Abstract
Respiratory epithelium is difficult to grow in vitro, as it requires a well-maintained polarizing air–liquid interface (ALI) to maintain differentiation. Traditional methods rely on permeable membrane culture inserts, which are difficult to work with and are ill-suited for the production of large numbers of cells, such as the quantities required for cell-based clinical therapies. Herein, we investigate an alternative form of culture in which the cells are placed on a porous substrate that is continuously rolled, such that the monolayer of cells is alternately submerged in media or apically exposed to air. Our prototype bioreactor is reliable for up to 21 days of continuous culture and is designed for scale-up for large-scale cell culture with continuous medium and gas exchange. Normal human bronchial epithelial (NHBE) cells were cultured on an absorbent substrate in the reactor for periods of 7, 14, and 21 days and were compared to static controls that were submerged in media. Quantification by immunohistochemistry and quantitative PCR of markers specific to differentiated respiratory epithelium indicated increased cilia, mucous production, and tight junction formation in the rolled cultures, compared to static. Together with scanning electron microscopy and paraffin histology, the data indicate that the intermittent ALI provided by the rolling bioreactor promotes a polarized epithelial phenotype over a period of 21 days.
Key words: Lung, Respiratory epithelium, Bioreactor, Air–liquid interface (ALI), Tissue engineering
INTRODUCTION
Respiratory epithelium is typically difficult to grow or maintain in a differentiated state in the laboratory (6). Even the most proliferative primary pulmonary epithelial cells rapidly change morphology and lose functionality, despite the use of finely tailored media (13). This is largely due to the lack of cell-polarizing forces in submerged culture (4,18). Without the influence of an air–liquid interface (ALI) or a normal basement membrane, epithelial cells in vitro grow haphazardly and devolve into amorphous, uncharacterized cell types (9,12). To enhance cell differentiation, respiratory epithelium has been cultured extensively using porous membrane culture well inserts (11,14,15,26). When used properly, the ALI provided by this culture device can coax respiratory epithelium into a differentiated state, characterized by mucin secretion, tight junction formation, and strong morphological polarity with apical cilia formation (9). However, the current design of such systems requires regular attention to monitor fluid levels and nutrient supply. Additionally, culture well inserts are not designed for large-scale culture, which is a requirement for a number of screening studies of respiratory cells or for therapeutic applications (6). Whole organ engineering by repopulating acellular lung matrices with pulmonary epithelium, for example, will require many hundreds of millions of differentiated and highly functional cell types. The feasibility of using decellularized lung matrices to produce implantable organs that exchange gas has recently been demonstrated (20,21), and scale-up to human-sized organs is being pursued (8,19). Therefore, a novel system that will enable the growth and differentiation of large numbers of these cells with relative ease is highly desirable (5,24).
With the exception of Grek and coworkers (10), who used hollow fibers to culture airway cells, previous studies have each attacked the problem of differentiating epithelium in vitro in the same way: by layering the cells on a permeable barrier that allows nutrient exchange with a fluid reservoir beneath the membrane (3,9,15,26). While approximately biomimetic, such designs can require extensive attention and manual labor to generate consistent results. The work of Macchiarini and colleagues (1,2,16), in which the interior of a tracheal scaffold was coated with respiratory epithelium via a rolling bioreactor, suggests that a rolling culture design might generate a functional ALI that is low maintenance and readily scalable.
The goal of this study was to design a bioreactor to expand and differentiate human respiratory epithelium in vitro. We hypothesized that an intermittent environmental cue of exposure to air might promote the polarization and differentiation of respiratory epithelium in vitro. We examined whether a specialized rotating culture apparatus could prove both effective and scalable. Our findings demonstrate that such a design is feasible and that it promotes appropriate respiratory cell differentiation.
MATERIALS AND METHODS
Bioreactor Design, Fabrication, and Assembly
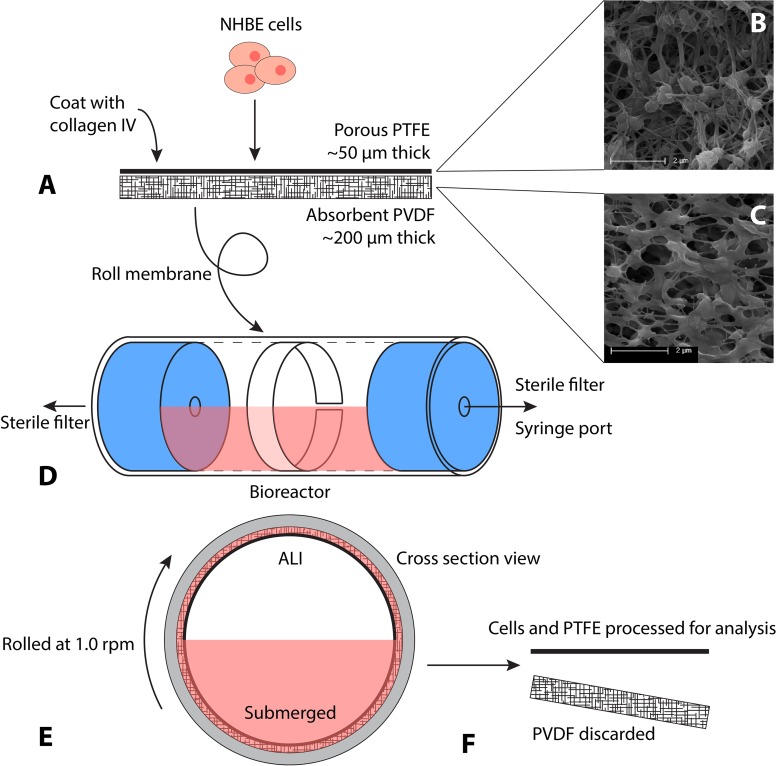
The bioreactor was designed to be inexpensive to produce, easy to assemble and to use, and to be readily adaptable for large-scale cell culture. Figure 1 details the bioreactor and experimental setup. Millipore’s Biopore Membrane (BGCM00010; EMD Millipore, Billerica, MA, USA) was used as a cell support. This product contains two components that adhere to one another unless separated with forceps (Fig. 1A). The upper layer, upon which cells were eventually seeded, is polytetrafluorethylene (PTFE), approximately 50 µm thick, with 0.4-μm pores (Fig. 1B). The lower layer, designed as a backing, is polyvinylidene fluoride (PVDF), approximately 200 µm thick, with 0.22-µm pores (Fig. 1C). These two components were treated as a single entity throughout culture. This allows the PVDF layer (BGCM00010; EMD Millipore) to act as an absorbent sponge for culture medium during ALI culture, supplying the above PTFE layer (BGCM00010; EMD Millipore) and attached cells with a medium reservoir for nutrient transfer (Fig. 1E) while elevated and exposed to air.
Figure 1.
Schematic showing setup and culture of constructs within bioreactor. Normal human bronchial epithelial (NHBE) cells are placed on the collagen-coated polytetrafluorethylene (PTFE) membrane bonded to an absorbent polyvinylidene fluoride (PVDF) sponge layer and allowed to adhere (A). (B and C) Scanning electron micrographs of the two construct layers, pictured without cells, showing their relative porosity and structure. Scale bars: 2 µm. This construct is rolled into a cylinder and placed inside the reactor vial where it aligns with the walls. The reactor is sealed, with ports to allow gas and media exchange (D). The reactor vial is half filled with media and rolled continuously throughout culture, generating intermittent air–liquid interface (ALI) and submerged conditions (E). When ready for cell harvesting and analysis, the construct is removed from the reactor, and the upper PTFE membrane with attached cells is processed (F).
The culture chambers consisted of a custom manufactured borosilicate glass tube, 38 mm outside diameter (OD), 32.3 mm inside diameter (ID), capped with two silicone stoppers (R-06298-14; Cole-Parmer, Vernon Hills, IL, USA). The length of these tubes was dependent on the desired culture surface area for a given experiment. A hole was bored through the center of each stopper to accommodate the insertion of Master-Flex L/S 16 tubing (ZW-06508-16; Cole-Parmer). A rotating, sealing Luer-lock (ZW-06464-90; Cole-Parmer) was then attached to one end of the vial to allow the chamber to rotate freely with a stopcock attached. Sterile syringe filters (EW-29705-50; Cole-Parmer) were attached to each end for periodic airflow, and a syringe port was attached to allow daily culture medium exchanges (Fig. 1D). This design maintains sterility over 3 weeks of rolling culture, with frequent medium changes and application of standard antibiotic agents (penicillin/streptomycin; 15140122; Gibco, Life Technologies, Grand Island, NY, USA). For large-scale application, the system can be adapted to accommodate continuous medium perfusion from a reservoir via a pump, which eliminates the need for daily medium and gas exchanges.
A custom system of rollers was designed and manufactured (Raredon Resources Inc., Florence, MA, USA) to rotate the reactor chambers at speeds ranging from 0 to 100 rpm. The rollers were designed to be modular, accommodating the simultaneous culture of up to 10 rolling chambers, with units easily added or removed as needed. The rollers proved reliable for continuous use in humidified air at 37°C.
The two-membrane construct was cut to fit the internal length and circumference of the culture chamber, was washed in 70% ethanol to sterilize it, and was rinsed twice with dH2O. The constructs were coated with collagen IV prior to cell seeding as previously described (6). Ten milligrams of human placental collagen type IV (C7521; Sigma-Aldrich, St. Louis, MO, USA) was dissolved in 20 ml dH2O with 40 µl of concentrated acetic acid (695092; Sigma-Aldrich) and incubated for 15 min at 37°C until fully dissolved. The solution was syringe filtered (PTFE 0.2 µm; 2915-08; Cole-Parmer) and stored in aliquots at −20°C. To coat the sterilized membranes, the stock solution was diluted 1:10 with sterile dH2O to yield a concentration of 0.05 mg/ml. The membranes were placed in a Petri dish (EW-06139-00; Cole-Parmer), and 1 ml of diluted collagen solution was applied to each 10-cm2 strip. The dish was transferred to an incubator and allowed to incubate at 37°C for at least 2 h or overnight.
NHBE Cell Culture
B-ALI-certified normal human bronchial/tracheal epithelial (NHBE) cells, from a 35-year-old male, were obtained from Lonza (CC-2540S; Lonza Group Ltd., Basel, Switzerland). The cells were plated onto T75 culture flasks (430641; Corning, Corning, NY, USA) and grown in B-ALI-growth medium (193516, with BulletKit 193515; Lonza). Once the cells reached >95% confluence, usually within 4–6 days, they were lifted from the flask with 3 ml of 0.05% trypsin-EDTA (25300-054; Gibco®). This cell suspension was resuspended in the appropriate volume of B-ALI-growth medium to generate a live cell concentration of approximately 2 × 106 cells/ml.
Approximately 2 million cells in 1 ml were seeded onto each 10-cm2 collagen IV-coated membrane, resulting in a cell density of 200,000 cells/cm2 (6). The constructs were incubated for 6 h to allow the cells to attach; the constructs were then completely submerged in B-ALI-growth medium and incubated for another 24 h. The membranes were then carefully rolled and inserted into presterilized reactor chambers under sterile conditions (Fig. 1). The reactor was sealed with the silicone caps, including preassembled rotating ports, stopcock, and attached filters and syringe port. The vials were filled with 15 ml of B-ALI-differentiation complete medium (193517, with BulletKit 193515; Lonza), corresponding to 1.5 ml of medium for every square centimeter of construct. The assembled reactors were placed on the custom roller and set at approximately 1 rpm. Bioreactors were maintained in humidified incubators at 37°C and 5% CO2.
A complete medium exchange was conducted daily via syringe ports. The chambers were then injected with 10 ml of incubator air through the sterile air filters on either end of the chamber.
All rolled constructs had a matched control, which was grown under static conditions in a Petri dish rather than in a rolled vial, to measure the impact of the intermittent ALI independently of other variables. Membrane preparation, cell lot and density, culture times, medium type, and medium volume per unit area were identical between rolled and static conditions. Assessments were taken immediately prior to insertion into the reactors, and after 7, 14, or 21 days of culture under either static or rolling conditions.
Primary Epithelial Cell Culture
All animal work was performed in accordance with AAALAC guidelines and was approved by the Yale Institutional Animal Care and Use Committee. Neonatal rats, 7–10 days old [Sprague–Dawley, green-fluorescent protein (GFP) modified, (CAG-EGFP) (CZ-004Osb) obtained from Masaru Okabe, Osaka University, Japan, SD-Tg] were euthanized via IP injection of sodium pentobarbital (150 mg/kg body weight, diluted appropriately for injection; 710101; Virbac Animal Health, Fort Worth, TX, USA). Lungs were harvested via a transverse incision below the costal margin, followed by a careful incision in the diaphragm to enter the thoracic cavity without damaging the lungs. Lungs, heart, and trachea were exposed and the trachea cannulated. The left and right atria were severed, and the lungs were perfused with heparin (100 U/ml; H4784; Sigma-Aldrich) in phosphate-buffered saline (PBS; 14190-144; Life Technologies, Gibco®) via the right ventricle until lungs were cleared of blood. Two milliliters of dispase (2 U/ml; D4818; Sigma-Aldrich) was injected into the lungs via the tracheal cannula, followed by 0.2–0.5 ml of 1% low melting point agarose (A9045; Sigma-Aldrich). Lungs were covered with ice for 2 min to allow the agarose to gel. The heart and trachea were removed, and the remaining lung tissue incubated in 1 ml/lung dispase solution for 45 min at room temperature. The tissue was then transferred to 7 ml of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-buffered Dulbecco’s modified Eagle’s medium (DMEM; 11330-032; Gibco) with 100 U/ml DNAse I (LS006333; Worthington Biochemical, Lakewood, NJ, USA) and teased apart with forceps to remove bronchi and airways, and minced with a scalpel. The cell suspension was then filtered through a series of cell strainers (22363549, 22363548, and 22363547; Fisher Scientific, Pittsburgh, PA, USA) and nylon mesh (20 μm pore size; NY20; Millipore). The filtrate was centrifuged at 130 × g for 8 min at 4°C. The pellet was then suspended and spread on a prewashed Petri dish, precoated with anti-cluster of differentiation 45 (CD45) and anti-CD32 antibodies for 24–48 h at 4°C (554875 and 550270, respectively; BD Pharmingen, Franklin Lakes, NJ, USA). After 2-h incubation at 37°C, the unbound cells were collected, spun down, and suspended in Lonza Small-Airway Epithelial Cell Growth Medium (SAGM™; CC-3118). CD45 was used to remove most immune cells such as macrophages. CD32 was also expressed on most immune cells. Separation of interstitial cells from epithelium was a result of the short plating/incubation time, as fibroblasts and smooth muscle cells attached much more rapidly than lung epithelium. This method of cell isolation was adapted from a previously reported isolation method for mouse type II cells (22). The remaining collected cells were seeded onto PTFE substrates within the bioreactors and on static control plates as described earlier. Samples were cultured for 7 days before being sacrificed for immunostaining.
Immunofluorescence
Constructs were extracted from the bioreactor and a portion fixed with 3.7% paraformaldehyde (PFA) fixative solution (BM-158; Boston BioProducts, Ashland, MA, USA) for 20 min, then transferred to 70% ethanol and stored until further use. To perform immunostaining, 1 cm × 0.5 cm pieces were cut from the whole, and the PTFE membranes were removed from the PVDF sponge layer (Fig. 1F). The PTFE layer is optically clear under fluorescent channels, allowing planar staining and imaging of the samples. The samples were laid on precleaned, uncharged slides, traced with a hydrophobic pen, and submerged in 0.2% Triton X-100 (Sigma-Aldrich) in PBS for 10 min. When staining for proliferating cells, antigen retrieval was performed using 75°C pH 6.0 citric acid buffer for 20 min (0.01 M sodium citrate dehydrate; 3646-10; Avantor Performance Materials, Center Valley, PA, USA, and 0.05% Tween-20; P7949-500; Sigma-Aldrich, in distilled water.) Slides were blocked in 5% bovine serum albumin (BSA; A7906; Sigma-Aldrich) and 0.75% glycine (G8898; Sigma-Aldrich) in PBS for 30 min, then incubated with primary antibody for 45 min, followed by an appropriate secondary antibody. The slides were rinsed again, stained with 4′,6-diamidino-2-phenylindole (DAPI; D1306; Life Technologies, Invitrogen™) in PBS for 1 min, and mounted using polyvinyl alcohol mounting medium with DABCO® (10981; Sigma-Aldrich). All steps were performed at room temperature unless otherwise noted.
Tight junctions were stained with an anti-zonula occludens-1 (ZO-1) antibody (339100 or 617300; Life Technologies, Invitrogen™) at a concentration of 1:100. Ciliated cells were identified using an antiacetylated tubulin antibody (T7451; Sigma-Aldrich) at a concentration of 1:3,000. Mucin-secreting cells were detected using an anti-mucin 5AC antibody (251768; Abbiotec, San Diego, CA, USA) at a concentration of 1:500. Cells undergoing apoptosis were tagged with an antiactive caspase 3 antibody (13847; Abcam, Cambridge, MA, USA) at a concentration of 1:500. Proliferating cell nuclei were marked with an antiproliferating cell nuclear antigen (PCNA) antibody (ab29; Abcam) at a concentration of 1:1,000. Surfactant protein-C was tagged with an antibody (AB3786; Millipore) at a concentration of 1:1,000, and keratin was tagged with an anti-pan-cytokeratin antibody (sc-8018; Santa Cruz Biotechnology, Santa Cruz, CA, USA) at a concentration of 1:50. Bound antibodies were tagged with either Alexa Fluor 405 or 488 goat anti-rabbit IgG (A31553 or A11034; Invitrogen) or Alexa Fluor 568 donkey anti-mouse IgG (A10037; Invitrogen), all at a concentration of 1:500.
Scanning Electron Microscopy (SEM)
Samples were removed from the bioreactor and fixed for 10 min with 2.5% paraformaldehyde/2.5% glutaraldehyde in 0.1 M sodium cacodylate (15949; Electron Microscopy Sciences, Hatfield, PA, USA). Samples were washed in deionized water for 5 min, and dehydrated using a series of graded ethanol washes (70%, 85%, 95%, 100%, 5 min each), before being submerged in hexamethyldisilazane (HMDS) (16700; Electron Microscopy Sciences) for 10 min. The membrane was removed from the HMDS, allowed to air dry at room temperature, and mounted on a standard SEM support. Samples were sputter coated with a model Auto108 sputter coater from Cressington Scientific Instruments, Ltd. (Watford, UK), using gold/palladium (Cressington 81003) and imaged using a model XL-30 ESEM-FEG microscope from FEI Company (Hillsboro, OR, USA).
Histology
Small portions of each sample were paraffin embedded for cross-sectioning by the Yale School of Medicine Histology Core. Ten-micrometer sections were cut every 100 µm through the sample, six times for each sample, and stained with hematoxylin and eosin (H&E; Sigma-Aldrich). Slides were imaged with a Zeiss Axioskop 2 microscope (Carl Zeiss AG, Oberkochen, Germany) at 40× magnification.
Quantitative Real-Time PCR
The RNeasy Mini Kit from Qiagen was used to extract total RNA following the manufacturer’s instructions. cDNA was synthesized with random hexamers as primers, using the SuperScript First-Strand Synthesis System according to manufacturer’s protocol (Invitrogen). Equal volume mixtures of the products were used as templates for PCR amplification. All reactions were performed in 25-μl volume with iQ™ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and 200 nM each of forward and reverse primers shown using iCyler and iQ software (Bio-Rad). Biological replicates for qRT-PCR n = 1.
The expression of mucin 5AC, ZO-1, forkhead box J1 (FOXJ1), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes by the NHBE cells was identified by qPCR using the following primers: mucin 5AC forward: ATTGCTATTATGCCCTGTGTA, mucin 5AC reverse: TGGTGGACGGACAGTCACT; ZO-1 forward: CGGGAAGTTACGTGGCGAAG, ZO-1 reverse: TCG GACAAAAGTCCGGGAAG; FOXJ1 forward: GCATA AGCGCAAACAGCCG, FOXJ1 reverse: TCGAAGAT GGCCTCCCAGT; GAPDH forward: GACAACAGCCTCAAGATCATCAG, GAPDH reverse: ATGGCATGGACTGTGGTCATGAG. Samples were run in triplicate (technical replicate n = 3). PCR conditions included an initial denaturation step of 4 min at 95°C, followed by 40 cycles of PCR consisting of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Average threshold cycle (Ct) values from the triplicate PCR reactions for a gene of interest (GOI) were normalized against the average Ct values for GAPDH from the same cDNA sample. Fold change of GOI transcript levels between sample A and sample B equals 2−ΔΔCt, where ΔCt = Ct(GOI) − Ct(GAPDH), and ΔΔCt = ΔCt(A) − ΔCt(B). Statistical significance for qRT-PCR was not calculated based on the recommendations of Rieu and Powers (23).
Quantitative Analysis
Imaging and quantification of immunostaining was performed using a Zeiss Axiovert zoom inverted microscope (Carl Zeiss AG), a Hamamatsu camera (Hamamatsu, Bridgewater, NJ, USA), and Volocity software (PerkinElmer, Waltham, MA, USA). For each sample, random locations were blindly selected by manipulating the movement stage with no visual inspection and images taken across all relevant excitation channels. This was repeated three times per sample to reduce sampling error. The image for each excitation channel per frame of view was then printed and manually counted. DAPI-stained nuclei were counted to yield a total cell number and positive protein expression counted to yield a percentage positive value for each sample. For ZO-1, individual closed polygons were counted as positive expression. For mucin 5AC, secretory focal points were counted as positive expression.
This technique yielded a mean of 761 cells counted per experimental sample, with a standard deviation of 432. At least three experimental replicates were performed for each time point of each culture condition (per sample, n ≥ 3), yielding a mean count per condition of 2,360 cells with a standard deviation of 972. A total of 9,633 cells were counted during ZO-1 quantification and 23,406 cells during mucin 5AC quantification. Standard deviation was calculated for each array of samples for a condition, and the mean and associated confidence interval (α = 0.05) were plotted for each data point. An independent two-sample single-tailed t-test, assuming unequal variances between the control and intermittent ALI conditions (Welch’s t-test), was used to calculate p values for each set of compared conditions at each time point.
RESULTS
Cell Proliferation and Viability
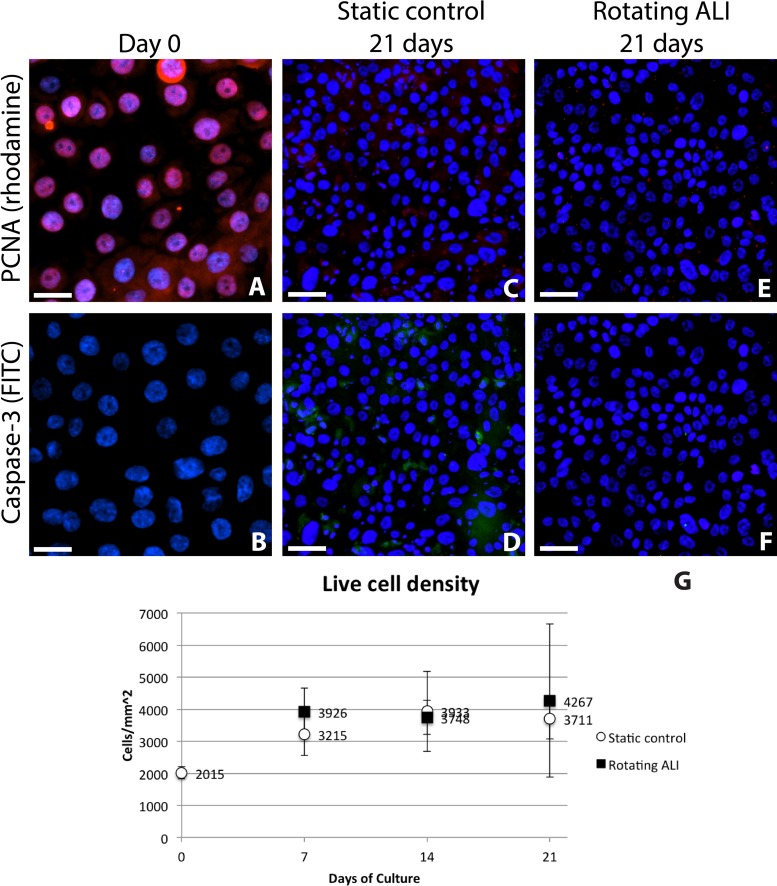
Cells at all day 0 time points, taken 6 h after cell seeding, stained positive for PCNA, as would be expected of recently proliferating cells, and displayed a live cell density of approximately 201,500 ± 19,000 cells/cm2 (Fig. 2). These day 0 samples showed very minimal cell death, with less than 1% of cells staining positive for caspase 3. Live cell density increased to approximately 350,000 cells/cm2 under both experimental conditions between day 0 and day 7; however, PCNA expression ceased by day 7 in the B-ALI-differentiation medium, and no further increase in cell density was observed (Fig. 2G). While occasional cell death occurred during culture, as shown in Figure 2D, most seeded cells remained viable for the duration of the experiment. Both sets of conditions were able to sustain a confluent sheet of cells for up to 3 weeks. No statistically significant difference in either PCNA or caspase 3 staining was observed between corresponding control and rotating ALI time points (p = 0.11 for day 7, p = 0.40 for day 14, and p = 0.34 for day 21) (Fig. 2G).
Figure 2.
Proliferating cell nuclear antigen (PCNA) and caspase 3 staining at day 0 and day 21 of culture for both conditions. Data are not shown for day 7 and day 14 time points, as the pattern of cell staining was consistent with that seen in the day 21 samples. (A, B) 6 h after seeding, the cells stained positive for PCNA [red; rhodamine (Alexa Fluor 568)] but not for caspase 3 [green; fluorescein isothiocyanate (FITC; Alexa Fluor 488)], indicating good cell attachment and viability on the constructs. After 21 days of culture, little difference was observed between the two experimental conditions, with both samples predominantly negative for both markers (C–F). Occasional cells did stain positive for caspase 3, as can be seen in green (D). Nuclei are stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). No significant differences were seen in the live cell density between static control and rotating ALI conditions (G). Scale bars: 25 µm (A, B); 50 µm (C–F).
Tight Junction Formation
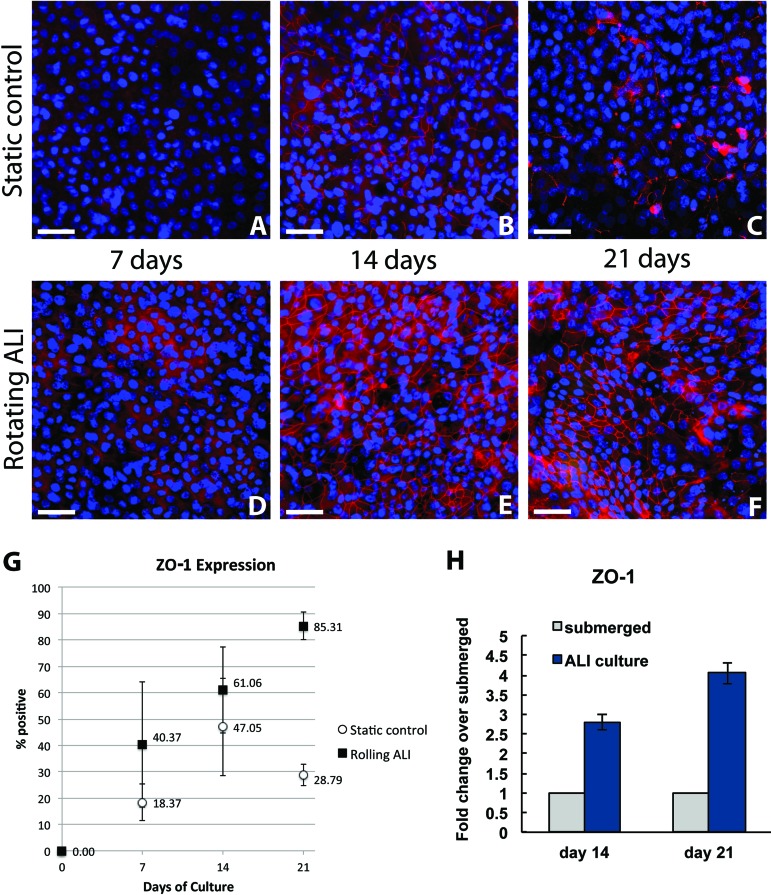
On immunostaining, fully formed tight junctions appear as bright fluorescent lines in a narrow focal plane surrounding the positive-staining cell. At day 7, 18.3 ± 6.8% of the static control population and 40.3 ± 23.8% of the rotating ALI population expressed ZO-1 tight junctions (p = 0.10). At day 14, 47.0 ± 18.5% of the static control population and 61.0 ± 16.4% of the rotating ALI population expressed ZO-1 tight junctions (p = 0.16). By day 21, however, 28.7 ± 4.0% of the static control population and 85.3 ± 5.1% of the rotating ALI population expressed ZO-1 tight junctions (p = 0.00005). Figure 3 shows the steady increase of ZO-1 expression by immunostaining in the rotating ALI cultures alongside the data for the static controls. At all time points, tight junctions formed during intermittent ALI culture (Fig. 3D–F) displayed a higher degree of visual organization and regularity than those formed in the static control populations (Fig. 3A–C). qPCR showed an increase in ZO-1 expression in NHBE cells cultured in the rotating ALI bioreactor compared to the cells cultured in static condition (Fig. 3H). In addition, the mRNA levels for ZO-1 increased over the time from day 14 to day 21 in rotating ALI samples.
Figure 3.
Tight junction formation. Images showing staining for zonula occludens-1 (ZO-1; red) tight junctions in static control and rotating ALI cultures (A–F). Note the initial formation of a tight junction network in static cultures and subsequent deterioration by day 21 (A–C) compared to the steady increase in junction density and organization over time in the rotating ALI cultures (D–F). Nuclei are stained with DAPI (blue). Quantitative assessment of cell ZO-1 expression from immunostaining confirmed this trend and is expressed as percentage of cells staining positive, with 95% confidence intervals (G). Expression of ZO-1 at day 14 and day 21 in NHBE cells cultured in rotating ALI culture system compared to static (submerged) controls (H).
Mucin Secretion
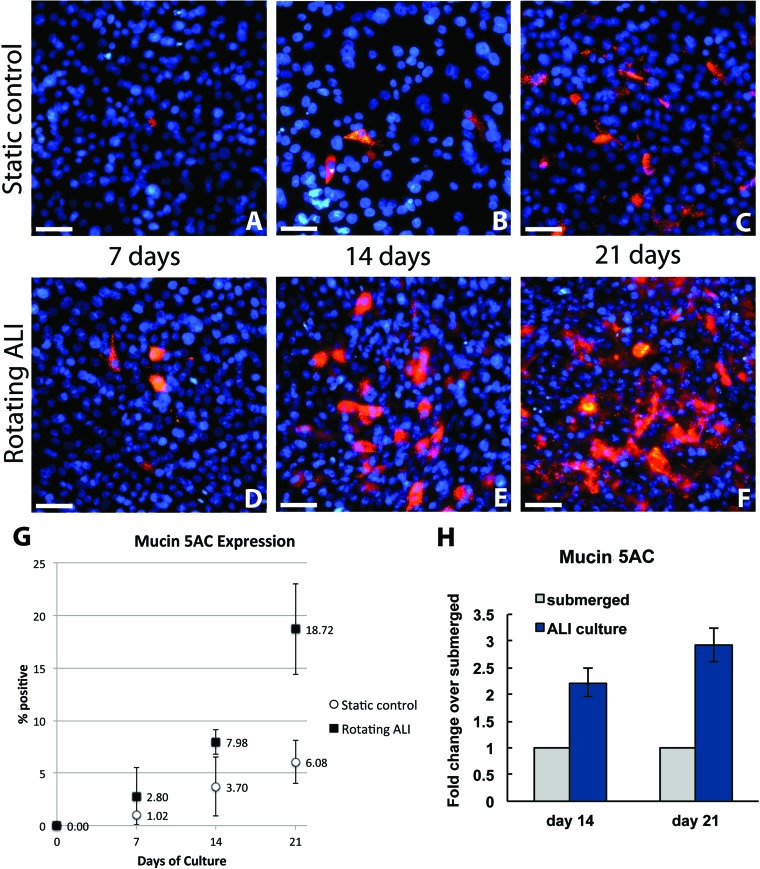
Mucin appears as brightly fluorescent vesicles in the cytoplasm and as extracellular secretions localized around the positive staining cell. At day 7, 1.0 ± 0.2% of the static control population and 2.8 ± 2.7% of the rotating ALI population expressed mucin 5AC (p = 0.16). By day 14, 3.7 ± 2.8% of the static control population and 7.9 ± 1.1% of the rotating ALI population expressed mucin 5AC (p = 0.03). At day 21, 6.0 ± 2.0% of the static control population and 18.7 ± 4.3% of the rotating ALI population expressed mucin 5AC (p = 0.009). At all time points, the rotating ALI condition showed visually higher levels of mucin secretion than the static control (Fig. 4A–F). Mucin 5AC expression in the rotated cultures sometimes manifested as patches of cells covered with a thin continuous layer of mucous (Fig. 4F), a feature that was not observed in the corresponding static controls (Fig. 4C). qPCR confirmed the trends observed by immunocytochemistry, with the 21-day rotating ALI sample displaying higher mucin 5AC expression compared to the static control samples and mucin 5AC expression increasing over time from day 14 to day 21 (Fig. 4H).
Figure 4.
Mucin 5AC expression. Representative images of mucin 5AC expression (red) for each condition at 7, 14, and 21 days of culture (A–F). Both experimental conditions showed an increase in expression over time, but the rate was significantly higher for the rotating ALI cultures, as can be seen by comparing (C) and (F). Nuclei are stained with DAPI (blue). Quantitative measure of mucin expression over time for both experimental conditions, expressed as percentage of cells staining positive, with 95% confidence intervals (G). RT-PCR analysis of mucin 5AC expression in NHBE cells cultured in rotating ALI system at day 14 and day 21 compared to the cells cultured in static conditions (H).
Cilia Formation
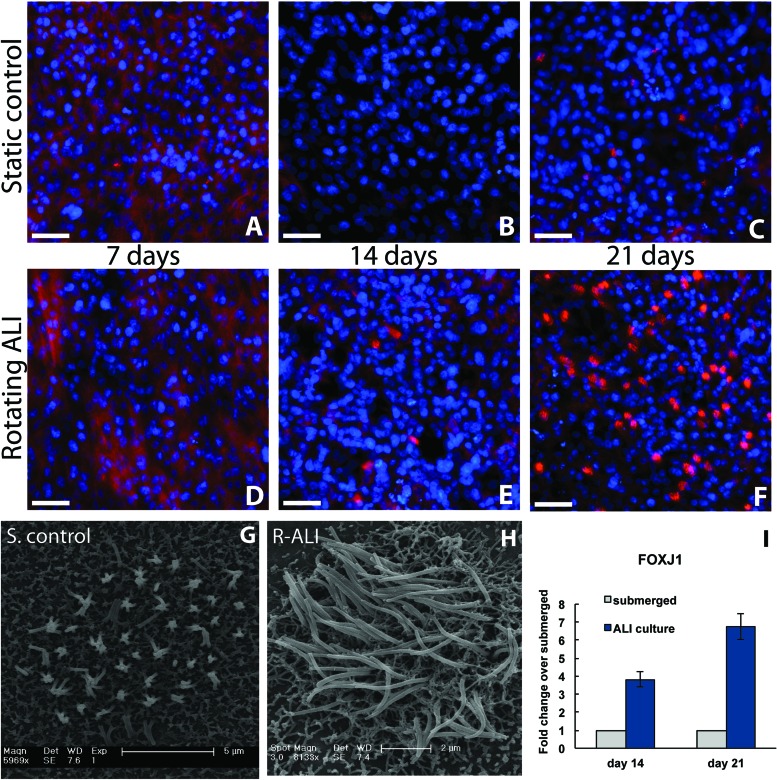
Acetylated tubulin is expressed weakly in the cytoplasm of nonpolarized NHBE cells and can be seen by immunostaining in the 7-day time point of both conditions (Fig. 5). This cytoplasmic staining decreases or disappears with further differentiation, and ciliated patches subsequently appear as brightly fluorescent clusters of rods over cells. Cilia formation appeared to be dependent on cell density, with tightly packed areas displaying the highest degree of cilia expression and organization. Cilia were first observed at day 14 of ALI culture (Fig. 5E) and were seen by day 21 of ALI culture across wide swaths of tissue (Fig. 5F). No cilia were detected in the static controls at either 7- or 14-day time points (Fig. 5A, B). qPCR for the ciliated cell marker, FOXJ1, confirmed these observations, with higher expression of FOXJ1 for 14-day rotating ALI cultures and 21-day rotated cultures compared to static controls at day 14 and day 21. In addition, the expression of FOXJ1 increased over the time from day 7 to day 21 (Fig. 5I). As can be seen in Figure 5C, occasional cells in the static control group showed cilia formation at day 21, which testifies to the ability of the medium to induce differentiation independent of environmental cues. Cilia seen in the static controls were sparse and appeared stunted and disorganized, which aligns with previous findings (4). SEM images confirmed that the ciliated cells present in the rotating ALI cultures (Fig. 5H) were more developed than those seen in the static controls (Fig. 5G), reaching approximately 5 µm in length.
Figure 5.
Cilia development. Immunocytochemistry for each experimental condition across all relevant time points (A–F). Acetylated tubulin can be found in the cytoplasm of nonpolarized NHBE cells (red; A, D). This expression disappears after 2 weeks of culture in the differentiation-inducing media (B, C). Fully formed cilia stain strongly for acetylated tubulin, as can be seen in (E, F). Nuclei are stained with DAPI (blue). Scanning electron micrographs comparing the highest degree of ciliation observed in the day 21 static controls (G) and in the day 21 rotating ALI cultures (H). Forkhead box J1 (FOXJ1) gene expression in NHBE cells cultured in rotating ALI system at day 14 and day 21 compared to the cells cultured under static conditions (I). Scale bars: 50 µm (A–F); 5 µm and 2 µm (G and H), respectively.
Histology
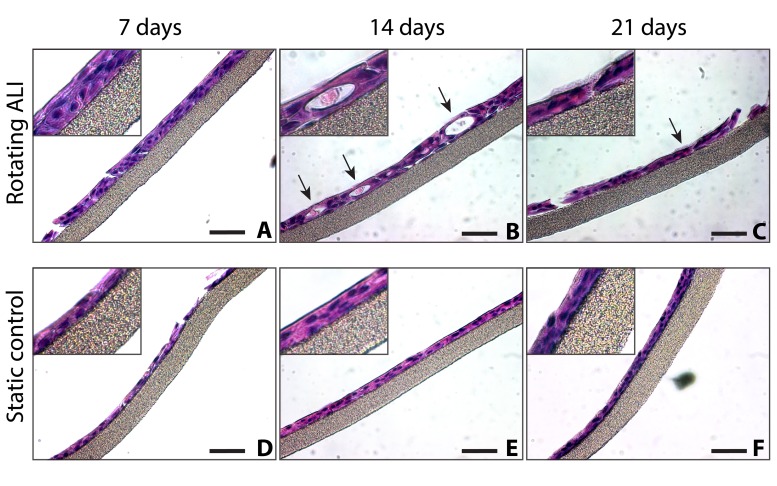
Transverse paraffin sections were taken to provide a more complete view of the tissue and to confirm the above findings (Fig. 6). Secretory cells were observed by day 14 in rotating ALI samples, as can be seen in Figure 6B (indicated with arrows). Identifiable cilia could occasionally be observed in the day 21 rotating ALI samples, shown in Figure 6C (indicated with arrow, magnified in inset). Secretory cells were rarely found in the corresponding static control time points, and cilia were never observed by H&E staining, though planar immunostaining did show some degree of ciliation in these samples (Fig. 5C). No consistent difference was observed between the experimental groups at day 7.
Figure 6.
Paraffin sections. Shown is the PTFE membrane with apically attached cells, without PVDF backing, cut perpendicular to membrane plane. Little difference was observed between the experimental conditions at day 7 (A, D). Goblet cells appeared by day 14 in the rotating cultures, indicated with arrows (B). An intermittent “brush border” indicating ciliation was occasionally observed by day 21 in the rotating cultures, indicated with arrows and magnified in inset (C). Little morphological development was witnessed over time in the static samples (D–F). Scale bars: 50 µm.
Primary Cell Culture
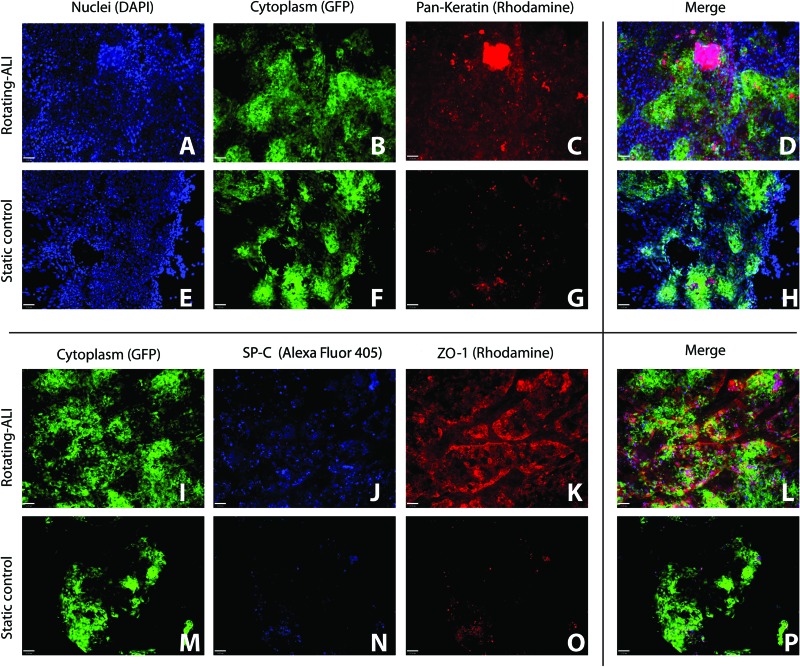
A preliminary experiment was performed to determine if the bioreactor could maintain primary lung epithelial cells in a differentiated state. GFP+ rat cells, enriched for alveolar epithelium, were cultured for 7 days under both experimental conditions. Cells were then fixed and stained for keratin, surfactant protein C (SP-C), and ZO-1 (Fig. 7). Cells cultured in the rotating bioreactor displayed greater keratin and ZO-1 expression than their submerged counterparts, which correlated with our findings from NHBE culture and suggested that the conditions within the reactor are selecting for cells with an epithelial phenotype. The cells also stained more positively for SP-C production when cultured within the reactor, compared to static culture, which suggests that the culture conditions might be appropriate for growing alveolar cell types as well as bronchial. This hypothesis has been confirmed by Ghaedi et al., who found that this reactor could be used to differentiate induced pluripotent stem cell-derived lung epithelial cells toward an alveolar type-I phenotype (7).
Figure 7.
Primary cell culture of green fluorescent protein-positive (GFP+) rat lung epithelial cells. Shown is immunostaining of freshly isolated GFP-modified rat pulmonary epithelium, enriched for alveolar cell types, at day 7 in both experimental conditions. (A–D) and (E–H) Immunostaining of keratin [red; rhodamine (Alexa Fluor 568)] in rolled and static cultures, respectively. Nuclei are stained with DAPI (blue). (I–L) and (M–P) Surfactant protein C (SP-C) (blue; Alexa Fluor 405) and ZO-1 [red; rhodamine (Alexa Fluor 568)] in rolled and static cultures, respectively. Cells cultured in the bioreactor displayed greater keratin expression, greater ZO-1 expression, and greater SP-C expression than their submerged counterparts, suggesting that the reactor might be a versatile tool for growing a broad array of epithelial subtypes. Scale bars: 53 µm.
Scalability and Adaptation for Bulk Production
The two-dimensional cultures of respiratory epithelium in these experiments were seeded at a density of 150,000–200,000 cells/cm2, which yields a confluent sheet of tightly packed cells (300,000–400,000 cells/cm2) similar to that found lining the airways in vivo. We found that a ratio of 1.5 ml of medium for every square centimeter of tissue, changed daily, was sufficient to support continuous culture. Assuming that eight 12-cm-diameter vials can be easily arranged along a linear meter, this type of system consequently allows for the seeding of 241,274 cm2 of construct, or approximately 48 billion cells, per cubic meter of culture space. Previous studies have estimated the number of epithelial cells in the nonalveolar portion of the lung to be approximately 10 billion (17), and in the alveolar region to be approximately 35 billion (25). Hence, the system can be adapted for large-scale laboratory and commercial cell production and differentiation, and might prove valuable for clinically scaled lung tissue engineering.
DISCUSSION
Current techniques for differentiating respiratory epithelium are dependent on permeable cell-insert technology, which was developed over 30 years ago by Whitcutt and colleagues (26). While cell-insert technology is well suited to the investigation of the impact of various drugs and other stimuli on cell behavior, it is not designed for use by the growing number of researchers who require large numbers of cells to engineer three-dimensional biological constructs or to grow tissue in vitro for clinical use. This report describes a low-maintenance bioreactor system that is capable of producing differentiated epithelial cells at scale, with little manual attention.
The current design of this system does not allow real-time imaging of the cells, a problem that might be overcome by finding an optically clear alternative to the PVDF sponge layer. Additionally, the culture vial dimensions, rotation speed, and ratio between submerged and ALI culture within the vial may not yet be optimized, and the tested set of conditions appeared to yield sparser cilia development than often reported for porous membrane inserts. Better results might be obtained by thickening the absorbent PVDF membrane and reducing the physical height of medium within the reactor, thereby altering the ratio of ALI to submerged culture time. It is important to note that the NHBE cells used in this experiment, sourced from Lonza, had been grown on plastic prior to storage, and had lost much of their native morphology by the time they were seeded into the reactor. The primary rat cells, however, were seeded almost immediately after isolation, and the data shown in Figure 7 strongly suggest that this reactor might support the expansion of primary epithelial cells without concurrent loss of differentiation.
Overall, the bioreactor provides a promising alternative to current methods for epithelial expansion. We have shown that by rolling a porous, absorbent construct alternately through media and air, we can generate an intermittent, functional ALI that promotes epithelial differentiation and that can induce human bronchial epithelial cells to express native markers. Cells cultured in this fashion display higher levels of tight junction formation, mucin secretion, and cilia formation compared to static cultures that were controlled for other variables. After 21 days of intermittent ALI culture in the rotating bioreactors, tight junction formation and mucous production is widespread, while groups of cells show well-developed cilia with an overall morphology similar to that found in vivo. With further refinement, this technique has potential for application in a variety of research areas, and it could accelerate research projects and clinical experiments that rely on the growth and differentiation of human tracheal, bronchial, and alveolar epithelium.
ACKNOWLEDGMENTS
This work was funded by 1U01HL111016-01 and R01HL098220-01. Thanks to Aaron Zimmerman and Jaclyn Shepard for helping to review the manuscript. L.E.N. has a research grant from United Therapeutics, Inc. L.E.N. has a financial interest in Humacyte, Inc., a regenerative medicine company. Humacyte did not fund these studies, and Humacyte did not affect the design, interpretation, or reporting of any of the experiments herein.
REFERENCES
- 1. Asnaghi M. A.; Jungebluth P.; Raimondi M. T.; Dickinson S. C.; Rees L. E.; Go T.; Cogan T. A.; Dodson A.; Parnigotto P. P.; Hollander A. P.; Birchall M. A.; Conconi M. T.; Macchiarini P.; Mantero S. A double-chamber rotating bioreactor for the development of tissue-engineered hollow organs: From concept to clinical trial. Biomaterials 30:5260–5269; 2009. [DOI] [PubMed] [Google Scholar]
- 2. Baiguera S.; Jungebluth P.; Burns A.; Mavilia C.; Haag J.; De Coppi P.; Macchiarini P. Tissue engineered human tracheas for in vivo implantation. Biomaterials 31:8931–8938; 2010. [DOI] [PubMed] [Google Scholar]
- 3. Bernacki S. H.; Nelson A. L.; Abdullah L.; Sheehan J. K.; Harris A.; Davis C. W.; Randell S. H. Mucin gene expression during differentiation of human airway epithelia in vitro. Am. J. Respir. Cell. Mol. 20:595–604; 1999. [DOI] [PubMed] [Google Scholar]
- 4. de Jong P. M.; van Sterkenburg M. A.; Hesseling S. C.; Kempenaar J. A.; Mulder A. A.; Mommaas A. M.; Dijkman J. H.; Ponec M. Ciliogenesis in human bronchial epithelial cells cultured at the air-liquid interface. Am. J. Respir. Cell Mol. Biol. 10(3):271–277; 1994. [DOI] [PubMed] [Google Scholar]
- 5. Fishman J. M.; De Coppi P.; Elliot M. J.; Atala A.; Macchiarini P. Airway tissue engineering. Expert Opin. Biol. Ther. 11(12):1623–1635; 2011. [DOI] [PubMed] [Google Scholar]
- 6. Fulcher M. L.; Gabriel S.; Burns K. A.; Yankaskas J. R.; Randell S. H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 107:183–206: 2005. [DOI] [PubMed] [Google Scholar]
- 7. Ghaedi G.; Mendez J. J.; Bove P. F.; Sivarapatna A.; Raredon M. S. B.; Niklason L. E. Alveolar epithelial differentiation of human induced pluripotent stem cells in a rotating bioreactor. Biomaterials 35:699–710; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gilpin S. E.; Guyette J. P.; Gonzalez X. R.; Asara J. M.; Mathisen D. J.; Vacanti J. P.; Ott H. C. Perfusion decellularization of human and porcine lungs: Bringing the matrix to clinical scale. J. Heart Lung Transplant, 33(3):298–308; 2014. [DOI] [PubMed] [Google Scholar]
- 9. Gray T. E.; Guzman K.; Davis C. W.; Abdullah L. H.; Nettesheim P. Mucociliary differentiation of serially passage normal human tracheobronchial epithelial cells. Am. J. Respir. Cell. Mol. Biol. 14:104–112; 1996. [DOI] [PubMed] [Google Scholar]
- 10. Grek C. L.; Newton D. A.; Qiu Y.; Wen X.; Spyropoulos D. D.; Baatz J. E. Characterization of alveolar epithelial cells cultured in semipermeable hollow fibers. Exp. Lung Res. 35:155–174; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hermanns M. I.; Unger R. E.; Kehe K.; Peters K.; Kirkpatrick C. J. Lung epithelial cell lines in coculture with human pulmonary microvascular endothelial cells: Development of an alveolo-capillary barrier in vitro. Lab. Invest. 84:736–752; 2004. [DOI] [PubMed] [Google Scholar]
- 12. Jorissen M.; Van der Schueren B.; Van den Berghe H.; Cassiman J. J. The preservation and regeneration of cilia on human nasal epithelial cells cultured in vitro. Arch. Otorhinolarygol. 246:308–314; 1989. [DOI] [PubMed] [Google Scholar]
- 13. Lechner J. F.; LaVeck M. A. A serum-free method for culturing normal human bronchial epithelial cells at clonal density. J. Tissue Cult. Meth. 9(2):43–48; 1985. [Google Scholar]
- 14. le Visage C.; Dunham B.; Flint P.; Leong K. W. Coculture of mesenchymal stem cells and respiratory epithelial cells to engineering a human composite respiratory mucosa. Tissue Eng. 10(9/10):1426–1435; 2004. [DOI] [PubMed] [Google Scholar]
- 15. Lin H.; Li H.; Cho H. J.; Bian S.; Roh H. J.; Lee M. K.; Kim J. S.; Chung S. J.; Shim C. K.; Kim D. D. Air-liquid interface (ALI) culture of human bronchial epithelial cell monolayers as an in vitro model for airway drug transport studies. J. Pharm. Sci. 96(2):341–350; 2007. [DOI] [PubMed] [Google Scholar]
- 16. Macchiarini P.; Jungebluth P.; Go T.; Asnaghi M. A.; Rees L. E.; Cogan T. A.; Dodson A.; Martorell J.; Bellini S.; Parnigotto P. P.; Dickinson S. C.; Hollander A. P.; Mantero S.; Conconi M. T.; Birchall M. A. Clinical transplantation of a tissue-engineered airway. Lancet 372:2023–2030; 2008. [DOI] [PubMed] [Google Scholar]
- 17. Mercer R. R.; Russell M. L.; Roggli V. L.; Crapo J. D. Cell number and distribution in human and rat airways. Am. J. Respir. Cell Mol. Biol. 10(6):613–624; 1994. [DOI] [PubMed] [Google Scholar]
- 18. Miller C.; George S.; Niklason L. Developing a tissue-engineered model of the human bronchiole. J. Tissue Eng. Regen. Med. 4:619–627; 2010. [DOI] [PubMed] [Google Scholar]
- 19. Nichols J. E. E.; Niles J.; Riddle M.; Vargas G.; Schilagard T.; Ma L.; Edward K.; Lafrancesca S.; Sakamoto J.; Vega S.; Ogedegbe R.; Mlcak R.; Deyo D.; Woodson L.; McQuitty C.; Lick S. Beckles D.; Melo E.; Cortiella J. Production and assessment of decellularized pig and human lung scaffolds. Tissue Eng. 19:2045–2062; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ott H. C.; Clippinger B.; Conrad C.; Schuetz C.; Pomerantseva I.; Ikonomou L.; Kotton D.; Vacanti J. P. Regeneration and orthotopic transplantation of a bioartificial lung. Nat. Med. 16(8):927–933; 2010. [DOI] [PubMed] [Google Scholar]
- 21. Petersen T. H.; Calle E. A.; Zhao L.; Lee E. J.; Gui L.; Raredon M. S. B.; Gavrilov K.; Yi T.; Zhuang Z. W.; Breuer C.; Herzog E.; Niklason L. E. Tissue-engineered lungs for in vivo implantation. Science 329(5991):538–541; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rice W. R.; Conkright J. J.; Na C. L.; Ikegami M.; Shannon J. M.; Weaver T. E. Maintenance of the mouse type II cell phenotype in vitro. Am. J. Physiol. Lung Cell Mol. Physiol. 283(2):L256–L264; 2002. [DOI] [PubMed] [Google Scholar]
- 23. Rieu I.; Powers S. J. Real-time quantitative RT-PCR: Design, calculations, and statistics. Plant Cell 21:1031–1033; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soleas J. P.; Paz A.; Marcus P.; McGuigan A.; Waddell T. K. Engineering airway epithelium. J. Biomed. Biotechnol. 2012:1–10; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stone K. C.; Mercer R. R.; Gehr P.; Stockstill B.; Crapo J. D. Allometric relationships of cell numbers and size in the mammalian lung. Am. J. Respir. Cell Mol. Biol. 6(2):235–243; 1992. [DOI] [PubMed] [Google Scholar]
- 26. Whitcutt M. L.; Adler K. B.; Wu R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell. Dev. B 24(5):420–428; 1988. [DOI] [PubMed] [Google Scholar]