Abstract
We have previously identified a zinc finger transcription factor, ZNF24 (zinc finger protein 24), as a novel inhibitor of tumor angiogenesis and have demonstrated that ZNF24 exerts this effect by repressing the transcription of VEGF in breast cancer cells. Here we focused on the role of ZNF24 in modulating the angiogenic potential of the endothelial compartment. Knockdown of ZNF24 by siRNA in human primary microvascular endothelial cells (ECs) led to significantly decreased cell migration and invasion compared with control siRNA. ZNF24 knockdown consistently led to significantly impaired VEGF receptor 2 (VEGFR2) signaling and decreased levels of matrix metalloproteinase-2 (MMP-2), with no effect on levels of major regulators of MMP-2 activity such as the tissue inhibitors of metalloproteinases and MMP-14. Moreover, silencing ZNF24 in these cells led to significantly decreased EC proliferation. Quantitative PCR array analyses identified multiple cell cycle regulators as potential ZNF24 downstream targets which may be responsible for the decreased proliferation in ECs. In vivo, knockdown of ZNF24 specifically in microvascular ECs led to significantly decreased formation of functional vascular networks. Taken together, these results demonstrate that ZNF24 plays an essential role in modulating the angiogenic potential of microvascular ECs by regulating the proliferation, migration, and invasion of these cells.— Jia, D., Huang, L., Bischoff, J., Moses, M. A. The endogenous zinc finger transcription factor, ZNF24, modulates the angiogenic potential of human microvascular endothelial cells.
Keywords: angiogenesis, transcriptional regulation, VEGF repression
Zinc finger protein 24 (ZNF24, also known as ZNF191, Zfp191, and KOX17) is a member of the Kruppel-like zinc finger transcription factor family (1). Its C terminus harbors 4 C2H2 zinc finger motifs that function as DNA binding domains (2), and its N terminus contains a SCAN domain that primarily serves as a dimerization domain in zinc finger proteins (3). ZNF24 is ubiquitously expressed during embryonic development, and its expression can be detected in every adult tissue examined (4, 5), indicating that its functions are essential in many different cell types. The importance of ZNF24 in regulating cellular functions has been revealed in part by the generation of ZNF24 knockout mice. Two independent studies have shown that knockout of ZNF24 leads to premature death at different time points of development (6, 7), indicating that ZNF24 plays an indispensable role in regulating key processes of organ development.
At the cellular level, ZNF24 has been implicated in regulating proliferation, differentiation, migration, and invasion of cells from different lineages. Overexpression of ZNF24 in neural progenitor cells maintains these cells in an actively proliferating state and inhibits neuronal differentiation (8). An important role of ZNF24 in regulating cell proliferation has been demonstrated during early embryonic development, where loss of ZNF24 leads to severely impaired proliferation of blastocysts (7). This could be one of the reasons why knockout of the ZNF24 gene leads to embryonic lethality. In the central nervous system, ZNF24 is required for the myelination function of differentiated oligodendrocytes (6). The function of ZNF24 in regulating cell migration and invasion has been primarily investigated in aortic vascular smooth muscle cells, where ZNF24 facilitates cell migration, which in turn contributes to the development of intimal hyperplasia after endovascular arterial injury (9).
In addition to regulating the function of normal cells, ZNF24 has also been shown to play confounding roles in key processes during cancer initiation and progression. Studies in our laboratory have shown that ZNF24 levels are significantly decreased in breast cancer and colon cancer tissues compared to normal tissues. It represses the transcription of VEGF, one of the principal proangiogenic factors, and therefore serves as a potent inhibitor of tumor angiogenesis (10, 11). Conversely, expression of ZNF24 is increased in hepatocellular carcinoma and is positively correlated with the growth of hepatocellular carcinoma cells (12).
Angiogenesis is a multistep process involving the degradation of basement membrane and extracellular matrix, EC proliferation, migration, invasion, and vessel maturation. A concert of pro- and antiangiogenic factors regulating these processes precisely controls angiogenesis temporally and spatially. These factors include angiogenic mitogens such as VEGF and bFGF (basic fibroblast growth factor), enzymes that degrade the extracellular matrix such as MMPs, and their endogenous inhibitors, TIMPs (13). To date, the function of ZNF24 in the endothelial compartment has not been studied. Our goal in this study was to determine whether ZNF24 plays an important role in the key process of EC proliferation, migration, and invasion using multiple human microvascular EC types, and whether ZNF24 expression is required for the formation of a functional vasculature in vivo.
MATERIALS AND METHODS
Cell culture and siRNA transfections
Human dermal microvascular endothelial cells (HMVEC-D) and human lung microvascular endothelial cells (HMVEC-L) were purchased from Lonza (Basel, Switzerland) and cultured in endothelial cell growth medium-2 for microvascular endothelial cells (EGM-2 MV; Lonza). Human mammary microvascular endothelial cells (HMVEC-M) were purchased from Sciencell Research Laboratories (Carlsbad, CA, USA) and cultured in endothelial cell (EC) medium (Sciencell). Bone marrow–derived mesenchymal progenitor cells (bmMPC) were isolated and cultured as described previously (14). The control nontargeting siRNA pools and siRNA pools targeting human ZNF24 were purchased from Thermo Fisher Scientific (Pittsburgh, PA, USA). Cells were transfected with siRNAs using the Dharmafect 1 reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Reverse transcription and quantitative PCR
RNA was collected using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and treated with DNase I (Qiagen). For PCR array analyses, RNA was isolated from HMVEC-D cells transfected with control siRNA or siZNF24. cDNA was synthesized using RT2 First Strand Kit (SABiosciences, Valencia, CA, USA). PCRs were performed using the RT2 Profiler PCR Cell Cycle Arrays (SABiosciences), and results were analyzed using the PCR Array Data Analysis Software (SABiosciences) according to the manufacturer’s instructions. For quantitative PCR (qPCR) analyses, cDNA was synthesized using the Superscript Vilo cDNA Synthesis Kit (Life Technologies, Grand island, NY, USA) and amplified using the following gene-specific primers: MMP2, forward, 5′-AAGTCTGGAGCGATGTGACC-3′, reverse, 5′-GAGTCCGTCCTTACCGTCAA-3′; TIMP1, forward, 5′-CTGTTGTTGCTGTGGCTGAT-3′, reverse, 5-TCTGGTTGACTTCTGGTGTCC-3′; TIMP2, forward, 5′-GGAAGTGGACTCTGGAAACG-3′, reverse, 5′-GGGGGCCGTGTAGATAAACT-3′; TIMP3, forward, 5′-CAGGACGCCTTCTGCAACT-3′, reverse, 5′-ATCTTGGTGAAGCCTCGGTA-3′; TIMP4, forward, 5′-TCTTCCCTCTGTGGTGTGAA-3′, reverse, 5′-AGGGCTCGATGTAGTTGCAC-3′; MMP14, forward, 5′-CCCCGTTGTCTCCTGCTC-3′, reverse, 5′-GCTGTGTGTGGGTACGTAGG-3′; CDKN3 (cyclin-dependent kinase inhibitor 3), forward, 5′-CGCCCAGTTCAATACAAACA-3′, reverse, 5′-GGAAGAGCACATAAACCGAGA-3′; CCND2 (cyclin D2), forward, 5′-GGACATCCAACCCTACATGC-3′, reverse, 5′-CCAAGAAACGGTCCAGGTAA-3′; and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), forward, 5′-AGCCACATCGCTCAGACAC-3′, reverse, 5′-AATGAAGGGGTCATTGATGG-3′. Values in all graphs represent mean ± sd from 3 independent experiments.
Immunoblotting analyses and ELISA
Whole-cell protein extracts were prepared by lysing the cells using a radioimmunoprecipitation assay (RIPA) buffer (Santa Cruz Biotechnology, Dallas, TX, USA) and were subjected to SDS-PAGE followed by immunoblotting analyses. Primary antibodies used were ZNF24 (R&D Systems, Minneapolis, MN, USA), phospho-VEGF receptor 2 (VEGFR2; Cell Signaling Technology, Danvers, MA), total VEGFR2 (Cell Signaling Technology), MMP-14 (EMD Millipore, Billerica, MA, USA) and GAPDH (EMD Millipore). Densitometry analyses of immunoblots were performed using Adobe Photoshop CS5 software (Adobe Systems, San Jose, CA, USA). ELISA assays were performed using Human Quantikine ELISA kits (R&D Systems) according to the manufacturer’s instructions as reported previously (15).
Migration and invasion assays
Migration assays were performed as described previously by us (16–18) using Costar 24-well Transwell plates (Corning, Tewksbury, MA, USA). Briefly, HMVEC-D and HMVEC-L cells transfected with control siRNA or siZNF24 were seeded at 25,000 cells per well in 100 μl of endothelial basal medium-2 (EBM-2; Lonza) in the upper chamber. EGM-2 MV or EBM-2 supplemented with 40 ng/ml VEGF (R&D Systems) and 1 μg/ml heparin (Stemcell Technologies, Vancouver, BC, Canada) was used in the bottom chamber as the chemoattractant. Cells were incubated at 37°C for 20 h before they were stained with Diff-Quik Stain Set (Siemens, Malvern, PA, USA). Cells remaining in the upper chamber were gently wiped off, and cells that had migrated to the bottom chamber were quantified using a microscope at ×200 (7 fields). To monitor differences in proliferation, cells were seeded at the same density in 24-well plates, cultured under the same conditions for 20 h, and counted using a Coulter counter. Fold changes in number of migrated cells were normalized against cell number differences in proliferation. Invasion assays were performed as previously described by us using Matrigel invasion chambers (Thermo Fisher Scientific) (17, 19). HMVEC-D and HMVEC-L cells transfected with control siRNA or siZNF24 were seeded at 50,000 cells per well in 500 μl of EBM-2 in the upper chamber. EGM-2 MV was used in the bottom chamber as the chemoattractant. Cells were incubated and quantified using the same methods described above for the migration assays. Values in all graphs represent mean ± sd from 3 independent experiments.
Proliferation and apoptosis assays
Proliferation assays were performed as previously described by us (16, 20–22). In brief, HMVEC-D, HMVEC-L, and HMVEC-M cells transfected with control siRNA or siZNF24 were serum starved overnight and seeded in EGM-2 MV at 5000 cells per well in triplicate in 48-well plates. Cell numbers were counted daily using a Coulter counter. Experiments were performed independently 3 times; a representative experiment is shown. Apoptosis assays were performed using the Dead Cell Apoptosis Kit (Life Technologies) according to the manufacturer’s instructions. Values represent mean ± sd from 3 independent experiments.
Substrate gel electrophoresis
MMP-2 enzymatic activity was detected using substrate gel electrophoresis as previously described (23–27). In brief, 33.3 μl of conditioned media or purified control MMP-2 (Merck Millipore, Billerica, MA) were mixed with 16.7 μl of buffer containing 4% SDS, 0.15 M Tris (pH 6.8), 20% glycerol, and 0.5% (w/v) bromphenol blue, then separated on a 10% SDS–acrylamide gel containing 0.1% (w/v) gelatin (Bio-Rad, Hercules, CA, USA). Gels were then incubated in a 2.5% Triton X-100 solution for 30 minutes at room temperature, followed by overnight incubation at 37°C in substrate buffer containing 50 mM Tris-HCl (pH 8), 5 mM CaCl2, and 0.02% NaN3, and stained with 0.5% Coomassie Brilliant Blue. For 4-aminophenylmercuric acetate (APMA) treatment, APMA was added to samples to a final concentration of 1 mM (28). Samples were then incubated for 2 hours at 37°C before gel electrophoresis.
In vivo vessel formation assays
Cell/Matrigel implant assays and microvessel density analyses were performed as described previously by us (14). In brief, 1 × 106 HMVEC-D cells transfected with control siRNA or siZNF24 were mixed with 1 × 106 bmMPC in 200 μl of phenol red–free growth factor–reduced Matrigel (BD Bioscience, San Jose, CA, USA), and subcutaneously injected into dorsal flanks of 6-week-old male nude mice (Massachusetts General Hospital, Boston, MA, USA). Each animal was injected at 2 sites with implants containing HMVEC-D cells transfected with either control siRNA or siZNF24. A total of 9 animals were injected. All animal studies were performed in accordance with the regulations of the Boston Children’s Hospital Institutional Animal Care and Use Committee. Animals were killed 7 days after injection, and implants were excised, fixed in 10% formalin overnight, and embedded in paraffin. Hematoxylin and eosin (H&E) staining was performed on 5 μm thick sections, and 10 random images were taken at ×400 for each implant. Microvessels were identified as lumenized structures containing erythrocytes, and microvessel density was quantified as number of vessels per square millimeter. Anti-human CD31 (Dako, Carpinteria, CA, USA) staining was performed on 5 μm thick sections, and 10 random images were taken at ×400 for each implant. Microvessels of human origin were identified as human CD31-positive vessels, and microvessel density was quantified as the number of vessels per square millimeter.
Statistical analyses
Values in all graphs represent mean ± sd or ± sem from 3 independent experiments unless otherwise stated. Unpaired 2-tailed Student’s t tests were used to compare mean differences in cell counts, mRNA, and protein levels between independent groups, as well as differences in microvessel density in the animal studies.
RESULTS
Knockdown of ZNF24 in microvascular ECs leads to decreased cell migration and attenuated VEGFR2 signaling
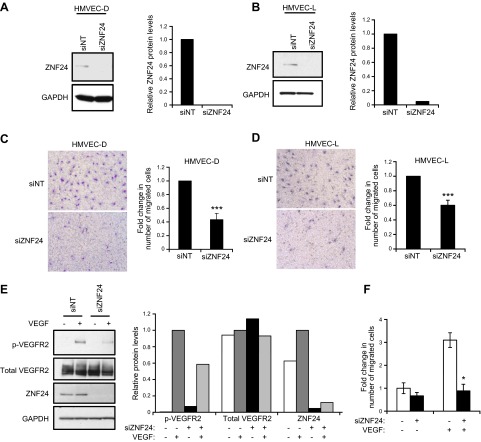
Previous studies have shown that ZNF24 plays essential roles in regulating key cellular processes of neural progenitor cells (8), breast and liver cancer cells (10–12), and vascular smooth muscle cells (9). However, the function of ZNF24 in the important endothelial compartment is completely unknown. To explore the function of ZNF24 in ECs, we first examined the effect of ZNF24 knockdown on EC migration utilizing modified Boyden chamber assays and full growth medium containing serum and a cocktail of proangiogenic factors as the chemoattractant. Transfection of human microvascular endothelial cells from skin (HMVEC-D) and from lung (HMVEC-L) with siZNF24 led to 100% and 95% decrease in endogenous ZNF24 levels, respectively (Fig. 1A, B). Knockdown of ZNF24 resulted in significantly decreased cell migration in both of these 2 cell types (Fig. 1C, D). To explore the mechanism by which endogenous ZNF24 facilitates microvascular EC migration, we examined the effect of ZNF24 knockdown on VEGFR2 signaling in these cells. Phosphorylation of VEGFR2 could not be detected when cells were untreated with VEGF (Fig. 1E, lanes 1 and 3). Upon VEGF stimulation, cells transfected with control siRNA exhibited robust phosphorylation of VEGFR2 (Fig. 1E, lane 2). This effect was, however, significantly attenuated in cells transfected with siZNF24 (Fig. 1E, lane 4), suggesting that knockdown of ZNF24 leads to decreased sensitivity to VEGF stimulation in these cells. We next asked whether ZNF24 knockdown has a similar effect on EC migration in response to VEGF. When using VEGF alone as the chemoattractant, cells transfected with siZNF24 exhibited significantly decreased cell migration compared with those transfected with control siRNA (Fig. 1F). These data suggest that endogenous ZNF24 is essential for the migration potential and VEGFR2 signaling in microvascular ECs.
Figure 1.
ZNF24 is essential for migration and VEGFR2 signaling of HMVEC cells. A and B) HMVEC-D (A) and HMVEC-L (B) cells were transfected with control siRNA or siZNF24, and protein levels were analyzed by immunoblotting and quantified by densitometry using Adobe Photoshop CS5. C and D) HMVEC-D (C) and HMVEC-L (D) cells were transfected with control siRNA or siZNF24, and cell migration was determined using EGM-2 MV as the chemoattractant. ***P < 0.001. E) HMVEC-D cells were transfected with control siRNA or siZNF24 and treated with or without VEGF. Protein levels were analyzed by immunoblotting and quantified by densitometry using Adobe Photoshop CS5. This experiment was conducted 3 times, and a representative experiment is shown. F) HMVEC-D cells were transfected with control siRNA or siZNF24, and cell migration was determined using VEGF as the chemoattractant. *P < 0.05.
Knockdown of ZNF24 in microvascular ECs leads to decreased cell invasion by down-regulating MMP-2
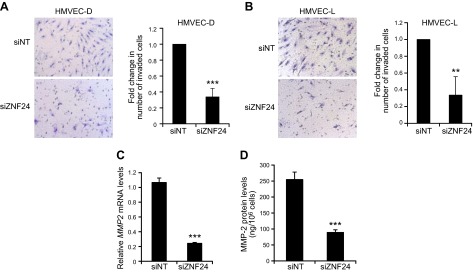
To explore the function of ZNF24 in regulating EC invasion, we conducted standard Matrigel invasion assays using HMVEC-D and HMVEC-L cells transfected with control siRNA or siZNF24. Knockdown of ZNF24 in these cells resulted in significantly decreased invasion of these cells (Fig. 2A, B), leading to the hypothesis that ZNF24 modulates the ability of ECs to degrade the extracellular matrix. To test this hypothesis, we determined the mRNA and protein levels of 2 key members of the matrix metalloproteinase (MMP) family, the gelatinases MMP-2 and MMP-9, which have been most strongly implicated in the development of the vasculature. Neither mRNA nor protein levels of MMP-9 could be detected in HMVEC-D cells (data not shown), whereas both mRNA and protein levels of MMP-2 were significantly decreased when ZNF24 was knocked down in these cells (Fig. 2C, D). Consistently, HMVEC-L cells transfected with siZNF24 exhibited significantly decreased MMP-2 mRNA and protein levels (Supplemental Fig. 1A, B). These results suggest that loss of endogenous ZNF24 contributes to decreased cell invasion possibly by down-regulating MMP-2 levels. Values in all graphs of qPCR analyses represent mean ± sd from 3 independent experiments.
Figure 2.
Knockdown of ZNF24 leads to decreased cell invasion and MMP-2 levels in HMVEC cells. A and B) HMVEC-D (A) and HMVEC-L (B) cells were transfected with control siRNA or siZNF24, and cell invasion was determined using EGM-2 MV as the chemoattractant. **P < 0.01; ***P < 0.001 C) HMVEC-D cells were transfected with control siRNA or siZNF24, and MMP2 mRNA levels were analyzed by qPCR. ***P < 0.001. Values in all graphs represent mean ± sd from 3 independent experiments. D) HMVEC-D cells were transfected with control siRNA or siZNF24, and MMP-2 protein levels in the conditioned media were analyzed by ELISA. ***P < 0.001.
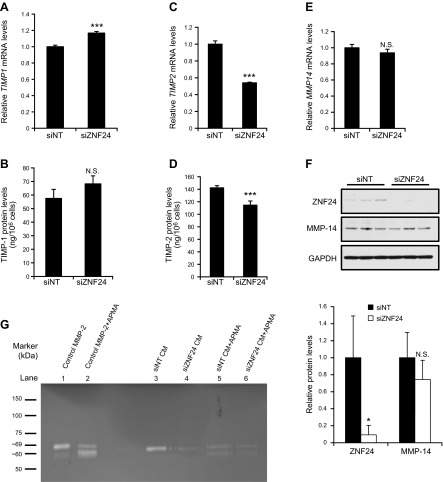
To determine whether down-regulation of MMP-2 protein levels further leads to decreases in its proteolytic activity, we first analyzed levels of TIMPs, which are endogenous inhibitors of MMPs (13, 29–31). Knockdown of ZNF24 led to a minor increase in TIMP1 mRNA levels and significant decreases in mRNA levels of TIMP2 and TIMP3, with no effect on levels of TIMP4 (Fig. 3A, C, and Supplemental Fig. 2). We further examined the protein levels of TIMP-1 and TIMP-2, which function as potent MMP-2 inhibitors. Protein levels of TIMP-1 did not exhibit significant changes when ZNF24 was knocked down in these cells (Fig. 3B), and protein levels of TIMP-2 were only marginally decreased when ZNF24 was knocked down (Fig. 3D). These results suggest that ZNF24 does not play a major role in regulating levels of endogenous inhibitors of MMP-2, TIMP-1 and TIMP-2. MMP-2 is secreted as a proenzyme which remains latent until the proteolytic removal of its prodomain by its activators (31). We examined the level of MMP-14, a key activator of MMP-2 (32). Knockdown of ZNF24 did not lead to significant changes in mRNA (Fig. 3E) or protein (Fig. 3F) levels of MMP-14. Taken together, these data suggest that endogenous ZNF24 does not contribute significantly to the regulation of major MMP-2 activators and inhibitors. As a result, decreased MMP-2 protein levels mediated by ZNF24 knockdown may in turn result in significantly lower levels of MMP-2 activity available for the degradation of the extracellular matrix.
Figure 3.
Knockdown of ZNF24 leads to decreased MMP-2 levels without affecting levels of major MMP-2 activity regulators. A, C, and E) HMVEC-D cells were transfected with control siRNA or siZNF24, and mRNA levels of TIMP1 (A), TIMP2 (C), and MMP14 (E) were analyzed by qPCR. ***P < 0.001; N.S., not significant. Values in all graphs represent mean ± sd from 3 independent experiments. B and D) HMVEC-D cells were transfected with control siRNA or siZNF24, and protein levels of TIMP-1 (B) and TIMP-2 (D) in the conditioned media were analyzed by ELISA. ***P < 0.001; N.S., not significant. F) HMVEC-D cells were transfected with control siRNA or siZNF24. Protein samples from 3 independent experiments were analyzed by immunoblotting and quantified by densitometry using Adobe Photoshop CS5. *P < 0.05; N.S., not significant. G) HMVEC-D cells were transfected with control siRNA or siZNF24. Conditioned media treated with or without APMA were analyzed by substrate gel electrophoresis for MMP-2 activity (lanes 3–6). Purified MMP-2 treated with or without APMA was used as control samples (lanes 1 and 2). This experiment was conducted 3 times, and a representative experiment is shown.
To test this hypothesis, we analyzed conditioned media from HMVEC-D cells transfected with control siRNA or siZNF24 using substrate gel electrophoresis. Knockdown of ZNF24 led to decreased MMP-2 levels in the conditioned media (Fig. 3G, lanes 3 and 4). To determine whether the MMP-2 detected in the conditioned media represents the latent proenzyme or its active form, we analyzed the conditioned media along with purified MMP-2 treated with or without APMA as controls. Pro-MMP-2 remains latent by forming an intramolecular complex between the cysteine residue in the propeptide domain and the zinc atom in the catalytic domain, which blocks the active site. Treatment of pro-MMP-2 with APMA, the standard activation reagent for MMPs, results in the disruption of this intramolecular complex, removal of the propeptide domain, and the subsequent conversion of pro-MMP-2 to its active form (33, 34). As shown in lane 1 (Fig. 3G), the molecular weight of the majority of the MMP-2 species in the control sample corresponds to pro-MMP-2 (top band). When treated with APMA (Fig. 3G, lane 2), the majority of the control MMP-2 protein was converted from the proform (top band) to the active form (bottom band), again demonstrating that the species in lane 1 represents the proform of MMP-2. APMA treatment of the conditioned media resulted in decreased intensity of the top band and increased intensity of the bottom band (Fig. 3G, lanes 5 and 6), suggesting that the proform of MMP-2 is present in the conditioned media of HMVEC-D cells. It is well appreciated that the activation of pro-MMP-2 requires its binding to TIMP-2 and subsequent formation of a ternary complex with membrane-bound MMP-14 (32, 35). In our 2-dimensional cell culture system, it is possible that pro-MMP-2 and TIMP-2 in the conditioned media remain free and unbound to MMP-14, therefore keeping the pro-MMP-2 in a latent state. In in vitro HMVEC cell invasion assays and in vivo assays, pro-MMP-2 can be activated through the interaction between the HMVEC cells and extracellular matrix (36, 37) and/or through their cross-talk with other cell types (38). Taken together, our results demonstrate that ZNF24 plays an important role in regulating the invasive activity of microvascular ECs, possibly by maintaining pro-MMP-2 levels that are available in the microenvironment for modulation when the degradation of the extracellular matrix is required. Values in all graphs of qPCR analyses represent mean ± sd from 3 independent experiments.
Microvascular EC proliferation is significantly inhibited when ZNF24 is knocked down
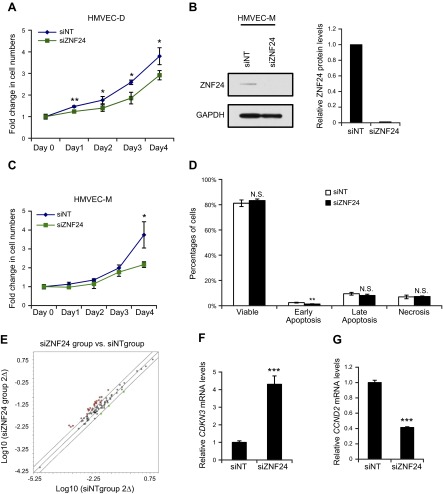
Proliferation of microvascular ECs is required for successful neovascularization. Because ZNF24 has been shown to regulate proliferation of several normal and malignant cell types (7–9), we explored whether knockdown of ZNF24 would have a significant impact on microvascular EC proliferation. HMVEC-D cells transfected with siZNF24 exhibited significantly decreased cell proliferation (Fig. 4A). Surprisingly, this inhibitory effect on cell proliferation mediated by ZNF24 knockdown was not observed in HMVEC-L cells (Supplemental Fig. 1C). Given that recent studies have revealed significant differences among microvascular ECs from different organs in their proteomic and gene expression profiles (39, 40), the lack of inhibitory effect mediated by siZNF24 on HMVEC-L cells could be due to changes in the expression levels of some of these genes that serve as ZNF24 downstream effectors in regulating cell proliferation. To determine whether the growth inhibitory effect of ZNF24 knockdown in HMVEC-D cells can also be observed in other microvascular EC types, we examined human microvascular ECs isolated from the mammary gland, HMVEC-M. Consistent with HMVEC-D cells, knockdown of ZNF24 in HMVEC-M cells led to significantly decreased proliferation (Fig. 4B, C). To confirm the decrease in cell numbers in siZNF24 group is indeed due to decreased cell proliferation rather than increased cell death, we examined the percentages of viable, early apoptotic, late apoptotic, and necrotic cells in siNT and siZNF24 groups by flow cytometry. Knockdown of ZNF24 did not lead to significant changes in the percentages of viable, late apoptotic, or necrotic cells (Fig. 4D). Knockdown of ZNF24 led to decreased percentage of early apoptotic cells, although in both the control siRNA group and the siZNF24 group the percentages of early apoptotic cells were extremely low (2.4% and 1.4%, respectively; Fig. 4D). Similar results were observed when these cells were stained with propidium iodide and analyzed by flow cytometry for the sub-G1 population (data not shown). We also analyzed protein extracts from HMVEC-D cells transfected with control siRNA or siZNF24 for levels of cleaved poly(ADP-ribose) polymerase, a marker for apoptosis. We did not detect cleaved poly(ADP-ribose) polymerase in either group (data not shown). Taken together, these results suggest that knockdown of ZNF24 does not have a significant effect on cell death, and that decreased cell numbers observed in siZNF24-transfected HMVEC cells are due to growth inhibition caused by ZNF24 knockdown. To identify cell cycle regulators that serve as downstream effectors of ZNF24 in regulating cell proliferation, we analyzed gene expression in HMVEC-D cells transfected with control siRNA or siZNF24 using the cell cycle qPCR array. Twenty-seven genes were differentially expressed between cells transfected with control siRNA or siZNF24 (Fig. 4E, and Supplemental Table 1A). These genes encode a network of proteins that are physically or functionally associated with each other to regulate the cell cycle. Six of these genes were also identified in another cell cycle qPCR array analysis of mammary epithelial cells transfected with control siRNA or siZNF24 (Supplemental Table 1A and data not shown). We verified the expression levels of these 6 genes using independent RNA samples, and all of these genes demonstrated changes in gene expression consistent with the qPCR array analyses (Fig. 4F, G and data not shown). Values in all graphs of qPCR analyses represent mean ± sd from 3 independent experiments. Studies have shown that among these 6 genes, increased levels of CDKN3 and decreased levels of CCND2 are associated with inhibition of cell cycle progression (41, 42), consistent with the effect mediated by ZNF24 knockdown. In summary, these data suggest that knockdown of ZNF24 in microvascular ECs inhibits cell proliferation by, at least in part, modulating levels of cell cycle regulators such as CDKN3 and CCND2.
Figure 4.
Knockdown of ZNF24 significantly inhibits HMVEC cell proliferation. A) HMVEC-D cells were transfected with control siRNA or siZNF24, and cell numbers were counted daily. *P < 0.05; **P < 0.01. B) HMVEC-M cells were transfected with control siRNA or siZNF24, and protein levels were analyzed by immunoblotting and quantified by densitometry using Adobe Photoshop CS5. C) HMVEC-M cells were transfected with control siRNA or siZNF24, and cell numbers were counted daily. *P < 0.05. D) HMVEC-D cells were transfected with control siRNA or siZNF24 and stained with annexin V and propidium iodide. Percentages of viable, early apoptotic, late apoptotic, and necrotic cells were analyzed by flow cytometry. **P < 0.01. N.S., not significant. E) Scatter plot showing the differential expression of cell cycle regulators in HMVEC-D cells transfected with siZNF24 compared to those transfected with control siRNA. Red dots indicate genes that are up-regulated by siZNF24 2-fold or more; green dots, genes down-regulated by siZNF24 2-fold or more; and black dots, genes not differentially expressed between the control siRNA group and the siZNF24 group. F, and G) HMVEC-D cells were transfected with control siRNA or siZNF24, and mRNA levels of CDKN3 (F) and CCND2 (G) were analyzed by qPCR. ***P < 0.001. Values in all graphs represent mean ± sd from 3 independent experiments.
The ability of microvascular ECs to form functional vascular networks in vivo is dependent on endogenous ZNF24 expression
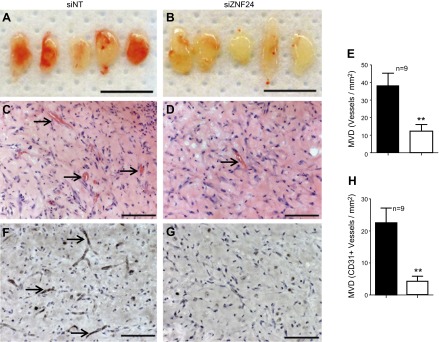
Our in vitro analyses using multiple microvascular ECs have demonstrated that knockdown of ZNF24 significantly inhibits migration, invasion and proliferation of these cells. To investigate the function of ZNF24 in the endothelial compartment in vivo, we examined the effect of ZNF24 knockdown on the ability of microvascular ECs to form new functional vascular networks in in vivo vessel formation assays. HMVEC-D cells were transfected with control siRNA or siZNF24, coimplanted subcutaneously with bmMPC in growth factor-reduced Matrigel on the back of 6 wk old nu/nu mice, and collected 7 d after injection as previously reported (14). Matrigel implants containing HMVEC-D cells transfected with control siRNA were well vascularized and perfused with erythrocytes (Fig. 5A, C), whereas those containing HMVEC-D cells transfected with siZNF24 formed fewer functional vessels (Fig. 5B, D). H&E staining revealed significantly lower number of lumenized vessels filled with erythrocytes in Matrigel implants of the siZNF24 group compared to those of the control siNT group (Fig. 5C, D). When quantified, knockdown of ZNF24 in HMVEC-D cells resulted in a 68% decrease in microvessel density (Fig. 5E). Anti-human CD31 staining confirmed that majority of these lumenized vessels were lined by HMVEC-D cells rather than host ECs (Fig. 5F, G). Consistent with H&E staining, the microvessel density of CD31 positive vessels exhibited significant decreases in siZNF24 group compared to the control group (Fig. 5H). These results lead to the conclusion that ZNF24 is essential for the ability of human microvascular ECs to form functional vascular networks in vivo.
Figure 5.
Loss of ZNF24 leads to significantly decreased functional vascular networks in vivo. HMVEC-D cells transfected with siNT (A, C, and F) or siZNF24 (B, D, and G) were coinjected with bmMPC in Matrigel, and implants were collected 7 days after injection. n = 9 for each group. A and B) Macroscopic view of representative Matrigel implants. Scale bars, 1 cm. C and D) Representative images of Matrigel implant sections stained with H&E. Arrows indicate lumenized blood vessels containing erythrocytes. Scale bars, 100 μm. E) Microvessel density was quantified by counting erythrocyte-filled vessels on the H&E staining. **P < 0.01. F, and G) Representative images of Matrigel implant sections stained with anti-human CD31. Arrows indicate blood vessels formed by HMVEC cells. Scale bars, 100 μm. H) Microvessel density of vessels formed by HMVEC cells was quantified by counting vessels positive for human CD31. **P < 0.01.
DISCUSSION
Recent studies have revealed the important functions of ZNF24 in regulating cell proliferation, differentiation, migration, and invasion, as well as tumor angiogenesis (8–12). Knockout of ZNF24 leads to embryonic lethality, suggesting that ZNF24 is essential for embryonic development (7). However, the mechanisms underlying the embryonic lethality are mostly unknown. In this study, we have shown for the first time that ZNF24 is an important and novel regulator in maintaining the angiogenic potential of microvascular ECs. Our data demonstrate that knockdown of ZNF24 expression resulted in decreased cell proliferation, possibly through up-regulation of cell cycle inhibitors such as CDKN3 and down-regulation of cell cycle activators such as CCND2. Knockdown of ZNF24 expression also led to decreased levels of VEGFR2 signaling and MMP-2 specifically, which in turn impaired the ability of these cells to migrate and invade. Consistent with our findings in vitro, microvascular ECs lacking ZNF24 expression exhibited significantly decreased ability to form functional vascular networks in vivo. Our results reveal that ZNF24 plays an important role in modulating cell proliferation and in sustaining proper levels of VEGFR2 signaling and MMP-2 in ECs, and thereby maintaining the angiogenic potential of these cells. We also identify a potential mechanism by which loss of ZNF24 gene leads to embryonic lethality at E7.5—that is, by compromising the formation of the vascular network in the developing embryo. Indeed, studies on genetically modified mouse models have demonstrated that partial or complete loss of VEGFR2 signaling by deletion of VEGF or VEGFR2 genes leads to embryonic lethality at time points similar to that of the ZNF24 knockout mice (43, 44). Although MMP2 knockout mice are viable and fertile, they exhibited significantly slower growth rate than their wild-type littermates and had impaired retinal angiogenesis (45, 46). These studies underlie the importance of VEGFR2 signaling and proper MMP-2 levels during the formation of the vasculature in embryonic development and during postnatal angiogenesis. Therefore, loss of the ZNF24 gene may lead to early lethality in part as a result of malfunctioning of these pathways.
ZNF24 has been shown to regulate Notch signaling in neural progenitor cells to inhibit neuronal differentiation in these cells (8). Because of the important role of Notch signaling in regulating angiogenesis (47), we analyzed the expression of genes involved in the Notch signaling pathway in HMVEC-D cells transfected with control siRNA or siZNF24 using the Notch signaling PCR array. Twenty genes exhibited differential expression between the control siRNA group and the siZNF24 group, including Notch ligands, Notch target genes, and genes involved in Notch receptor processing (Supplemental Table 1B). Although levels of Notch1 and Notch4, the predominant Notch receptors in the endothelium, did not exhibit significant changes when ZNF24 was knocked down, levels of prevalent Notch ligands in ECs, DLL4 and JAG1, were both down-regulated when ZNF24 was knocked down. ADAM10, a member of the ADAM family (A Disintegrin And Metalloproteinase), which is essential for the proteolytic cleavage of Notch receptor and subsequent activation of the Notch signaling pathway (48), was also down-regulated in HMVEC-D cells transfected with siZNF24. These data strongly suggest that knockdown of ZNF24 in HMVEC-D attenuates Notch signaling in these cells.
ECs isolated from different organs and under different disease conditions show significant heterogeneity in their morphology, functions, and molecular signatures (49). In our study, we observed distinct effects of ZNF24 knockdown in HMVEC cells isolated from different organs. Specifically, knockdown of ZNF24 led to decreased cell proliferation in HMVEC cells of dermal but not pulmonary origin. One possible reason for this interesting observation is that these HMVEC cells may express different levels of endogenous ZNF24 such that knockdown of endogenous ZNF24 in these HMVEC cell types may have different effects. However, immunoblotting analyses demonstrated that endogenous levels of ZNF24 did not exhibit significant differences among these HMVEC cell types (Supplemental Fig. 3). These data indicate that differences in the function of ZNF24 in regulating proliferation of these cell types may be due to variations in its ability to regulate its downstream target genes rather than to differences in expression levels of endogenous ZNF24. We have found that, unlike skin-derived HMVEC cells in which ZNF24 modulates levels of cell cycle regulators CDKN3 and CCND2, knockdown of ZNF24 in lung-derived HMVEC cells led to much less significant changes in CDKN3 levels and no changes in CCND2 levels (Supplemental Fig. 1D, E). Interestingly and importantly, recent studies using proteomic and transcriptomic analyses have revealed unique features of ECs from these 2 origins (39, 40). In particular, HMVEC cells isolated from skin overexpress fascin, an actin-bundling protein with important functions in the filopodia (40). Fascin has been shown to play a positive role in the regulation of cell proliferation in multiple cell lines (50, 51). It is possible that ZNF24 modulates the proliferation of HMVEC-D cells in part through regulating fascin. In HMVEC-L cells, however, this effect is lost due to low endogenous levels of fascin as a ZNF24 downstream effector. We also examined levels of β-catenin and cyclin D1, which have been shown to serve as downstream effectors of ZNF24 in regulating cell proliferation in hepatocellular carcinoma cells (12). Knockdown of ZNF24 in HMVEC cells did not lead to changes in β-catenin and cyclin D1 levels (data not shown), indicating that ZNF24 exerts similar function in regulating cell proliferation through distinct target genes in different cell types.
We have observed that knockdown of ZNF24 in HMVEC cells leads to significantly decreased VEGFR2 signaling by down-regulating the phosphorylation of VEGFR2 without affecting total VEGFR2 protein levels. The localization of VEGFR2 in lipid rafts of the EC membrane is essential for the dimerization and phosphorylation of the receptor and subsequent activation of the signaling pathway (52). Therefore, disruption of the localization of VEGFR2 in lipid rafts by cholesterol depletion or by modulating the expression of cholesterol homeostasis regulators can lead to impaired phosphorylation of VEGFR2 without affecting total VEGFR2 protein levels (53, 54). Combining our observations with the results from these studies, it is possible that knockdown of ZNF24 changes the localization of VEGFR2 on the EC membrane, thereby attenuating the phosphorylation of the receptor and its downstream signaling.
Our results have demonstrated that ZNF24 serves as a new positive regulator of the angiogenic potential of microvascular ECs. Interestingly, we observed the opposite effect of ZNF24 in human breast cancer cells, in which it functions as a potent inhibitor of tumor angiogenesis by repressing the transcription of VEGF (10, 11). One possible reason for the distinct functions of ZNF24 in different cell types is that similar to many other transcription factors, the function of ZNF24 may be context dependent. By binding to different target DNA sequences, these transcription factors may favor distinct cofactors, thereby activating or repressing the transcription of its unique downstream target genes in different cell types (55, 56). In ECs, ZNF24 regulates the mRNA levels of MMP2, CDKN3 and CCND2. To explore whether MMP2, CDKN3, and CCND2 are direct target genes for ZNF24 in HMVEC cells, we searched the proximal promoter regions of these genes for known ZNF24 binding sites: TCAT repeats (57), ATTAATT (12), and GCTTTCCATTT (11). There is no known ZNF24 binding site within the proximal promoter regions of these genes, indicating that ZNF24 may regulate the transcription of these genes indirectly. Our results have demonstrated that knockdown of ZNF24 leads to attenuated VEGF signaling and possibly attenuated Notch signaling in HMVEC cells. It has been shown that activated VEGF and Notch signaling leads to increased MMP2 expression and decreased CDKN3 expression, respectively (58, 59). Activated Notch signaling also leads to increased expression of c-Myc (60), which in turn up-regulates the transcription of CCND2 and promotes cell cycle progression (41). Therefore, in HMVEC cells, ZNF24 may directly regulate genes involved in VEGF and Notch signaling pathways, which then lead to changes in MMP2, CDKN3, and CCND2 gene expression. In breast cancer cells, ZNF24 directly represses the transcription of VEGF. It is possible that the target DNA sequences and chromatin environment of the promoter regions of these genes dictates different binding partners for ZNF24 in these cell types, thereby exerting cell type–specific functions. One interesting and important question remaining to be answered is whether ZNF24 can modulate the angiogenic potential of the tumor endothelium. Further studies using mouse and human ECs isolated from tumor tissues will fully unveil the complicated function of ZNF24 in angiogenesis.
Supplementary Material
Acknowledgments
Research reported in this study was supported by the Breast Cancer Research Foundation (New York, NY, USA) and the Advanced Medical Research Foundation (Brookline, MA, USA).
Glossary
- APMA
4-aminophenylmercuric acetate
- bmMPC
bone marrow–derived mesenchymal progenitor cell
- CCND2
cyclin D2
- CDKN3
cyclin-dependent kinase inhibitor 3
- EBM-2
endothelial basal medium-2
- EC
endothelial cell
- EGM-2 MV
endothelial cell growth medium-2
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HMVEC
human microvascular endothelial cell
- HMVEC-D
human dermal microvascular endothelial cell
- HMVEC-L
human lung microvascular endothelial cell
- HMVEC-M
human mammary microvascular endothelial cell
- MMP
matrix metalloproteinase
- qPCR
quantitative PCR
- TIMP
tissue inhibitor of matrix metalloproteinases
- VEGFR2
VEGR receptor 2
- ZNF24
zinc finger protein 24
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Han Z. G., Zhang Q. H., Ye M., Kan L. X., Gu B. W., He K. L., Shi S. L., Zhou J., Fu G., Mao M., Chen S. J., Yu L., Chen Z. (1999) Molecular cloning of six novel Krüppel-like zinc finger genes from hematopoietic cells and identification of a novel transregulatory domain KRNB. J. Biol. Chem. 274, 35741–35748 [DOI] [PubMed] [Google Scholar]
- 2.Wang H., Sun R., Liu G., Yao M., Fei J., Shen H. (2008) Characterization of the target DNA sequence for the DNA-binding domain of zinc finger protein 191. Acta Biochim. Biophys. Sin. (Shanghai) 40, 704–710 [PubMed] [Google Scholar]
- 3.Edelstein L. C., Collins T. (2005) The SCAN domain family of zinc finger transcription factors. Gene 359, 1–17 [DOI] [PubMed] [Google Scholar]
- 4.Khalfallah O., Faucon-Biguet N., Nardelli J., Meloni R., Mallet J. (2008) Expression of the transcription factor Zfp191 during embryonic development in the mouse. Gene Expr. Patterns 8, 148–154 [DOI] [PubMed] [Google Scholar]
- 5.Prost J. F., Nègre D., Cornet-Javaux F., Cortay J. C., Cozzone A. J., Herbage D., Mallein-Gerin F. (1999) Isolation, cloning, and expression of a new murine zinc finger encoding gene. Biochim. Biophys. Acta 1447, 278–283 [DOI] [PubMed] [Google Scholar]
- 6.Howng S. Y., Avila R. L., Emery B., Traka M., Lin W., Watkins T., Cook S., Bronson R., Davisson M., Barres B. A., Popko B. (2010) ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev. 24, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Chen X., Yang H., Wang S., Guo B., Yu L., Wang Z., Fu J. (2006) The zinc finger transcription factor 191 is required for early embryonic development and cell proliferation. Exp. Cell Res. 312, 3990–3998 [DOI] [PubMed] [Google Scholar]
- 8.Khalfallah O., Ravassard P., Lagache C. S., Fligny C., Serre A., Bayard E., Faucon-Biguet N., Mallet J., Meloni R., Nardelli J. (2009) Zinc finger protein 191 (ZNF191/Zfp191) is necessary to maintain neural cells as cycling progenitors. Stem Cells 27, 1643–1653 [DOI] [PubMed] [Google Scholar]
- 9.Lv L., Zhang J., Wang P., Meng Q., Liang W., Zhang L. (2014) Zinc finger protein 191 deficiency attenuates vascular smooth muscle cell proliferation, migration, and intimal hyperplasia after endovascular arterial injury. J. Vasc. Surg. 59, 500–509 [DOI] [PubMed] [Google Scholar]
- 10.Harper J., Yan L., Loureiro R. M., Wu I., Fang J., D’Amore P. A., Moses M. A. (2007) Repression of vascular endothelial growth factor expression by the zinc finger transcription factor ZNF24. Cancer Res. 67, 8736–8741 [DOI] [PubMed] [Google Scholar]
- 11.Jia D., Hasso S. M., Chan J., Filingeri D., D’Amore P. A., Rice L., Pampo C., Siemann D. W., Zurakowski D., Rodig S. J., Moses M. A. (2013) Transcriptional repression of VEGF by ZNF24: mechanistic studies and vascular consequences in vivo. Blood 121, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G., Jiang S., Wang C., Jiang W., Liu Z., Liu C., Saiyin H., Yang X., Shen S., Jiang D., Zhou P., Han D., Hu X., Yi Q., Yu L. (2012) Zinc finger transcription factor 191, directly binding to β-catenin promoter, promotes cell proliferation of hepatocellular carcinoma. Hepatology 55, 1830–1839 [DOI] [PubMed] [Google Scholar]
- 13.Harper J., Moses M. A. (2006) Molecular regulation of tumor angiogenesis: mechanisms and therapeutic implications. EXS (96), 223–268 [DOI] [PubMed] [Google Scholar]
- 14.Melero-Martin J. M., De Obaldia M. E., Kang S. Y., Khan Z. A., Yuan L., Oettgen P., Bischoff J. (2008) Engineering robust and functional vascular networks in vivo with human adult and cord blood–derived progenitor cells. Circ. Res. 103, 194–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith E. R., Zurakowski D., Saad A., Scott R. M., Moses M. A. (2008) Urinary biomarkers predict brain tumor presence and response to therapy. Clin. Cancer Res. 14, 2378–2386 [DOI] [PubMed] [Google Scholar]
- 16.Fernández C. A., Moses M. A. (2006) Modulation of angiogenesis by tissue inhibitor of metalloproteinase-4. Biochem. Biophys. Res. Commun. 345, 523–529 [DOI] [PubMed] [Google Scholar]
- 17.Roy R., Rodig S., Bielenberg D., Zurakowski D., Moses M. A. (2011) ADAM12 transmembrane and secreted isoforms promote breast tumor growth: a distinct role for ADAM12-S protein in tumor metastasis. J. Biol. Chem. 286, 20758–20768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., McNeish B., Butterfield C., Moses M. A. (2013) Lipocalin 2 is a novel regulator of angiogenesis in human breast cancer. FASEB J. 27, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J., Bielenberg D. R., Rodig S. J., Doiron R., Clifton M. C., Kung A. L., Strong R. K., Zurakowski D., Moses M. A. (2009) Lipocalin 2 promotes breast cancer progression. Proc. Natl. Acad. Sci. USA 106, 3913–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez C. A., Roy R., Lee S., Yang J., Panigrahy D., Van Vliet K. J., Moses M. A. (2010) The anti-angiogenic peptide, loop 6, binds insulin-like growth factor-1 receptor. J. Biol. Chem. 285, 41886–41895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy R., Moses M. A. (2012) ADAM12 induces estrogen-independence in breast cancer cells. Breast Cancer Res. Treat. 131, 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foradori M. J., Chen Q., Fernandez C. A., Harper J., Li X., Tsang P. C., Langer R., Moses M. A. (2014) Matrilin-1 is an inhibitor of neovascularization. J. Biol. Chem. 289, 14301–14309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moses M. A., Wiederschain D., Loughlin K. R., Zurakowski D., Lamb C. C., Freeman M. R. (1998) Increased incidence of matrix metalloproteinases in urine of cancer patients. Cancer Res. 58, 1395–1399 [PubMed] [Google Scholar]
- 24.Fernández C. A., Yan L., Louis G., Yang J., Kutok J. L., Moses M. A. (2005) The matrix metalloproteinase-9/neutrophil gelatinase-associated lipocalin complex plays a role in breast tumor growth and is present in the urine of breast cancer patients. Clin. Cancer Res. 11, 5390–5395 [DOI] [PubMed] [Google Scholar]
- 25.Pories S. E., Zurakowski D., Roy R., Lamb C. C., Raza S., Exarhopoulos A., Scheib R. G., Schumer S., Lenahan C., Borges V., Louis G. W., Anand A., Isakovich N., Hirshfield-Bartek J., Wewer U., Lotz M. M., Moses M. A. (2008) Urinary metalloproteinases: noninvasive biomarkers for breast cancer risk assessment. Cancer Epidemiol. Biomarkers Prev. 17, 1034–1042 [DOI] [PubMed] [Google Scholar]
- 26.Coticchia C. M., Curatolo A. S., Zurakowski D., Yang J., Daniels K. E., Matulonis U. A., Moses M. A. (2011) Urinary MMP-2 and MMP-9 predict the presence of ovarian cancer in women with normal CA125 levels. Gynecol. Oncol. 123, 295–300 [DOI] [PubMed] [Google Scholar]
- 27.Roy R., Louis G., Loughlin K. R., Wiederschain D., Kilroy S. M., Lamb C. C., Zurakowski D., Moses M. A. (2008) Tumor-specific urinary matrix metalloproteinase fingerprinting: identification of high molecular weight urinary matrix metalloproteinase species. Clin. Cancer Res. 14, 6610–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herron G. S., Werb Z., Dwyer K., Banda M. J. (1986) Secretion of metalloproteinases by stimulated capillary endothelial cells. I. Production of procollagenase and prostromelysin exceeds expression of proteolytic activity. J. Biol. Chem. 261, 2810–2813 [PubMed] [Google Scholar]
- 29.Moses M. A., Sudhalter J., Langer R. (1990) Identification of an inhibitor of neovascularization from cartilage. Science 248, 1408–1410 [DOI] [PubMed] [Google Scholar]
- 30.Fernández C. A., Butterfield C., Jackson G., Moses M. A. (2003) Structural and functional uncoupling of the enzymatic and angiogenic inhibitory activities of tissue inhibitor of metalloproteinase-2 (TIMP-2): loop 6 is a novel angiogenesis inhibitor. J. Biol. Chem. 278, 40989–40995 [DOI] [PubMed] [Google Scholar]
- 31.Roy R., Zhang B., Moses M. A. (2006) Making the cut: protease-mediated regulation of angiogenesis. Exp. Cell Res. 312, 608–622 [DOI] [PubMed] [Google Scholar]
- 32.Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., Goldberg G. I. (1995) Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 270, 5331–5338 [DOI] [PubMed] [Google Scholar]
- 33.Stetler-Stevenson W. G., Krutzsch H. C., Wacher M. P., Margulies I. M., Liotta L. A. (1989) The activation of human type IV collagenase proenzyme. Sequence identification of the major conversion product following organomercurial activation. J. Biol. Chem. 264, 1353–1356 [PubMed] [Google Scholar]
- 34.Van Wart H. E., Birkedal-Hansen H. (1990) The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. USA 87, 5578–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emmert-Buck M. R., Emonard H. P., Corcoran M. L., Krutzsch H. C., Foidart J. M., Stetler-Stevenson W. G. (1995) Cell surface binding of TIMP-2 and pro-MMP-2/TIMP-2 complex. FEBS Lett. 364, 28–32 [DOI] [PubMed] [Google Scholar]
- 36.Théret N., Lehti K., Musso O., Clément B. (1999) MMP2 activation by collagen I and concanavalin A in cultured human hepatic stellate cells. Hepatology 30, 462–468 [DOI] [PubMed] [Google Scholar]
- 37.Stanton H., Gavrilovic J., Atkinson S. J., d’Ortho M. P., Yamada K. M., Zardi L., Murphy G. (1998) The activation of ProMMP-2 (gelatinase A) by HT1080 fibrosarcoma cells is promoted by culture on a fibronectin substrate and is concomitant with an increase in processing of MT1-MMP (MMP-14) to a 45 kDa form. J. Cell Sci. 111, 2789–2798 [DOI] [PubMed] [Google Scholar]
- 38.Lozito T. P., Jackson W. M., Nesti L. J., Tuan R. S. (2014) Human mesenchymal stem cells generate a distinct pericellular zone of MMP activities via binding of MMPs and secretion of high levels of TIMPs. Matrix Biol. 34, 132–143 [DOI] [PubMed] [Google Scholar]
- 39.Chi J. T., Chang H. Y., Haraldsen G., Jahnsen F. L., Troyanskaya O. G., Chang D. S., Wang Z., Rockson S. G., van de Rijn M., Botstein D., Brown P. O. (2003) Endothelial cell diversity revealed by global expression profiling. Proc. Natl. Acad. Sci. USA 100, 10623–10628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dib H., Chafey P., Clary G., Federici C., Le Gall M., Dwyer J., Gavard J., Tamas N., Bussone G., Broussard C., Camoin L., Witko-Sarsat V., Tamby M. C., Mouthon L. (2012) Proteomes of umbilical vein and microvascular endothelial cells reflect distinct biological properties and influence immune recognition. Proteomics 12, 2547–2555 [DOI] [PubMed] [Google Scholar]
- 41.Bouchard C., Thieke K., Maier A., Saffrich R., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M. (1999) Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 18, 5321–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gyuris J., Golemis E., Chertkov H., Brent R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803 [DOI] [PubMed] [Google Scholar]
- 43.Carmeliet P., Ferreira V., Breier G., Pollefeyt S., Kieckens L., Gertsenstein M., Fahrig M., Vandenhoeck A., Harpal K., Eberhardt C., Declercq C., Pawling J., Moons L., Collen D., Risau W., Nagy A. (1996) Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380, 435–439 [DOI] [PubMed] [Google Scholar]
- 44.Shalaby F., Rossant J., Yamaguchi T. P., Gertsenstein M., Wu X. F., Breitman M. L., Schuh A. C. (1995) Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66 [DOI] [PubMed] [Google Scholar]
- 45.Itoh T., Ikeda T., Gomi H., Nakao S., Suzuki T., Itohara S. (1997) Unaltered secretion of beta-amyloid precursor protein in gelatinase A (matrix metalloproteinase 2)-deficient mice. J. Biol. Chem. 272, 22389–22392 [DOI] [PubMed] [Google Scholar]
- 46.Ohno-Matsui K., Uetama T., Yoshida T., Hayano M., Itoh T., Morita I., Mochizuki M. (2003) Reduced retinal angiogenesis in MMP-2-deficient mice. Invest. Ophthalmol. Vis. Sci. 44, 5370–5375 [DOI] [PubMed] [Google Scholar]
- 47.Hofmann J. J., Iruela-Arispe M. L. (2007) Notch signaling in blood vessels: who is talking to whom about what? Circ. Res. 100, 1556–1568 [DOI] [PubMed] [Google Scholar]
- 48.Bozkulak E. C., Weinmaster G. (2009) Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 29, 5679–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aird W. C. (2012) Endothelial cell heterogeneity. Cold Spring Harb Perspect Med 2, a006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Y., Li A., Faller W. J., Libertini S., Fiorito F., Gillespie D. A., Sansom O. J., Yamashiro S., Machesky L. M. (2013) Fascin 1 is transiently expressed in mouse melanoblasts during development and promotes migration and proliferation. Development 140, 2203–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie J. J., Xu L. Y., Zhang H. H., Cai W. J., Mai R. Q., Xie Y. M., Yang Z. M., Niu Y. D., Shen Z. Y., Li E. M. (2005) Role of fascin in the proliferation and invasiveness of esophageal carcinoma cells. Biochem. Biophys. Res. Commun. 337, 355–362 [DOI] [PubMed] [Google Scholar]
- 52.Labrecque L., Royal I., Surprenant D. S., Patterson C., Gingras D., Béliveau R. (2003) Regulation of vascular endothelial growth factor receptor-2 activity by caveolin-1 and plasma membrane cholesterol. Mol. Biol. Cell 14, 334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noghero A., Perino A., Seano G., Saglio E., Lo Sasso G., Veglio F., Primo L., Hirsch E., Bussolino F., Morello F. (2012) Liver X receptor activation reduces angiogenesis by impairing lipid raft localization and signaling of vascular endothelial growth factor receptor-2. Arterioscler. Thromb. Vasc. Biol. 32, 2280–2288 [DOI] [PubMed] [Google Scholar]
- 54.Fang L., Choi S. H., Baek J. S., Liu C., Almazan F., Ulrich F., Wiesner P., Taleb A., Deer E., Pattison J., Torres-Vázquez J., Li A. C., Miller Y. I. (2013) Control of angiogenesis by AIBP-mediated cholesterol efflux. Nature 498, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arvey A., Agius P., Noble W. S., Leslie C. (2012) Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 22, 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hutchins A. P., Diez D., Takahashi Y., Ahmad S., Jauch R., Tremblay M. L., Miranda-Saavedra D. (2013) Distinct transcriptional regulatory modules underlie STAT3’s cell type–independent and cell type–specific functions. Nucleic Acids Res. 41, 2155–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albanèse V., Biguet N. F., Kiefer H., Bayard E., Mallet J., Meloni R. (2001) Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum. Mol. Genet. 10, 1785–1792 [DOI] [PubMed] [Google Scholar]
- 58.Rodrigues M., Xin X., Jee K., Babapoor-Farrokhran S., Kashiwabuchi F., Ma T., Bhutto I., Hassan S. J., Daoud Y., Baranano D., Solomon S., Lutty G., Semenza G. L., Montaner S., Sodhi A. (2013) VEGF secreted by hypoxic Müller cells induces MMP-2 expression and activity in endothelial cells to promote retinal neovascularization in proliferative diabetic retinopathy. Diabetes 62, 3863–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanda T., Li X., Gutierrez A., Ahn Y., Neuberg D. S., O’Neil J., Strack P. R., Winter C. G., Winter S. S., Larson R. S., von Boehmer H., Look A. T. (2010) Interconnecting molecular pathways in the pathogenesis and drug sensitivity of T-cell acute lymphoblastic leukemia. Blood 115, 1735–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palomero T., Lim W. K., Odom D. T., Sulis M. L., Real P. J., Margolin A., Barnes K. C., O’Neil J., Neuberg D., Weng A. P., Aster J. C., Sigaux F., Soulier J., Look A. T., Young R. A., Califano A., Ferrando A. A. (2006) NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 103, 18261–18266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.