Abstract
The mammalian heart expresses two myosin heavy chain genes (Myh6 and Myh7), which are major components of the thick filaments of the sarcomere. We have determined that a third myosin heavy chain, MYH7B, is also expressed in the myocardium. Developmental analysis shows Myh7b expression in cardiac and skeletal muscle of Xenopus, chick and mouse embryos, and in smooth muscle tissues during later stages of mouse embryogenesis. Myh7b is also expressed in the adult human heart. The promoter region of the Myh7b gene shows remarkable similarity between diverse species, suggesting that transcriptional control mechanisms have been conserved. Using luciferase reporter analysis in rat cardiomyocytes, it can be shown that MEF2, GATA and E-box regulatory elements are essential for efficient expression of the Myh7b gene. In addition two conserved elements that do not correspond to consensus binding sites for known transcription factors are also essential for full transcriptional activity of the Myh7b reporter. Finally, the Myh7b gene shows a transcriptional response similar to Myh6 in response to cardiac hypertrophy.
Keywords: Myosin heavy chain, cardiac development, hypertrophy, promoter analysis, somites, smooth muscle
INTRODUCTION
Myosin heavy chains (MYHs) are essential components of the thick filaments of striated muscle. MYH proteins contain almost 2000 amino acid residues and have MWs of about 220 kDa. Approximately 1300 residues of the C-terminal region form the linear helical domain of the protein and the helical domains of two MYH proteins intertwine to produce a rigid coiled structure. The 700 amino acid N-terminal region forms a globular head containing the ATP catalytic site and the actin-binding domain. Hundreds of these MYH molecules assemble into the thick filament of mature muscle, with the globular heads protruding to contact the actin thin filaments.
Due to their critical role in contractility, MYH genes have been extensively studied and the structure, genomic organization, developmental expression and transcriptional regulation of these genes are well documented (reviewed in Morkin, 2000). Although a variety of different myosin motor proteins are found in diverse cell types, the thick filaments of mammalian heart muscle are generally considered to contain just two conventional MYH proteins, MYH6 and MYH7 (Mahdavi et al., 1982; Gordon et al., 2000; Morkin, 2000). These proteins show distinct developmental expression profiles and distributions in the adult heart. In humans, the MYH7 is the predominant isoform expressed in the ventricular chambers while MYH6 is preferentially expressed in the atrial chambers.
In the mammalian genome, the Myh6 and Myh7 genes are tandemly linked and are believed to have arisen through a gene duplication event (Yamauchi-Takihara et al., 1989; Gulick et al., 1991). The two genes have evolved to show alterations in primary sequence and also differences in expression during development and in the adult heart. The duplication event that generated Myh6 and Myh7 in mammals is not evident in the genomes of other vertebrates (e.g. birds, fish, Amphibia) (Desjardins et al., 2002) and a different set of MHC genes is expressed in cardiac muscle of these animals. For example, in the chicken at least three myosin heavy chain genes, MYH6, vMHC and ssMHC are expressed in the heart (Gonzalez-Sanchez and Bader, 1985; Yutzey et al., 1994; Machida et al., 2002). The mammalian orthologue of ssMHC has been assigned the official name, MYH7b (Desjardins et al., 2002). While recent publications have shown that Myh7b is expressed in a range of mouse muscle tissues (van Rooij, et al., 2009) and in a subset of fibers in the extra-ocular muscles (Rossi et al., 2010) very little is known about developmental expression of Myh7b particularly during heart development.
We have carried out developmental studies showing that Myh7b is expressed in a range of muscle tissues in Xenopus, chick and mouse embryos. Furthermore, Myh7b transcripts are expressed at significant levels in the mature human heart. Transcriptional analysis indicates that MEF2, GATA and E-box binding elements are required for efficient cardiomyocyte specific expression of Myh7b, but we also demonstrate the importance of additional regulatory elements that do not correspond to known transcription factor binding sites. Finally, despite distinct differences in transcription regulatory machinery, the Myh7b gene responds to hypertrophic stimuli in a manner equivalent to Myh6.
MATERIALS AND METHODS
In situ hybridization
Whole mount in situ hybridization analysis of Myh7b expression in Xenopus, chicken and mouse embryos was performed using standard procedures (Wilkinson, 1992). Digoxigenin-labeled, antisense Myh7b RNA probes were prepared from PCR amplified Xenopus tropicalis and mouse Myh7b sequences (CX999202 and NM_001085378) inserted into pGEM-Teasy (Promega). Chick MYH7B EST (BU144006) was linearized with Not I and transcribed with T3 RNA polymerase. In situ hybridization to tissue sections of mouse embryos and chick heart was carried out using the method of Grapin-Botton et al. (2001).
RT-PCR analysis
The presence of MYH7B transcripts in adult human tissues was assayed by RT-PCR using a commercially available cDNA panel (OriGene). One μl of each cDNA sample was used as template in radioactive RT-PCR that included 0.3 μCi of α-32P per reaction. RT-PCR cycle number was determined to assure the reaction was in the linear range of amplification. PCR samples were separated on non-denaturing 5% acrylamide gels, dried and then exposed to X-ray film. Human beta-actin primers supplied with the cDNA panel were used as a loading control.
Western Blot Analysis
Mouse tissues were excised, rinsed in ice-cold phosphate buffered saline (PBS), and frozen rapidly in liquid N2. The tissues were homogenized in buffer (pH 6.8) containing 8M urea, 2 M thiourea, 3% SDS, 0.05 M Tris-Cl and 50% glycerol v/v. Tissue homogenates were separated on a 4-15% Mini-Protean TGX precast gel (Bio-RAD), and blotted on 0.2 μm nitrocellulose (Whatman Protran, Whatman, Germany). After transfer the membrane was blocked for 30 min at 37°C in 2% BSA/PBS, incubated for 1h in either monoclonal anti-myosin tail (MF20) antibodies (Bader et al., 1982) or affinity purified rabbit anti-myosin 7B-AP diluted in blitz buffer [150 mM NaCl, 4%BSA, 10mM NaPO4 (pH 7.3-7.5), 1mM EDTA, 20% Triton X-100]. For the competition assay the antibody was competed with the antigenic peptides QRHLERALEERRRQEE and EEQAGRDEEQRLAAEL at a 1:50 antibody to peptide ratio. After washes, the membrane was incubated for 45 min with either peroxidase-conjugated donkey anti- rabbit or -donkey anti-mouse (jacksonImmunoResearch) diluted 1:15,000 in blitz. Supersignal West Pico Chemiluminescent substrate (Thermo Scientific, IL) was used to visualize reaction product.
MYH7B immunocytochemistry
Rabbit immune serum was generated against the synthetic peptide sequence QRHLERALEERRRQEE (corresponding to residues 1457-1472 of the mouse MYH7B sequence) by Strategic Biosolutions (Windham, ME). Polyclonal antibody was isolated from serum by affinity purification and assayed by protein blotting using standard techniques. Mouse cardiac muscle myofibrils were isolated and washed as described (Knight and Trinick 1982). Myofibrils were fixed in 3% paraformaldehyde/PBS for 20 min, permeabilized in 0.2% Triton X-100/PBS and blocked in 2% BSA/1% normal donkey serum/PBS. Myofibrils were probed with affinity purified rabbit anti-MYH7B and monoclonal α-ACTININ EA-53 (Sigma, MO) antibodies, followed by incubation in Texas red-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc., PA) and AlexaFluor 488-conjugated goat anti-mouse IgG (Molecular Probes, OR). Stained samples were mounted with AquaPolymount (Polysciences, Inc., PA) and analyzed on an Olympus model IX70 using a 100x objective (1.35 NA). Micrographs were recorded as digital images on a Photometrics CCD camera (model Series 300).
Computational Analysis
The syntenic relationship of the Myh7b gene in selected vertebrate genomes was analyzed using Metazome (http://www.metazome.net) and Ensembl (http://www.ensembl.org) genomic analysis resources. Alignment of 5’ flanking sequences of Myh7b genes from different species (Fig. 6) was carried out using MacVector. Transcription factor binding site searches were performed using TESS (Schug, 2008) and rVista (Loots and Ovcharenko, 2003).
Figure 6. High sequence conservation in the of proximal promoter regions of the human, mouse, chick and Xenopus Myh7b genes.
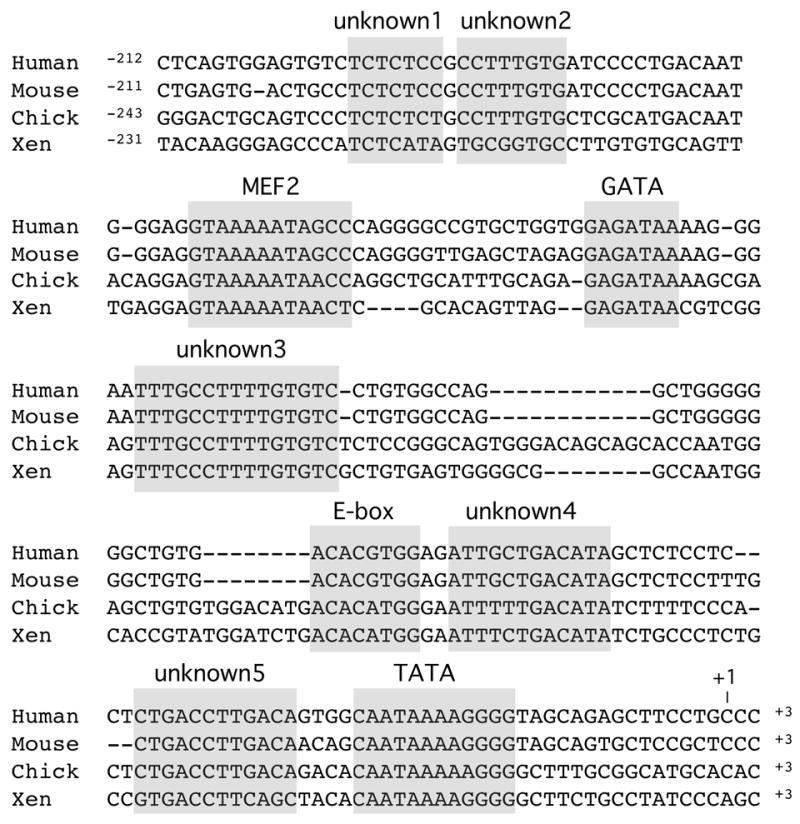
Elements which show conservation between all four species are boxed and spaces introduced to maximize the alignment are indicated by (-). In addition to consensus binding sites for MEF2, GATA, E-box and TATA factors, at least 5 other conserved elements are present. These have been named Unknowns 1-5 as indicated. Putative transcription start site is indicated as +1.
Mutational analysis of the Myh7b promoter
A DNA fragment containing the mouse Myh7b proximal promoter and first intron (1029 bp) was PCR amplified from mouse genomic DNA and inserted into the pGL3 luciferase reporter plasmid (Promega). Inverse PCR was used to individually mutate each of the conserved regions shown in Fig. 6. Fetal rat ventricular cardiomyocytes were isolated as described previously (Gustafson et al., 1987) and maintained in 12 well plates. Half μg of the appropriate Myh7b reporter was transected using SuperFect transfection reagent (Qiagen). Luciferase reporter gene activity was measured in a TD-20/20 luminometer (Turner Designs) and normalized to 0.1 μg of a co-transfected CMV-β-galactosidase reporter construction (pCMVβ; Clontech) according to the instructions supplied with the Dual-Light Combined Reporter Gene Assay System kit (Applied Biosystems).
PMA treatment and thoracic aortic banding
After 3 day in primary culture rat ventricular myocytes were treated with PMA (phorbol 12-myristate 13-acetate) at a final concentration of 0.1 μM for 24 hours, as described by Allo et al. 1991. Male mice (C57BL6; 6 to 8 weeks old) were subjected to pressure overload by thoracic aortic banding (TAB) as previously described by Rockman et al. (1991). Following PMA or TAB treatment, RNA was extracted, cDNA prepared and RT-PCR analysis carried out using directions provided with the iQ SYBR Green Supermix (Bio-Rad). Ornithine decarboxylase (Odc) transcript levels were used as a loading control. Real-time PCR analysis was carried out using the Rotor-Gene 3000 system (Corbett).
RESULTS
Identification of vertebrate orthologues of Myh7b
In a survey of sequences expressed during early heart development of Xenopus we identified transcripts encoding the myosin heavy chain protein variant, Myh7b. This is the orthologue of chicken slow skeletal myosin-2, which is known to be expressed in cardiac muscle (Machida et al., 2002). Examination of synteny shows that the MYH7B gene is present at an identical location in the human, mouse, chick and Xenopus genomes (Fig. 1). Note that, although this myosin gene has been given the name, Myh7b, it is more ancient than Myh7 and quite distinct in primary sequence (Desjardins et al., 2002). For example, the human MYH7B protein sequence is 79% identical to chicken MYH7B at the amino acid level but only 67% and 68% identical to the human MYH6 and MYH7 sequences respectively.
Figure 1. Syntenic arrangement of Myh7b orthologues.

Alignment of Myh7b genes from different species using the Ensembl database shows syntenic arrangement of adjacent genes, thereby allowing unambiguous identification of the Myh7b genes as orthologues. Identical genes are shown with the same texture pattern. ACSS2 = acetyl-Coenzyme A synthetase 1; GSS = glutathione synthetase; TRPC4AP = transient receptor potential cation channel 4, associated protein: GGTL3; gamma-glutamyltransferase-like 3
Developmental expression of Myh7b
Using whole mount in situ hybridization, we analyzed Myh7b expression during development of Xenopus, chick and mouse embryos. In Xenopus, Myh7b transcripts were first detected very weakly in the somites of the neurula embryo (Fig. 2A). During subsequent development, Myh7b expression became more prominent in the somites, particularly surrounding the nuclei that are aligned at the center of the somite bundle (Keller, 2000). Starting at the early tailbud stages, Myh7b was clearly visible in the developing heart (Fig. 2B-D) and this expression persisted into the adult (data not shown). In the chick embryo, MYH7B expression is visible in the developing heart, starting at approximately HH12, and at later stages is also visible in the somites (Fig. 2E, F). Using in situ hybridization to tissue sections, we observed MYH7B transcripts apparently evenly distributed in all regions of the myocardium of the D19.5 chick heart (Fig. 2G). However, expression was not present in non-muscle tissues of the heart, including the developing valves or the epicardium (Fig. 2H,I). In sections through the adult heart, in situ staining appears to be concentrated in the perinuclear region, which has previously been reported for transcripts encoding contractile proteins (Aigner and Pette, 1990; Russell and Dix, 1992).
Figure 2. Developmental expression of myh7b in Xenopus and chick.
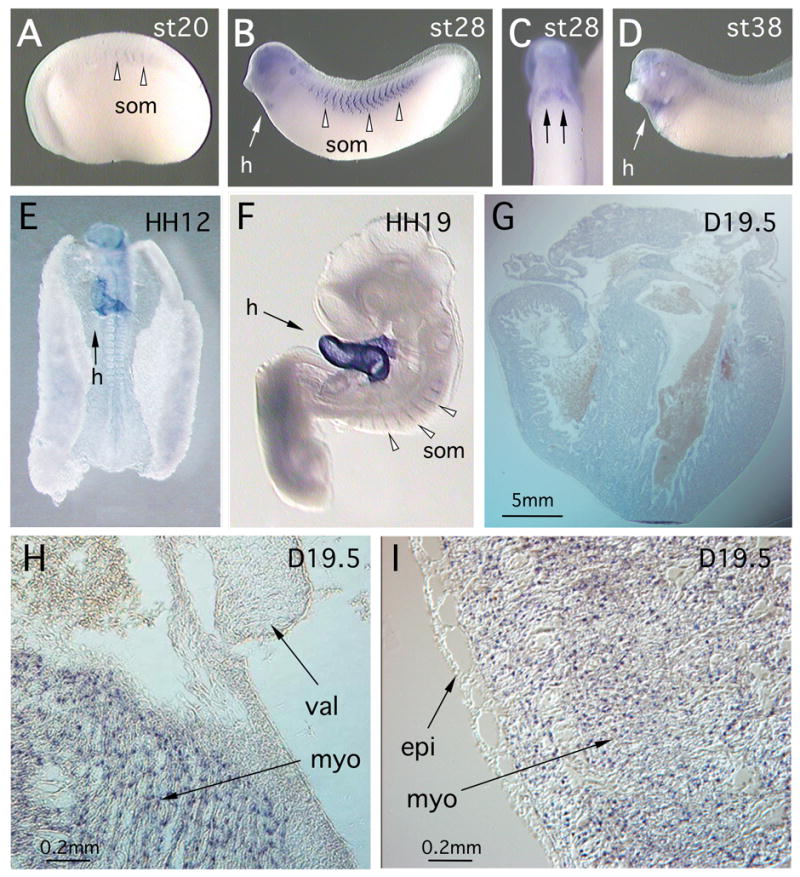
In situ hybridization analysis was used to detect myh7b transcripts. (A) myh7b expression was first detected in the somites of the neurula stage frog embryo, (B) At the early tailbud stage, transcripts were observed in developing heart tissue (h) and in the perinuclear region of somite cells (som – arrowheads). (C) Ventral view of the frog embryo showed expression in the paired heart patches (arrows) prior to fusion at the ventral midline. (D). During later development, myh7b expression persisted in the heart (h) but was reduced in the somites (E) Chick embryo at HH12, showing MYH7B expression in the folding heart tube (h). (F). During later development, transcripts persisted in the heart (h) and became visible in the somites (arrowheads). (G) Longitudinal section through the heart of a D19.5 chick embryo. MYH7B transcripts were detected throughout the myocardial layer. (H). Magnified view of atrio-ventricular septal region of D19.5 heart, showing expression of MYH7B in myocardium (myo) but absence from developing valve tissue (val). (I). Magnified view of the surface of the D19.5 heart, showing absence of MYH7B expression in the epicardial tissue layer (epi).
In the mouse, Myh7b transcripts were first detected at E8.5 (Fig. 3A) and by E10.5 expression was clearly visible throughout the developing myocardium and in the somites, particularly in more posterior regions (Fig. 3B). This early expression pattern was more or less identical to that observed in Xenopus and chick embryos. To characterize mouse Myh7b expression during later developmental stages, we used in situ hybridization to tissue sections. In E11.5 and E12.5 embryos, Myh7b transcripts were clearly visible in the developing heart, throughout the myocardial layer of the atrial and ventricular chambers (Fig. 3C, D). Myh7b transcripts were detected in the somites of E12.5 embryos (Fig. 3E) and in the psoas muscles of later stage embryos (Fig. 3F). At E14.5 Myh7b expression was also observed in the smooth muscle layer of the larger blood vessels, including the descending aorta (Fig. 3F) and in the smooth muscle layer of the small intestine (Fig. 3G). These results indicate that mouse Myh7b is expressed in cardiac muscle and, apparently at lower levels, in skeletal and smooth muscle tissues.
Figure 3. Developmental expression of Myh7b in the mouse.
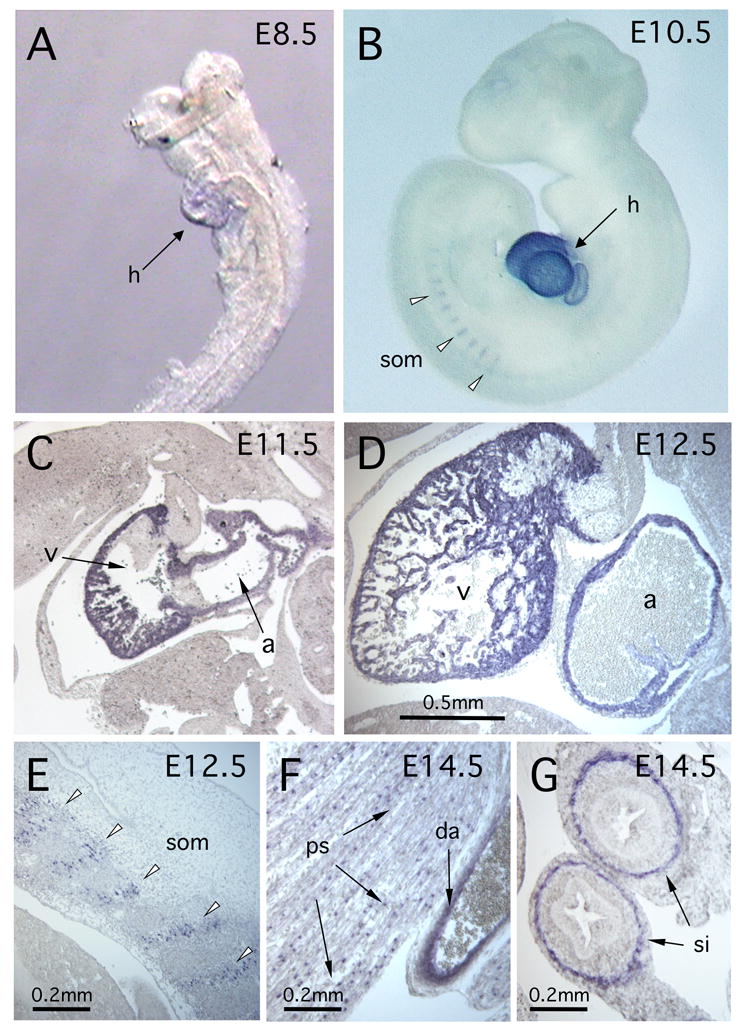
In situ hybridization was used to detect Myh7b transcripts. (A) E8.5 embryo showing Myh7b expression in the folding heart tube (h). (B) In the E10.5 embryo Myh7b transcripts persisted in the heart (h) and were also visible in the developing somites (som - arrowheads). (C) Section in situ hybridization to E11.5 embryo showing Myh7b expression in the myocardial tissue of the developing heart. Atrium (a) and ventricle (v) are indicated. (D) Myh7b expression persisted throughout atrial and ventricular tissues of the heart at E12.5. Atrium (a) and ventricle (v) are indicated. (E) Section through E12.5 embryo showing Myh7b transcripts in the somites (arrowheads). (F) Myh7b transcripts were detected in the psoas muscle (ps) and in the smooth muscle layer of the descending aorta (da) of an E14.5 embryo. (G) Section through the gut of an E14.5 embryo showing Myh7b expression in the smooth muscle of the small intestine (si).
To demonstrate that MYH7B protein is also expressed in the heart, we generated a specific antibody using a peptide sequence that is conserved in both human and mouse MYH7B but which is absent from the MYH6 and MYH7 proteins (Fig. 4A). When the MYH7B antibody was used in protein blot experiments, a single band corresponding to MYH7B protein was detected in extracts from the mouse heart and extra-ocular muscle (Fig. 4B). No signal was detected in spleen extracts. Extra-ocular muscle was included in these experiments because MYH7B is reported to be expressed in this tissue (Rossi et al., 2010). Specificity of detection was confirmed by blocking with the peptide antigen (lane H+Pep in Fig. 4B). The protein detected by MYH7B antibody was identical in size to that detected by the pan-myosin heavy chain antibody, MF20 (Bader et al., 1982). Immuno-staining of isolated mouse cardiac myofibrils revealed a pattern consistent with incorporation of MYH7B protein into thick filaments (Fig. 4C). We conclude from these experiments that MYH7B protein is expressed in the mouse heart and assembles into myofilaments.
Figure 4. Antibody detection of MYH7B protein in mouse tissues.
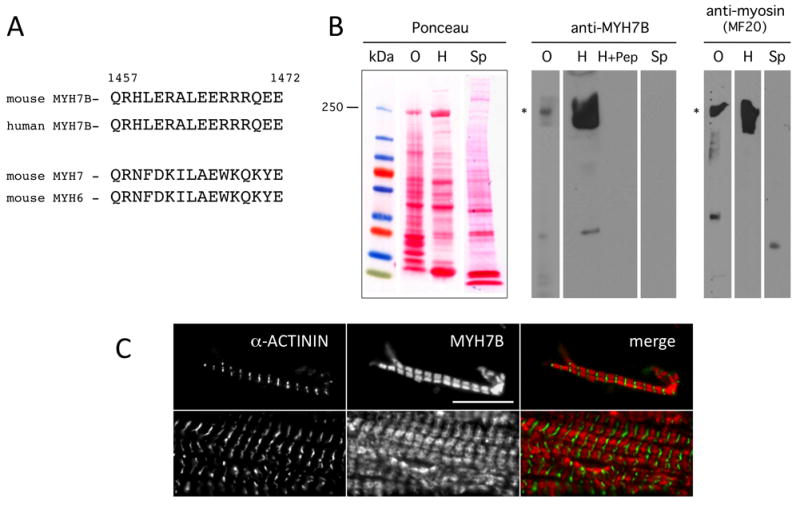
(A) Alignment of the protein sequences of human and mouse MYH7B (top) and mouse MYH7 and MYH6 (bottom). The MYH7B sequence is perfectly conserved between mouse and human but is quite distinct from mouse MYH7 or MYH6 sequences. The MYH7B peptide sequence indicated was used as an antigen for antibody preparation. (B) Protein blot detection of MYH7B in adult mouse tissues. Ponceau-S staining shows the appearance of protein extracts used in these studies. The second panel shows that anti-MYH7B antibody detected a single prominent band of approximately 200 kDa (indicated by asterisk) in the ocular muscle and heart muscle lanes. This band was eliminated by blocking with the peptide antigen (lane labeled H+Pep). The third panel shows staining with the pan-myosin heavy chain antibody, MF20. The major band (asterisk) is the same size at that detected by the anti-MYH7B antibody. (C) Immunohistological detection of α-ACTININ and MYH7B in isolated cardiac myofibrils. α-ACTININ marks the Z-disk and MYH7B shows localization to the thick fibrils. All immunohistological images are presented at the same scale. Scale bar is 10 microns.
The MYH7B gene is expressed in adult human heart
Searches of human EST databases reveal 20 entries corresponding to transcripts from the MYH7B gene, indicating that human MYH7B is transcriptionally active. To better characterize MYH7B expression we used RT-PCR to detect transcripts in commercial RNA samples from a range of human tissues. MYH7B is expressed at high levels in the adult heart and apparently at much lower levels in skeletal muscle, kidney and brain (Fig. 5). While independent microarray studies of normal human tissues have also reported MYH7B expression in kidney and brain (GeneNote) it is possible that the very low levels of signal in the kidney and brain lanes in Fig. 5 represent contamination from the overloaded heart sample.
Figure 5. MYH7B is expressed at relatively high levels in the human heart.
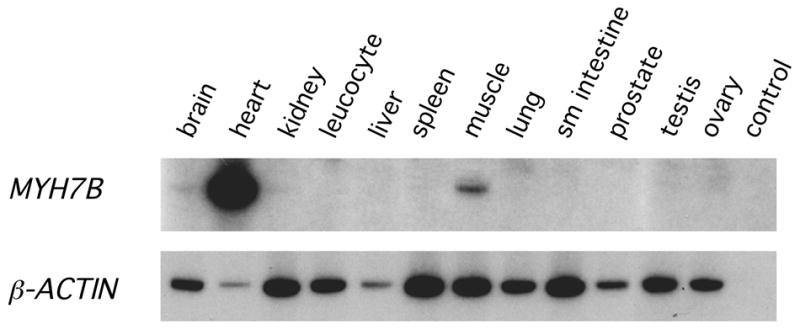
(A) RT-PCR analysis of MYH7B sequences was carried out using a range of human tissues, as indicated at the top of the figure. The same samples were assayed for β-ACTIN as a measure of total input material. Note that MYH7B expression is much greater in heart than in any other human tissue examined.
Conservation of Myh7b promoter sequences
Using genome data we have aligned and compared the promoter regions of the MYH7B genes from human, mouse, chicken and Xenopus (Fig. 6). This sequence alignment reveals a remarkable degree of sequence conservation between species, especially in the proximal promoter region, suggesting that transcription regulatory mechanisms may be conserved. Apart from the region upstream of the transcription start site, our computer analysis detected no other regions of extended sequence similarity between Xenopus and other vertebrates, either further 5’ to the gene or within the first two introns. We observe that MEF2 and GATA binding sites are conserved and that both of these elements are known to function in regulation of cardiac genes (reviewed in Bruneau, 2002; Nemer and Nemer, 2003). However, the Myh7b promoter does not contain the SRE, NKE or M-CAT elements (SRF factor, Nkx2-5 and NTEF-1 binding sites respectively) that are present in the MyH6 and MyH7 promoters. More surprisingly we found that five highly conserved elements, labeled as unknown1-5 (Fig. 6) do not correspond to conserved consensus binding sites for known transcription factors. This raises the possibility that novel factors may be involved in regulation of Myh7b.
Transcriptional regulation of Myh7b expression
To directly examine the importance of the evolutionarily conserved elements for Myh7b expression, we analyzed the activity of the Myh7b regulatory regions in primary rat cardiomyocte cells and in transgenic embryos (Fig. 7). For these studies we inserted a fragment of the mouse Myh7b gene containing 655 bp of sequence upstream of the transcription start site (including all of the conserved elements shown in Fig. 6) plus first intron sequences upstream of the luciferase reporter gene (Fig. 7A). Transfection into rat cardiomyocytes or mouse L929 cells showed that Myh7b regulatory regions increased luciferase activity over the minimal TK promoter in cardiomyocytes but not in the fibroblast cell line (Fig. 7B), indicating that Myh7b regulatory regions conferred some tissue specificity. Second, the possible importance of first intron sequences for gene regulation was tested by transfecting luciferase reporter constructions, with or without the first intron, into rat cardiomyocytes. As shown in Fig. 7C, sequences in the first intron did not appear to play a significant role in reporter expression levels in the cardiac cells. Next, to determine whether the reporter constructions contained all regulatory elements necessary for cardiac expression, we carried out transgenic studies using the Xenopus transgenesis system. When a construction containing the mouse Myh7b 5’ regulatory elements plus first intron (Fig. 7A, #2) was used to generate transgenic frog embryos, expression of the EGFP reporter was clearly visible in the developing heart (Fig. 7D-F). This indicates that sequences contained within the 1029 bp of the construction are sufficiently conserved to allow expression of the mouse Myh7b promoter in the Xenopus heart.
Figure 7. The proximal promoter region of the mouse Myh7b gene is sufficient to direct cardiac-specific reporter expression.
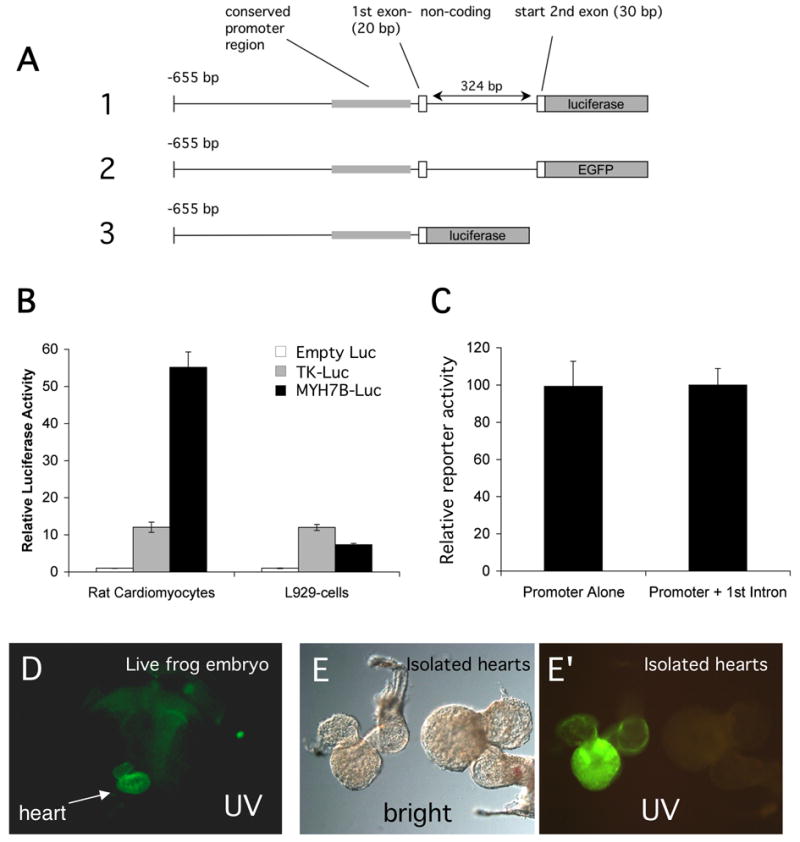
(A) Diagram showing regions of mouse genomic DNA included in luciferase and EGFP reporter constructions. The shaded box corresponds to the region of conserved sequence illustrated in Fig. 5. The first exon of the Myh7b gene is non-coding. (B) Luciferase reporter constructions with or without first exon sequences (#1 and 3 respectively in A), were transfected into rat ventricular cardiomyocytes and reporter activity was assayed. Results indicate that first intron sequences do not contribute to total levels of reporter activity. (C) Luciferase reporter constructions (number 1 and controls) were transfected into rat ventricular cardiomyocytes or into mouse L929 fibroblasts and assayed for reporter activity. The Myh7b promoter is significantly more active in cardiomyocytes than in fibroblast cells. (D-F). The mouse Myh7b promoter is sufficient to direct cardiac expression of an EGFP reporter in the Xenopus embryo. (D) Ventral view of living Xenopus embryo under UV light, showing EGFP expression in the developing heart. (E and F) Bright field and UV light images respectively of hearts isolated from transgenic Xenopus embryos. The heart from Myh7b transgenic embryo (left) shows EGFP expression throughout the atrial and ventricular myocardium compared to the non-transgenic control (at right).
To investigate the transcriptional regulatory activity of the individual conserved elements in the Myh7b regulatory region, we carried out mutagenic analysis using the Myh7b-luciferase reporter construction including the first intron as a template (#1 in Fig. 7A). Mutagenized constructions were then transfected into rat cardiomyocytes and luciferase activity measured (Fig. 8). As expected, these experiments demonstrated that the GATA, MEF2 and E-box binding sites were important for full transcriptional activity of the promoter. However, we also observed that the elements Un4 and Un5 were essential for full transcriptional activity of the Myh7b promoter (Fig. 8). Indeed the individual Un4 and Un5 elements were second only to the MEF2 site in importance for reporter gene expression. When both the Un4 and Un5 sites were mutated, reporter activity in cardiomyocytes was reduced to less than 20% of wild type levels. Mutation of the conserved elements Un1 and Un2 resulted in very little change in luciferase activity (Fig. 8). This may be because they function to regulate Myh7b transcription in other types of tissue, for example smooth or skeletal muscle cells, or because they mediate Myh7b response to altered physiological conditions.
Figure 8. Mutational analysis of the Myh7b gene regulatory region.
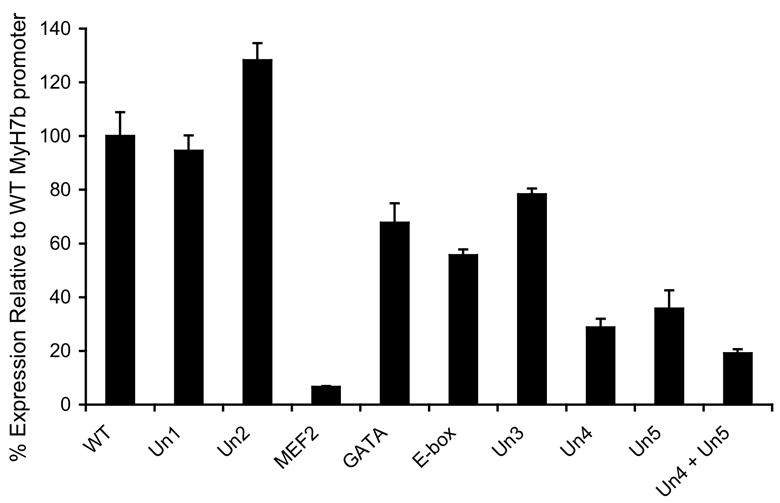
Wild type and mutant constructions were assayed for luciferase reporter activity after transfection into rat ventricular cardiomyocytes. Conserved elements were scrambled to eliminate possible binding targets either individually or in combination. Reporter activity of mutant constructions is shown relative to the wild type control. Bars (+/- SE) represent average of triplicate measurements.
Myh7b expression is regulated in cardiac hypertrophy
During cardiac hypertrophy the ratio of expression of the two myosin heavy chain isoforms expressed in the heart undergoes a dramatic shift. In rodents, expression of the MYH6 isoform is reduced whereas the MYH7 isoform increases (Nadal-Ginard and Mahdavi, 1989; Chien et al., 1991). Since Myh7b is transcribed in the heart, it would be of interest to determine whether transcription of this isoform also responded to cardiac hypertrophy. For initial studies, rat ventricular cardiomyocytes were treated for 24 hours with phorbol 12-myristate 13-acetate (PMA), which functions as an activator of protein kinase C signaling and stimulates a cardiac hypertrophy response (Dunnmon et al., 1990). After PMA treatment gene expression levels were compared to controls using quantitative PCR analysis (Fig. 9A). As expected from previous studies, Myh6 transcript levels decreased and Myh7 and Atrial natriuretic factor (Anf) levels increased (Nadal-Ginard and Mahdavi, 1989; Buttrick et al., 1994). In response to PMA treatment, expression of Myh7b decreased to approximately 25% of control levels. This response is similar to Myh6, rather than Myh7. To validate the results obtained with PMA, we have examined Myh7b expression in an in vivo model, where hypertrophy was induced in mouse hearts by aortic banding (Rockman et al., 1991; Tatsuguchi et al., 2007). As shown in Fig. 9B, Myh7b expression levels decreased when compared to sham operated controls, again consistent with the response of the Myh6 gene.
Figure 9. Myh7b expression is reduced in response to cardiac hypertrophy.
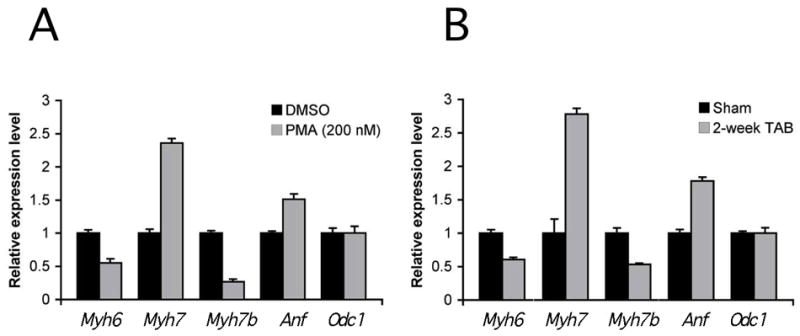
(A) Neonatal rat ventricular cardiomyocytes were treated with the protein kinase C activator, PMA (200 nM) for 24 hours and expression of endogenous genes was determined using quantitative RT-PCR. Control cells were treated with DMSO carrier. Transcript levels in PMA treated cells are presented relative to controls. Transcripts for ornithine decarboxylase-1 (Odc1) served as loading controls. Transcripts levels for Myh7 and Anf increased, and levels for Myh6 decreased. Bars (+/- SE) represent averages of triplicate experiments. (B). Mice were subjected to aortic banding for 2 weeks to induce a hypertrophic response. Transcript levels were then measured using quantitative RT-PCR. Sham operated animals served as controls and Odc1 transcripts were measured as loading controls.
DISCUSSION
Myh7b expression during development
Using gene synteny we have unambiguously identified the orthologues of the Myh7b gene in the mouse, human, chick and Xenopus genomes. The Myh7b gene has been analyzed in some detail at the structural level (Desjardins et al., 2002), but information on its expression is sporadic. At present, expression data indicate that Myh7b is expressed in slow tonic fibers of extra-ocular muscles (Rossi et al., 2010) and in a subset of mouse embryonic tissues (van Rooij et al., 2009).
We have used in situ hybridization to examine developmental expression of Myh7b in Xenopus, chick and mouse embryos. These experiments indicate that Myh7b expression in the developing heart and somites is conserved in each of these evolutionarily distinct organisms. In the Xenopus embryo, Myh7b expression in the heart is first detected at approximately the same time as other differentiation markers, when myocardial precursor cells are present in two symmetrical patches of tissue on either side of the ventral midline (Fig. 2C). In chick and mouse, Myh7b transcripts were not detected until the heart tube stage (Fig. 2E, 3A). This represents a slight delay relative to the other cardiac myosin genes, which are strongly expressed prior to heart tube formation (Lyons et al., 1990; Yutzey et al., 1994). Using in situ hybridization to tissue sections we were able to examine Myh7b expression during later stages of heart development. In both chick and mouse (Fig. 2H, I and Fig. 3C, D), we observed Myh7b expression throughout the myocardium. In addition, Myh7b was expressed in the smooth muscle tissue of the small intestine and also in the walls of major blood vessels. Therefore, Myh7b is expressed in at least a subset of cardiac, skeletal and smooth muscle tissues. Earlier studies showed that MYH7B is expressed in extra-ocular muscles in the mouse (Rossi et al, 2010), but we did not observe this expression domain in our in situ hybridizatin analysis, most likely because expression was not activated at the developmental stages examined. In order to demonstrate that protein was indeed synthesized from Myh7b transcripts, we raised antibodies against a unique sequence of mammalian MYH7B and used this to detect a single band in mouse heart and extra-ocular muscle tissue (Fig. 4).
Although limited in scope, RT-PCR analysis of human tissues (Fig. 5) showed that MYH7B transcripts are present in a range of adult organs. Expression levels are highest in the myocardium, followed by skeletal muscle, with much lower levels observed in kidney and brain. The tissue distribution in human organs is broadly consistent with the tissue distribution in the mouse embryo. One exception is that Myh7b transcripts are present in the mouse small intestine but absent from human intestine (compare Fig. 3G and Fig. 5). This may be due to expression differences between species or to loss of Myh7b expression in adult gut tissues.
One of our original reasons for studying Myh7b was the possibility that the expression reported in the conduction system of the chick heart (Machida et al., 2002) would also be observed in other vertebrates. However, our experimental results do not reproduce the expression pattern reported for chick. The previous studies indicated that MYH7B transcripts were not detectable until late stages of heart development (approximately 19 days after fertilization) and then only in the Purkinje fibers of the conduction system, but not myocardium (Machida et al., 2002). In our studies of chick embryos, expression of MYH7B was detected as early as 2 days after fertilization (HH12 - approximately 48 h, Fig. 2E) and transcripts were visible throughout all myocardial tissues (Fig. 2G-I). We note that the pattern of MYH7B expression that we observed in chicken is very similar to the expression patterns detected in frog and mouse. At present we do not have an explanation for this apparent discrepancy.
Transcriptional regulation of Myh7b in the heart
Examination of the proximal promoter regions of Myh7b genes from different species shows a remarkable degree of sequence conservation (Fig. 6) suggesting that transcriptional control mechanisms have been evolutionarily conserved. Like many other cardiac-expressed genes, the Myh7b regulatory region contains binding sites for GATA, MEF2 and E-box family transcription factors (Morkin, 2000). Mutational analysis in primary rat cardiomyocytes indicates that each of these binding sites is required for maximal expression from the Myh7b promoter, with the MEF2 binding site playing the most important role (Fig. 8). The importance of the MEF2, E-box and GATA elements for cardiac expression, suggests some conservation of regulatory mechanisms between these distantly related genes.
Analysis of the other conserved elements in the Myh7b promoter revealed that the regions called Un4 and Un5 are also required for maximum expression. Analysis using several different transcription factor search programs failed to identify a high confidence consensus binding site for either of these elements. Nevertheless, the fact that these elements were required for efficient expression of the Myh7b reporter in cardiomyocytes suggests that further study of these sites and identification of binding proteins may be justified.
Based on analysis of protein sequences, previous studies suggest that MYH7B may belong to the slow class of myosin heavy chains (Desjardins et al., 2002). In this respect, MYH7B is likely to be more closely related to MYH6 than to MYH7 at the functional level. During cardiac hypertrophy, we observe that expression of both Myh6 and Myh7b is reduced relative to control transcripts while, as expected, Myh7 is upregulated (Fig. 8). Again, this suggests that Myh7b may have more functional similarity to Myh6 than to Myh7.
Evolutionary conservation of Myh7b expression
During embryonic development, Xenopus, chick and mouse show extremely similar patterns of Myh7b expression in the developing heart and somites (Figs. 2, 3). In all organisms and at all developmental stages, the expression of Myh7b in the heart appears to be much lower than for other cardiac myosin transcripts at the equivalent stage. The question therefore is whether MYH7B serves some specific function in regulation of cardiac physiologic function. On one hand, the levels of Myh7b expression are low and the protein is unlikely to comprise more than a small fraction of the total myosin protein in the thick filaments. At this point it is unclear whether the small amount of MYH7B protein contributes to regulation of cardiac muscle function. On the other hand, expression of Myh7b is maintained with the same developmental pattern in animals that have been evolving independently for hundreds of millions of years (Kumar and Hedges, 1998), suggesting that, at least under some conditions, expression of Myh7b provides a selective advantage.
Acknowledgments
We would like to thank Da-Zhi Wang (University of North Carolina) for providing the RNA from hypertrophic hearts and Charles Ordahl for advice on Myh7b expression in the somites. A.S.W. is the recipient of a Postdoctoral Fellowship award from the American Heart Association. P.A.K. is the Allan C. Hudson and Helen Lovaas Endowed Professor of the Sarver Heart Center at the University of Arizona College of Medicine. This work was supported by the Sarver Heart Center and by the NHLBI of the NIH, grants HL083146 to CCG and HL74184 and AHA-0950064G to PAK.
References
- Aigner S, Pette D. In situ hybridizaton of slow myosin heavy chain mRNA in normal and transforming rabbit muscles with the use of a nonradioactively labeled cRNA. Histochemistry. 1990;95:11–18. doi: 10.1007/BF00737222. [DOI] [PubMed] [Google Scholar]
- Allo SN, McDermott PJ, Carl LL, Morgan HE. Phorbol ester stimulation of protein kinase C activity and ribosomal DNA transcription. J Biol Chem. 1991;266:22003–22009. [PubMed] [Google Scholar]
- Bader D, Masaki T, Fischman DA. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–70. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res. 2002;90:509–519. doi: 10.1161/01.res.0000013072.51957.b7. [DOI] [PubMed] [Google Scholar]
- Buttrick PM, Kaplan M, Leinwand LA, Scheuer J. Alterations in gene expression in the heart after chronic pathological and physiological loads. J Mol Cell Cardiol. 1994;26:61–67. doi: 10.1006/jmcc.1994.1008. [DOI] [PubMed] [Google Scholar]
- Chien KR, Knowlton KU, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- Desjardins PR, Burkman JM, Shrager JB, Allmond LA, Stedman HH. Evolutionary implications of three novel members of the human sarcomeric myosin heavy chain gene family. Mol Biol Evol. 2002;19:375–393. doi: 10.1093/oxfordjournals.molbev.a004093. [DOI] [PubMed] [Google Scholar]
- Dunnmon PM, Iwaki K, Henderson SA, Sen A, Chien KR. Phorbol esters induce immediate-early genes and activate cardiac gene transcription in neonatal rat myocardial cells. J Mol Cell Cardiol. 1990;22:901–910. doi: 10.1016/0022-2828(90)90121-h. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sanchez A, Bader D. Characterization of a myosin heavy chain in the conductive system of the adult and developing chicken heart. J Cell Biol. 1985;100:270–275. doi: 10.1083/jcb.100.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Majithia AR, Melton DA. Key events of pancreas formation are triggered in gut endoderm by ectopic expression of pancreatic regulatory genes. Genes Dev. 2001;15:444–454. doi: 10.1101/gad.846001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick J, Subramanian A, Neumann J, Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- Keller R. The origin and morphogenesis of amphibian somites. Curr Top Dev Biol. 2000;47:183–246. doi: 10.1016/s0070-2153(08)60726-7. [DOI] [PubMed] [Google Scholar]
- Knight PJ, Trinick JA. Preparation of myofibrils. Methods Enzymol. 1982;85(Pt B):9–12. doi: 10.1016/0076-6879(82)85004-0. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lyons GE, Ontell M, Cox R, Sassoon D, Buckingham M. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J Cell Biol. 1990;111:1465–1476. doi: 10.1083/jcb.111.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of trancription factor binding sites. Nucleic Acids Res. 2003;32:W217–221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S, Noda S, Takao A, Nakazawa M, Matoka R. Expression of slow skeletal myosin heavy chain-2 gene in Purkinje fiber cells in chick heart. Biol Cell. 2002;94:389–399. doi: 10.1016/s0248-4900(02)00010-2. [DOI] [PubMed] [Google Scholar]
- Mahdavi V, Periasamy M, Nadal-Ginard B. Molecular characterization of two myosin heavy chain genes expressed in the adult heart. Nature. 1982;297:659–664. doi: 10.1038/297659a0. [DOI] [PubMed] [Google Scholar]
- Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc Res Tech. 2000;50:522–531. doi: 10.1002/1097-0029(20000915)50:6<522::AID-JEMT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Nadal-Ginard B, Mahdavi V. Molecular basis of cardiac performance. Plasticity of the myocardium generated through protein isoform switches. J Clin Invest. 1989;84:1693–1700. doi: 10.1172/JCI114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemer G, Nemer M. Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and –6. Dev Biol. 2003;254:131–148. doi: 10.1016/s0012-1606(02)00026-x. [DOI] [PubMed] [Google Scholar]
- Rockman HA, Ross RS, Harris AN, Knowlton KU, Steinhelper ME, Field LJ, Ross J, Jr, Chien KR. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci USA. 1991;88:8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi AC, Mammucari C, Argentini C, Reggiani C, Schiaffino S. Two novel/ancient myosins in mammalian skeletal muscles: MYH14/7b and MYH15 are expressed in extraocular muscles and muscle spindles. J Physiol. 2010;588:353–364. doi: 10.1113/jphysiol.2009.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell B, Dix DJ. Mechanisms for intracellular distribution of mRNA: in situ hybridization studies in muscle. Am J Physiol. 1992;262:C1–8. doi: 10.1152/ajpcell.1992.262.1.C1. [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics. 2008;Chapter 2(Unit 2.6) doi: 10.1002/0471250953.bi0206s21. [DOI] [PubMed] [Google Scholar]
- Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–73. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. Whole mount in situ hybridization of vertebrate embryos. In: Wilkinson DG, editor. Situ Hybridization: A Practical Approach. Oxford: IRL Press; 1992. pp. 75–83. [Google Scholar]
- Yamauchi-Takihara K, Sole MJ, Liew J, Ing D, Liew CC. Characterization of human cardiac myosin heavy chain genes. Proc Natl Acad Sci USA. 1989;86:3504–3508. doi: 10.1073/pnas.86.10.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutzey KE, Rhee JT, Bader D. Expression of the atrial-specific myosin heavy chain AMHC1 and the establishment of anteroposterior polarity in the developing chicken heart. Development. 1994;120:871–883. doi: 10.1242/dev.120.4.871. [DOI] [PubMed] [Google Scholar]


