Abstract
Integrative and conjugative elements (ICEs) of the ICESa2603 family have been isolated from several species of Streptococcus spp.; however, the comparative genomic and evolutionary analyses of these particular ICEs are currently only at their initial stages. By investigating 13 ICEs of the ICESa2603 family and two ICESa2603 family-like ICEs derived from diverse hosts and locations, we have determined that ICEs comprised a backbone of 30 identical syntenic core genes and accessory genes that were restricted to the intergenic sites or the 3′-end of the non-conserved domain of core genes to maintain its function. ICESa2603 family integrase IntICESa2603 specifically recognized a 15-bp att sequence (TTATTTAAGAGTAAC) at the 3′-end of rplL, which was highly conserved in genus Streptococcus. Phylogenetic analyses suggest that extensive recombination/insertion and the occurrence of a hybrid/mosaic in the ICESa2603 family were responsible for the significant increase in ICE diversity, thereby broadening its host range. Approximately 42.5 and 38.1% of the tested Streptococcus suis and Streptococcus agalactiae clinical isolates respectively contained ICESa2603 family Type IV secretion system (T4SS) genes, and 80.5 and 62.5% of which also respectively carried intICESa2603, indicating that ICESa2603 family is widely distributed across these bacteria. Sequencing and conjugation transfer of a novel sequence type ST303 clinical S. suis isolate HB1011 demonstrated that the 89K-subtype ICESsuHB1011 retained its transferrable function, thereby conferring tetracycline and macrolide resistance.
Keywords: integrative and conjugative elements, antimicrobial resistance, horizontal gene transfer, ICESa2603 family, erm(B), tet(O), Streptococcus suis, Streptococcus agalactiae
Introduction
Integrative and conjugative elements (ICEs) are self-transmissible mobile genetic elements (MGEs) that primarily reside in the host cell's chromosome, yet have the ability to be transferred between cells by conjugation (Burrus et al., 2002; Burrus and Waldor, 2004). ICEs are considered as mosaic elements with both phage- and plasmid-like features that can integrate into and replicate with the host cell's chromosomes similar to those observed in bacteriophages, as well as transfers, via conjugation in plasmids. The number of identified ICEs continues to increase with the exponential expansion of sequenced complete genomes (Te Poele et al., 2008; Wozniak and Waldor, 2010). Guglielmini et al (Guglielmini et al., 2011) previously described the prevalence and diversity of ICEs by conducting bioinformatics analysis of clustered conjugative apparatus modules in various chromosomal locations. Based on this definition, 18% of sequenced prokaryotic genomes contain at least one ICE, implying that the role of ICEs in horizontal gene transfer is more important than previously conceived.
An ICE typically comprises three distinct modular structures that mediate its integration and excision, conjugation, and regulation. In addition, ICEs contain genes that confer specific phenotypes that are related to its existence in its hosts such as resistance to antibiotics and heavy metals. Therefore, ICEs are important vectors for the horizontal transfer of genetic information, thereby facilitating rapid bacterial evolution (Bi et al., 2011; Rodriguez-Blanco et al., 2012). The SXT/R391 family is the most extensively studied ICE family that was initially identified in Vibrio cholerae O139 (Waldor et al., 1996). To date, more than 40 members of this family have been identified in different clinical and environmental Vibrio species, as well as related gammaproteobacteria (Mata et al., 2011; Rodriguez-Blanco et al., 2012). The SXT/R391 family shares a core structure containing 52 conserved genes and a common chromosomal integration site at the 5′-end of the prfC gene, with other parts all integrating into 3′-ends of distinct element-specific tRNA gene loci (Wozniak et al., 2009; Wozniak and Waldor, 2010). All known elements contain variable DNAs that are inserted into specific positions of the backbone (hot spots) and encoding various beneficial traits such as antibiotic resistant determinants including but not limited to floR, strBA, sul2, dfrA1, dfr18, tetA′, kanR, and an emerging extended-spectrum cephalosporin resistance gene blaCMY−2 (Harada et al., 2010). SetR, a homolog of the phage λ repressor CI, derepresses the master transcriptional activators required for SXT transfer, including the int and tra operons (Daccord et al., 2012). Beaber et al. (2004) have shown that DNA-damaging agents mitomycin C and quinolinones which induce the host SOS response, increase the transfer of SXT by several hundred times. The horizontal dissemination of antibiotic resistance genes through ICEs under selective pressure poses new challenges to the use of antimicrobial agents. Extensive knowledge of SXT/R391 family ICEs prompted us to explore the genetic features of other family ICEs.
Comparative analysis of the genomes of different isolates of Streptococcus agalactiae has revealed that nearly 2/3 of its regions of diversity in the isolates consist of ICEs or ICE-like elements (Brochet et al., 2008), which include two of the largest families, namely, Tn916 and ICESa2603. The Tn916 family is extensively distributed across various organisms, including important human pathogens such as Enterococcus faecalis, Clostridium difficile, Staphylococcus aureus, and Streptococcus spp. (Roberts and Mullany, 2011). Furthermore, its transfer mechanism has been well studied (Roberts and Mullany, 2009). In contrast, the ICESa2603 family, although identified in various Streptococcus species, including S. agalactiae and Streptococcus suis, has not been comprehensively investigated. In the current study, we first comparatively analyzed 13 ICEs of the ICESa2603 family and two ICESa2603 family-like ICEs to perform an in-depth genetic characterization of the core genes that are present in all members of this ICE family. Next, we investigated the potential accessory functions encoded by the variable DNA harbored by these mobile elements as well as the conserved att site for integration and the oriT site for origination transfer. Phylogenetic analyses of the 30 core genes allowed us to classify the 15 ICEs were into four subgroups. Furthermore, the distribution of these ICEs was determined in S. suis and S. agalactiae. Finally, an experimental transfer of a swine origin 89K-like subgroup transferable ICE, ICESsuHB1011, was introduced. It is worth noting that isolates with erm(B)- and tet(O)-carrying transferable 89K-subtype ICEs were detected in different swine and bovine farms of China, thereby disseminating tetracycline and macrolide resistance genes and contributing to human pathogenesis (Tang et al., 2006).
Materials and methods
Bacterial strains and genomic DNA extraction
A total of 73 S. suis isolates from swine (including one S. suis strain from ATCC) and 21 S. agalactiae isolates from bovine (including one S. agalactiae strain from ATCC) were used in the present study (Table S1). Isolates were routinely grown overnight on tryptic soya broth or agar plates supplemented with 5% calf serum at 37°C. The bacterial culture was centrifuged (10,000 g for 5 min at room temperature), and the pellets were harvested and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) supplemented with lysozyme (5 mg/mL) and incubated for 30 min at 37°C. Genomic DNA was prepared using a Bacteria DNA kit (Omega, Norcross, GA, USA), following the manufacturer's instructions. The extracted DNA was used as template for PCR and sequencing.
In silico identification of the ICESa2603 family
To identify and compare the ICESa2603 family ICEs, the reference genomes of ICESa2603 family were obtained from NCBI (Table 1). ICESa2603, ICESde3396, ICESdy12394-1, ICESpa43144-1, ICESthJIM8232-1, ICESsu32457, ICESsuBM407-2, and ICESsuSC84 were previously classified as members of the ICESa2603 family (Bi et al., 2011; http://db-mml.sjtu.edu.cn/ICEberg/index.php). ICESa09mas018883, ICESsu05ZYH33-1 (also designated as 89K according to its size), ICESsu98HAH33-1, ICESsuD9, ICESsuSS12, and ICESsuHB1011 were grouped into this family because these encode the integrase gene that is closely related to intICESa2603 and share significant sequence alignment and syntenic core structure. ICESsuT15 and ICESluvan were identified as ICESa2603 family-like ICEs because these encoded a serine recombinase (SR) family integrase gene instead of the tyrosine family integrase gene, intICESa2603.
Table 1.
Properties of the ICESa2603 family ICEs.
| ICE name | Host strain | Accession | Size (bp) | % GC | Origin | References |
|---|---|---|---|---|---|---|
| ICESa2603 | S. agalactiae 2603V/R | AE009948.1 | 54349 | 38 | United States | Tettelin et al., 2002 |
| ICESde3396 | S. dysgalactiae subsp. equisimilis NS3396 | EU142041.1 | 63668 | 31 | Australia, 2001 | Davies et al., 2005, 2009 |
| ICESdy12394-1 | S. dysgalactiae subsp. equisimilis ATCC12394 | CP002215.1 | 50568 | 39 | United States, 1939 | Suzuki et al., 2011 |
| ICESpa43144-1 | S. pasteurianus ATCC43144 | AP012054.1 | 58476 | 37 | United States | Lin et al., 2011 |
| ICESthJIM8232-1 | S. thermophilus JIM8232 | FR875178.1 | 50501 | 37 | France, 2002 | Delorme et al., 2011 |
| ICESa09mas018883 | S. agalactiae 09mas018883 | HF952104.1 | 46903 | 37 | – | Zubair et al., 2013 |
| ICESsu32457 | S. suis 32457 | FR823304.2 | 54879 | 50.5 | Italy, 2007 | Palmieri et al., 2012 |
| ICESsu05ZYH33-1 | S. suis 05ZYH33 | CP000407.1 | 88851 | 37 | China, 2005 | Chen et al., 2007 |
| ICESsu98HAH33-1 | S. suis 98HAH33 | CP000408.1 | 89154 | 37 | China, 1998 | Chen et al., 2007 |
| ICESsuSC84 | S. suis SC84 | FM252031.1 | 89165 | 37 | China, 2005 | Holden et al., 2009 |
| ICESsuBM407-2a | S. suis BM407 | FM252032.1 | 80320 | 36 | Vietnam, 2004 | Holden et al., 2009 |
| ICESsuD9 | S. suis D9 | CP002641.1 | 55989 | 39.8 | China | Zhang et al., 2011 |
| ICESsuSS12 | S. suis SS12 | CP002640.1 | 64284 | 38.2 | China | Zhang et al., 2011 |
| ICESsuT15a,b | S. suis T15 | CP006246.1 | 71412 | 36.3 | – | – |
| ICESluvanb | S. lutetiensis 5-F9 | HE963029.1 | 94189 | 44 | Netherlands | Bjorkeng et al., 2013 |
| ICESsuHB1011c | S. suis HB1011 | – | – | – | China, 2010 | This study |
The SNF2 family protein was absent (ICESsuBM407-2) or partially absent (ICESsuT15).
ICESsuT15 and ICESluvan with a serine recombinase (SR) family integrase were referred as ICESa2603 family-like ICEs.
ICESsuHB1011 sequence was not complete.
Phylogenetic analyses
The ICESa2603 family and family-like ICEs were aligned using ClustalW (Larkin et al., 2007) with default settings. Nucleotide and amino acid conservations were assessed using the appropriate BLAST algorithms. MAUVE 2.4 (Darling et al., 2004) and ACT software (Carver et al., 2005) were used to identify core genes. The core genes of each ICE were further aligned with the corresponding genes of ICESa2603 to calculate nucleic acid percentage identity (Figure 1A).
Figure 1.
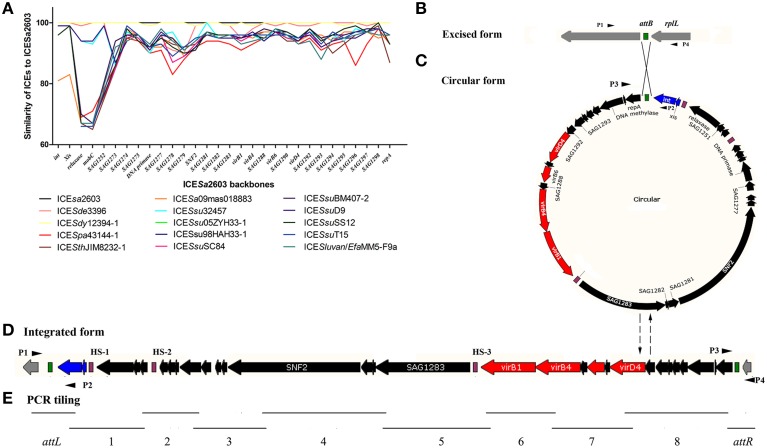
Core genes identity (%) of the ICESa2603 family and schematic diagram of the integration and excision forms. The attB (B), attICE (C), attL, and attR (D) sites are shown as green rectangles. The core genes are shown by navy (int and xis), red (T4SS genes), and black (other core genes). The HS sites are shown as purple rectangles. (A); Core genes identity (%) of each ICE of ICESa2603 family to that of ICESa2603. (B–D); Schematic diagram of the integration and excision forms of the ICESa2603 family. (E); Schematic diagram of PCR tiling to detect the presence of ICEs fragments.
To obtain an overview of the relationship of the ICESa2603 family and family-like ICEs and its possible evolution, phylogenetic trees were generated from the alignment of whole core genes or each core gene using the neighbor-joining method with bootstrapping (N = 1000) parameters by thesoftware, MEGA6 (Tamura et al., 2013) and FigTree v1.4.2.
Evaluation of the att and oriT sites
To identify the putative att sites, genomic sequences of the ICE-carrying strains were analyzed for the presence of directly repeated sequences flanking the ICEs. The ICESa2603 family-like ICESsuT15, where there are no conserved directly repeated sequences presented, the putative att site was predicted by comparing the junction sequence with S. suis P1/7.
The predicted oriT site of ICESsu05ZYH33-1 was first identified by Li et al. (2011). We determined that the oriT site is conserved in all other ICESa2603 family members. MEGA 6 (Tamura et al., 2013) and BioEdit v7 were used to align and mapthe oriT site the members of the ICESa2603 family.
PCR and sequencing
To determine the presence of ICESa2603 family ICEs in S. suis and S. agalactiae, primers for the ICESa2603 core genes were designed and used in PCR analysis (Table S2). The integrated form and the extrachromosomal circular form of the ICEs of S. suis and S. agalactiae were detected by using combination primers, P1–P4 (Figures 1B–D and Table S2). PCR was also performed to detect the presence of ICE fragments in S. suis and S. agalactiae by PCR tiling assay as described previously (Sitkiewicz et al., 2011) using specific oligonucleotide primers (Figure 1E and Table S2). The products of a representative S. suis strain, HB1011, with the ICEs backbone as determined by PCR tiling were sequenced by Genewiz, Inc. (Suzhou, Jiangsu, China).
Antimicrobials and susceptibility tests
Tetracycline, erythromycin, streptomycin, and rifampin (Sigma, St. Louis, MO, USA) were used in the present study. MICs were determined according to the guidelines described by the Clinical and Laboratory Standards Institute (CLSI, 2010).
Conjugation experiments
S. suis BAA-853 and S. agalactiae G9, which were susceptible to tetracycline and erythromycin, were used to generate a rifampin- and streptomycin-resistant phenotype by using the stepwise-induced method with sub-MICs of rifampin and streptomycin (Haenni et al., 2010). In the mating experiments, the above two induced strains (tetracycline- and erythromycin-susceptible but rifampin- and streptomycin-resistant) were used as recipients and S. suis HB1011 (tetracycline- and erythromycin-resistant but rifampin- and streptomycin- susceptible) with the ICESsuHB1011 was utilized as donor. Filter mating assays were performed as previously described (Li et al., 2011). The transconjugant was further confirmed by PCR, sequencing, and MLST typing.
Results
General features of the ICESa2603 family
A list of 13 ICEs of the ICESa2603 family and two ICESa2603 family-like ICEs from the NCBI complete genome database were analyzed and compared in this study (Table 1). An ICE that encodes an integrase gene closely related to intICESa2603, defined as having >60% gene or protein homology, and has significant sequence alignment (60% nucleic acid identity of core genes) and syntenic core structure were classified as a member of ICESa2603 family (Bi et al., 2011; Figure 2A) (http://db-mml.sjtu.edu.cn/ICEberg/index.php). Although ICESsuT15 and ICESluvan contained the backbone sequences of the ICESa2603 family, its integrase genes belonged to the SR family instead of the tyrosine family site-specific integrase of intICESa2603, and these were referred to as ICESa2603 family-like ICEs (Figure 2B). The strains, which were originally isolated from different countries around the world, belonged to six species of the Streptococcus spp. (Table 1).
Figure 2.
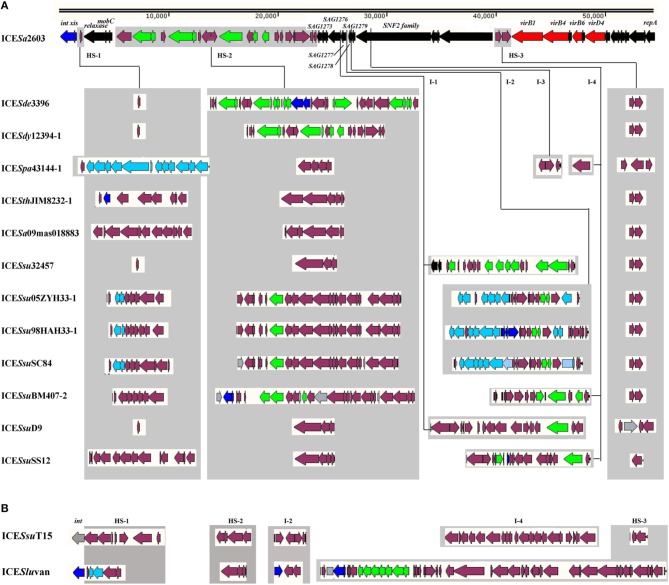
Genomic organization of the ICESa2603 family. HS-1, HS-2, and HS-3 were the hotspots of ICESa2603 family, which were presented in all ICEs. I-1, I-2, I-3, and I-4 could also receive additional genes in part of the ICEs. Genes are shown in different colors: black, ICEs core genes; navy, int and xis; azure, nisin operon genes; red, T4SS genes; green, antibiotic and heavy metal resistance genes; purple, other cargo genes. (A); ICESa2603 family ICEs. (B); ICESa2603 family-like ICEs.
The ICEs of the ICESa2603 family were first compared by using the MAUVE 2.4 (Darling et al., 2004) and ACT software (Carver et al., 2005) to visualize conserved and variable regions. All ICEs, with sizes ranging from 46,903 bp (ICESa09mas018883) to 89,165 bp (ICESsuSC84), shared a common conserved structure and had variable regions (Table 1 and Figure 2A). The syntenic core structure of the ICESa2603 family backbone (33 kb) contained 30 conserved core genes (Figures 1A, 2). There were three sites within the conserved ICESa2603 family structure for inserting variable DNAs in all of the ICEs (Figure 2). The three sites were named as HS-1, HS-2, and HS-3 (i.e., hotspots) for ICE acquisition of new genes of resistance to antibiotics and heavy metals. The three hotspots were located in the intergenic region of the ICE backbone, suggesting that the acquisition of these variable DNA regions did not interrupted core ICE gene. Additional DNA was inserted into the 3′-end non-conserved region of gene SAG1277 (Gene ID: 1014084), SAG1278 (Gene ID: 1014085), SAG1279 (Gene ID: 1014086), or SNF2 (Gene ID: 1014087) of some ICEs (Figure 2). The four inserted sites were designated as I-1, I-2, I-3, and I-4, respectively. The function of SAG1277, SAG1278, and SAG1279 remains unclear.
The ICE core genes
The present study has identified 30 core genes in 13 ICEs, which comprised the recombination module, the conjugation module, and various other genes of unknown functions. Integrases were members of the tyrosine recombinase family that contained a signature R·· H·· RH·· Y active site residue within its C-terminal catalytic domain. ICESa2603 family integrase IntICESa2603 mediates site-specific recombination between identical attICE and attB sites and integration into the 3′-end of 50S ribosomal subunit protein L7/L12 gene (rplL) (Haenni et al., 2010), which was determined to be conserved in all Streptococcus spp. (Table S3).
The conjugative transfer module of the ICESa2603 family was grouped into two parts clustered. The first cluster comprised 3 genes (SAG1250, SAG1251, and SAG1252 in S. agalactiae 2603V/R), which were annotated as Tn5252 orf4 relaxase, Tn5252 orf9 mobC, and Tn5252 orf10, respectively. Protein SAG1250 showed similarities to various prokaryotic DNA relaxases that covalently bind to and nick the DNA at the origin of transfer (oriT) with the help of the auxiliary MobC to initiate DNA processing (Lanka and Wilkins, 1995), which include ORF4 relaxase (AAC98434.1) of conjugative transposon Tn5252 (52% identity and 72% similarity) and MobA (AAQ55244.1) of S. aureus plasmid pC223 (22% identity and 41% similarity). Protein SAG1251, which is located adjacent to relaxase, shared 57% identity and 79% similarity to Tn5252 ORF9 MobC (AAC98435.1) and 27% identity and 50% similarity to MobC (AAQ55243.1) of S. aureus plasmid pC223. The other part of the conjugation module belonged to a type IV secretion system (T4SS). The VirD4 coupling protein (40% identity to Agrobacterium tumefaciens VirD4), which functions as a substrate receptor, binds to the ICE DNA-relaxase complex (relaxosome) and presents it to the mating pair formation (Mpf) channels. Compared to Agrobacterium tumefaciens Vir T4SS (Christie, 1997), which are assembled from virB1–VirB11, only VirB1 (transglycosylase, 18% identity), VirB4 (ATPase, 41% identity), and VirB6 (inner-membrane protein, 12% identity) have been identified among the Mpf channels of the ICESa2603 family (Figure 2).
Additional conserved genes were located adjacent to the T4SS genes. Upstream of the virD4 gene were genes coding for cytosine DNA methylase (SAG1297), replication initiation factors (SAG1299), and other unknown conserved genes, whereas downstream of the cluster were genes of the SNF2 family (SAG1280), DNA primase (SAG1276), and other unknown conserved genes.
Variable DNA regions
In addition to conserved core genes in all ICEs analyzed, variable DNA regions were also detected in the ICE backbone sequences (Figure 2). Most of the variable DNA sequences were inserted at three intergenic hotspots (HS), namely, HS-1, HS-2, and HS-3. The DNA content of various ICEs in HS was highly identical. For example, ICESa2603, ICESde3396, ICESdy12394-1, ICESsu32457, and ICESsuD9 had identical contents in HS-1. The DNA contents in HS-2 were more variable (only ICESsu32457 and ICESsuD9 shared identical genes), whereas in HS-3, these were more conserved (only ICESpa43144-1, ICESsuD9, and ICESsuSS12 varied from the other ICEs). There were other exogenous DNA insertion sites (I), namely, I-1, I-2, I-3, and I-4 in at the 3′-end of the non-conserved region of genes SAG1277, SAG1278, SAG1279, and SNF2 in some ICEs, respectively. For example, ICESsu32457 and ICESsuD9 had exogenous DNAs in I-1, whereas ICESsu05ZYH33-1, ICESsu98HAH33-1, ICESsuSC84, ICESsuT15, and ICESluvan had variable DNAs in I-2. Additional DNA inserts were identified at the I-3 site in ICESpa43144-1 and at the I-4 site in ICESpa43144-1, ICESsuBM407-2, ICESsuSS12, and ICESsuT15.
Other notable accessory genes in this family were identified as resistance genes for antibiotics and heavy metals, intact or remnant lantibiotic biosynthesis operon, and other MGEs such as IS elements and Tn916-like elements (Figure 2). The antibiotic and heavy metal resistance genes in ICEs are presented in Table 2. ICESa2603, ICESde3396, and ICESdy12394-1 carry heavy metal resistance genes, whereas ICEs from S. suis mostly contain macrolide resistance gene erm(B) and/or tetracycline resistance gene tet(M) or tet(O). ICESpa43144-1, ICESsu05ZYH33-1, ICESsu98HAH33-1, ICESsuSC84, and ICESluvan had remnant nisin ORFs, in which a two-component signal transduction system nisK/nisR, is essential for full virulence of highly invasive S. suis serotype 2 (Li et al., 2008; Wang et al., 2014). In the first four ICEs, remnant nisin ORFs were presented in two variable regions at the insertion sites of HS-1 and I-2 (Figure 2A).
Table 2.
Attachment site analysis and resistance profile of the ICESa2603 family.
| ICEs name | % Identity to Int2603 | % Identity to rplL2603 | attB (3) | Putative att sites | Resistance profile |
|---|---|---|---|---|---|
| ICESa2603 | 100 | 100 | rplL | TTATTTAAGAGTAAC | Hg, Cd, Cu |
| ICESde3396 | 100 | – | – | – | Cd, Cu, As |
| ICESdy12394-1 | 100 | 84 | rplL | TTATTTAAGAGTGAT | Cd, Cu |
| ICESpa43144-1 | 96 | 84 | hypothetical protein | TTATTTAAGAGTAAC | |
| ICESthJIM8232-1 | 96 | 88 | rplL | TTATTTAAGAGTAAC | |
| ICESa09mas018883 | 81 | 99 | rplL | TTATTTAAGAGTAAC | |
| ICESsu32457 | 99 | –a | rplL | TTATTTAAGAGTAAC | erm(B), aadE, aphA, tet(40), tet(O/W/32/O) |
| ICESsu05ZYH33-1 | 96 | 84 | rplL | TTATTTAAGAGTAAC | tet(M), aadE |
| ICESsu98HAH33-1 | 96 | 84 | rplL | TTATTTAAGAGTAAC | tet(M), aadE |
| ICESsuSC84 | 96 | 84 | rplL | TTATTTAAGAGTAAC | tet(M), aadE |
| ICESsuBM407-2 | 96 | 84 | rplL | TTATTTAAGAGTAAC | tet(M), tet(L), tet(O), erm(B), aadE, cat |
| ICESsuD9 | 99 | 84 | rplL | TTATTTAAGAGTAAC | tet(O), erm(B) |
| ICESsuSS12 | 96 | 84 | rplL | TTATTTAAGAGTAAC | tet(O), erm(B) |
| ICESsuT15b | – | 84 | SSU0877 in S. suis P1/7 | CCTCTTATGTCAAGTAACTG (attL) CATTATTATGACACAATCCC (attR) | |
| ICESluvanb | – | –a | rumA | CACGTGGAGTGCGTAGTGTT (attL) TTCTCAAGGACCAGACAACA (attR) | van resistance operon |
| ICESsuHB1011c | 98 | – | rplL | TTATTTAAGAGTAAC | tet(O), erm(B) |
The rplL sequences were not indicated.
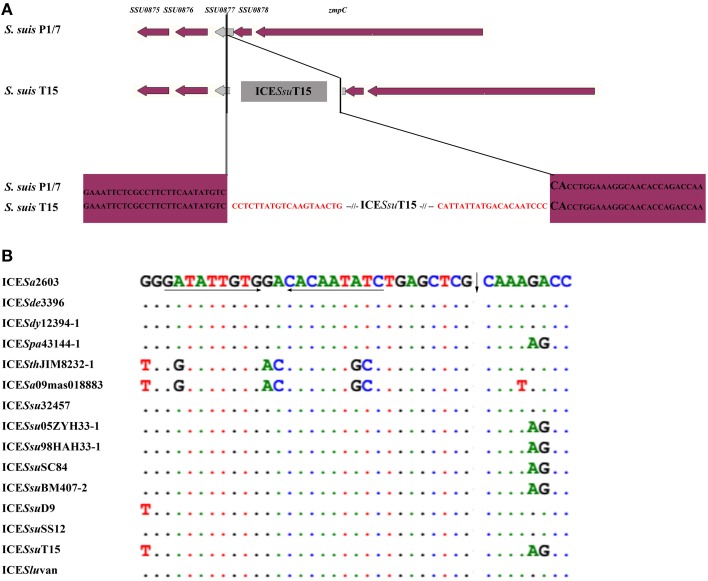
The ICESsuT15 and ICESluvan's Int belonging to serine recombinase (SR) family shared low homology with the ICESa2603 tyrosine family site-specific integrase. ICESluvan was inserted in rum, and the att site was experimentally confirmed by Bjorkeng et al. (2013). ICESsuT15 was inserted in the 344 sites of SSU0877 in S. suis P1/7, the putative att site was predicted, and the Int of ICESsuT15 and ICESluvan may recognize CA dinucleotide in certain region of chromosome sites.
ICESsuHB1011 sequence was not complete and the rplL sequences were not known.
MGEs were also found to be inserted in HS or I sites. For example, IS116 and IS4 family IS elements were inserted in the I-4 region of ICESpa43144-1 and ICESsuD9, respectively (Figure 2A). Moreover, a Tn916-like element was inserted in HS-2 of ICESsu05ZYH33-1, ICESsu98HAH33-1, ICESsuSC84, and ICESsuBM407-2 (Figure 2A). A Tn1549-like element was inserted in I-4 of ICESluvan, thereby forming a mosaic ICE ((Bjorkeng et al., 2013); Figure 2B). The integrase of ICESluvan, which belongs to theSR family instead of the tyrosine recombinase family of IntICESa2603, resulted in the alteration of the insertion site, thus generating a new ICE (ICESa2603 family-like ICE).
The att site and oriT site
ICEs of the ICESa2603 family were found to specially integrate at the 3′-end of the rplL gene of Streptococcus spp., by recognizing the att sequence, TTATTTAAGAGTAAC (Table 2). Although different identities of rplL gene were observed, the att sequence (15 bp from the 3′-end of the rplL gene) appeared more conserved in Streptococcus spp. (Table 2 and Table S3), thus providing the sequence basis for its distribution among members of the family. ICESsuT15 and ICESluvan with Int and belonging to SR instead of tyrosine family integrase of ICESa2603 family were assumed to have a different att site. As shown in Figure 3A, there were no consensus insertion sequences have been identified in ICESsuT15.
Figure 3.
att site of ICESsuT15 (A) and oriT site of the ICESa2603 family (B). (A); ORFs in purple represent the chromosome genes while in gray shows the integraing site of ICESsuT15, the putative att sequence was indicated in red. (B); the oriT site of the ICESa2603 were listed in the top, the bases different from ICESa2603 were indicated. Arrows under the sequences represent the locations of inverted repeats, and vertical arrows show the nick sites.
A ~200 bp highly conserved region (92–100% identity) highlighted in the visual comparison with Mauve 2.4 (Darling et al., 2004) is considered to be the putative the oriT region. The left adjacent region was SAG1252 (Tn5252 orf10, 65–100% identity) gene that showed relatively low identity and the right adjacent region was a variable DNA hotspot site. In all ICEs except for ICESthJIM8232-1 and ICESa09mas018883, the oriT site contained a 9-nt inverted repeat sequence, GATATTGTG, which served as a binding site for relaxase, and the precise nickase cleavage site was predicted to reside at position 5′-CTCG/CAAA (Figure 3B).
Phylogenetic analyses of the ICE core genomes
To analyze the evolution of the ICE core genomes, we first compared the identity of each ICE core genes to the corresponding ICESa2603 genes (Figure 1A). Most ICE core genes exhibited a >90% identity at the nucleotide level. On the other hand, the less conserved genes were located at the junction between core genes and variable region. For example, the DNA processing genes (SAG1250–SAG1252) of the conjugative transfer module between HS-1 and HS-2 were much less conserved (65–72%) than other core genes, whereas the oriT site remained highly conserved.
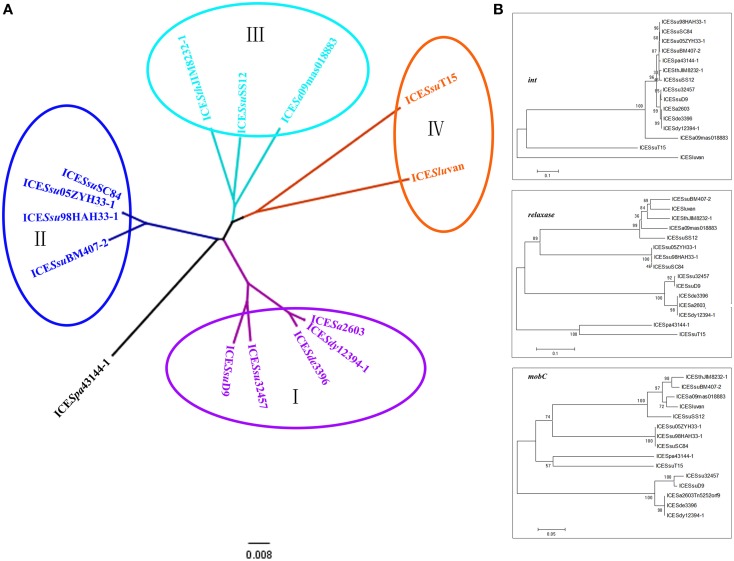
Next, we generated phylogenetic trees based on all the 30 core genes (Figure 4A) or individual representative genes (Figure 4B). The ICEs were then further divided into four subgroups: ICESa2603 family groups I–III (ICESa2603-like, 89K-like, and ICESthJIM8232-1-like) and ICESa2603 family-like group IV. Interestingly, these subgroups also showed variable features: The ICEs of group IV could be distinguished from that of other groups based on the int, only one regulatory gene (SAG1249) was found in HS-1 region of group I ICEs (ICESa2603-like); and a transposon Tn916 was inserted into HS-2 region of group II ICEs, thereby generating mosaic ICEs (Figures 2, 4A).
Figure 4.
Phylogenetic tree of ICESa2603 family. (A), phylogenetic tree generated from 30 core genes. Four subgroups (I–IV) can be classified; (B), phylogenetic tree of int, relaxase, and mobC.
To gain more insights into the evolution of individual core genes, we created phylogenetic trees for each core gene. However, most core genes were determined to be highly conserved, thus making it difficult to generate a reliable tree. Hence, we concentrated on the less conserved genes and selected int, relaxase, and mobC for phylogenetic analyses. Figure 4B shows the two subgroups of int: the tyrosine recombinase family int genes, which were highly conserved and present in all ICESa2603 families (88–100% identity), and the SR family int genes that were present in ICESa2603 family-like ICEs. According to the tree of relaxase and mobC, all ICEs in group I were aligned together and shared ~94% similarity, whereas other ICEs generated distinct branching patterns (with < 80% similarity).
Distribution of members of the ICESa2603 family in S. suis and S. agalactiae
To evaluate the presence of ICEs in S. suis isolates from pigs and S. agalactiae isolates from cows in China, several core ICEs genes were detected (Table 3 and Table S1). Of the 73 S. suis isolates screened, 31 were determined to contain ICEs based on the presence of T4SS genes (virB1, virB4, virB6, and virD4), in which 80.5% (25/31) also carried intICESa2603. Similarly, 38.1% of the tested S. agalactiae isolates harbored T4SS genes, amongst which 62.5% (5/8) were intICESa2603-positive. Different primer pairs P1–P4 were employed and identified the integration/excision form of ICESa2603 family in the present study (Table 3). Table 3 shows that 88% (22/25) of the ICESa2603 family strains were excised from the chromosomes, thereby forming circular intermediates prior to its conjugative transfer into recipient cells.
Table 3.
Distribution of ICESa2603 family core genes in S. suis and S. agalactiae.
To determine whether the ICEs were intact, PCR tiling was performed. Ten overlapping fragments were amplified using the long-PCR method (Figure 1E and Table S2). Table 4 and Table S1 showed that almost all fragments of ICE core regions (fragments 4, 6, 7, and 8) could be amplified in macrolide- and tetracycline-resistant S. suis, whereas other fragments (1, 2, 3, 5) linking the variable regions (HS-1–HS-3 and I-1–I-4) were more variable. PCR tiling also detected various ICE subgroups. For example, the strain with fragments 2–8, but without fragments attL, 1, and attR, belonged to subgroup IV (ICESa2603 family-like); Furthermore, subgroup I could be distinguished from other subgroups based on the size of fragment 1.
Table 4.
Analyses of ICESa2603-family fragments in part of macrolide and tetracycline resistant S. suis by PCR tiling assay.
| Fragmentsa | attL | 1 | 2 | 3b | 4 | 5 | 6 | 7 | 8 | attR |
|---|---|---|---|---|---|---|---|---|---|---|
| HB1004 | + | + | - | - | + | + | + | + | + | + |
| HB1006 | - | - | - | - | + | + | + | + | + | + |
| HB1011c | + | + | + | - | + | + | + | +b | + | + |
| HB1012 | + | + | - | - | + | + | + | + | + | + |
| HB1013 | - | - | - | - | + | + | + | - | - | - |
| NJ4 | + | - | - | - | + | + | + | + | - | - |
| JS07002 | + | + | - | - | + | - | + | + | + | + |
| JS07015 | + | + | - | - | + | + | + | + | + | + |
| SC05017 | - | - | - | - | + | + | + | + | + | - |
| SS2-XY | + | - | - | - | + | + | + | + | + | + |
| YY060816 | - | - | - | - | + | + | + | + | + | - |
The PCR tiling assay and fragments can be seen in Figure 1E.
Failure of amplifying this fragment, probably due to too long additional DNA segments inserted in the 4 regions (I-1 to I-4)
The fragment 7 in HB1011 was ~2 kb larger than ICESa2603 core region.
ICESsuHB1011, a swine origin 89K-like subgroup transferable ICEs
Because ICEs play an important role in the horizontal transmission of antibiotic resistance, we selected a swine-derived clinical macrolide- and tetracycline-resistant strain HB1011 (ST303, a new ST type) for subsequent conjugation experiments. The transfer of macrolide and tetracycline resistance from HB1011 to S.suis BAA-853 (ST1) was confirmed by sequencing of the PCR products (Figures S1, S2) and MLST typing. The MICs of erythromycin and tetracycline and PCR products of erm(B) and tet(O) in conjugant JH-1 confirmed that erythromycin and tetracycline resistance genes erm(B) and tet(O) were indeed transferred (Table S4). We further sequenced the tiling fragments of ICESsuHB1011, which is an 89K-like ICE (Figure 5). The results showed that erm(B) and tet(O) genes were inserted into the SNF2 gene (I-4, Figure 1).
Figure 5.
ORF map of ICESsuHB1011 to the 89K PAI (ICESsu05ZYH33-1) of S. suis 05ZYH33. 89K PAI (ICESsu05ZYH33-1) was proved to be a pathogenicity island response for the human S. suis Outbreak in Sichuan, China in 2005. Genes are shown in different colors: black, ICEs core genes; navy, int and xis; azure, nisin operon genes; red, T4SS genes; green, antibiotic and heavy metal resistance genes.
Discussion
ICEs are self-transmissible MGEs that encode the machinery for conjugation, as well as intricate regulatory systems to control excision from chromosomes and its conjugative transfer (Wozniak and Waldor, 2010). ICEs can be considered as mosaic elements that present the combined features of plasmids and bacteriophages. Similar to plasmids, ICEs transfer via conjugation and in parallel to phages, ICEs integrate into and replicate with its host chromosome (Burrus and Waldor, 2004). ICEs often carry accessory genes that encode for antibiotic resistance, virulence factors and various other functions. These are therefore important vectors for the horizontal dissemination of genetic information, thereby facilitating rapid bacterial evolution.
ICESa2603 is a ~54 kb ICE that was first discovered in S. agalactiae 2603V/R (Tettelin et al., 2002). It has the capacity to retain its features of transferability to S. agalactiae, S. dysgalactiae subsp dysgalactiae, and S. uberis (Haenni et al., 2010). Subsequently, ICESa2603-like ICEs have been detected in clinical isolates of Streptococcus spp. from both human and animal species from Europe and Asia (Table 1). Therefore, it is likely that the members of the ICESa2603 family have been distributed and have the ability to undergo horizontal transmission in Streptococcus spp. The number of ICESa2603 family ICEs and its host range may therefore further expand as additional research studies are currently being conducted. However, comparative genomics of ICESa2603 family ICEs and its evolution have not been reported. Here, we first comparatively analyzed 13 ICEs of the ICESa2603 family. The results showed that the ICESa2603 family has a backbone of 30 core genes that encode the core functions of integration/excision, conjugation and regulation.
The integration/excision modules of ICESa2603 consisted of IntICESa2603 (SAG1247) and XisICESa2603 (SAG1248). IntICESa2603 belongs to the tyrosine based site-specific recombinases. This family of integrase originated from bacterial phages and conjugate transposons. One member is the integrase from Bacillus subtilis conjugative transposon ICEBs1, which mediates integration into the 3′-ends of tRNA-Leu loci (Lee et al., 2007). IntICESa2603 shares 23% identity with IntICEBs1, but mediate site-specific recombination between identical attICE and attB sites and the integration into the 3′-end of the 50S ribosomal subunit protein L7/L12 gene (Haenni et al., 2010). The attB sites (TTATTTAAGAGTAAC) of ICESa2603 family is conserved in Streptococcus spp. and even Enterococcus faecium and Enterococcus faecalis, which provided the sequence basis for the horizontal transfer of this family. To date, members of the ICESa2603 family has been identified in six species of Streptococcus spp. (Table 2). The ICESa2603 family-like ICEs, share 28 of 30 core genes with ICESa2603 family ICEs, which contain the SR family integrase, although a low level of identity was observed between ICESluvan and ICESsuT15 (Figures 1A, 4B). A study on another SR IntTn5397 has shown that the insertion site contains a central GA dinucleotide; however, no other consensus insertion sequences have been identified (Wang and Mullany, 2000). Based on the experimental transfer of ICESluvan (Bjorkeng et al., 2013) and the analysis of ICESsuT15 (Figure 3A), we believe that Int of ICESluvan and ICESsuT15 recognize the CA dinucleotide in certain regions of chromosomal sites. The ICESluvan is capable of transmission, which provides an illustration that acquisition of a new integration/excision module (most likely via recombination) generates new types of ICEs (here we termed as ICESa2603 family-like ICEs). Xis, the adjacent recombination directionality factor (RDF), both facilitates excision and inhibits integration. It binds to the ends of the integrated element adjacent to Int binding sites. Deletion of the xis gene of ICESsu05ZYH33-1 demonstrated that xis stimulates, but is not essential for ICESsu05ZYH33-1 excision from chromosomes (Li et al., 2011).
The conjugation module of ICESa2603 family is similar with that of plasmids. When ICESa2603 transfers, a relaxase (SAG1250) first binds to and excises circular ICE DNA at oriT sites (Li et al., 2011). Thereafter, with the help of a coupling protein, MobC (SAG1251), the relaxase-DNA complex is presented to type IV coupling protein (T4CP) and the Mpf complex. Finally, the T4CPMpf complex transfers the ICE DNA to the receipt cell. The T4SS complex includes genes encoding for proteins similar to A. tumefaciens VirB1, VirB4, VirB6, and VirD4. Zhang et al. reported that these T4SS gene clusters were conserved both in composition and order in the genomes of various Streptococcal species and thus collectively described these as a Type–IVC secretion system (Zhang et al., 2012). Knockout of two key components genes (i.e., virD4 and virB4) of the Type–IVC secretion system not only abolished its transferability but also eliminated the lethality of the highly virulent strain and impaired its ability to trigger a host immune response in infected mouse models (Li et al., 2011; Zhao et al., 2011). These results suggest that T4SS has retained its ancestral function as a conjugation module while its capacity to translocate effector protein(s) to the cell surface or into eukaryotic target cells undergoes evolution.
Our understanding of the mechanism underlying the regulation module of the ICESa2603 family is limited. Comparative analysis of the 30 core genes of ICESa2603 family ICEs has shown that no common regulators were conserved in all ICEs of this family. Instead, a gene (SAG1249) encoding a Cro/CI family transcriptional regulator was detected in all experimentally proven transferable ICEs, despite not being classified as core genes in the present study. This regulator possibly regulates ICE gene expression and ICE transfer (Beaber and Waldor, 2004). Recently, Xu et al. described a two-component system, NisK/NisR, in subgroup II (89K-subgroup, Figure 4A) ICEs that contributes to the virulence of S. suis serotype 2 (Xu et al., 2014). It is tempting to know whether this two-component system is involved in the regulation of transfer. A bacteriophage abortive infection (Abi) system, AbiE, which assists bacteria from being killed by bacteriophages, has been recently identified as a type IV toxin-antitoxin system in ICESa2603 (SAG1284 and SAG1285) (Chopin et al., 2005; Dy et al., 2014). Dy et al. (2014) demonstrated that the AbiE system enables plasmid maintenance when these were introduced into pUC19. It is possible that the abiE operon and native promoter enable the maintenance of the circular form of ICESa2603, thereby increasing the frequency for ICE transfer.
Phylogenetic and comparative analyses have shown that most of the core genes of the ICESa2603 family exhibited 91–100% identity at the nucleotide level (Figure 1A). No detectable difference in the degree of conservation of most core genes was observed, which suggests equal selective pressures on core genes of the ICEs. However, some genes (i.e., Int, xis, relaxase, mobC, and SAG1278) exhibited lower degrees of conservation. The difference in the identity of the Int and xis genes, as earlier described, was probably generated by recombination. The relaxase and mobC genes can be divided into four subgroups (Figure 4B). The relaxase and mobC genes exhibited a high level of identity within subgroups (>80%), but a low identity between subgroups (65–80%). These results suggest that the different subgroups of relaxase and mobC genes (DNA processing module) might have also been generated by recombination of the entire DNA processing module. The relative low level of conservation of SAG1278, in which of the 3′-end is often the insertion site (I-2), might have been ascribed to the insertion of accessory genes, as indicated by the lower level of identity of this gene in each ICE. These results also suggest that individual core genes are exposed to different evolutionary pressures.
BLAST analysis of the core conjugation and integration module proteins of the ICESa2603 family showed that ICESa2603 shared some similarity with the Streptococcus pneumoniae conjugative transposon Tn5252 (Alarcon-Chaidez et al., 1997). Based on limited knowledge, we propose that ICEs were disseminated across a diverse range of hosts, thereby resulting in exposure to different selective stress. To survive stress, cargo genes encoding antibiotic resistance, nitrogen fixation, virulence factors, and various other functions were introduced into the HS and I sites. In the case of the ICESa2603 family, insertion of variable DNAs might have disrupted the core regulatory module gene (SAG1249), but another regulatory module gene could be functioned to compensate and restore stability in the chromosome (Li et al., 2008, 2011). Another benefit is that recombination of the core module increases the adaptability and diversity of ICEs or MGEs. Examples include the ICESa2603 family-like ICEs, ICESluvan, in which only the conjugation module intTyr–xis was replaced by SR family intSR yet retained its transferability (Bjorkeng et al., 2013). A very recent study (Marini et al., 2015) has shown that mating between S. suis 32457 (donor) and S. agalactiae 2603V/R (recipient) yields a hybrid ICE, ICESa2603/ICESsu32457, and interestingly, could be transferred across various Streptococcus pyogenes strains. These findings strongly support the concept that recombination plays an important role in horizontal gene transfer and generates novel ICEs or even new types of MGEs. These findings and the above mentioned recombination/insertion/hybrid events further broaden range of ICE diversity. The mosaic patterns of the ICESa2603 family consisted of genes from groups A, B, and G Streptococcus organisms, Faecalibacterium, enterococci, and possibly Listeria innocua and Enterococci faecalis, indicating that recombination and mobilization events are key factors in the assembly of these families. The exchange of integration modules among ICEs or between ICEs and other integrating MGEs (e.g., phages) could potentially create new ICEs with altered insertion site specificities or modified host ranges.
Figure 4A shows that all four subgroups of the ICESa2603 family were detected in S. suis, which is indicative of its important role in the transmission of these types of ICEs of S. suis. Screening of ICESa2603 core genes showed that 42.5% of swine-derived S. suis isolates contained ICE T4SS genes and 80.5% of these belong to ICESa2603 family. These results suggest that S. suis isolates harbor ICESa2603 family ICEs that are extensively distributed in swine in China, which has become a challenge to the hog industry and even a threat to human health.
89K-like subgroup ICEs (ICESsu98HAH33-1 and ICESsu05ZYH33-1) were detected in two S. suis isolates and have caused two extensive human outbreaks of streptococcal toxic shock–like syndrome (STSLS) in China in 1998 and 2005 (Tang et al., 2006). Further studies (Li et al., 2008; Zhao et al., 2011; Zhong et al., 2014) have demonstrated that 89K is a pathogenicity island (PAI). Li et al. (2008) earlier showed that a two-component signal transduction system (SalK/SalR) is essential for full virulence of highly invasive S. suis serotype 2. SalK/SalR, which is also the regulatory module of the nisin lantibiotic operon, was inserted into the HS-1, and possibly acts as a regulator, together with SAG1249. Furthermore, knockout of three key components (virB1, virB4, and virD4) of T4SS significantly reduces but does not abolish the virulence of S. suis in a mouse model (Zhao et al., 2011; Zhong et al., 2014). These findings indicate that T4SS harbored by 89K PAI contributes to the development of STSLS and mediates the horizontal transfer of 89K. However, those types of ICEs did not contain genes that encode resistance to macrolides and tetracycline. In the present study, we observed that almost all of the macrolide- and tetracycline-resistant S. suis also contained T4SS genes, indicating that ICEs play an important role in spreading macrolide and tetracycline resistance in S. suis. The isolated strain HB1011, which was determined to harbor ICESsuHB1011 that carried erm(B) and tet(O), which was indicative of its multiple functions, including horizontal transfer resistance and pathogenicity. In addition, other strains of S. agalactiae and S. dysgalactiae subsp. equisimilis whose ICEs harbored T4SS that was similar to 89K did not cause STSLS. Therefore, it is of great importance to identify factors and mechanisms by which the T4SS of 89K mediates pathogenicity. It has been proposed that ICESa2603 family T4SS might have retained its ancestral function as a mobile DNA element, but have also evolved its capacity to translocate effector protein(s) to the cell surface or into eukaryotic target cells (Bhatty et al., 2013).
Conclusions
In summary, the structures of the ICESa2603 family ICEs in isolates of different streptococcal species are clearly illustrated in this study by comparative analyzing 13 ICEs of the ICESa2603 family and two ICESa2603 family-like ICEs. Screening of ICESa2603 core genes shows that ICESa2603 family ICEs are distributed widely in S. suis and S. agalactiae in China and mainly belong to the 89K-subtype family ICEs. Furthermore, the spread of the ICEs containing resistance genes erm(B) and tet(O) among S. suis strains by conjugation had a profound effect on the horizontal transmission of antibiotic resistance to macrolides and tetracyclines. Further research is needed to characterize the mechanisms involved in triggering ICEs transfer intra- and inter-species.
Author contributions
JH, DG, and LW developed the concept and designed experiments. JH, YL, KS, and LG performed the experiments and collected the data. JH, YL, and JK conducted all bioinformatics analyses. JH and LW prepared the manuscript. All authors have contributed to, seen and approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The study was partially supported by the National Natural Science Foundation of China (No. 31572567), Natural Science Foundation of Jiangsu Province of China (No. BK2012771), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The authors wish to thank Professor Shile Huang from Louisiana State University, Health Sciences Center in USA for reviewing the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00055
References
- Alarcon-Chaidez F., Sampath J., Srinivas P., Vijayakumar M. N. (1997). TN5252: a model for complex streptococcal conjugative transposons. Adv. Exp. Med. Biol. 418, 1029–1032. 10.1007/978-1-4899-1825-3_242 [DOI] [PubMed] [Google Scholar]
- Beaber J. W., Hochhut B., Waldor M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. 10.1038/nature02241 [DOI] [PubMed] [Google Scholar]
- Beaber J. W., Waldor M. K. (2004). Identification of operators and promoters that control SXT conjugative transfer. J. Bacteriol. 186, 5945–5949. 10.1128/JB.186.17.5945-5949.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatty M., Laverde Gomez J. A., Christie P. J. (2013). The expanding bacterial type IV secretion lexicon. Res. Microbiol. 164, 620–639. 10.1016/j.resmic.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D., Xu Z., Harrison E. M., Tai C., Wei Y., He X., et al. (2011). ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 40, D621–D626. 10.1093/nar/gkr846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkeng E. K., Hjerde E., Pedersen T., Sundsfjord A., Hegstad K. (2013). ICESluvan, a 94-kilobase mosaic integrative conjugative element conferring interspecies transfer of VanB-type glycopeptide resistance, a novel bacitracin resistance locus, and a toxin-antitoxin stabilization system. J. Bacteriol. 195, 5381–5390. 10.1128/JB.02165-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet M., Couve E., Glaser P., Guedon G., Payot S. (2008). Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J. Bacteriol. 190, 6913–6917. 10.1128/JB.00824-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V., Pavlovic G., Decaris B., Guedon G. (2002). Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46, 601–610. 10.1046/j.1365-2958.2002.03191.x [DOI] [PubMed] [Google Scholar]
- Burrus V., Waldor M. K. (2004). Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155, 376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G., Parkhill J. (2005). ACT: the artemis comparison tool. Bioinformatics 21, 3422–3423. 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- Chen C., Tang J. Q., Dong W., Wang C. J., Feng Y. J., Wang J., et al. (2007). A glimpse of streptococcal toxic shock syndrome from comparative genomics of S-suis 2 chinese isolates. PLoS ONE 2:e315. 10.1371/journal.pone.0000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M. C., Chopin A., Bidnenko E. (2005). Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 8, 473–479. 10.1016/j.mib.2005.06.006 [DOI] [PubMed] [Google Scholar]
- Christie P. J. (1997). Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2010). Performance standards for antimicrobial susceptibility testing; Twentieth informational supplement, in CLSI Document M100-S20, ed Cockerill F. R. (Wayne, PA: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- Daccord A., Mursell M., Poulin-Laprade D., Burrus V. (2012). Dynamics of the SetCD-regulated integration and excision of genomic islands mobilized by integrating conjugative elements of the SXT/R391 family. J. Bacteriol. 194, 5794–5802. 10.1128/JB.01093-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. R., Shera J., Van Domselaar G. H., Sriprakash K. S., McMillan D. J. (2009). A novel integrative conjugative element mediates genetic transfer from group G Streptococcus to other {beta}-hemolytic Streptococci. J. Bacteriol. 191, 2257–2265. 10.1128/JB.01624-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. R., Tran T. N., McMillan D. J., Gardiner D. L., Currie B. J., Sriprakash K. S. (2005). Inter-species genetic movement may blur the epidemiology of streptococcal diseases in endemic regions. Microbes Infect. 7, 1128–1138. 10.1016/j.micinf.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Delorme C., Bartholini C., Luraschi M., Pons N., Loux V., Almeida M., et al. (2011). Complete genome sequence of the pigmented Streptococcus thermophilus strain JIM8232. J. Bacteriol. 193, 5581–5582. 10.1128/JB.05404-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy R. L., Przybilski R., Semeijn K., Salmond G. P., Fineran P. C. (2014). A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 42, 4590–4605. 10.1093/nar/gkt1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J., Quintais L., Garcillan-Barcia M. P., De La Cruz F., Rocha E. P. (2011). The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222. 10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni M., Saras E., Bertin S., Leblond P., Madec J. Y., Payot S. (2010). Diversity and Mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. dysgalactiae subsp dysgalactiae, and S. uberis. Appl. Environ. Microbiol. 76, 7957–7965. 10.1128/AEM.00805-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S., Ishii Y., Saga T., Tateda K., Yamaguchi K. (2010). Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob. Agents Chemother. 54, 3545–3550. 10.1128/AAC.00111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T. G., Hauser H., Sanders M., Thi H. N., Cherevach I., Cronin A., et al. (2009). Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS ONE 4:e6072. 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E., Wilkins B. M. (1995). DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64, 141–169. 10.1146/annurev.bi.64.070195.001041 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lee C. A., Auchtung J. M., Monson R. E., Grossman A. D. (2007). Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 66, 1356–1369. 10.1111/j.1365-2958.2007.06000.x [DOI] [PubMed] [Google Scholar]
- Li M., Shen X., Yan J., Han H., Zheng B., Liu D., et al. (2011). GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79, 1670–1683. 10.1111/j.1365-2958.2011.07553.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wang C., Feng Y., Pan X., Cheng G., Wang J., et al. (2008). SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS ONE 3:e2080. 10.1371/journal.pone.0002080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin I. H., Liu T. T., Teng Y. T., Wu H. L., Liu Y. M., Wu K. M., et al. (2011). Sequencing and comparative genome analysis of two pathogenic Streptococcus gallolyticus subspecies: genome plasticity, adaptation and virulence. PLoS ONE 6:e20519. 10.1371/journal.pone.0020519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini E., Palmieri C., Magi G., Facinelli B. (2015). Recombination between Streptococcus suis ICESsu32457 and Streptococcus agalactiae ICESa2603 yields a hybrid ICE transferable to Streptococcus pyogenes. Vet. Microbiol. 178, 99–104. 10.1016/j.vetmic.2015.04.013 [DOI] [PubMed] [Google Scholar]
- Mata C., Navarro F., Miro E., Walsh T. R., Mirelis B., Toleman M. (2011). Prevalence of SXT/R391-like integrative and conjugative elements carrying blaCMY-2 in Proteus mirabilis. J. Antimicrob. Chemother. 66, 2266–2270. 10.1093/jac/dkr286 [DOI] [PubMed] [Google Scholar]
- Palmieri C., Magi G., Mingoia M., Bagnarelli P., Ripa S., Varaldo P. E., et al. (2012). Characterization of a Streptococcus suis tet(O/W/32/O)-carrying element transferable to major streptococcal pathogens. Antimicrob. Agents Chemother. 56, 4697–4702. 10.1128/AAC.00629-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. P., Mullany P. (2009). A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17, 251–258. 10.1016/j.tim.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Roberts A. P., Mullany P. (2011). Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 35, 856–871. 10.1111/j.1574-6976.2011.00283.x [DOI] [PubMed] [Google Scholar]
- Rodriguez-Blanco A., Lemos M. L., Osorio C. R. (2012). Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrob. Agents Chemother. 56, 2619–2626. 10.1128/AAC.05997-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitkiewicz I., Green N. M., Guo N., Mereghetti L., Musser J. M. (2011). Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC Microbiol. 11:65. 10.1186/1471-2180-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Lefebure T., Hubisz M. J., Pavinski Bitar P., Lang P., Siepel A., et al. (2011). Comparative genomic analysis of the Streptococcus dysgalactiae species group: gene content, molecular adaptation, and promoter evolution. Genome Biol. Evol. 3, 168–185. 10.1093/gbe/evr006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Wang C., Feng Y., Yang W., Song H., Chen Z., et al. (2006). Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e151. 10.1371/journal.pmed.0030151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Poele E. M., Bolhuis H., Dijkhuizen L. (2008). Actinomycete integrative and conjugative elements. Antonie Van Leeuwenhoek 94, 127–143. 10.1007/s10482-008-9255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Masignani V., Cieslewicz M. J., Eisen J. A., Peterson S., Wessels M. R., et al. (2002). Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U.S.A. 99, 12391–12396. 10.1073/pnas.182380799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor M. K., Tschape H., Mekalanos J. J. (1996). A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178, 4157–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Mullany P. (2000). The large resolvase TndX is required and sufficient for integration and excision of derivatives of the novel conjugative transposon Tn5397. J. Bacteriol. 182, 6577–6583. 10.1128/JB.182.23.6577-6583.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Gao Y., Teng K., Zhang J., Sun S., Zhong J. (2014). Restoration of bioactive lantibiotic suicin from a remnant lan locus of pathogenic Streptococcus suis serotype 2. Appl. Environ. Microbiol. 80, 1062–1071. 10.1128/AEM.03213-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R. A., Fouts D. E., Spagnoletti M., Colombo M. M., Ceccarelli D., Garriss G., et al. (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. 10.1371/journal.pgen.1000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R. A., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- Xu J., Fu S., Liu M., Xu Q., Bei W., Chen H., et al. (2014). The two-component system NisK/NisR contributes to the virulence of Streptococcus suis serotype 2. Microbiol. Res. 169, 541–546. 10.1016/j.micres.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Zhang A., Yang M., Hu P., Wu J., Chen B., Hua Y., et al. (2011). Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genomics 12:523. 10.1186/1471-2164-12-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Rong C., Chen C., Gao G. F. (2012). Type-IVC secretion system: a novel subclass of type IV secretion system (T4SS) common existing in gram-positive genus Streptococcus. PLoS ONE 7:e46390. 10.1371/journal.pone.0046390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Liu G., Li S., Wang M., Song J., Wang J., et al. (2011). Role of a type IV-like secretion system of Streptococcus suis 2 in the development of streptococcal toxic shock syndrome. J. Infect. Dis. 204, 274–281. 10.1093/infdis/jir261 [DOI] [PubMed] [Google Scholar]
- Zhong Q., Zhao Y., Chen T., Yin S., Yao X., Wang J., et al. (2014). A functional peptidoglycan hydrolase characterized from T4SS in 89K pathogenicity island of epidemic Streptococcus suis serotype 2. BMC Microbiol. 14:73. 10.1186/1471-2180-14-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubair S., De Villiers E. P., Fuxelius H. H., Andersson G., Johansson K. E., Bishop R. P., et al. (2013). Genome sequence of Streptococcus agalactiae strain 09mas018883, isolated from a Swedish cow. Genome Announc. 1:e00456-13. 10.1128/genomeA.00456-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.