Abstract
Bladder cancer is a common malignant disease, with non–muscle-invasive bladder cancer (NMIBC) representing the majority of tumors. This cancer subtype is typically treated by transurethral resection. In spite of treatment, up to 70% of patients show local recurrences. Intravesical BCG (Bacillus Calmette-Guerin) immunotherapy has been widely used to treat NMIBC, but it fails to suppress recurrence of bladder tumors in up to 40% of patients. Therefore, the development of prognostic markers is needed to predict the progression of bladder cancer and the efficacy of intravesical BCG treatment. This study demonstrates the effectiveness of an E2F4 signature for prognostic prediction of bladder cancer. E2F4 scores for each sample in a bladder cancer expression dataset were calculated by summarizing the relative expression levels of E2F4 target genes identified by ChIP-seq, and then the scores were used to stratify patients into good- and poor-outcome groups. The molecular signature was investigated in a single bladder cancer dataset and then its effectiveness was confirmed in two meta-bladder datasets consisting of specimens from multiple independent studies. These results were consistent in different datasets and demonstrate that the E2F4 score is predictive of clinical outcomes in bladder cancer, with patients whose tumors exhibit an E2F4 score >0 having significantly shorter survival times than those with an E2F4 score <0, in both non–muscle-invasive, and muscle-invasive bladder cancer. Furthermore, although intravesical BCG immunotherapy can significantly improve the clinical outcome of NMIBC patients with positive E2F4 scores (E2F4>0 group), it does not show significant treatment effect for those with negative scores (E2F4<0 group).
Implications
The E2F4 signature can be applied to predict the progression/recurrence and the responsiveness of patients to intravesical BCG immunotherapy in bladder cancer.
Introduction
Bladder cancer is the fourth most common tumor type among males and the ninth leading cause of cancer in the United States (1). Worldwide, over 430,000 new cases occurred in 2012 (2). More than 90% of bladder cancers are urothelial cell carcinoma, with the other 10% comprised of squamous cell carcinoma, adenocarcinoma, sarcoma, and small cell carcinoma (3). The majority of bladder tumors are non–muscle-invasive that are papillary and confined to the urothelial mucosa (Ta) or to the lamina propria (T1), whereas the remaining are muscle-invasive (T2–T4; ref. 4). Patients with Ta and T1 tumors are typically treated by transurethral resection, and in some cases, intravesicular chemo- and immunotherapy (5). In particular, Bacillus Calmette-Guerin (BCG) is the most effective intravesical immunotherapy for treating early-stage bladder cancer (6). Unfortunately, in spite of treatment, up to 70% of patients show local recurrences, and the frequency of recurrences has a significant effect on the quality of life for patients (3). For this reason, prognostic markers for predicting those bladder tumors most likely to recur or progress are needed to assist the decision of treatment strategy and the schedule of cystoscopy follow-up.
Prognostic markers are helpful for the clinicians to select the most appropriate therapeutic treatment. Conventional clinical factors such as tumor stage, tumor grade, and histopathologic features have been used for predicting the prognostic outcome of cancer patients (7). Later, single molecular factors, including mRNA abundance, protein abundance, mutation of a specific gene, and DNA methylation status, were suggested as prognostic markers (8–10). More recently, the wide application of genomewide technologies (e.g., microarray) facilitated the development of prognostic multigene assays (11–14). An example of this, Oncotype DX, a commercial assay, utilizes a 21-gene signature to predict the risk of distant recurrence in estrogen receptor (ER)-positive breast cancers and their responsiveness to chemotherapy (15). A large number of gene signatures like this have been proposed for prognosis prediction in different cancer types.
In bladder cancer, a few signatures have been proposed for prognosis prediction. Dyrskjot and colleagues have identified a gene signature for predicting recurrence frequency in non–muscle-invasive tumors (16), and a gene signature for predicting disease progression (17, 18). Lee and colleagues reported an expression signature of E2F1 that is predictive of superficial to invasive progression of bladder tumors (19). Kim and colleagues have identified genetic signatures that are associated with disease progression in patients with non–muscle-invasive bladder cancer (NMIBC) (20). In addition, Sjodahl and colleagues defined five molecular subtypes of bladder cancer based on gene expression profiles of tumor samples (21). These subtypes show distinct clinical outcomes and differ with respect to the expression and mutation frequency of certain cancer-related genes, such as FGFR3, PIK3CA, and TP53. In general, prognostic signature genes are selected by optimizing supervised predictive models in a training dataset and then validating in test datasets (22). Because of “the curse of dimensionality” (i.e., the number of genes is much larger than the number of samples), most of the selected genes are “passengers” rather than “driving genes” that are functionally related to prognosis (23). As a consequence, many of signatures show reduced or no predictive power in independent datasets (24). One strategy to address this issue is to incorporate prior knowledge into prognosis models.
Transcription factors play critical roles in tumor development, progression, and metastasis (25–28). We have previously developed a method to identify transcriptional regulatory programs that are predictive of cancer prognosis (29, 30). A regulatory program consists of transcription factors and its target genes identified by ChIP-chip or ChIP-seq experiments (31, 32). Given a cancer gene expression dataset, we apply a method called Binding Association with Sorted Expression (BASE; ref. 33) to infer the regulatory activities of the transcription factors in all samples, and then examine their correlation with clinical outcomes of patients using the Cox regression model (30). In contrast to multigene prognostic signatures identified by supervised models, regulatory programs are defined on the basis of prior knowledge learned from ChIP-seq/chip data. We have previously identified an E2F4 regulatory program that showed robust power in predicting the survival of breast cancer patients (29). E2F4 is a member of the E2F transcription factor family, which plays critical roles in cell-cycle progression and differentiation (34). Mutated E2F4 has been reported in various tumors (35, 36). Abnormal expression or mutation of E2F4 causes the malfunction of cell-cycle controls and results in malignant tumors (36). Moreover, transgenic mice with overexpressed E2F4 develop tumors, indicating that E2F4 is an oncogene (37).
In this study, we examine the efficacy of the E2F4 program in predicting progression of bladder cancer. Our analyses indicate that E2F4 scores differ significantly between bladder tumor and normal bladder samples, between superficial and invasive primary tumors, and in primary tumor samples at different stages and grades. The E2F4 scores in primary tumor samples are predictive to the progression of patients, particularly, in NMIBC. Furthermore, we find that E2F4 scores can be used to predict the treatment effect of intravesical therapy. Finally, we validate the effectiveness of the E2F4 signature for progression prediction in two meta-bladder cancer datasets.
Materials and Methods
Datasets
Five bladder cancer gene expression datasets were analyzed in this study. All of them were available from the GEO (Gene Expression Omnibus) database, with accession numbers: GSE13507 (256 samples), GSE1827 (80 samples), GSE19915 (160 samples), GSE31684 (93 samples), and GSE32894 (308 samples). In the GSE13507 dataset, 10, 58, 165, and 23 samples are from normal bladder tissues, normal bladder tissue surrounding bladder tumors, primary bladder tumors, and recurrent bladder tumors, respectively. We converted probeset expression into gene expression for all datasets. For genes with multiple probesets, the probeset with the highest average intensity in all samples was selected to represent the corresponding genes.
The ChIP-seq datasets for E2F4 were downloaded as wig files from previous publications, providing genome-wide occupation of E2F4 in GM06900 (38), HeLa, and K562 (39) cell lines. We used the probabilistic method TIP (Target Identification from Profiles; ref. 40) to identify E2F4 target genes in each cell line using a threshold of false discovery rate (FDR < 0.01). Genes shared in the three cell lines were then identified, resulting in an E2F4 core geneset with 199 genes.
Preparation of meta-bladder datasets
We generated two meta-bladder cancer datasets that contain samples with matched gene expression profiles and survival information to further test the performance of our predictor. The first meta-dataset includes a total of 482 primary bladder tumor samples from three one-channel datasets, GSE13507, GSE31684, and GSE32894 (20, 21, 41). All of the samples were renormalized by quantile normalization to have the same distribution at the gene level (42). Then, expression values were log transformed and gene-wise median normalization was performed to convert the data into relative expression values. After median normalization, the median expression values in the 482 samples for all genes were zero. The second meta-dataset includes a total 240 primary bladder tumor samples from two two-channel arrays, GSE1827 and GSE19915 (4). The dataset contains the relative expression values (log ratios) of genes against a reference sample (RNA pooled from 10 human cell lines). No additional processing was performed for this meta-dataset.
Calculation of E2F4 scores
Given a bladder cancer dataset or a meta-dataset, we applied an algorithm called BASE to infer E2F4 activity in all of the samples (33). The BASE algorithm sorts genes based on their relative expression levels in a sample, and then summarizes the distribution of the E2F4 target genes in the ranked gene list using a nonlinear random walk-based method. For each sample, BASE gives rise to an E2F4 score. A positive E2F4 score indicates that E2F4 targets tend to be highly expressed in the ranked gene list, implying high E2F4 activity in the sample. Conversely, a negative E2F4 score indicates that E2F4 targets tend to be lowly expressed in the ranked gene list, and therefore implying low E2F4 activity in the sample. In general, the E2F4 scores follow a bimodal distribution with two peaks on the positive and negative sides, respectively.
Statistical analysis
To investigate the effectiveness of E2F4 program for predicting prognosis, bladder cancer samples were dichotomized into E2F4>0 and E2F4<0 groups. Kaplan–Meier survival curves were derived from the Cox proportional hazard models (43). The difference between the survival curves of the two groups was compared with significance being estimated by using the log-rank test. Multivariate Cox proportional hazards models were used to examine the prognostic ability of the E2F4 program in the presence of potential confounding factors, including age, grade, and stage. For these models, E2F4 was treated as a binary variable, with 0 and 1 representing the E2F4<0 and E2F4>0 groups, respectively. The grade and stage variables were treated as integers, with stage ranging from 0 to 4 to represent Ta, T1, T2, T3, and T4 samples. Analyses were performed in R using the “survival” package. Specifically the “survfit” function was called to create Kaplan–Meier survival curvess, and the “survdiff” function was called to compare the difference between two survival curves.
Results
Overview of our analysis
Figure 1 shows a schematic diagram of our analysis. Given a gene expression dataset for a number of bladder tumor samples, we applied a method called BASE to infer the regulatory activities of E2F4 (denoted as E2F4 scores) in these samples. The E2F4 scores were calculated on the basis of the expression of a core set of E2F4 target genes identified from ChIP-seq experiments. When target genes exhibit a trend to be highly expressed in a sample, BASE results in a positive E2F4 score, indicating high E2F4 activity in this sample. Conversely, when target genes are lowly expressed, BASE results in a negative E2F4 score, indicating low E2F4 activity in the corresponding sample. The core E2F4 target genes represent a set of genes that are regulated by E2F4 in a non–tissue-specific manner. They were identified as the E2F4 targets shared in multiple human cell lines (K562, GM12878, and HeLa) defined from ChIP-seq data.
Figure 1.
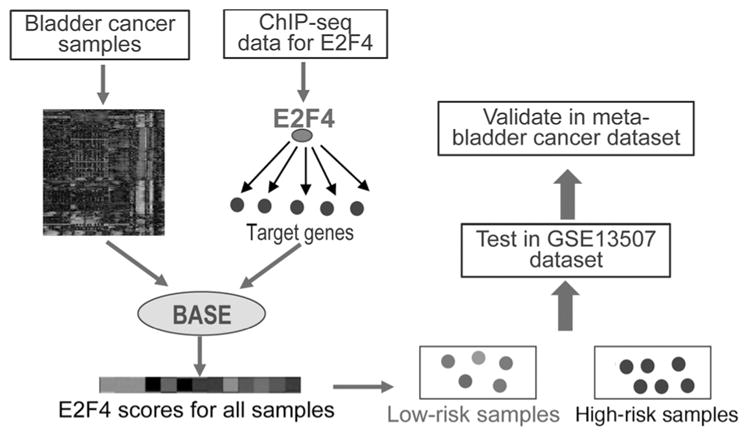
Schematic diagram of our analysis. The BASE algorithm is used to calculate E2F4 scores in bladder cancer samples based on the expression of E2F4 target genes.
Bladder tumor samples were then stratified into high-risk (E2F4>0) and low-risk (E2F4<0) groups based on their E2F4 scores. The survival times of the two groups were compared to examine whether E2F4 scores are predictive of bladder cancer prognosis. We first tested the E2F4 program for survival prediction in the GSE13507 dataset that contained expression profiles for normal and tumorous bladder samples (20). Different survival times were tested, including overall survival time (OS), cancer-specific survival time (CSS), recurrence-free survival time (RFS), and progression-free survival time (PFS). Then we validated our findings in two meta-bladder datasets that combined samples from multiple experiments using a one-channel platform and a two-channel platform, respectively.
E2F4 scores in different subsets of bladder samples
First, we compared the E2F4 activities in different subsets of samples contained in the GSE13507 dataset, which included normal bladder samples from nondiseased individuals, pathologically normal tissue surrounding bladder tumors, primary bladder tumor samples, and recurrent bladder tumor samples. As expected, the E2F4 scores are significantly higher in tumor samples (primary and recurrent) than in normal bladder samples (normal and surrounding; P = 2E–17, Wilcoxon rank sum test). As shown in Fig. 2A, 53% of primary (88/165) and 73% of recurrent tumor samples (16/23) have positive E2F4 scores, whereas the majority of normal samples have negative E2F4 scores: 86% of surrounding (50/58) and 100% normal bladder samples. Compared with the normal samples, surrounding samples show slightly higher E2F4 scores (P = 0.02; Wilcoxon rank sum test), suggesting these “surrounding” bladder samples might be contaminated with tumor cells. Compared with primary tumor samples, the recurrent tumor samples show higher E2F4 scores (P = 0.03; Wilcoxon rank sum test). The primary tumor samples were collected from patients with or without recurrence during follow-up. As shown in Fig. 2B, the primary tumor samples from recurrent patients have larger fraction of positive E2F4 scores than those from nonrecurrent patients (58% vs. 36%; P = 0.02, Fisher exact test), However, primary and recurrent tumor samples from recurrent patients do not exhibit significant difference in their E2F4 scores (Fig. 2C, P > 0.05, Wilcoxon rank sum test).
Figure 2.
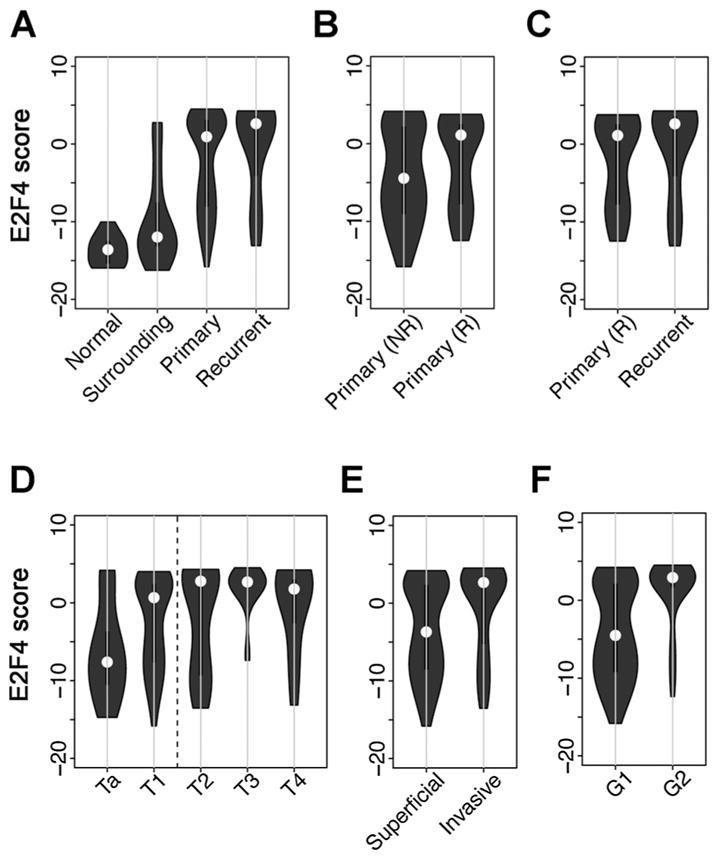
The distribution of E2F4 scores in different subsets of samples. A, normal bladder tissues, normal bladder surrounding bladder tumors, primary bladder tumors, and recurrent bladder tumors. B, primary bladder tumors from patients with and without recurrence. C, primary and recurrent bladder tumors from recurrent patients. D, different tumor stages. E, superficial (Ta and T1, also called NMIBC) and invasive bladder tumors (T2–4, also called MIBC). F, grade G1 and G2.
The primary tumor samples in this dataset are from different stages that include 24 Ta, 80 T1, 31 T2, 19 T3, and 11 T4 samples. Figure 2D shows the distribution of E2F4 scores in these five stage groups. As shown, the E2F4 scores demonstrate an increasing trend from Ta to T4. When superficial samples (Ta and T1) and invasive samples (T2–T4) were compared, we observed a significant difference in their E2F4 scores (P = 0.0007; Wilcoxon rank sum test) as shown in Fig. 2E. In addition, when primary tumor samples with different grade were compared, the G2 group showed significantly higher E2F4 scores than the G1 group (Fig. 2F, P = 8E–9, Wilcoxon rank sum test). Taken together, these results suggest that E2F4 scores of samples are highly correlated with their clinical factors, such as tumor stage, grade, and the recurrence of patients.
E2F4 program is predictive of survival of bladder cancer patients
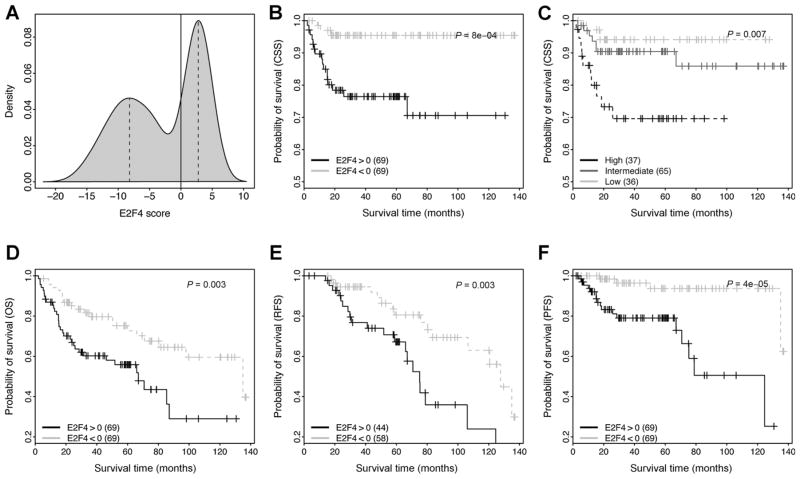
Next, we focused on the primary bladder tumor samples of the GSE13507 dataset, and utilized their E2F4 scores to predict patient survival. As the survival of patients can be complicated by the treatment to them, we excluded samples from patients treated with systemic chemotherapy, resulting in 138 primary samples. Fig. 3A shows that E2F4 scores have a bimodal distribution with positive and negative peaks, which enable us to stratify patients in two different ways. First, we simply divided patients into positive (E2F4>0) and negative (E2F4<0) groups (Fig. 3B). As shown, the E2F4>0 group shows significantly shorter CSS time than the E2F4<0 group (P = 0.0008). At the median follow-up time (40 months), 23% of E2F4>0 patients but only 4% E2F4<0 patients died from cancer. Second, we determined the E2F4 scores at the positive and the negative peaks (see dashed lines in Fig. 3A) and used them as the cut-off values to divide patients into high-, intermediate-, and low-risk groups (Fig. 3C). As shown, the three groups show a significant difference in their CSS times (P = 0.007).
Figure 3.
Application of E2F4 program to prognosis prediction in bladder cancer. A, the distribution of E2F4 scores in primary bladder tumor samples. B, stratification of patients into E2F4>0 and E2F4<0 groups, and then compare their CSS curves. C, stratification of patients into high, intermediate and low groups based on their E2F4 scores, and then comparing their CSS curves. The scores at the two dashed lines in (A) are used as cut-off values for defining groups. D, comparison of the overall survival (OS) curves between the E2F4>0 and E2F4<0 groups. E, comparison of the RFS curves between the E2F4>0 and E2F4<0 groups. Only NMIBC samples are used in this analysis. F, comparison of the PFS curves between the E2F4>0 and E2F4<0 groups. Note that only patients without systemic chemotherapy treatment are included in these analyses.
In addition to CSS time, we also tested the capacity of the E2F4 program for predicting OS (Fig. 3D), RFS (Fig. 3E, for NMIBC only) and PFS (Fig. 3F) of patients. As shown, E2F4 scores are predictive of all these measures of survival, with the highest accuracy achieved for PFS of patients. Moreover, we repeated the same analyses using all of the 165 primary tumor samples (i.e., without filtering out systemic chemotherapy– treated patients), and obtained similar results (Supplementary Fig. S1). Finally, we performed a multivariate Cox proportional hazards regression analysis to test for the association between E2F4 class (E2F4>0 or E2F4<0) and PFS when controlling for potential confounding factors, including age, grade, and stage. We applied this analysis to the 102 NMIBC samples that had not been treated by systemic chemotherapy, and found that E2F4 class remained significant (P = 0.02). This suggests that the E2F4 program provides additional information to traditional clinical variables when predicting PFS.
Application of E2F4 program to NMIBC and MIBC
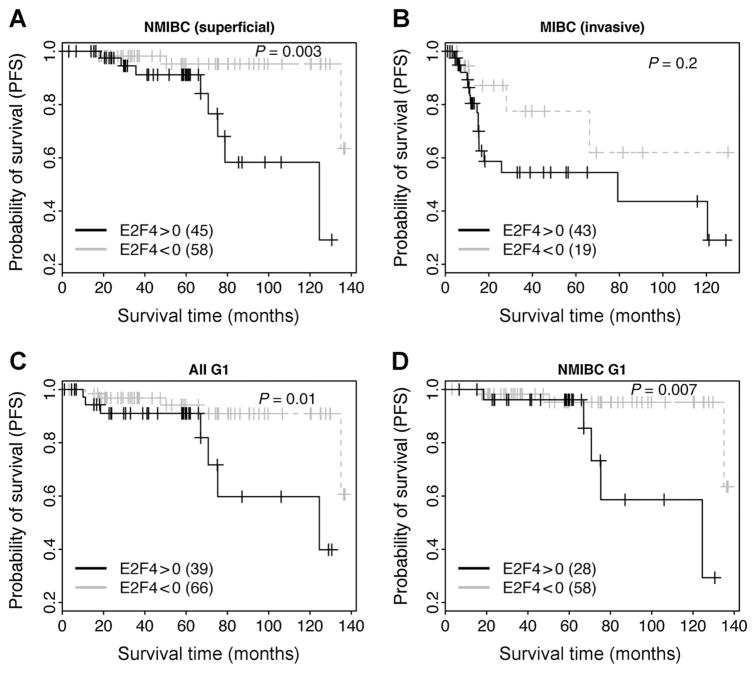
In the 165 primary bladder tumor samples, 103 are NMIBC (non–muscle-invasive at Ta or T1 stages, also called superficial tumor) and 52 are MIBC (muscle invasive at T2, T3, or T4 stages). After excluding systemic chemotherapy–treated patients, we obtained 102 NMIBC and 36 MIBC samples. We investigated the effectiveness of the E2F4 program for predicting PFS in both subtypes. Our results indicated that the program is valid in both NMIBC (Fig. 4A) and MIBC (Fig. 4B). It is known that tumor grade is correlated with patient survival, and we have shown in Fig. 2F that E2F4 scores were significantly different between G1 and G2 samples. Thus, we next tested the E2F4 program in 93 G1 samples without systemic chemotherapy treatment. As shown, E2F4>0 patients showed significantly shorter PFS times than E2F4<0 patients in all G1 samples (Fig. 4C) as well as in the NMIBC G1 samples (Fig. 4D).
Figure 4.
Application of E2F4 program to prognosis prediction in NMIBC and MIBC samples. The survival curves of E2F4>0 and E2F4<0 groups are compared in NMIBC (A), MIBC (B), grade G1 (C), and grade G1 NMIBC (D) samples. Note that only patients untreated by systemic chemotherapy are included in this analysis.
Similar results were identified when all primary tumor samples (with and without systemic chemotherapy) were used for above analysis (Supplementary Figure S2). However, lower predictive powers of the E2F4 program were observed for MIBC samples. The dataset contains 52 and 36 MIBC samples respectively before and after systemic chemotherapy exclusion. When all of the 52 MIBC samples were used, we no longer observed a significant difference in survival between the E2F4>0 and the E2F4<0 groups (P = 0.2; Supplementary Fig. S2B) in spite of more samples being included in Fig. 4B. This indicates that systemic chemotherapy does have an effect on the progression of patients and including treated samples complicates the prognostic analyses.
Application of E2F4 program to predicting intravesical therapy effectiveness in NMIBC
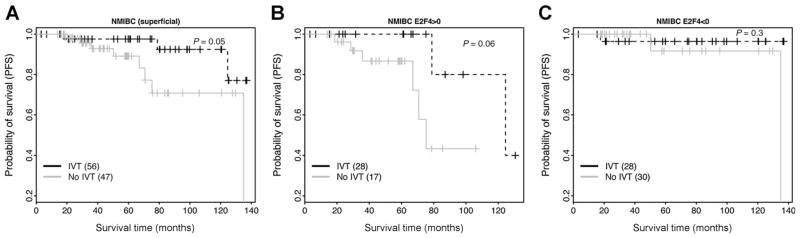
Some of the NMIBC samples in GSE13507 dataset received one cycle of intravesical BCG immunotherapy (IVT). A comparison between the IVT-treated and the IVT-untreated groups showed significantly longer PFS times of the former group, suggesting that, overall, NMIBC patients can benefit from IVT (Fig. 5A). We then examined whether the E2F4 signature can be used to predict the treatment effect of IVT. Specifically, we stratified the NMIBC patients into E2F4>0 and E2F4<0 groups. Survival analyses indicate that IVT can extend the survival times of the patients in the E2F4>0 group (Fig. 5B), but not of the patients in the E2F4<0 group (Fig. 5C). For the E2F4<0 group, all patients showed good prognosis with or without IVT. This suggests that applying IVT treatment to this group may not benefit patients. Thus, the E2F4 program might be used as a predictive marker for determining the effectiveness of IVT in treating NMIBC.
Figure 5.
E2F4 program is predictive of the efficacy of intravesical BCG immunotherapy in NMIBC. A, the survival curves of intravesical therapy-treated and untreated groups are compared all in A, E2F4>0 (B), and E2F4<0 NMIBC samples (C). IVT: intravesical BCG immunotherapy.
E2F4 scores in bladder cancer molecular subtypes
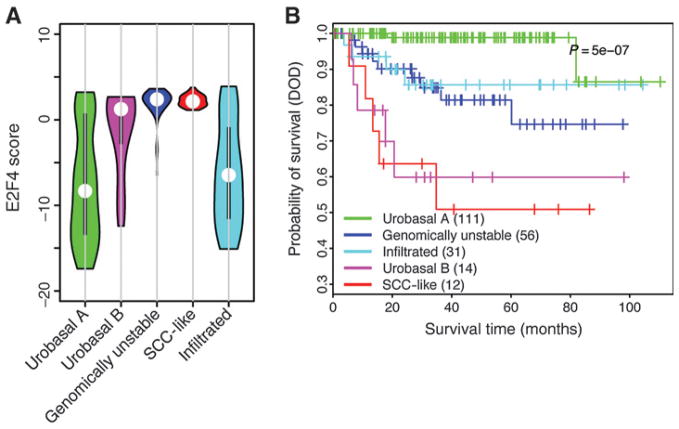
On the basis of gene expression profiles, bladder tumor samples can be classified into five different molecular subtypes: urobasal A, genomically unstable, urobasal B, squamous cell carcinoma-like (SCC-like), and an infiltrated class of tumors (21). These molecular subtypes showed distinct survival patterns. We calculated the E2F4 scores of samples from the GSE32894 dataset, in which the molecular subtypes of samples were carefully defined (Fig. 6A). We found that the urobasal A samples tend to have lower E2F4 scores, consistent with their good prognosis (Fig. 6B), whereas the SCC-like samples have highest E2F4 scores, which is known to be associated with poor prognosis. The other subtype with poor prognosis, urobasal B, also showed relatively high E2F4 scores. The infiltrated subtype showed intermediate prognosis, and samples of this subtype have intermediate E2F4 score. The genomically unstable subtype is associated with intermediate prognosis; however, we found that samples of this subtype tend to have high E2F4 scores. This suggests that prognosis is not fully determined by the proliferation of cells captured by the E2F4 program. It is also affected by some other factors, such as genome stability.
Figure 6.
The distribution of E2F4 scores in different molecular subtypes of bladder cancer. A, E2F4 score distribution. B, survival curves of different molecular subtypes.
Validation of E2F4 program in meta-bladder cancer datasets
To validate the findings obtained from the GSE13507 dataset, we investigated the effectiveness of the E2F4 program for progression prediction in two meta-bladder datasets, containing 482 and 240 samples measured by one-channel and two-channel arrays, respectively. To include as many samples as we could, we examined the OS time, for which data were available for the majority of samples.
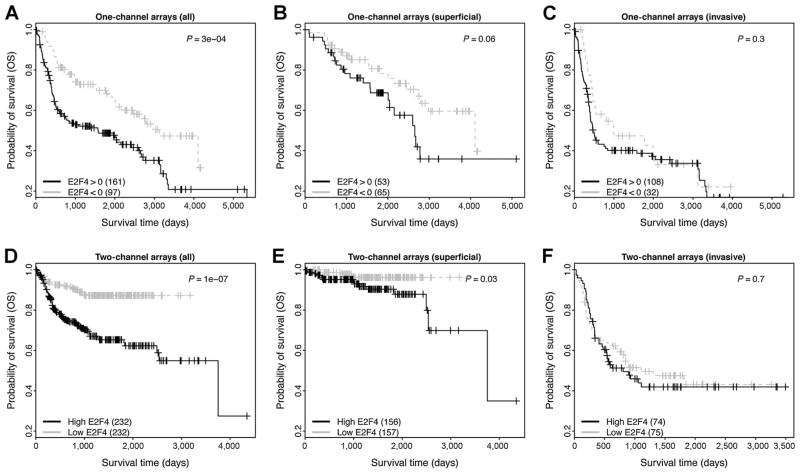
The capacity of the E2F4 program for predicting the OS time of patients was validated in both meta-datasets (Fig. 7). As shown, in the one-channel metadata the E2F4>0 group shows significantly shorter survival times than the E2F4<0 groups with P=3E–4 (Fig. 7A, log-rank test). Similarly, in the two-channel metadata the high-E2F4 group shows significantly shorter survival times than the low-E2F4 groups with P = 1E-7 (Fig. 7D, log-rank test). When we further divided samples into NMIBC (superficial) and MIBC (invasive), we found that the E2F4 program was more effective for predicting overall survival of NMIBC than MIBC samples. As shown, the program results in P = 0.06 (Fig. 7B) and P = 0.03 (Fig. 7E) in the two meta-datasets, respectively. For MIBC samples, the two groups stratified on the basis of E2F4 scores are not significantly different in their OS times (P > 0.1; Fig. 7C and F). This is caused by the fact that (i) OS time is less bladder cancer related than PFS time, and thus more difficult to predict (Fig. 3D and F); and (ii) some MIBC patients have been treated by chemotherapy, which complicates the analysis. It should be noted that due to the majority of samples in the two-channel metadata having negative E2F4 scores, we stratified samples for this dataset using the median E2F4 scores as the threshold. In all the two-channel arrays used in this analysis, RNA pooled from ten human cell lines was used as the reference. Thus, negative E2F4 scores indicate relatively lower E2F4 activities in bladder tumors with respect to the pooled RNA reference.
Figure 7.
Validation of E2F4 program in two meta-bladder cancer datasets. The survival curves of E2F4>0 and E2F4<0 groups are compared all in A, superficial (NMIBC; B) and invasive (MIBC; C) samples from the one-channel metadata, as well as from the two-channel metadata (D–F).
Discussion
In this analysis, we demonstrated the effectiveness of an E2F4 program for predicting prognosis of bladder cancer. We showed that samples with high E2F4 scores tend to have a poorer prognosis than those with low E2F4 scores. This trend is observed in both NMIBC and MIBC samples. The E2F4 program consists of 199 shared target genes of E2F4 that are identified on the basis of ChIP-seq data in three human cell lines: K562, GM06900 and HeLa. These genes are regulated by E2F4 in all three of the cell lines, representing a core non–tissue-specific target gene set. Although none of the cell lines are a bladder cancer cell line, we assume they can be used to infer the E2F4 regulatory activities in bladder tumor samples. Nevertheless, we would expect to improve this analysis if bladder tissue–specific E2F4 target genes are identified (e.g., from ChIP-seq in bladder cancer cell lines) and used for activity inference.
Our results support that the E2F4 program is predictive of the treatment effect of intravesical BCG immunotherapy. In intravesical therapy, drugs are directly inserted into the bladder rather than oral administration or injection into a vein. IVT can be either immunotherapy or chemotherapy depending on which drugs are used (44). As drugs delivered in this way mainly affect the cells inside of the bladder lining, intravesical therapy is used only for superficial bladder cancers (NMIBC, Ta and T1). BCG, is the most effective intravesical immunotherapy for treating early-stage bladder cancer (6). The exact mechanism by which BCG prevents bladder cancer recurrence is unknown, but it is thought that the presence of live but attenuated bacteria in the bladder might trigger a localized immune reaction that leads to clearance of residual cancer cells (45). It has been reported that intravesical BCG immunotherapy is only effective in less than 2 of 3 of the cases with NMIBC (46). Strikingly, our results indicates that the E2F4>0 patients but not E2F4<0 patients would benefit from intravesical BCG immunotherapy. Thus, the E2F4 program might be used as a predictive marker to predict whether intravesical BCG immunotherapy can benefit a specific NMIBC patient.
We find that prognostic analysis might be complicated by treatment. In GSE13507 dataset, 26 of 62 MIBC patients were treated by systemic chemotherapy (47). When only untreated MIBC samples were used for prognostic analysis, the E2F4>0 group showed shorter PFS time than the E2F4<0 group with P = 0.06 (Fig. 4B, log-rank test). In contrast, when all MIBC samples were used, a less significant difference was observed (P = 0.2) in spite of higher statistical power (more samples) in the analysis. This is presumably caused by the correlation between E2F4 scores and chemotherapy treatment effect. The E2F4>0 patients are associated with shorter survival time, meanwhile, they are more sensitive to chemotherapy than the E2F4<0 patients. As a consequence, for patients treated with chemotherapy, survival times of the E2F4>0 group will be extended more significantly than the E2F4<0 group, which obscures the difference between the two groups. This also explains why we did not observe a significant difference between the E2F4>0 and E2F4<0 groups in both of the two meta-bladder cancer datasets–chemotherapy- treated samples were included in the analyses due to the lack of treatment information for the majority of samples.
Going forward, we believe the E2F4 program has the potential to be applied in a clinical setting. Given a new bladder cancer sample, gene expression profile can be measured by RT-PCR, microarray, or RNA-seq experiment. After matching the distribution with a predefined control profile by quantile normalization, the E2F4 score in this sample can be calculated and used for prognostic prediction. The test can be further improved by selecting a subset of E2F4 targets that is highly correlated with E2F4 activity so that E2F4 scores can be calculated solely based on expression of these genes rather than the whole gene expression profiles.
In summary, we have defined an E2F4 program based on ChIP-seq and microarray expression data. The program is predictive of prognosis of patients in both NMIBC and MIBC. The program can also be used to predict the sensitivity of NMIBC patients to intravesical BCG immunotherapy.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by the American Cancer Society Research Grant, #IRG-82-003-30, the Centers of Biomedical Research Excellence (COBRE) grant GM103534, the National Center For Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001086, and by the start-up funding package provided to C. Cheng by the Geisel School of Medicine at Dartmouth College.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: C. Cheng
Development of methodology: C. Cheng
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): C. Cheng
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): C. Cheng, F.S. Varn, C.J. Marsit
Writing, review, and/or revision of the manuscript: C. Cheng, F.S. Varn, C.J. Marsit
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): C. Cheng
Study supervision: C. Cheng
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2015;65(5):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Pasin E, Josephson DY, Mitra AP, Cote RJ, Stein JP. Superficial bladder cancer: an update on etiology, molecular development, classification, and natural history. Rev Urol. 2008;10:31–43. [PMC free article] [PubMed] [Google Scholar]
- 4.Lindgren D, Frigyesi A, Gudjonsson S, Sjodahl G, Hallden C, Chebil G, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70:3463–72. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 5.National comprehensive cancer network (NCCN) NCCN Guidelines for Treatment of Cancer. http://www.nccn.org/professionals/physician_gls/pdf/bladder.pdf.
- 6.Alexandroff AB, Jackson AM, O’Donnell MA, James K. BCG immunotherapy of bladder cancer: 20 years on. Lancet. 1999;353:1689–94. doi: 10.1016/S0140-6736(98)07422-4. [DOI] [PubMed] [Google Scholar]
- 7.Hayes DF, Trock B, Harris AL. Assessing the clinical impact of prognostic factors: when is “statistically significant” clinically useful? Breast Cancer Res Treat. 1998;52:305–19. doi: 10.1023/a:1006197805041. [DOI] [PubMed] [Google Scholar]
- 8.Su SF, de Castro Abreu AL, Chihara Y, Tsai Y, Andreu-Vieyra C, Daneshmand S, et al. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladder cancer recurrence. Clin Cancer Res. 2014;20:1978–89. doi: 10.1158/1078-0432.CCR-13-2637. [DOI] [PubMed] [Google Scholar]
- 9.Park HS, Park WS, Bondaruk J, Tanaka N, Katayama H, Lee S, et al. Quantitation of Aurora kinase A gene copy number in urine sediments and bladder cancer detection. J Natl Cancer Inst. 2008;100:1401–11. doi: 10.1093/jnci/djn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sapre N, Anderson PD, Costello AJ, Hovens CM, Corcoran NM. Gene-based urinary biomarkers for bladder cancer: an unfulfilled promise? Urol Oncol. 2014;32:e9–17. doi: 10.1016/j.urolonc.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 2002;41:154–61. [PubMed] [Google Scholar]
- 13.Habel LA, Shak S, Jacobs MK, Capra A, Alexander C, Pho M, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006;8:R25. doi: 10.1186/bcr1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–34. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 15.Ravdin PM, Siminoff LA, Davis GJ, Mercer MB, Hewlett J, Gerson N, et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 2001;19:980–91. doi: 10.1200/JCO.2001.19.4.980. [DOI] [PubMed] [Google Scholar]
- 16.Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–6. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 17.Dyrskjot L, Zieger K, Kruhoffer M, Thykjaer T, Jensen JL, Primdahl H, et al. A molecular signature in superficial bladder carcinoma predicts clinical outcome. Clin Cancer Res. 2005;11:4029–36. doi: 10.1158/1078-0432.CCR-04-2095. [DOI] [PubMed] [Google Scholar]
- 18.Dyrskjot L, Zieger K, Real FX, Malats N, Carrato A, Hurst C, et al. Gene expression signatures predict outcome in non-muscle-invasive bladder carcinoma: a multicenter validation study. Clin Cancer Res. 2007;13:3545–51. doi: 10.1158/1078-0432.CCR-06-2940. [DOI] [PubMed] [Google Scholar]
- 19.Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB, et al. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol. 2010;28:2660–7. doi: 10.1200/JCO.2009.25.0977. [DOI] [PubMed] [Google Scholar]
- 20.Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS, Jeong P, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9:3. doi: 10.1186/1476-4598-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sjodahl G, Lauss M, Lovgren K, Chebil G, Gudjonsson S, Veerla S, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18:3377–86. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- 22.Catto JW, Abbod MF, Wild PJ, Linkens DA, Pilarsky C, Rehman I, et al. The application of artificial intelligence to microarray data: identification of a novel gene signature to identify bladder cancer progression. Eur Urol. 2010;57:398–406. doi: 10.1016/j.eururo.2009.10.029. [DOI] [PubMed] [Google Scholar]
- 23.Kern SE. Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer Res. 2012;72:6097–101. doi: 10.1158/0008-5472.CAN-12-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldron L, Haibe-Kains B, Culhane AC, Riester M, Ding J, Wang XV, et al. Comparative meta-analysis of prognostic gene signatures for late-stage ovarian cancer. J Natl Cancer Inst. 2014;106(5):dju049. doi: 10.1093/jnci/dju049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risolino M, Mandia N, Iavarone F, Dardaei L, Longobardi E, Fernandez S, et al. Transcription factor PREP1 induces EMT and metastasis by controlling the TGF-beta-SMAD3 pathway in non-small cell lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111:E3775–84. doi: 10.1073/pnas.1407074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh JE, Toniolo PA, Frank DA. Targeting transcription factors: promising new strategies for cancer therapy. Curr Opin Oncol. 2013;25:652–8. doi: 10.1097/01.cco.0000432528.88101.1a. [DOI] [PubMed] [Google Scholar]
- 27.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–9. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 28.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–94. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 29.Khaleel SS, Andrews EH, Ung M, Direnzo J, Cheng C. E2F4 regulatory program predicts patient survival prognosis in breast cancer. Breast Cancer Res. 2014;16:486. doi: 10.1186/s13058-014-0486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu M, Liu CC, Cheng C. REACTIN: regulatory activity inference of transcription factors underlying human diseases with application to breast cancer. BMC Genomics. 2013;14:504. doi: 10.1186/1471-2164-14-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aparicio O, Geisberg JV, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Cell Biol. 2004 Sep;Chapter 17(Unit 17.7) doi: 10.1002/0471143030.cb1707s23. [DOI] [PubMed] [Google Scholar]
- 32.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 33.Cheng C, Yan X, Sun F, Li LM. Inferring activity changes of transcription factors by binding association with sorted expression profiles. BMC Bioinformatics. 2007;8:452. doi: 10.1186/1471-2105-8-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowland BD, Bernards R. Re-evaluating cell-cycle regulation by E2Fs. Cell. 2006;127:871–4. doi: 10.1016/j.cell.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Schwemmle S, Pfeifer GP. Genomic structure and mutation screening of the E2F4 gene in human tumors. Int J Cancer J Int du Cancer. 2000;86:672–7. doi: 10.1002/(sici)1097-0215(20000601)86:5<672::aid-ijc11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 36.Souza RF, Yin J, Smolinski KN, Zou TT, Wang S, Shi YQ, et al. Frequent mutation of the E2F-4 cell cycle gene in primary human gastrointestinal tumors. Cancer Res. 1997;57:2350–3. [PubMed] [Google Scholar]
- 37.Wang D, Russell JL, Johnson DG. E2F4 and E2F1 have similar proliferative properties but different apoptotic and oncogenic properties in vivo. Mol Cell Biol. 2000;20:3417–24. doi: 10.1128/mcb.20.10.3417-3424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee BK, Bhinge AA, Iyer VR. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res. 2011;39:3558–73. doi: 10.1093/nar/gkq1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng C, Min R, Gerstein M. TIP: a probabilistic method for identifying transcription factor target genes from ChIP-seq binding profiles. Bioinformatics. 2011;27:3221–7. doi: 10.1093/bioinformatics/btr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riester M, Taylor JM, Feifer A, Koppie T, Rosenberg JE, Downey RJ, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18:1323–33. doi: 10.1158/1078-0432.CCR-11-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 43.Cox DR. Regression Models and Life-Tables. J Roy Statist Soc Ser B. 1972;34:187–220. [Google Scholar]
- 44.Shelley MD, Mason MD, Kynaston H. Intravesical therapy for superficial bladder cancer: a systematic review of randomised trials and meta-analyses. Cancer Treat Rev. 2010;36:195–205. doi: 10.1016/j.ctrv.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Naoe M, Ogawa Y, Takeshita K, Morita J, Iwamoto S, Miyazaki A, et al. Bacillus Calmette-Guerin-pulsed dendritic cells stimulate natural killer T cells and gammadeltaT cells. Int J Urol. 2007;14:532–8. doi: 10.1111/j.1442-2042.2006.01697.x. discussion 8. [DOI] [PubMed] [Google Scholar]
- 46.Zlotta AR, Fleshner NE, Jewett MA. The management of BCG failure in non-muscle-invasive bladder cancer: an update. Can Urol Assoc J. 2009;3:S199–205. doi: 10.5489/cuaj.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta S, Mahipal A. Role of systemic chemotherapy in urothelial urinary bladder cancer. Cancer Control. 2013;20:200–10. doi: 10.1177/107327481302000308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.