Abstract
The existence of many highly similar genes in the lymphocyte receptor gene loci makes them difficult to investigate, and the determination of phased “haplotypes” has been particularly problematic. However, V(D)J gene rearrangements provide an opportunity to infer the association of Ig genes along the chromosomes. The chromosomal distribution of H chain genes in an Ig genotype can be inferred through analysis of VDJ rearrangements in individuals who are heterozygous at points within the IGH locus. We analyzed VDJ rearrangements from 44 individuals for whom sufficient unique rearrangements were available to allow comprehensive genotyping. Nine individuals were identified who were heterozygous at the IGHJ6 locus and for whom sufficient suitable VDJ rearrangements were available to allow comprehensive haplotyping. Each of the 18 resulting IGHV|IGHD|IGHJ haplotypes was unique. Apparent deletion polymorphisms were seen that involved as many as four contiguous, functional IGHV genes. Two deletion polymorphisms involving multiple contiguous IGHD genes were also inferred. Three previously unidentified gene duplications were detected, where two sequences recognized as allelic variants of a single gene were both inferred to be on a single chromosome. Phased genomic data brings clarity to the study of the contribution of each gene to the available repertoire of rearranged VDJ genes. Analysis of rearrangement frequencies suggests that particular genes may have substantially different yet predictable propensities for rearrangement within different haplotypes. Together with data highlighting the extent of haplotypic variation within the population, this suggests that there may be substantial variability in the available Ab repertoires of different individuals.
A major gap in our knowledge of the human genome is the lack of phased genomic data describing the chromosomal associations of different nucleotide sequences (1). Phased (or “haplotype”) data bring additional power to the investigation of species evolution (2) and to the exploration of relationships between human populations (3). Such data are of particular importance where the chromosomal location of genes could be of physiological consequence, as is the case, for example, with the rearrangeable genes that encode the highly variable receptors of B cells and T cells of the immune system.
The mammalian immune system has the ability to respond to almost any Ag to which it is exposed because of the incredible diversity of lymphocyte receptor molecules. BCR diversity is made possible by multiple sets of highly similar genes that recombine to form functional VDJ gene rearrangements encoding the Ig H chain and VJ gene rearrangements encoding the Ig L chain (4). Ig gene recombination is an intrachromosomal event, and knowledge of gene associations would facilitate understanding of the generation of diversity. Yet we have very little phased data relating to the Ig gene loci. Although both the HapMap project (5) and the 1000 Genomes Project (6) have identified numerous polymorphisms in these loci, the association of so many highly similar sequences on each chromosome prevents the inference of haplotypes using conventional linkage disequilibrium-based phasing methods.
The H chain variable gene (IGHV) locus was first mapped in the 1990s (7), but the difficulties associated with the sequencing of so many similar genes and pseudogenes has meant that a complete sequence of the locus has only been reported once (8), and it was this sequence that was incorporated into the first two published versions of the human genome (9, 10). The sequence is not a true haplotype, for it was assembled from contigs derived from three lymphoblastoid cell lines (8). A complete description of the H chain diversity gene (IGHD) locus has also only been published once (11), and a handful of early reports described a number of distinct haplotypes encoding the six genes of the shorter joining gene (IGHJ) locus (12–14). More recently, it has been suggested that some of the apparent allelic variants that defined these IGHJ haplotypes may have been misidentified as a consequence of sequencing errors (15).
High-throughput sequencing has recently been applied to the study of human Ig (16–18) and TCR genes (19). Although such technology can be used for the direct investigation of germline genes (18), in this study we report how the analysis of amplified H chain VDJ gene rearrangements can be used to efficiently infer phased data covering all rearrangeable genes in the H chain V region gene loci. Despite the challenges involved in the identification of the germline genes that contribute to some VDJ rearrangements, in large data sets each rearrangement provides independent evidence of the presence of particular IGHV, IGHD, and IGHJ genes on one chromosome. Together, thousands of rearrangements can also provide evidence for the absence of particular genes, for deletion polymorphisms have been reported to occur in the IGHV (20) and IGHD (21) loci. Ig gene haplotypes, including deletion polymorphisms, can therefore be inferred, and in this study we describe the inferred diplotypes of nine individuals.
The IgH gene locus is revealed as a locus of considerable variability. Not only are the 18 haplotypes described in this study all unique, but the extent of the differences between the haplotypes suggests the existence of many additional haplotypes. This variation is likely to result in substantial individual differences in repertoires of available Abs.
Materials and Methods
Sequence generation and data set preparation
Ig gene sequence data sets were generated from samples of human peripheral blood that were obtained under a Stanford University Institutional Review Board-approved protocol. Donors were of unknown ethnicity and were recruited from the San Francisco Bay Area. PBMCs were isolated by centrifugation of diluted blood layered over Hypaque 1077 (Sigma-Aldrich), and purified DNA template was prepared either by column purification (Qiagen, Valencia, CA) or by magnetic bead-based isolation (Magnapure, Roche Diagnostics Corporation, Indianapolis, IN). PCR was performed using BIOMED-2 primers to which were added 59 sequencing elements, and unique 6-, 7-, or 10-nucleotide sample “barcodes” were added to the IGHJ primers as previously described (17). To facilitate pooling, bulk sequencing, and later determination of the sample source of each sequence, production of amplicons for later sequencing using the 454 Titanium sequencer also used additional 10-nucleotide sample barcodes that were added to the IGHV primers (17). Analysis of many of the resulting amplicons has previously been described in two separate reports (16, 17). For this study, longer amplicons were also produced using the same IGHJ primer in conjunction with BIOMED2 IGHV framework 1 (FR1) primers as reported in Table I (22). PCR amplifications were performed, then amplicons were pooled and purified as previously described (17). High-throughput amplicon pyrosequencing data were then generated using either 454 FLX chemistry or Titanium chemistry (Roche, Branford, CT).
Table I.
IGHV FR1-region family-specific forward primers used in PCR amplifications of rearranged Ig gene sequences
| IGHV Family | Sequences (5′ to 3′) |
|---|---|
| IGHV1 | GGCCTCAGTGAAGGTCTCCTGCAAG |
| IGHV2 | GTCTGGTCCTACGCTGGTGAAACCC |
| IGHV3 | CTGGGGGGTCCCTGAGACTCTCCTG |
| IGHV4 | CTTCGGAGACCCTGTCCCTCACCTG |
| IGHV5 | CGGGGAGTCTCTGAAGATCTCCTGT |
| IGHV6 | TCGCAGACCCTCTCACTCACCTGTG |
Sequences were sorted into individual data sets based on the presence of perfect matches to sample barcodes, IGHV primers, and to the first three bases of the IGHJ common primer.
Determination of genotypes
The sets of germline IGHV, IGHD, and IGHJ genes that collectively define an individual's genotype were determined by analysis of VDJ gene data sets. The sets of sequences were first aligned to a set of germline genes using Vmatch, a general sequence alignment utility (23). The gene set included both the UNSWIg IGHV germline repertoire (24) (http://www.ihmmune.unsw.edu.au/unswig.php) and the IMGT repertoire of functional genes and pseudogenes (25). Levels of mutations in the IGHV genes were reviewed, and where large numbers of alignments to a particular gene carried shared mismatches, further investigations were carried out to identify putative unreported polymorphisms, as previously described (16). For each 454 sequence, the five germline IGHV sequences or putative IGHV polymorphisms that aligned with the least mismatches and longest alignment length were selected as candidate progenitor sequences for the rearrangement. In some cases, it was not possible to distinguish between two possible contributing alleles, because the critical nucleotides that distinguish them are located upstream of the IGHV primers that had been used for sequence amplification. In such cases, if a contending allele had been highlighted previously as being of dubious provenance (24), it was excluded from further consideration. Maximum likelihood genotyping was then carried out to find the set of alleles that had the highest probability of resulting in the observed sequence set. Where a single allele provided the best alignment for all instances of that gene, it was immediately assigned to the final genotype.
We defined G as a genotype for a specific gene and S (S = {s1, s2,..., si}) as the set of sequences that were derived from this specific gene. For each gene, the maximum likelihood genotype was defined as
| (1) |
where Pr(S|G) is the probability of the sequence set S given a genotype G.
Most genes are normally present as one or two alleles in an individual genotype; however, in cases of gene duplication, there may appear to be three or even four alleles of the same gene. For each IGHV gene, the potential genotypes were the various sets of up to four alleles ({g1, g2, ..., gn},1 ≤ n ≤ 4) identifiable from the draft genotype. The probability Pr(si|gn) of sequence si given the allele gn can be estimated taking into account the position of apparent mutations in the sequence and their sequence context, as previously implemented in iHMMune-align (26).
Therefore, Pr(si|G) was estimated as:
| (2) |
Assuming, initially, that all alleles of a given gene are equally likely to appear in a genotype,
| (3) |
Therefore according to Eqs. 1, 2, and 3,
| (4) |
A preliminary analysis of the IGH sequences was then performed using the partitioning utility iHMMune-align (26) to identify the germline IGHD segments that were most likely to have been incorporated into each rearrangement. By reference to the iHMMune-align output, duplicate sequences, clonally related sequences and chimeric sequences, and sequences containing insertions or deletions were then identified and removed from the data sets. For each individual, the IGHD genotype was then defined by the presence or absence of each gene in the iHMMune-align partitioning results using IGHD gene-dependent rules. Three members of the IGHD1 gene family (IGHD1-1, IGHD1-7, and IGHD1-20) were not included in the genotypes because these genes are short and highly similar and therefore cannot be identified with sufficient certainty. Similarly, the IGHD7-27 gene was excluded because of its very short length. The IGHD2-2 gene was included in the genotyping, but no attempt was made to identify the three rarely distinguishable IGHD2-2 alleles. No attempt was made to distinguish between IGHD5-5 and IGHD5-18, as these genes share identical coding regions. Finally, the IGHD4-11, IGHD1-14, and IGHD6-25 genes were excluded as they are apparently not rearrangeable (27). Rules based on specific sequence motifs were used to discriminate between alleles for the IGHD2-8, IGHD2-21, IGHD3-3, IGHD3-10, and IGHD3-16 genes. For other IGHD genes, inclusion in the final genotype was based on the presence or absence of satisfactory alignments in the iHMMune-align output.
Genotyping of IGHJ genes was performed using a strategy similar to that of IGHV genotyping. The 39 end of each VDJ sequence was aligned against the IGHJ repertoire using Vmatch (23). After determination of the draft IGHJ genotype, the maximum likelihood genotyping method was applied to ambiguous sequences using a simplified estimation of Pr(si|gn) based on the number of mutations observed in their IGHV region alignment.
Inference of haplotypes
Genotypes were examined, and individuals who were heterozygous at the IGHJ6 loci were identified. Data sets for each donor were checked to ensure that they contained the minimum number of sequences required to give a 95% probability of being able to detect the presence of an IGHVor IGHD gene within a haplotype with an average rearrangement frequency of 0.5%. This was determined as follows:
| (5) |
where N is the required number of sequences, θ is the desired probability of observing the gene, and f = 0.005 × (the proportion of the total data set utilizing the least frequently observed IGHJ6 allele).
For suitable individuals, N varied between 4,289 and 10,272. VDJ sequences from these individuals were repartitioned with iHMMune-align using customized germline repertoires based on the genotype of each individual. The two IGH haplotypes of each individual were then determined by examining the number of instances in which an IGHV or IGHD gene associated with each of the IGHJ alleles of the heterozygous IGHJ6 locus. Genes and allelic variants were only included in this analysis if at least 10 VDJ rearrangements were identified that appeared to include that gene or allele.
On the assumption that a proportion of the apparent IGHV and IGHD gene sequences identified within VDJ rearrangements would be incorrectly associated with an IGHJ6 allele, two hypotheses were considered: that each germline gene or allelic variant was either found on one or on both chromosomes. Where x sequences were assigned to chromosome A, y sequences were assigned to chromosome B, and x ≥ y, the probabilities associated with each hypothesis were calculated as follows:
| (6) |
| (7) |
where n = x + y is the total number of alignments to the gene, ε is the error rate of assignments to chromosome A, and p is the overall proportion of sequences in the data set involving rearrangement of chromosome A.
The error rate was set by reference to published evaluations of the performance of the iHMMune-align utility (28). Where a single allele was present in the genotype, ε was set as 0.02, and where multiple alleles were present, ε was set as 0.1. The likelihood ratio (LR) was then calculated to determine the most likely explanation of the results, as follows:
| (8) |
where LR1 was >20, the more likely hypothesis was confirmed. Because the presence of the gene within the individual's genotype had been independently determined, where LR1 was <20, it was accepted that the gene would likely be present on at least one chromosome. The likelihood ratio of the probabilities that the gene was either present on both chromosomes or only on chromosome B was then calculated as follows:
| (9) |
where the value of this ratio was >20, the presence of the gene on chromosome A was confirmed, while the presence or absence of the gene on chromosome B remained uncertain.
Unrearranged IGH locus genomic DNA sequencing
Genomic DNA was prepared from PBMC samples using column purification kits (Qiagen) and was treated with RNase A prior to genomic library construction. DNA was fragmented using fragmentase enzyme (New England Biolabs) and size selected with agarose gel electrophoresis and gel purification column kits (Qiagen). Fragmented DNA was end-repaired to form blunt-ended fragments using Klenow DNA Polymerase I, T4 DNA Polymerase, and T4 Polynucleotide Kinase (New England Biolabs), then linkered with Illumina linkers for single-end nondirectional sequencing. Agarose gels were used for size selection of linkered fragments of 150- to 400-bp size, and fragments were purified using a gel purification column kit (Qiagen). Twenty-five cycles of PCR were carried out on the linkered libraries with linker-specific primers and Phusion polymerase (New England Biolabs), and the final library was gel purified. Sequencing of each genomic fragment library with single-end 36 base reads was carried out in a single lane of an Illumina Genome Analyzer IIx sequencer.
Genomic DNA sequence analysis
Single-end 36 base genomic DNA reads were aligned to the HG19 version of the reference human genome using the Needleman–Wunsch algorithm implemented in the Novoalign alignment software (Novocraft). Reads aligning to multiple sites in the genome were excluded from further analysis. Reads aligning to unique positions in the genome were evaluated further, after collapsing identical reads to minimize bias from PCR amplification of the genomic libraries. Collapsed reads aligning to the predicted region of D segment deletion (spanning from IGHD3-3 to IGHD2-8) were counted and were compared with the total count of reads aligning to the rest of the genome. Statistical testing for the significance of the proportion of reads in the region of putative deletion compared with the number of reads over the rest of the genome, comparing the sample SBKN14 (predicted to contain the deletion) with a control sample, was calculated using χ2 testing and Fisher's exact test.
Results
Sets of VDJ rearrangements were considered from 44 individuals for whom there were sufficient unique sequences to allow comprehensive IGHV, IGHD, and IGHJ genotyping. The sequence data sets for these individuals averaged 4240 VDJ rearrangements. Sequences containing no mismatches to germline genes composed 47.5% of the data sets, and most of these sequences were therefore likely derived from IgM+/IgD+ naive B cells. This general lack of mutation aided genotype determination. The defined genotypes included between 39 and 55 distinct, functional germline IGHV sequences. A number of individuals were seen who appear to carry homozygous deletion polymorphisms involving IGHV genes, including IGHV4-30-2, IGHV3-30-3, IGHV4-30-4, IGHV4-39, IGHV4-b, and IGHV5-a. Deletions of the contiguous genes IGHV1-8 and IGHV3-9 were also inferred in several individuals. As many as seven rearrangeable pseudogenes were seen in a single genotype, and rearrangeable pseudogenes included humIGHV177, humIGHV181, IGHV1-14, IGHV1-17, IGHV3-19, IGHV3-22, IGHV3-41, IGHV3-47, IGHV3-52, and IGHV4-55. IGHD geno-types included between 15 and 23 distinct sequences, and apparent homozygous IGHD deletion polymorphisms were also seen in the IGHD locus. The partitioning of ~5% of sequences failed to identify a candidate IGHD gene that met the stringent requirements of iHMMune-align for IGHD gene identification (26).
Little diversity was seen among the IGHJ genotypes, although 15 individuals were heterozygous at the IGHJ6 locus. Although sequence data sets from every such individual can be used to define at least partial haplotypes, the inference of phased haplotypes was restricted to data sets that included at least 4200 unique rearrangements. This ensured that resulting haplotypes included many loci and that these loci could all be analyzed using multiple sequence reads. Nine individuals had sufficient IGHJ6-utilizing sequences for such comprehensive haplotyping. In these individuals, IGHJ6-utilizing sequences accounted for between 29.7 and 50.1% of all sequences.
Rearranged VDJ sequences were analyzed to determine the most likely haplotypes. A number of likely IGHD gene deletion polymorphisms were highlighted. Because of the difficulty of aligning rearranged sequences against the germline IGHD repertoire, the data sets that included these likely deletions, as well as any other rare IGHJ/IGHD pairings, were carefully reviewed. In one individual (AL13), a handful of sequences suggested the presence of the IGHD4-4 gene in a haplotype. In other respects, this region of the haplotype matched two other haplotypes that shared a deletion of the IGHD3-3, IGHD4-4, IGHD6-6, IGHD1-7, and IGHD2-8 genes. (The presence or absence of IGHD5-5 could not be determined because its sequence is identical to the IGHD5-18 gene.) Review of the IGHD4-4 alignments suggests that they were all in error, for they were all short and mutated. Apparent deletion polymorphisms involving single genes (IGHD2-8, IGHD3-9, and IGHD5-24) were also reviewed and confirmed.
The absence of D segments in sets of rearranged IGH VDJ sequences could, in theory, be the result of sequence variants preventing rearrangement of these segments, or could be due to deletion removing these regions of the genome entirely. To evaluate further for the presence of a homozygous germline genomic deletion of the region of the IGH locus containing D segments IGHD3-3 to IGHD2-8, whole genome shotgun sequencing was performed on DNA from an individual predicted to have the D3-3 to D2-8 deletion and on a control sample. Supplemental Fig. 1 shows that in subject SBKN14, zero reads aligned to the region of predicted deletion, whereas multiple reads aligned to this region in the control sample. χ2 testing and Fisher's exact test for the significance of the proportion of reads in the region of predicted deletion compared with the rest of the genome gave p values of 2 × 10−6 (χ2) and 7 × 10−7 (Fisher's exact test).
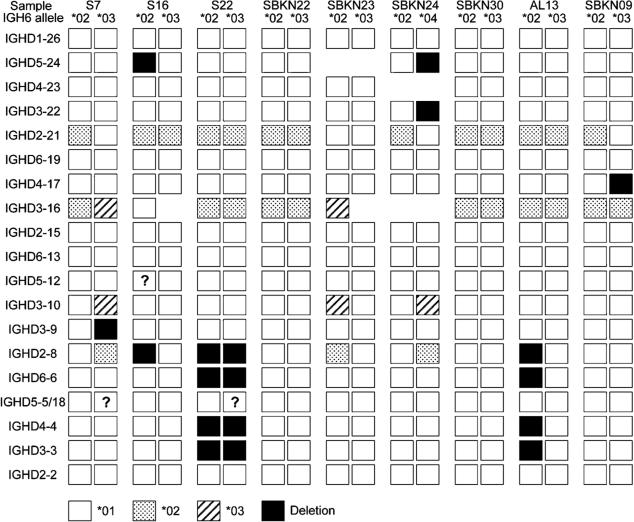
The diversity of the 18 IGHD haplotypes is represented in Fig. 1. Without consideration of loci where the allelic variant of the gene could not be determined with certainty, seven different inferred haplotypes were seen, including one haplotype that was identified nine times. IGHD haplotype analysis for one individual is presented as Table II, and analysis for all individuals is available as Supplemental Table I.
FIGURE 1.
Partial diplotypes of the IGHD locus, as determined for nine individuals. Only functional IGHD genes are shown. Genes that are highly similar (IGHD1-1, IGHD1-7, and IGHV1-20), and the very short IGHD7-27 gene are not shown, as these genes could not be distinguished with certainty in all individuals. Genes that are not rearrangeable (IGHD1-14, IGHD4-11, and IGHD6-25) are also omitted. IGHD5-5 and IGHD5-18 are identical sequences, and therefore one or both of the genes may be present where shown. Alleles are coded as shown in the key. Deletion polymorphisms are shown in black. Where a gene that was present in the genotype could not be identified in a haplotype with certainty, it is indicated as “?.”
Table II.
Sequence counts of IGHD genes and allelic variants associated with IGHJ6 alleles in VDJ rearrangements, and LRs for the inference of the presence of IGHD genes within the diplotypes of a representative individual
| Sequence Counts |
Presence of IGHD Genes on Chromosomes as Determined by LRs |
||||
|---|---|---|---|---|---|
| IGHJ6*02 | IGHJ6*03 | Chromosome 1a | Chromosome 2 | LR1 | |
| IGHD2-2b | 97 | 199 | ✓ | ✓ | 1.47E+71 |
| IGHD3-3*01 | 124 | 119 | ✓ | ✓ | 4.09E+129 |
| IGHD4-4/11*01 | 8 | 23 | ✓ | ✓ | 3.79E+03 |
| IGHD5-5/18*01 | 9 | 31 | ✓ | ✓ | 1.75E+02 |
| IGHD6-6*01 | 51 | 50 | ✓ | ✓ | 5.32E+54 |
| IGHD2-8*01 | 0 | 11 | x | ✓ | 2.62E+03 |
| IGHD2-8*02 | 9 | 0 | NDc | ND | |
| IGHD3-9*01 | 26 | 12 | ✓ | ✓ | 6.17E+09 |
| IGHD3-10*01 | 3 | 41 | x | ✓ | 3.15E+10 |
| IGHD3-10*p03 | 82 | 4 | ✓ | x | 2.10E+14 |
| IGHD5-12*01 | 30 | 13 | ✓ | ✓ | 1.45E+10 |
| IGHD6-13*01 | 92 | 43 | ✓ | ✓ | 2.32E+35 |
| IGHD2-15*01 | 84 | 21 | ✓ | ✓ | 2.32E+35 |
| IGHD3-16*02 | 0 | 4 | ND | ND | |
| IGHD3-16*p03 | 28 | 0 | ✓ | x | 5.88E+05 |
| IGHD4-17*01 | 48 | 16 | ✓ | ✓ | 6.50E+09 |
| IGHD6-19*01 | 38 | 23 | ✓ | ✓ | 4.37E+21 |
| IGHD2-21*01 | 30 | 18 | ✓ | ✓ | 7.46E+16 |
| IGHD3-22*01 | 65 | 28 | ✓ | ✓ | 6.17E+21 |
| IGHD4-23*01 | 9 | 4 | ✓ | ✓ | 1.52E+03 |
| IGHD5-24*01 | 6 | 1 | ND | ND | |
| IGHD1-26*01 | 44 | 32 | ✓ | ✓ | 1.84E+32 |
Chromosome 1 is the IGHJ6*02-defined chromosome.
Data are shown for all readily identifiable and rearrangeable IGHD genes. No attempt was made to distinguish between the highly similar IGHD2-2 alleles.
No attempt was made to assign a gene or allelic variant to a chromosome where fewer than 10 VDJ rearrangements appeared to include that sequence.
x, Absent; ✓, present.
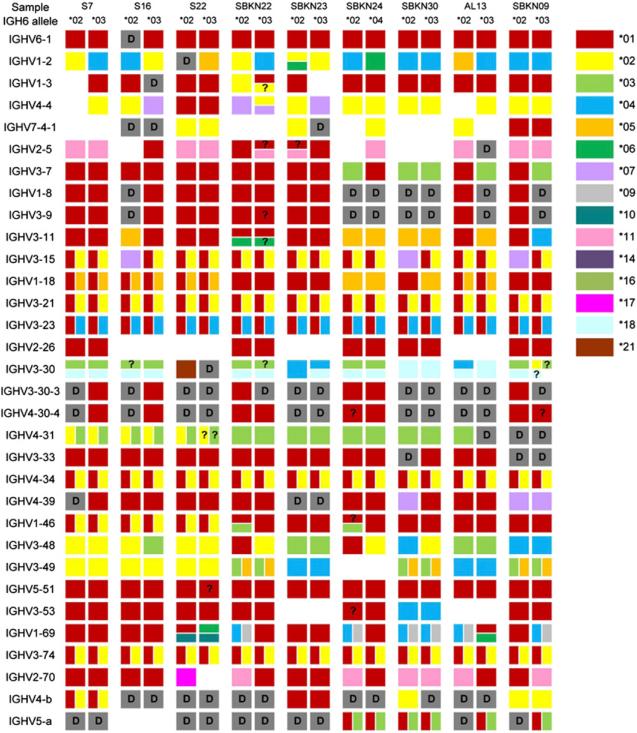
Analysis of IgH rearrangements as described earlier was then used to define the presence or absence of IGHV genes in each haplotype. A new putative variant of IGHV1-18 was identified. We have named this putative allele IGHV1-18*p05, and it is presented as Supplemental Table II. Subsequent investigation of dbSNP showed that the polymorphism has been reported by the 1000 Genomes Project (rs72695948). The diplotype of one individual is presented for the 3′ region of the genome, from IGHV6-1 to IGHV1-18, as Table III. One of the haplotypes shown appears to lack the contiguous IGHV1-8 and IGHV3-9 genes. The diversity of the 18 unique IGHV haplotypes is illustrated in the partial haplotypes of Fig. 2. Statistical analysis could not determine the presence or absence of some genes with certainty for all haplotypes, and these uncertainties are also indicated in Fig. 2. Additional associations across the IGHV locus are also available for all individuals as Supplemental Table III. These haplotypes include between 35 and 46 functional genes. In total, 54 IGHV loci were included in the inferred haplotypes, including seven loci where the data were indicative of duplications of previously reported genes. Such apparent duplications were seen for IGHV1-2, IGHV3-11, IGHV3-30, IGHV1-46, IGHV4-59, IGHV3-64, and IGHV1-69.
Table III.
Sequence counts of IGHV genes and allelic variants associated with IGHJ6 alleles in VDJ rearrangements, and LRs for the inference of the presence of IGHV genes and allelic variants within the diplotypes of a representative individual
| Sequence Counts |
Presence of IGHV Genes on Chromosomes as Determined by LRs |
||||
|---|---|---|---|---|---|
| IGHJ6*02 | IGHJ6*03 | Chromosome 1a | Chromosome 2 | LR1 | |
| IGHV6-1*01b | 2 | 15 | x | ✓ | 6.60E+01 |
| IGHV1-2*02 | 2 | 28 | x | ✓ | 1.63E+06 |
| IGHV1-2*04 | 20 | 0 | ✓ | x | 5.82E+04 |
| IGHV1-3*01 | 42 | 3 | ✓ | x | 2.62E+07 |
| IGHV4-4*02 | 16 | 1 | ✓ | x | 1.35E+03 |
| IGHV2-5*01 | 0 | 12 | x | ✓ | 1.89E+03 |
| IGHV2-5*p11 | 2 | 0 | NDc | ND | |
| IGHV3-7*01 | 18 | 22 | ✓ | ✓ | 4.47E+18 |
| IGHV1-8*01 | 1 | 28 | x | ✓ | 1.84E+07 |
| IGHV3-9*01 | 1 | 13 | x | ✓ | 4.12E+02 |
| IGHV3-11*01 | 1 | 14 | x | ✓ | 1.28E+03 |
| IGHV3-15*01/02 | 0 | 26 | x | ✓ | 1.25E+07 |
| IGHV3-15*07 | 29 | 6 | ✓ | x | 6.63E+02 |
| IGHV1-18*01/p05 | 30 | 30 | ✓ | ✓ | 1.41E+33 |
Chromosome 1 is the IGHJ6*02-defined chromosome.
Data are shown for functional IGHV genes at the 3′ end of the IGHV locus.
No attempt was made to assign a gene or allelic variant to a chromosome where fewer than 10 VDJ rearrangements appeared to include that sequence.
x, Absent; ✓, present.
FIGURE 2.
Partial diplotypes of the IGHV locus, as determined for nine individuals. Only functional IGHV genes are shown. Rarely identified genes with published rearrangement frequencies of <0.5% (IGHV3-13, IGHV3-20, IGHV1-24*01, IGHV4-30-2, IGHV3-43, IGHV1-45, IGHV1-58, IGHV3-64, IGHV3-66, IGHV3-72, IGHV3-73, and IGHV1-f) are also omitted (22). IGHV4-59 and IGHV4-61 are also omitted as these genes could not be distinguished with certainty in all individuals. Alleles are color-coded as shown in the key. Where a gene that was present in the genotype could not be identified in a haplotype with certainty, it is indicated as “?.” Where a gene or allelic variant was known to be present in a genotype but insufficient sequences were available to allow confident haplotyping, no data are recorded. Ambiguities (e.g., *01 or *02) are indicated by vertically split cells, and duplications are shown by horizontally split cells. Apparent deletion polymorphisms are shown as gray cells and are marked “D.”
Apparent deletion polymorphisms were inferred at the following IGHV loci: IGHV1-8, IGHV3-9, IGHV3-30, IGHV4-30-2, IGHV3-30-3, IGHV4-30-4, IGHV4-31, IGHV3-33, and IGHV4-39. In most cases, deletion of IGHV1-8 was associated with deletion of IGHV3-9, and this polymorphism was seen in 7 of the 18 haplotypes. Twelve different haplotypes included deletions of between one and four of the six contiguous functional genes from IGHV3-30 to IGHV3-33. Without consideration of uncertainties, eight distinct patterns of deletions were seen. Unmapped genes are generally rearranged at frequencies that preclude haplotyping, but IGHV4-b and IGHV5-a rearrange at higher frequencies and were also seen to be absent from many inferred haplotypes.
In many cases, although likelihood ratios suggested the absence of a gene from a chromosome, small numbers of sequences were seen that appeared to associate the “deleted” gene with that chromosome. Review of these data sets showed that these aberrant sequences were typically highly mutated. It is likely that these mutations led to a misidentification of the IGHV gene or allele in the rearranged sequences. For example, the data set generated from one individual included 120 sequences that used the IGHV3-30 gene and could be associated with the chromosome bearing IGHJ6*02. A further 13 sequences appeared to link the IGHV3-30 gene with the alternate chromosome. These sequences were all highly mutated. It is likely that these rearrangements do not involve the IGHV3-30 gene, but rather involve the highly similar IGHV3-33 gene. Mutations made some of the many IGHV3-33 sequences align more closely to the germline IGHV3-30 gene.
Inferred gene duplications were also supported by mutation analysis. If an apparent duplication was inferred as a consequence of misidentification of IGHV alleles, we would expect to see that a high proportion of these sequences were mutated. This was not the case.
Rearrangement frequencies were analyzed, and IGHD gene rearrangement frequencies were particularly variable. For example, IGHD5-12 was present in 0.4–5.0% and IGHD2-2 in 4.5–28.0% of all rearrangements of single chromosomes. IGHV gene rearrangement frequencies were less variable, and these genes were usually present in between 0.5 and 2% of all rearrangements. In contrast, a single allele of the IGHV1-69 gene was present in 18.0% of rearrangements of one chromosome, and two IGHV1-69 alleles were responsible for 20.6% of all rearrangements of another chromosome that carried an evident IGHV1-69 gene duplication. Much of the variability that was seen appears to result from differing rearrangement frequencies of the different allelic variants of the IGHV genes. This was most clearly seen in the case of the IGHV7-4-1 and IGHV1-3 genes. Although the IGHV7-4-1*01 allele was detected in the genotype of six individuals, it was only present at a high enough frequency to allow haplotyping in one individual, where this allele was present in 1.1% of all rearrangements of a single chromosome. The alternative IGHV7-4-1*02 allele was detected in five haplotypes, and it was seen in between 2.3 and 7.5% (mean: 3.9%) of rearrangements of those chromosomes. Similarly, the IGHV1-3*01 allele was present in 14 haplotypes and was seen in between 2.5 and 5.6% (mean: 3.7%) of rearrangements of those chromosomes. The alternative IGHV1-3*02 allele was present in three genotypes, but was only present at a high enough frequency to be identified within one haplotype, where it was associated with 0.7% of rearrangements of that chromosome. No significant differences in rearrangement frequencies were detected between unmutated and highly mutated sequences.
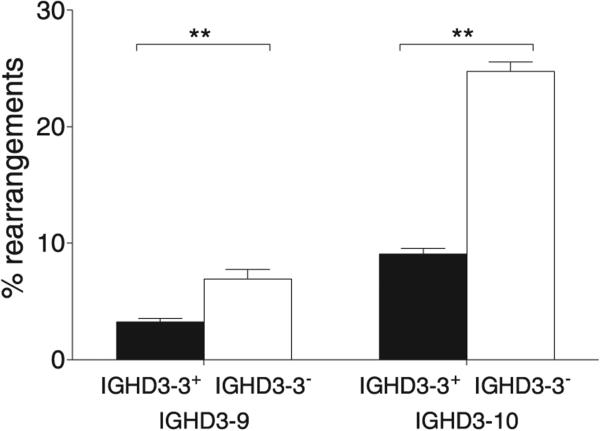
Additional variability appeared to be associated with partial haplotypes, and this was particularly evident for genes of the IGHD locus. For example, as shown in Fig. 3, the frequency of rearrangements utilizing the IGHD3-9*01 and IGHD3-10*01 genes was significantly higher on chromosomes that lacked the genes of the IGHD3-3 to IGHD2-8 locus (Mann–Whitney U test: p < 0.01 in both cases).
FIGURE 3.
Mean rearrangement frequencies and SEMs of the IGHD3-9 and IGHD3-10 genes on chromosomes that carry the full set of 5′ IGHD genes (+) and on chromosomes where the genes from IGHD3-3 to IGHD2-8 appear to be deleted (−). Significant differences are indicated: **p < 0.01.
Discussion
The IgH V region gene locus contains in the order of 120 highly similar functional genes and pseudogenes interspersed among >700 repetitive elements that make up almost half of the locus (8). This has made the study of the chromosomal associations of these genes very difficult. The HapMap project and the 1000 Genomes Project have identified many variant sequences, but neither project is capable of generating comprehensive phased data for the locus. A recent report on the 1000 Genomes Project shows that the sequencing is either too short or too shallow for reliable reconstruction of the IGHV locus (6). In addition, both projects use libraries prepared from EBV-immortalized lymphoblastoid cell lines. By definition, these cells have lost genes from the Ig loci through gene rearrangement. Analysis of data from the HapMap project, for the identification of deletion polymorphisms, did not include the IGH, IGK, and IGL loci for this reason (5). However, although assembly of contigs for the locus still remains problematic, long-read 454 pyrosequencing is now being applied to the study of the Ab repertoire (17).
In 2010, we published an analysis of large data sets of H chain VDJ rearranged gene sequences amplified from the DNA of 12 individuals (16). This study examined the genotype of the IGHV, IGHD, and IGHJ loci for each of the individuals, highlighting substantial differences in the germline IGHV repertoires as a result of allelic variation and varying levels of heterozygosity at different gene loci. We also were able to infer likely gene deletion and duplication polymorphisms. In the current study, the greater clarity that comes from haplotype analysis has allowed us to confirm and extend these initial findings. Although the size of the data sets and the rate of errors generated during pyrosequencing precluded proper analysis of those Ig genes that are rearranged at very low frequency, most genes could be included in the analysis. Those genes that could not be properly analyzed make a relatively small contribution to the overall Ab repertoires of these individuals. As sequencing costs and pyrosequencing error rates are both falling, the future generation of larger relatively error-free data sets will likely allow the confident inference, within a haplotype, of even these rarely used genes.
Our earlier study led to the inference of the existence of 14 previously unreported IGHV polymorphisms, as well as a new IGHD allele and a new IGHJ allele (16). In this study, using IGHV FR1 primers, we have been able to identify an additional putative polymorphism. These sequences have all been named as putative alleles by the inclusion of the descriptor “p” in their proposed allele names. This unofficial naming and reporting is important, for the existence of these alleles will not be accepted by the WHO/IUIS/IMGT Nomenclature Subcommittee for Igs (IG) and TCRs (TR), who stipulate that new allelic variants must be identified as unrearranged genomic sequences. Thousands of VDJ rearrangements now stand as evidence of the existence of these putative alleles, and data from the dbSNP provide additional evidence in support of their existence. The principles governing the acceptance of allelic variants by the nomenclature subcommittee may therefore require revision, given that rapid confirmation of the existence of these new polymorphisms by the stipulated genomic screening is unlikely.
The common heterozygosity that we see at Ig gene loci undoubtedly contributes to repertoire diversity. Although many allelic variants differ from one another by just a single amino acid, even such small differences can give rise to Abs with quite different binding properties (29). Susceptibility to Haemophilus influenzae type b disease, for example, has been associated with allelic variation in the κ L chain genes. It is also now clear that the apparent heterozygosity that can be seen in genotypes is often a consequence of the carriage of multiple “alleles” on a single chromosome. Such duplication of Ig genes has been reported previously. By employing RFLP analysis with sequence-specific oligonucleotide probes, Sasso and colleagues (30) identified two separate loci for sequences that are now identified as IGHV3-30 and IGHV4-28, as well as for IGHV1-69 (31). They also claimed there can be multiple copies of the IGHV3-23 sequence on a single chromosome (32), and others have reported duplication of IGHV4-31 (33). Although it has not been possible to confirm duplication of IGHV3-23 in this study, as all sequences seen were identical to the IGHV3-23*01 sequence, duplications were seen for IGHV3-30 and IGHV1-69. The apparent duplicated IGHV3-30 sequences may actually be amplifications of the IGHV3-30-5 locus, as sequences that map to this locus have been shown to be identical to IGHV3-30 sequences (34). Duplications of IGHV1-2, IGHV3-11, and IGHV4-59, which have not previously been reported, were also seen.
Gene deletion polymorphisms were also inferred in the current study, and in many cases, homozygous deletions were inferred that were supported by the analysis of genotypes. In our earlier genotyping study, we were able to infer that one individual carried a homozygous deletion polymorphism of a number of contiguous IGHD genes (16). In the absence of phased data, however, single copy deletion polymorphisms are impossible to detect by such analysis. Very limited phased data for the IGHV locus were generated in the early 1990s, leading to the inference of an IGHV4-4 deletion polymorphism (35). The reporting of a complete sequence of the IGHV locus in 1998 also revealed the existence of indels involving the unmapped genes IGHV1-f, IGHV4-b, and IGHV5-a (8).
Additional IGHV deletion polymorphisms have recently been revealed. Chimge and colleagues (20) have constructed IGHV haplotypes from single sperm, using multiplex PCR followed by microarray detection using IGHV gene-specific probes. They reported that two individuals of five carried heterozygous deletions of the IGHV4-39 gene. They have also reported deletions of IGHV4-61 (36) and of IGHV7-4-1, IGHV1-8, and IGHV3-9 (37). Recently, they amplified DNA sequence tags to define haplotypes for the complex region from IGHV3-30 to IGHV3-33, where a five-gene indel has been reported (34), and confirmed common deletions within this locus (33). However, a limitation of these studies is that they are only able to define haplotypes by the presence or absence of genes.
The haplotyping technique reported in the current study now reveals the full extent of diversity within the IGH locus because of its ability to define haplotypes that include the identification of allelic variants of each gene. This is perhaps seen most strikingly in our ability to define the haplotypes of the IGHV3-30 to IGHV3-33 region. Twelve different partial haplotypes were seen for this section of the IGHV locus, and these involved as many as four apparent gene deletion polymorphisms. We inferred these and other likely deletion polymorphisms from the absence of certain VDJ rearrangements. It is possible that the data reflect an incompatibility of some IGHV and IGHJ allelic combinations, rather than the absence of particular genes. It is also possible that epigenetic variation blocks recombination of some genes in some individuals. Whereas this study reports “functional haplotypes,” the fact that identical deletions were inferred in different individuals and the fact that most of these apparent deletion polymorphisms have been previously reported lead us to the working hypothesis that the deletions are not merely functional but are physical. In the case of the inferred deletion of the contiguous IGHD genes from IGHD3-3 to IGHV2-8, whole genome shotgun sequencing demonstrates that this dramatic deletion is a physical one.
The value of haplotyping goes beyond the deliverable of a more complete description of the genes of the Ig gene loci. Haplotyping also brings clarity to the study of Ig gene rearrangement frequencies. Variability in recombination frequencies of different genes has been attributed either to variations in recombination signal sequences (38) or to variation in the accessibility of genes through chromatin remodeling (39). Although variation in RSS sequences undoubtedly contributes to unequal utilization of IGHV genes (40), the hypothesis that recombination is affected by chromosomal position and context is also well supported (41).
In this study, rearrangement frequencies were shown to vary between alleles. This variation was seen even where analysis of genomic sequencing data, including the original report of the locus (8), shows such alleles to be associated with identical RSS (data not shown). Rearrangement frequencies also appear to vary according to the genomic context of a gene, such that rearrangement frequencies were most similar among haplotypes that were most similar. This was most dramatically seen in the very high rearrangement frequencies for IGHD3-9 and IGHD3-10 on chromosomes that appear to lack the IGHD3-3 to IGHD2-8 genes. The IGHD3-9 and IGHD3-10 genes are immediately downstream of this deletion. This elevation in the rearrangement frequencies would not be expected to “plug” a hole in the repertoire, created by the absence of IGHD3-3, for although these genes are from the same IGHD gene family, they encode strikingly different amino acid sequences. It therefore may be more likely to be a reflection of changes in the noncoding sequences that flank these genes, as a consequence of the gene deletions, and of the roles of these noncoding sequences in the regulation of gene rearrangement.
Before the development of modern sequencing technologies, when investigations of genomic differences were still very difficult, it was suggested that enormous variability of the Ig gene loci could exist within the human population (42). Analysis of thousands of VDJ rearrangements in this study now clearly demonstrates that this is true. This study has shown that the functional IGH haplotypes that encode the Ig H chain are incredibly diverse, and that this diversity includes a surprisingly high frequency of deletion and duplication polymorphisms. Where genes are present, dramatic variations are also seen in rearrangement frequencies, particularly with the genes of the IGHD locus. Together, this variation suggests that the repertoire of VDJ rearrangements may vary substantially between individuals. It may well be that individual variation in the genes of the Ig loci, and resulting repertoire variation, emerge as important contributors to individual variation in immunocompetency and individual susceptibility to Ab-mediated immunopathology. If the contribution of the locus to health and disease is to be understood, haplotype variation and repertoire variation within the population must now be explored.
Supplementary Material
Abbreviations used in this article
- FR1
framework 1
- LR
likelihood ratio
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Tewhey R, Bansal V, Torkamani A, Topol EJ, Schork NJ. The importance of phase information for human genomics. Nat. Rev. Genet. 2011;12:215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nievergelt CM, Libiger O, Schork NJ. Generalized analysis of molecular variance. PLoS Genet. 2007;3:e51. doi: 10.1371/journal.pgen.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 5.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, Altshuler DM. International HapMap Consortium. 2006. Common deletion polymorphisms in the human genome. Nat. Genet. 38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 6.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook GP, Tomlinson IM, Walter G, Riethman H, Carter NP, Buluwela L, Winter G, Rabbitts TH. A map of the human immunoglobulin VH locus completed by analysis of the telomeric region of chromosome 14q. Nat. Genet. 1994;7:162–168. doi: 10.1038/ng0694-162. [DOI] [PubMed] [Google Scholar]
- 8.Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 10.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 11.Corbett SJ, Tomlinson IM, Sonnhammer ELL, Buck D, Winter G. Sequence of the human immunoglobulin diversity (D) segment locus: a systematic analysis provides no evidence for the use of DIR segments, inverted D segments, “minor” D segments or D-D recombination. J. Mol. Biol. 1997;270:587–597. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]
- 12.Mattila PS, Schugk J, Wu H, Mäkelä O. Extensive allelic sequence variation in the J region of the human immunoglobulin heavy chain gene locus. Eur. J. Immunol. 1995;25:2578–2582. doi: 10.1002/eji.1830250926. [DOI] [PubMed] [Google Scholar]
- 13.Rabbitts TH. The nineteenth Colworth Medal Lecture. The human immunoglobulin genes. Biochem. Soc. Trans. 1983;2:119–126. [PubMed] [Google Scholar]
- 14.Ravetch JV, Siebenlist U, Korsmeyer S, Waldmann T, Leder P. Structure of the human immunoglobulin m locus: characterization of embryonic and rearranged J and D genes. Cell. 1981;27:583–591. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- 15.Lee CE, Jackson KJ, Sewell WA, Collins AM. Use of IGHJ and IGHD gene mutations in analysis of immunoglobulin sequences for the prognosis of chronic lymphocytic leukemia. Leuk. Res. 2007;31:1247–1252. doi: 10.1016/j.leukres.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Boyd SD, Gaëta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J. Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, et al. Measure ment and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 1: 12ra23. 2009 doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Jackson KJ, Gaeta B, Pomat W, Siba P, Sewell WA, Collins AM. Genomic screening by 454 pyrosequencing identifies a new human IGHV gene and sixteen other new IGHV allelic variants. Immunogenetics. 2011;63:259–265. doi: 10.1007/s00251-010-0510-8. [DOI] [PubMed] [Google Scholar]
- 19.Venturi V, Quigley MF, Greenaway HY, Ng PC, Ende ZS, McIntosh T, Asher TE, Almeida JR, Levy S, Price DA, et al. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J. Immunol. 2011;186:4285–4294. doi: 10.4049/jimmunol.1003898. [DOI] [PubMed] [Google Scholar]
- 20.Chimge NO, Pramanik S, Hu G, Lin Y, Gao R, Shen L, Li H. Determination of gene organization in the human IGHV region on single chromosomes. Genes Immun. 2005;6:186–193. doi: 10.1038/sj.gene.6364176. [DOI] [PubMed] [Google Scholar]
- 21.Zong SQ, Nakai S, Matsuda F, Lee KH, Honjo T. Human immunoglobulin D segments: isolation of a new D segment and polymorphic deletion of the D1 segment. Immunol. Lett. 1988;17:329–333. doi: 10.1016/0165-2478(88)90006-5. [DOI] [PubMed] [Google Scholar]
- 22.van Dongen JJ, Langerak AW, Brüggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, García-Sanz R, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 23.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Jackson KJL, Sewell WA, Collins AM. Many human immunoglobulin heavy-chain IGHV gene polymorphisms have been reported in error. Immunol. Cell Biol. 2008;86:111–115. doi: 10.1038/sj.icb.7100144. [DOI] [PubMed] [Google Scholar]
- 25.Lefranc MP. Nomenclature of the human immunoglobulin heavy (IGH) genes. Exp. Clin. Immunogenet. 2001;18:100–116. doi: 10.1159/000049189. [DOI] [PubMed] [Google Scholar]
- 26.Gaëta BA, Malming HR, Jackson KJL, Bain ME, Wilson P, Collins AM. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 27.Lee CEH, Gaëta B, Malming HR, Bain ME, Sewell WA, Collins AM. Reconsidering the human immunoglobulin heavy-chain locus: 1. An evaluation of the expressed human IGHD gene repertoire. Immunogenetics. 2006;57:917–925. doi: 10.1007/s00251-005-0062-5. [DOI] [PubMed] [Google Scholar]
- 28.Jackson KJ, Boyd S, Gaëta BA, Collins AM. Benchmarking the performance of human antibody gene alignment utilities using a 454 sequence dataset. Bioinformatics. 2010;26:3129–3130. doi: 10.1093/bioinformatics/btq604. [DOI] [PubMed] [Google Scholar]
- 29.Liu L, Lucas AH. IGH V3-23*01 and its allele V3-23*03 differ in their capacity to form the canonical human antibody combining site specific for the capsular polysaccharide of Haemophilus influenzae type b. Immunogenetics. 2003;55:336–338. doi: 10.1007/s00251-003-0583-8. [DOI] [PubMed] [Google Scholar]
- 30.Sasso EH, Willems van Dijk K, Bull A, van der Maarel SM, Milner EC. VH genes in tandem array comprise a repeated germline motif. J. Immunol. 1992;149:1230–1236. [PubMed] [Google Scholar]
- 31.Sasso EH, Willems van Dijk K, Bull AP, Milner EC. A fetally expressed immunoglobulin VH1 gene belongs to a complex set of alleles. J. Clin. Invest. 1993;91:2358–2367. doi: 10.1172/JCI116468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasso EH, Buckner JH, Suzuki LA. Ethnic differences of polymorphism of an immunoglobulin VH3 gene. J. Clin. Invest. 1995;96:1591–1600. doi: 10.1172/JCI118198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pramanik S, Cui X, Wang HY, Chimge NO, Hu G, Shen L, Gao R, Li H. Segmental duplication as one of the driving forces underlying the diversity of the human immunoglobulin heavy chainvariable gene region. BMC Genomics. 2011;12:78. doi: 10.1186/1471-2164-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter G, Tomlinson IM, Cook GP, Winter G, Rabbitts TH, Dear PH. HAPPY mapping of a YAC reveals alternative haplotypes in the human immunoglobulin VH locus. Nucleic Acids Res. 1993;21:4524–4529. doi: 10.1093/nar/21.19.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin EK, Matsuda F, Nagaoka H, Fukita Y, Imai T, Yokoyama K, Soeda E, Honjo T. Physical map of the 3′ region of the human immunoglobulin heavy chain locus: clustering of autoantibody-related variable segments in one haplotype. EMBO J. 1991;10:3641–3645. doi: 10.1002/j.1460-2075.1991.tb04930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui X, Li H. Determination of gene organization in individual haplotypes by analyzing single DNA fragments from single spermatozoa. Proc. Natl. Acad. Sci. USA. 1998;95:10791–10796. doi: 10.1073/pnas.95.18.10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pramanik S, Li H. Direct detection of insertion/deletion polymorphisms in an autosomal region by analyzing high-density markers in individual spermatozoa. Am. J. Hum. Genet. 2002;71:1342–1352. doi: 10.1086/344713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roch FA, Hobi R, Berchtold MW, Kuenzle CC. V(D)J recombination frequency is affected by the sequence interposed between a pair of recombination signals: sequence comparison reveals a putative recombinational enhancer element. Nucleic Acids Res. 1997;25:2303–2310. doi: 10.1093/nar/25.12.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Semin. Immunol. 2010;22:353–361. doi: 10.1016/j.smim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Feeney AJ, Tang A, Ogwaro KM. B-cell repertoire formation: role of the recombination signal sequence in non-random V segment utilization. Immunol. Rev. 2000;175:59–69. [PubMed] [Google Scholar]
- 41.Bates JG, Cado D, Nolla H, Schlissel MS. Chromosomal position of a VH gene segment determines its activation and inactivation as a substrate for V(D)J recombination. J. Exp. Med. 2007;204:3247–3256. doi: 10.1084/jem.20071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milner EC, Hufnagle WO, Glas AM, Suzuki I, Alexander C. Polymorphism and utilization of human VH Genes. Ann. N. Y. Acad. Sci. 1995;764:50–61. doi: 10.1111/j.1749-6632.1995.tb55806.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.