Abstract
Background
Dysregulation of miR-9 is a common feature of many types of cancers, including oral squamous cell carcinoma (OSCC). However, whether the expression level of serum miR-9 is changed in patients with OSCC remains unknown.
Material/Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to examine the expression level of serum miR-9 in OSCC patients, oral leukoplakia (OLK) patients, and healthy volunteers, then we evaluated the association between serum miR-9 expression level and clinical outcome of OSCC patients.
Results
The expression level of serum miR-9 was significantly downregulated in patients with OSCC or OLK in comparison with healthy controls (P<0.01). Serum miR-9 expression level was associated with various clinicopathological parameters, including T stage (P=0.013), lymph node metastasis (P=0.002), and TNM stage (P=0.007). In addition, the OSCC patients in the low serum miR-9 expression group had poorer overall survival rate (P=0.022) and disease-free survival rate (P=0.004) compared with those in the high serum miR-9 expression group. Multivariate analysis showed that serum miR-9 was an independent prognostic factor for OSCC.
Conclusions
Serum miR-9 was downregulated in patients with OSCC and patients with OLK. In addition, low serum miR-9 was correlated with poor prognosis of OSCC, indicating miR-9 might play a tumor suppressive role in OSCC and can serve as a promising biomarker for this deadly disease.
MeSH Keywords: Biological Markers; Leukoplakia, Oral; MicroRNAs; Mouth Neoplasms; Prognosis; Serum
Background
Oral squamous cell carcinoma (OSCC) is the most common malignancy of the oral cavity, accounting for approximately 90% of oral neoplasms [1,2]. Despite aggressive treatment modalities, this malignant disease remains a leading cause of cancer-related deaths worldwide [3]. The overall 5-year survival rate of OSCC, which is less than 50%, has not changed much in the past decades [4]. As OSCC is generally asymptomatic at an early stage, most cases are diagnosed at the advanced stage, when the treatment options are limited and the clinical outcome is rather poor. Screening and an early detection are effective methods to solve this problem. Unfortunately, highly sensitive and specific biomarkers for OSCC are still not available for clinical use and exploring novel molecular markers is urgently needed.
MicroRNAs are a family of small, non-coding RNAs (approximately 21–25 nt in length) with the ability to regulate protein output via silencing the expression of targeted genes at the posttranscriptional level [5,6]. MiRNAs play important roles in regulation of most cellular processes, such as cell proliferation, differentiation, migration, and survival [7,8]. Deregulation of miRNAs is closely linked to the pathological mechanisms of many human diseases, including cancer [9,10]. MiRNAs can not only regulate the expression levels of oncogenes and tumor suppressor genes to influence the progression of cancer, but also might directly function as oncogenes or tumor suppressor genes. MiRNAs have been shown to be involved in tumorigenesis of OSCC. Tu et al. reported that miR-372 and miR-373 were up-regulated in OSCC tissues. In addition, overexpression of these 2 miRNAs was closely correlated with nodal metastasis, lymphovascular invasion, and poor survival, indicating that miR-372 and miR-373 might act as oncogenes in OSCC and promote its progression [11]. MiR-181a was down-regulated in OSCC and enforced expression of miR-181a can inhibit tumor cell behavior, suggesting that miR-181a might play a tumor suppressive role in OSCC [12].
Dysregulation of miR-9 has been shown to participate in the carcinogenesis of many types of cancers, including OSCC [13–15]. Although previous studies have demonstrated that miR-9 is significantly down-regulated in OSCC tissues and cell lines and might function as a tumor suppressor in OSCC [13], whether the expression level of serum miR-9 is changed in patients with OSCC has until now remained unknown. Therefore, the aim of this study was to evaluate the serum miR-9 expression level in OSCC patients and its potential clinical values.
Material and Methods
Study population and clinical specimens
The study was approved by the Ethics Committee of the Affiliated Hospital of Binzhou Medical College and written informed consent was obtained from all the participants: 104 OSCC patients, 30 OLK patients, and 40 healthy volunteers. Study subjects were recruited from patients who received treatment at the Department of Oral Surgery, the Affiliated Hospital of Binzhou Medical College and all cases were pathologically confirmed by at least 2 pathologists. In addition, subjects had not received any kind of therapy before serum sample collection. The clinical data of OSCC patients are summarized in Table 1.
Table 1.
The association between serum miR-9 expression and clinicopathological characteristics of OSCC.
| Parameters | Group | Total | Serum miR-9 | P | |
|---|---|---|---|---|---|
| Low | High | ||||
| Gender | Female | 43 | 20 | 23 | 0.918 |
| Male | 61 | 29 | 32 | ||
| Age | ≤60 | 36 | 21 | 15 | 0.095 |
| >60 | 68 | 28 | 40 | ||
| Smoking status | Non-smoker | 55 | 29 | 26 | 0.225 |
| Smoker | 49 | 20 | 29 | ||
| Tumor site | Tongue | 34 | 14 | 20 | 0.398 |
| Non-tongue | 70 | 35 | 35 | ||
| T stage | T1–T2 | 66 | 25 | 41 | 0.013 |
| T3–T4 | 38 | 24 | 14 | ||
| Lymph node metastasis | No | 63 | 22 | 41 | 0.002 |
| Yes | 41 | 27 | 14 | ||
| TNM stage | I–II | 59 | 21 | 38 | 0.007 |
| III–IV | 45 | 28 | 17 | ||
| Grade | Well/moderate | 71 | 29 | 42 | 0.060 |
| Poor | 33 | 20 | 13 | ||
Real-time PCR
Up to 4-ml blood samples were drawn from subjects and then the specimens were centrifuge at 3500g at room temperature for 5 min. Serum was transferred into RNA-free EP tubes and stored at −80°C until use. The Qiagen miRNeasy kit (Qiagen, Hilden, Germany) was used to isolate total RNA from serum samples according to the manufacturer’s protocol. Then the miScript SYBR Green PCR Kit (Qiagen) was used to generate cDNA and the subsequent PCR amplification. The PCR reaction was performed using a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). RNU6B was used as internal control for normalization of data, and the expression level of serum miR-9 was calculated and determined using the 2−ΔΔCT method. Experiments were performed in triplicate. The primers for miR-9 were: sense 5′-CGGAGATCTTTTCTCTCTTCACCCTC-3′, antisense 5′-CAAGAATTCGCCCGAACCAGTGAG-3′.
Statistical analysis
Because the data were not normally distributed, the expression levels of serum miR-9 in patients with OLK, patients with OSCC, and healthy volunteers were analyzed using the Kruskal-Wallis test. The chi-square test was used to evaluate the correlation between serum miR-9 expression level and clinical parameters of OSCC. The association between serum miR-9 and overall survival/disease-free survival was examined using the Kaplan-Meier method and log-rank test. Univariate and multivariate analysis were performed to identify the independent risk factors for OSCC. All data were analyzed using SPSS v21.0 software (SPSS Inc., Chicago, IL, USA). P values<0.05 were regarded as statistically significant.
Results
The expression level of serum miR-9 in patients with OSCC and patients with OLK
The expression levels of serum miR-9 in patients with OSCC and those with OLK were significantly decreased compared with healthy controls (P<0.01). In addition, OLK patients had a higher serum miR-9 expression level than OSCC patients (P<0.05) (Figure 1).
Figure 1.
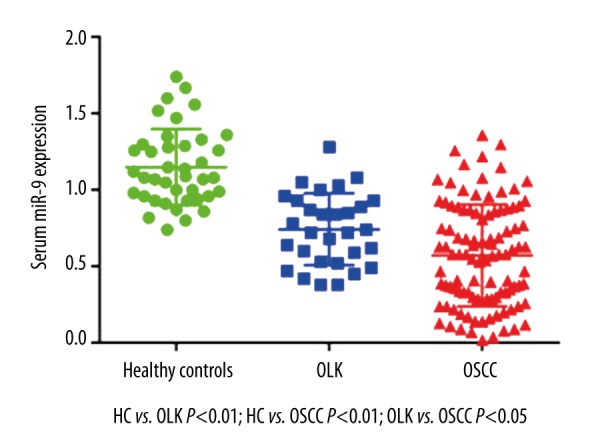
Expression level of serum miR-9 in OLK and OSCC. Serum miR-9 expression level was significantly reduced in patients with OSCC and patients with OLK in comparison with healthy volunteers (P<0.01). In addition, OLK patients had a higher serum miR-9 expression level than OSCC patients (P<0.05).
The association between serum miR-9 expression level and clinical parameters of OSCC
The average expression level of serum miR-9 was first evaluated in healthy volunteers, and then the relative expression level of serum miR-9 in all OSCC patients was determined by comparing it with the average expression level of serum miR-9 in healthy controls. The median expression level of serum miR-9 (0.58 fold) was used to divide the OSCC patients into a high serum miR-9 expression group and a low serum miR-9 expression group. The chi-square test showed that the expression level serum miR-9 was associated with T stage (P=0.013), lymph node metastasis (P=0.002), and TNM stage (P=0.007). However, it was not correlated with sex (P=0.918), age (P=0.095), smoking status (P=0.225), tumor site (P=0.398), or tumor grade (P=0.060) (Table 1).
OSCC patients in the low serum miR-9 expression group had poorer overall survival and disease-free survival
The survival analysis revealed that OSCC patients in the low serum miR-9 expression group had poorer overall survival (P=0.022) (Figure 2) and disease-free survival rates (P=0.004) than the patients in the high serum miR-9 expression group (Figure 3).
Figure 2.
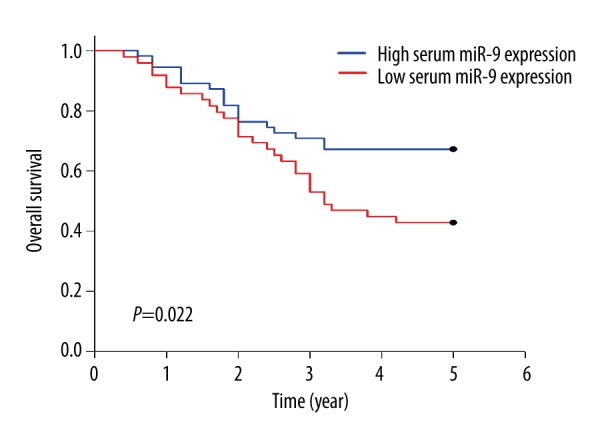
Association between serum miR-9 and overall survival. The overall survival rate of OSCC patients in the low serum miR-9 expression group was 42.86%, which was significantly lower than in the patients (67.27%) in the high serum miR-9 expression group (P=0.022).
Figure 3.
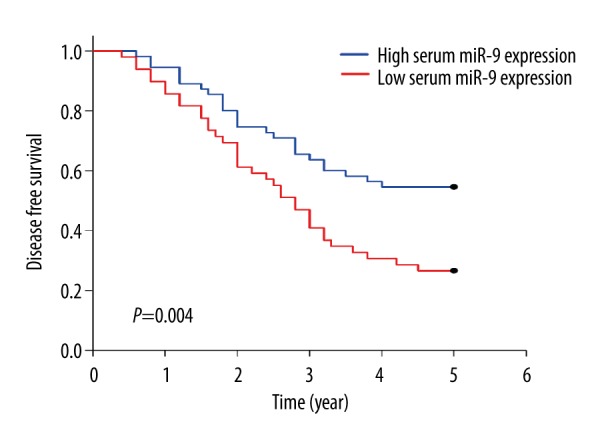
Correlation between serum miR-9 and disease-free survival. OSCC patients in the low serum miR-9 expression group had a poorer disease-free survival compared with those in the high serum miR-9 expression group (26.53% vs. 54.55%; P=0.004).
Serum miR-9 was an independent risk factor for OSCC
For overall survival, univariate analysis showed that T stage (P=0.038), lymph node metastasis (P=0.006), TNM stage (P=0.026) and serum miR-9 expression level (P=0.015) were significant prognostic factors for OSCC. All the above clinical parameters were also independent risk factors for OSCC (Table 2)
Table 2.
Univariate and multivariate analysis of serum miR-9 expression in OSCC overall survival.
| Parameters | Overall survival | |||
|---|---|---|---|---|
| Univariate OR (95%CI) | P | Multivariate OR (95%CI) | P | |
| Gender (Male vs. Female) | 1.256 (0.310–1.816) | 0.546 | ||
| Age (>60 vs. ≤60) | 1.413 (0.524–2.362) | 0.389 | ||
| Smoking status (Smoker vs. Non-smoker) | 1.108 (0.497–1.544) | 0.732 | ||
| Tumor site (Tongue vs. Non-tongue) | 0.867 (0.426–1.219) | 0.507 | ||
| T stage (T3–T4 vs. T1–T2) | 2.614 (1.318–4.354) | 0.038 | 2.961 (1.384–4.732) | 0.041 |
| Lymph node metastasis (Yes vs. No) | 4.390 (2.065–8.697) | 0.006 | 6.127 (2.546–10.873) | 0.008 |
| TNM stage (III–IV vs. I–II) | 3.017 (1.623–5.783) | 0.026 | 3.720 (1.856–6.028). | 0.024 |
| Grade (Poor vs. Well/moderate) | 1.935 (0.974–2.840) | 0.058 | ||
| Serum miR-9 expression (Low vs. High) | 3.621 (1.892–6.983) | 0.015 | 4.054 (1.980–7.862) | 0.019 |
In disease-free survival, T stage (P=0.043), lymph node metastasis (P=0.005), TNM stage (P=0.031), and serum miR-9 expression level (P=0.011) were significant prognostic factors in univariate analysis. Lymph node metastasis (P=0.006), TNM stage (P=0.040), and serum miR-9 expression level (P=0.018) were independent risk factors (Table 3).
Table 3.
Univariate and multivariate analysis of serum miR-9 expression in OSCC disease free survival.
| Parameters | Overall survival | |||
|---|---|---|---|---|
| Univariate OR (95%CI) | P | Multivariate OR (95%CI) | P | |
| Gender (Male vs. Female) | 1.484 (0.581–2.308) | 0.345 | ||
| Age (>60 vs. ≤60) | 1.208 (0.348–1.560) | 0.620 | ||
| Smoking status (Smoker vs. Non-smoker) | 1.639 (0.394–2.836) | 0.196 | ||
| Tumor site (Tongue vs. Non-tongue) | 0.940 (0.621–1.388) | 0.765 | ||
| T stage (T3–T4 vs. T1–T2) | 2.631 (1.378–4.293) | 0.043 | 2.738 (0.985–3.952) | 0.058 |
| Lymph node metastasis (Yes vs. No) | 5.829 (2.571–10.686) | 0.005 | 6.521 (2.524–11.630) | 0.006 |
| TNM stage (III–IV vs. I–II) | 3.487 (1.492–5.363) | 0.031 | 3.295 (1.407–4.259) | 0.040 |
| Grade (Poor vs. Well/moderate) | 2.282 (0.954–3.473) | 0.062 | ||
| Serum miR-9 expression (Low vs. High) | 4.420 (1.813–7.362) | 0.011 | 4.592 (1.924–8.271) | 0.018 |
Discussion
In the present study, our results showed that the expression level of serum miR-9 was significantly down-regulated in patients with OSCC or OLK, indicating that miR-9 might be involved in regulating the initiation and progression of OSCC. Detecting serum miR-9 expression level in patients with oral pre-cancer might help screen and identify the high risk population that has the potential to develop into OSCC. In addition, low serum miR-9 expression level was associated with advanced stage and poor prognosis of OSCC, and serum miR-9 was an independent risk factor for OSCC. Based on our study results, MiR-9 probably acts as a tumor suppressor in OSCC and this finding is consistent with previous reports. Yu et al. showed that miR-9 was under-expressed in OSCC tissues and oral cancer cell lines. In addition, overexpression of miR-9 could suppress the proliferative capacity of oral cancer cells both in vitro and in vivo. MiR-9 might have a tumor-suppressive role by downregulating the expression of CXC chemokine receptor 4 via the Wnt/β-catenin signaling pathway [13]. Curcumin, a phytochemical derived from the rhizome of Curcuma longa, has demonstrated antitumor activity in many types of tumors. Xiao et al. recently reported that curcumin inhibited proliferation of oral cancer cells by upregulating miR-9 expression and inhibiting Wnt/β-catenin signaling [16], indicating that the regulatory role of miR-9 in OSCC closely interacts with Wnt/β-catenin signaling. In addition to OSCC, miR-9 has also been shown to suppress tumorigenesis in various types of cancers. Lu et al. demonstrated that the expression of miR-9 was reduced in nasopharyngeal carcinoma (NPC) tissues and cell lines, and the reduction of miR-9 in NPC specimens was correlated with clinical stage and metastasis. Moreover, ectopic expression of miR-9 could significantly suppress proliferation, migration, and invasive capacity of NPC cell lines, indicating that miR-9 might be a negative regulator of NPC progression [17]. Emmrich et al. revealed that miR-9 was downregulated in patients with acute myeloid leukemia (AML). However, overexpression of miR-9 inhibited growth and induced monocytic differentiation in specific AML cell lines but not in all types of AML cells investigated, indicating the tumor suppressive role of miR-9 was strictly cell context-dependent [18]. Similarly, Wan et al. showed that miR-9 was downregulated in human gastric adenocarcinoma and that enforced expression of miR-9 could inhibit proliferation of cancer cells both in vitro and in vivo. Moreover, the tumor-related gene NF-kappaB1was identified as a downstream targeted gene of miR-9 in gastric cancer [19].
Although miR-9 can suppress tumorigenesis in some types of cancers, it might also act as an oncogene. Wu et al. reported that the expression level of miR-9 was significantly upregulated in gliomas tissues compared with normal adjacent tissues. In addition, miR-9 expression level was closely associated with WHO grade, Karnofsky performance score, and overall survival, indicating that miR-9 may have an important role in tumor progression in human gliomas [20]. Zhu et al. showed that the expression level of miR-9 was higher in colorectal cancer (CRC) with distant metastasis than that in non-metastasis CRC. Overexpression of miR-9 changed the morphological appearance of colorectal and enhanced the motility, suggesting miR-9 might be crucial for CRC metastasis [21].
Several reasons might explain the contradictory role of miR-9 in different types of cancers. Firstly, the concrete function of miR-9 may be cancer-dependent. Because a single miRNA can target a number of downstream genes, the concrete microenvironment context might influence which targeted genes will be activated or suppressed. It is common to observe the phenomenon that a specific miRNA function as an oncogene in a certain type of cancer while it acts as a tumor suppressor in another type of cancer. Secondly, the concrete function of miR-9 might be cell-dependent. MiR-9 is a tumor suppressor in pediatric AML with t(8;21) [18]. However, it was upregulated in patients with mixed-lineage leukemia-rearranged AML compared with the controls and could promote the progression of this specific type of AML [22]. The genetic changes in the leukemia cells might be responsible for the opposite role of miR-9 in different subtypes of AML.
Conclusions
The expression level of serum miR-9 was downregulated in patients with OLK and those with OSCC. Low serum miR-9 was associated with advanced stage and poor prognosis of OSCC. Collectively, our data demonstrate that miR-9 is a tumor suppressor in OSCC and can serve as a potential therapeutic target to treat this malignant disease.
Footnotes
Conflict of interest
None declared.
Source of support: Departmental sources
References
- 1.Stewart BW, Greim H, Shuker D, et al. Defence of IARC monographs. Lancet. 2003;361(9365):1300. doi: 10.1016/S0140-6736(03)13003-6. [DOI] [PubMed] [Google Scholar]
- 2.Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008;87(1):14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Nagler RM. Molecular aspects of oral cancer. Anticancer Res. 2002;22(5):2977–80. [PubMed] [Google Scholar]
- 5.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 6.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14(8):475–88. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 7.Clark EA, Kalomoiris S, Nolta JA, et al. Concise review: MicroRNA function in multipotent mesenchymal stromal cells. Stem Cells. 2014;32(5):1074–82. doi: 10.1002/stem.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meza-Sosa KF, Pedraza-Alva G, Pérez-Martínez L. microRNAs: key triggers of neuronal cell fate. Front Cell Neurosci. 2014;8:175. doi: 10.3389/fncel.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–33. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan H, Lu H, Wang X, et al. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu HF, Chang KW, Cheng HW, et al. Upregulation of miR-372 and -373 associates with lymph node metastasis and poor prognosis of oral carcinomas. Laryngoscope. 2015 doi: 10.1002/lary.25464. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Shin KH, Bae SD, Hong HS, et al. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun. 2011;404:896–902. doi: 10.1016/j.bbrc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- 13.Yu T, Liu K, Wu Y, et al. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/β-catenin signaling pathway. Oncogene. 2014;33:5017–27. doi: 10.1038/onc.2013.448. [DOI] [PubMed] [Google Scholar]
- 14.Sondermann A, Andreghetto FM, Moulatlet AC, et al. MiR-9 and miR-21 as prognostic biomarkers for recurrence in papillary thyroid cancer. Clin Exp Metastasis. 2015;32:521–30. doi: 10.1007/s10585-015-9724-3. [DOI] [PubMed] [Google Scholar]
- 15.Xie D, Shang C, Zhang H, et al. Up-regulation of miR-9 target CBX7 to regulate invasion ability of bladder transitional cell carcinoma. Med Sci Monit. 2015;21:225–30. doi: 10.12659/MSM.893232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao C, Wang L, Zhu L, et al. Curcumin inhibits oral squamous cell carcinoma SCC-9 cells proliferation by regulating miR-9 expression. Biochem Biophys Res Commun. 2014;454:576–80. doi: 10.1016/j.bbrc.2014.10.122. [DOI] [PubMed] [Google Scholar]
- 17.Lu J, Luo H, Liu X, et al. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis. 2014;35:554–63. doi: 10.1093/carcin/bgt354. [DOI] [PubMed] [Google Scholar]
- 18.Emmrich S, Katsman-Kuipers JE, Henke K, et al. miR-9 is a tumor suppressor in pediatric AML with t(8;21) Leukemia. 2014;28:1022–32. doi: 10.1038/leu.2013.357. [DOI] [PubMed] [Google Scholar]
- 19.Wan HY, Guo LM, Liu T, et al. Regulation of the transcription factor NF-kappaB1 by microRNA-9 in human gastric adenocarcinoma. Mol Cancer. 2010;9:16. doi: 10.1186/1476-4598-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Wang L, Li G, et al. Increased expression of microRNA-9 predicts an unfavorable prognosis in human glioma. Mol Cell Biochem. 2013;384:263–68. doi: 10.1007/s11010-013-1805-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhu L, Chen H, Zhou D, et al. MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Med Oncol. 2012;29:1037–43. doi: 10.1007/s12032-011-9975-z. [DOI] [PubMed] [Google Scholar]
- 22.Chen P, Price C, Li Z, et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci USA. 2013;110:11511–16. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]


