies.201200158. [DOI] [PubMed] [Google Scholar]
- 23.Schaefer M, Pollex T, Hanna K, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–5. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33(18):2020–39. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studte P, Zink S, Jablonowski D, et al. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol Microbiol. 2008;69(5):1266–77. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- 26.Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. 2014;14(10):662–72. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- 27.Xu B, Tong N, Li JM, Zhang ZD, Wu HF. ELAC2 polymorphisms and prostate cancer risk: a meta-analysis based on 18 case-control studies. Prostate Cancer Prostatic Dis. 2010;13(3):270–7. doi: 10.1038/pcan.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YC, Giovannucci E, Kraft P, Hunter JD. Sequence variants of elaC homolog 2 (Escherichia coli) (ELAC2) gene and susceptibility to prostate cancer in the Health Professionals Follow-Up Study. Carcinogenesis. 2008;29(5):999–1004. doi: 10.1093/carcin/bgn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramcharan SK, Lip GY, Stonelake PS, Blann AD. Angiogenin outperforms VEGF, EPCs and CECs in predicting Dukes’ and AJCC stage in colorectal cancer. Eur J Clin Invest. 2013;43(8):801–8. doi: 10.1111/eci.12108. [DOI] [PubMed] [Google Scholar]
- 30.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22(20):2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37(19):6575–86. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iizuka S, Oridate N, Nashimoto M, Fukuda S, Tamura M. Growth inhibition of head and neck squamous cell carcinoma cells by sgRNA targeting the cyclin D1 mRNA based on TRUE gene silencing. PLoS One. 2014;9(12):e114121. doi: 10.1371/journal.pone.0114121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10(12):1798–806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–9. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Saikia M, Krokowski D, Guan BJ, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287(51):42708–25. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21(2):368–79. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161(4):790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280(52):42744–9. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 40.Folkman J, Klagsbrun M. Angiogenic factors. Science. 1987;235(4787):442–7. doi: 10.1126/science.2432664. [DOI] [PubMed] [Google Scholar]
- 41.Qin L, Bromberg-White JL, Qian CN. Opportunities and challenges in tumor angiogenesis research: back and forth between bench and bed. Adv Cancer Res. 2012;113:191–239. doi: 10.1016/B978-0-12-394280-7.00006-3. [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Hu GF. Angiogenin-mediated ribosomal RNA transcription as a molecular target for treatment of head and neck squamous cell carcinoma. Oral Oncol. 2010;46:648–53. doi: 10.1016/j.oraloncology.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshioka N, Wang L, Kishimoto K, Tsuji T, Hu GF. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc Natl Acad Sci USA. 2006;103:14519–24. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savitskaya YA, Rico G, Linares L, et al. Circulating natural IgM antibodies against angiogenin in the peripheral blood sera of patients with osteosarcoma as candidate biomarkers and reporters of tumorigenesis. Biomark Cancer. 2010;2:65–78. doi: 10.4137/BIC.S6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhahbi JM. 5′tRNA Halves: the next generation of immune signaling molecules. Front Immunol. 2015;6:74. doi: 10.3389/fimmu.2015.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DI. Deep sequencing of serum small RNAs identifies patterns of 5′tRNA half and YRNA fragment expression associated with breast cancer. Biomark Cancer. 2014;6:37–47. doi: 10.4137/BIC.S20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t’Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40(18):9272–85. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao H, Bojanowski K, Ingber DE, et al. New role for tRNA and its fragment purified from human urinary bladder carcinoma conditioned medium: inhibition of endothelial cell growth. J Cell Biochem. 1999;76(1):109–17. doi: 10.1002/(sici)1097-4644(20000101)76:1<109::aid-jcb11>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 49.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613–23. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsumoto K, Bay BH. Significance of the Y-box proteins in human cancers. J Mol Genet Med. 2005;1(1):11–7. doi: 10.4172/1747-0862.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanov P, O’Day E, Emara MM, Wagner G, Lieberman J, Anderson P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111(51):18201–6. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi EW, Nayak LV, Bates PJ. Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 2010;38(5):1623–35. doi: 10.1093/nar/gkp1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959–68. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849(7):861–70. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584(2):265–71. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zywicki M, Bakowska-Zywicka K, Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res. 2012;40(9):4013–24. doi: 10.1093/nar/gks020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Telonis AG, Loher P, Honda S, et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015;6(28):24797–822. doi: 10.18632/oncotarget.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speer J, Gehrke CW, Kuo KC, Waalkes TP, Borek E. tRNA breakdown products as markers for cancer. Cancer. 1979;44(6):2120–3. doi: 10.1002/1097-0142(197912)44:6<2120::aid-cncr2820440623>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 59.Martens-Uzunova ES, Jalava SE, Dits NF, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31(8):978–91. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 60.Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;27:2. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.t’Hoen PA, Friedländer MR, Almlöf J, et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31(11):1015–22. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 62.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100(20):1432–8. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kern SE. Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer Res. 2012;72(23):6097–101. doi: 10.1158/0008-5472.CAN-12-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maute RL, Schneider C, Sumazin P, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1404–9. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruggero K, Guffanti A, Corradin A, et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol. 2014;88(7):3612–22. doi: 10.1128/JVI.02823-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mishima E, Inoue C, Saigusa D, et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J Am Soc Nephrol. 2014;25(10):2316–26. doi: 10.1681/ASN.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanada T, Weitzer S, Mair B, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495(7442):474–80. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karaiskos S, Naqvi AS, Swanson KE, Grigoriev A. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol Direct. 2015;10:51. doi: 10.1186/s13062-015-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10(4):553–63. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]