Abstract
This study aimed to investigate whether single nucleotide polymorphisms (SNPs) of five NLR family genes (NOD1, NOD2, NLRP1, NLRP3 and CIITA) are associated with Behcet’s disease (BD) in a Chinese Han population. The study was carried out in 950 BD patients and 1440 controls for 19 SNPs in the selected NLR genes. In the first-stage study, significantly decreased frequencies of the CIITA//rs12932187 C allele (Pc = 1.668E-02) and NOD1//rs2075818 G allele (Pc = 4.694E-02) were found in BD patients as compared to controls . After performing a second stage validation study and combination of data we confirmed the association of CIITA//rs12932187 and NOD1//rs2075818 with BD. In CIITA//rs12932187, the frequencies of the CC genotype and C allele were significantly lower in BD than in controls (Pc = 3.331E-06; Pc = 6.004E-07, respectively). In NOD1//rs2075818, the GG genotype and G allele showed significantly decreased frequencies in BD patients when compared to controls (Pc = 1.022E-02; Pc = 6.811E-05, respectively). Functional experiments showed that carriers with the CC genotype in CIITA//rs12932187 had a lower CIITA mRNA expression level and an enhanced IL-10 secretion as compared to GG and CG carriers. This study provides evidence that the CIITA and NOD1 gene are involved in the susceptibility to Behcet’s disease.
Behçet’s disease (BD) is a multifactorial disease which presents with oral aphthae, genital ulcerations, ocular inflammation, skin lesions and a pathognomonic pathergy test1. The etiology of BD is largely unknown, but cumulative evidence suggests that an excessive T-cell mediated inflammatory response is associated with disease activity2. Previous studies revealed that a number of genetic factors are involved in disease susceptibility, such as STAT3, STAT4, TLR2, miR-182, FAS and CD40 genes3,4,5,6,7,8,9.
BD is associated with important morbidity of which the intraocular inflammation may lead to serious visual handicap. The disease is currently being treated with corticosteroids and a variety of immunosuppressive agents. Further knowledge of the inflammatory pathways involved in the disease process may lead to the development of new drugs to target these disorders. One of the approaches currently used includes the analysis of the association of these diseases with gene polymorphisms of proteins involved in the immune or inflammatory response. Since BD may be triggered by an infectious process we focused on gene polymorphisms associated with the microbial immune response8.
Nucleotide-binding domain and leucine-rich repeat containing (NLRs) including at least 22 known proteins, exist in the cytosol and play an important role in the recognition of microbial products10. They are characterized by three structural domains: a NACHT- domain for oligomerization and activation of the NLRs, an LRR domain at the C-terminus which is responsible for recognition of microbial patterns, and a protein–protein interaction domain at the N-terminus that could be formed of a pyrin (PYD)-, caspase (CARD) or a baculo-virus inhibitor of apoptosis repeat (BIR) domain, triggering the signal transduction cascade10.
Few NLRs have been well characterized thus far, however, more recent studies demonstrate that variation in NLRs genes are associated with autoimmune or inflammatory disease. NOD2 was the first identified CD (Crohn’s disease) susceptibility gene11 and variations of NOD1 have been shown to confer risk to inflammatory bowel diseases (IBD) and CD12,13. NLRP3 variations have also been found to be associated with several autoimmune diseases including neonatal-onset multi-system inflammatory disease (NOMID), Muckle-Wells syndrome (MWS) and familial cold urticaria (FCU)14,15,16. NLRP1 has been found to confer risk to autoimmune rheumatoid arthritis (RA), Addison’s disease, type I diabetes and vilitigo17,18,19. Variation in CIITA was also found to be related to a number of autoimmune diseases such as RA, myocardial infarction and multiple sclerosis (MS)20,21.
On the basis of these previous studies, we conducted this research to investigate whether polymorphisms of NLR family genes including NOD1, NOD2, NLRP1, NLRP3 and CIITA gene were associated with BD.
Results
Clinical Features
Nineteen SNPs in five selected NLR genes (NOD1, NOD2, NLRP1, NLRP3 and CIITA) were genotyped successfully and all SNPs did not deviate from the Hardy-Weinberg equilibrium in controls. The case group comprised 186 women and 764 men, and the average age of the BD patients was 33.0031 ± 8.4 years. The healthy control cohort consisted of 1440 subjects (321 women, 1119 men), in which the average age was 35.9 ± 11.2 years. There were no statistical differences in age and sex between cases and controls (P > 0.05). The demographics and clinical features of BD patients enrolled in the study are summarized in Table 1.
Table 1. Clinical Features, Age and Gender Distribution in Controls as well as Patients with Ocular BD.
| Clinical Features | Total | % |
|---|---|---|
| Ocular BD Patients | 950 | |
| Age at onset, year ± SD | 33.1 ± 8.4 | |
| Female | 950 | 19.6 |
| Male | 755 | 80.4 |
| Uveitis | 950 | 100 |
| Oral ulcer | 950 | 100 |
| Genital ulcer | 466 | 49.1 |
| Skin lesions | 570 | 60.0 |
| Pathergy reaction | 231 | 24.3 |
| Hypopyon | 202 | 21.3 |
| Arthritis | 151 | 15.9 |
| Controls | 1440 | |
| Age at onset, year ± SD | 35.9 ± 11.2 | |
| Female | 321 | 22.3 |
| Male | 1119 | 77.7 |
SD = standard deviation.
BD = Behcet disease.
Genotype Results
In first stage study, the frequency of the CIITA//rs12932187 C allele (Pc = 1.668 × 10−2, OR = 0.713, 95% CI = 0.591–0.861) and NOD1//rs2075818 G allele (Pc = 4.694E-02, OR = 0.698, 95% CI = 0.562–0.868) were decreased in BD patients compared to controls (Table 2). The other seventeen SNPs did not show a significant association with BD (Supplementary Table S1). In the second stage, we tested another set of 566 BD patients and 870 healthy controls to confirm the result of the first stage study. After combination of the data, the frequencies of CC genotype and C allele in CIITA//rs12932187, were significantly decreased in the BD patients (Pc = 3.331 × 10−6, OR = 0.617, 95% CI = 0.519–0.735; Pc = 6.004 × 10−7, OR = 0.709, 95% CI = 0.629–0.799, respectively). In NOD1//rs2075818, the frequencies of the GG genotype and G allele were also decreased in the BD patients (Pc = 1.022E-02, OR = 0.536, 95% CI = 0.386–0.745; Pc = 6.811 × 10−5, OR = 0.720, 95% CI = 0.629–0.824, respectively).
Table 2. Polymorphisms of NOD1//rs2075818 and CIITA//rs12932187 in Behcet’s Disease.
| SNPs | Stage | Genotype | Cases, No. |
Controls, No. |
P Value | Pc Value | OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|---|
| Allele | (Frequency) | (Frequency) | |||||||
| rs12932187 (CIITA) | Firsta | CC | 113 | (0.298) | 233 | (0.405) | 8.212E-04 | 4.927 E-02 | 0.625 (0.474 to 0.824) |
| CG | 204 | (0.538) | 280 | (0.486) | 0.115 | NS | 1.232 (0.950 to 1.598) | ||
| GG | 62 | (0.164) | 63 | (0.109) | 0.015 | NS | 1.593 (1.093 to 2.323) | ||
| C | 430 | (0.567) | 746 | (0.648) | 4.169E-04 | 1.668E-02 | 0.713 (0.591 to 0.861) | ||
| G | 328 | (0.433) | 406 | (0.352) | 4.169E-04 | 1.668E-02 | 1.402 (1.162 to 1.691) | ||
| Replicationb | CC | 170 | (0.304) | 359 | (0.416) | 1.878E-05 | 1.120E-02 | 0.612 (0.489 to 0.767) | |
| CG | 298 | (0.533) | 408 | (0.473) | 0.028 | NS | 1.270 (1.026 to 1.573) | ||
| GG | 91 | (0.163) | 95 | (0.110) | 0.004 | NS | 1.570 (1.152 to 2.140) | ||
| C | 638 | (0.571) | 1126 | (0.653) | 9.586E-06 | 3.835E-04 | 0.706 (0.605 to 0.824) | ||
| G | 480 | (0.429) | 598 | (0.347) | 9.586E-06 | 3.835E-04 | 1.417 (1.214 to 1.653) | ||
| Combinedc | CC | 283 | (0.302) | 592 | (0412) | 5.552E-08 | 3.331E-06 | 0.617 (0.519 to 0.735) | |
| CG | 502 | (0.535) | 688 | (0.478) | 0.007 | NS | 1.255 (1.064 to 1.480) | ||
| GG | 153 | (0.163) | 158 | (0.110) | 1.693E-04 | 1.016E-05 | 1.579 (1.243 to 2.006) | ||
| C | 1068 | (0.569) | 1872 | (0.651) | 1.501E-08 | 6.004E-07 | 0.709 (0.629 to 0.799) | ||
| G | 808 | (0.431) | 1004 | (0.349) | 1.501E-08 | 6.004E-07 | 1.411 (1.252 to 1.589) | ||
| rs2075818 (NOD1) | First | CC | 235 | (0.618) | 299 | (0.525) | 0.004 | NS | 1.469 (1.128 to 1.913) |
| CG | 130 | (0.342) | 217 | (0.381) | 0.226 | NS | 0.846 (0.645 to 1.109) | ||
| GG | 15 | (0.040) | 54 | (0.094) | 0.001 | NS | 0.393 (0.218 to 0.707) | ||
| C | 600 | (0.789) | 825 | (0.724) | 1.173E-03 | 4.694E-02 | 1.432 (1.152 to 1.780) | ||
| G | 160 | (0.211) | 315 | (0.276) | 1.173E-03 | 4.694E-02 | 0.698 (0.562 to 0.868) | ||
| Replicationb | CC | 328 | (0.588) | 431 | (0.502) | 0.002 | NS | 1.416 (1.142 to 1.756) | |
| CG | 193 | (0.346) | 341 | (0.397) | 0.052 | NS | 0.803 (0.644 to 1.002) | ||
| GG | 37 | (0.066) | 87 | (0.101) | 0.023 | NS | 0.630 (0.422 to 0.940) | ||
| C | 849 | (0.761) | 1203 | (0.700) | 4.29E-04 | 1.715E-02 | 1.361 (1.146 to 1.617) | ||
| G | 267 | (0.239) | 515 | (0.300) | 4.29E-04 | 1.715E-02 | 0.735 (0.619 to 0.872) | ||
| Combinedc | CC | 563 | (0.601) | 730 | (0.511) | 1.941E-05 | 1.164E-03 | 1.438 (1.217 to 1.699) | |
| CG | 323 | (0.344) | 558 | (0.390) | 0.023 | NS | 0.820 (0.691 to 0.973) | ||
| GG | 52 | (0.055) | 141 | (0.099) | 1.703E-04 | 1.022E-02 | 0.536 (0.386 to 0.745) | ||
| C | 1449 | (0.772) | 2028 | (0.710) | 1.703E-06 | 6.811E-05 | 1.389 (1.214 to 1.589) | ||
| G | 427 | (0.228) | 830 | (0.290) | 1.703E-06 | 6.811E-05 | 0.720 (0.629 to 0.824) | ||
NS = no significant difference; OR = odds ratio; Pc = P value with Bonferroni correction.
SNP = single nucleotide polymorphism.
aFirst stage (stage 1), case-to-control ratio: 380:576; bReplication stage (stage 2), case-to-control ratio: 559:862; cCombined stage(a+b), case-to-control ratio: 939:1438.
mRNA level and downstream inflammatory factors
Because of the significant association of CIITA//rs12932187 and NOD1//rs2075818 with BD, we tested the expression of NOD1 and CIITA in PBMCs obtained from healthy individuals with known genotypes of the two SNPs. Real-time PCR did not show a detectable association between the various genotypes and the expression of NOD1 and CIITA when testing unstimulated PBMCs (Supplementary Fig. S1 and Supplementary Fig. S3). Following stimulation by LPS, carriers with the CC genotype in CIITA//rs12932187 had a lower mRNA expression of CIITA compared with the GG or CG genotype carriers (P = 0.004, Fig. 1). anti-CD3/anti-CD28 stimulation did not affect CIITA expression (Supplementary Fig. S2) and no effect on NOD1 mRNA expression was observed for the various rs2075818 genotypes by either normal or stimulated PBMCs (Supplementary Fig. S4 and Supplementary Fig. S5).
Figure 1. The influence of various rs12932187 genotypes on CIITA mRNA expression after stimulation with LPS.
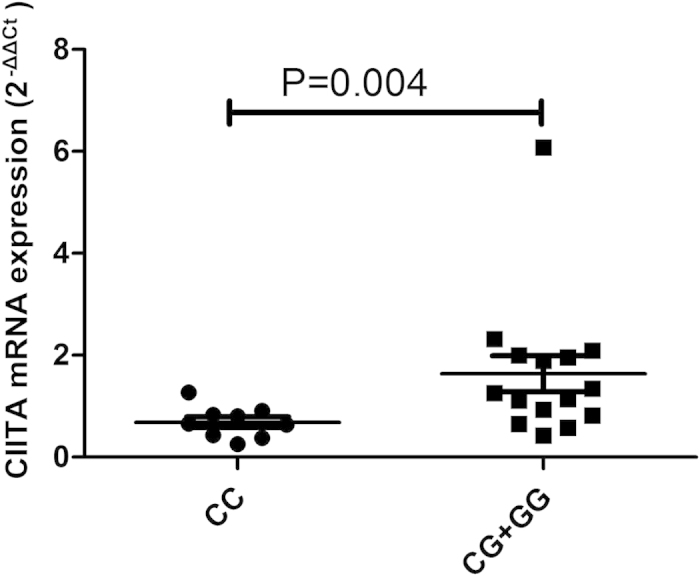
The expression of CIITA in PBMCs treated with LPS. PBMCs were obtained from healthy individuals with diverse genotypes of CIITA//rs12932187.
Since the different genotypes of CIITA//rs12932187 had an effect on CIITA mRNA expression, we decided to investigate whether the different genotypes influenced the cytokine response of PBMCs following LPS stimulation. We measured the PBMC expression levels of IL-6, IL-8, IL-10, IL-1β, TNF-α and MCP-1 by ELISA. These cytokines have all been shown to play a role in the development of BD as shown by earlier studies22,23. Carriers of the CC genotype had a higher secretion level of IL-10 as compared to GG and CG carriers (P = 0.017, Fig. 2). No significant effect on secretion levels of IL-6, IL-8, IL-1β, TNF-α and MCP-1 was found (Supplementary Fig. S6–S10).
Figure 2. The influence of various rs12932187 genotypes on secretion of IL-10 after stimulation with LPS.
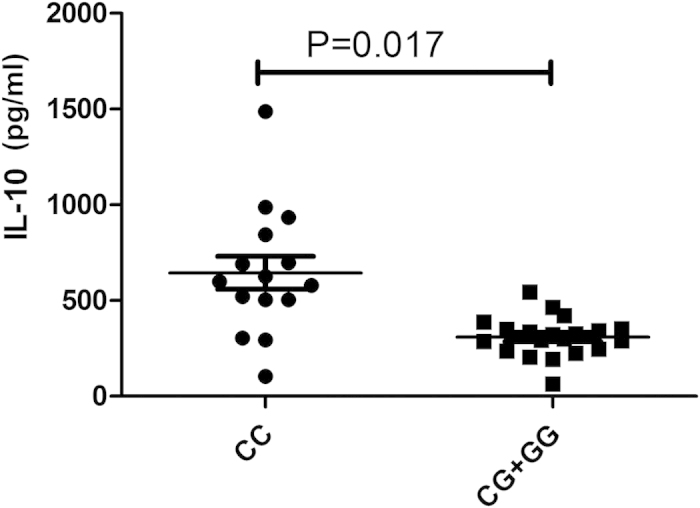
The production of IL-10 in PBMCs obtained from healthy genotype controls. PBMCs were treated with LPS.
Discussion
In the present study, we investigated the associations of 19 SNPs in NOD1, NOD2, NLRP1, NLRP3 and CIITA with BD in a Chinese Han population. Two SNPs, rs12932187 of CIITA and rs2075818 of NOD1 contributed to the genetic susceptibility of BD. Functional studies showed that carriers of the CC genotype of CIITA//rs12932187 had a lower CIITA mRNA expression level and an increased IL-10 secretion by PBMCs as compared to GG and CG carriers.
CIITA acts as a transcriptional coactivator and has been associated with various inflammatory and autoimmune diseases24,25. CIITA mediates activated immune responses and its deficiency has been shown to cause Type II Bare lymphocyte syndrome (BLS)26. Variability in the CIITA gene has also been reported to be associated with several autoimmune and inflammatory diseases such as myocardial infarction (MI), rheumatoid arthritis (RA), type I diabetes (T1D) and multiple sclerosis (MS)23,24. A case-control study was performed in 1320 MS cases and 1363 independent healthy controls and the results showed that CIITA//rs4774 was associated with MS, particularly in DRB1*1501(+) cases27. Another study showed that the two SNPs rs4774 and rs6498122 of CIITA were associated with oral lichen planus (OLP)28. CIITA//rs8048002 was found to be associated with RA in a Swedish cohort29. CIITA//rs12932187 and rs11074938 were found to be susceptibility markers of nasal passages inflammation in asthma patients in a Japanese population30. In our study, only the CIITA//rs12932187 G allele and GG genotype were associated with BD risk. CIITA has been shown to function not only as a transcriptional regulator of MHC genes, but is also a transcriptional regulator of over 60 immunologically important genes, including IL-4, IL-10 and a number of thyroid-specific genes24,25. A study in CIITA-deficient (CIITA(−/−)) mice showed that CIITA negatively regulates the expression of IL-10 by DCs, which supports the findings in humans as presented here31. IL-10 is considered an immune regulatory cytokine which controls innate and adaptive immune responses32. Low IL-10 serum levels have been reported in Asian patients with BD33. The functional tests we performed showed that carriers with the CC genotype of CIITA//rs12932187 had a lower CIITA mRNA expression level and an enhanced IL-10 secretion as compared to GG and CG carriers. The protective effect of the C allele and CC genotype concerning BD development could thus be explained by the fact that these individuals produce more anti-inflammatory IL-10 in response to a microbial stimulus than carriers of the G allele. Further studies are needed to support this hypothesis.
NOD1 has been characterized as a critical regulator of innate immunity. Various studies have reported the association between NOD1 gene variants and autoimmune disease10. The NOD1//rs2075818 G allele was found to decrease the risk of CD and rs2907748 AA and AG genotypes showed a decreased frequency in UC13. These findings are in agreement with our study and could be due to the fact that BD as well as these inflammatory bowel disease are considered as an autoinflammatory disease caused by an aberrant response to a microbial agent. We were not able to detect a functional explanation for the association with NOD1//rs2075818.
NLRP1 and NLRP3 have been shown to play an important role in the processing of pivotal pro-inflammatory cytokines such as IL-1β and IL-1814,16. Gene variants of NLRP3 have been shown to be associated with Psoriatic Juvenile Idiopathic Arthritis in a Caucasian population34. Moreover, genetic variants of NLRP1 were observed to confer risk for the development of vitiligo35. Nevertheless, our study did not find an association between NLRP1 and NLRP3 and BD in a Chinese Han population. NOD2, that was already identified as a CD-susceptibility gene11, was not associated with BD.
Our study has some limitations. Since the subjects in our study were all Chinese Han, the conclusions of the study are only valid to the Chinese Han population and should be studied and replicated in other ethnic groups. Furthermore, all the BD patients in this study were recruited from ophthalmology departments and a selection bias in our patient population may be present. Whether our findings can be generalized to other uveitis entities is not known and deserves further study. We did test the SNPs described in this study on uveitis patients with Vogt Koyanagi Harada syndrome but did not observe statistically significant associations (data not shown).
In conclusion, this study for the first time reports an association of CIITA//rs12932187 and NOD1//rs2075818 with susceptibility to BD in a Chinese Han population. A functional variant of CIITA//12932187 was shown to regulate CIITA expression and IL-10 production.
Materials and Methods
Study population
In the first stage of this study, a total of 384 BD patients and 576 controls were enrolled to identify disease susceptibility loci in the family of NLR genes. In the second (confirmatory) stage, another set of 566 BD patients and 864 controls were added to replicate the susceptible SNPs found in the first stage study.
All blood samples were enrolled at the Zhongshan Ophthalmic Center (Guangzhou, China) and the First Affiliated Hospital of Chongqing Medical University (Chongqing, China) from November 2006 to February 2015. The diagnosis of BD patients is based on the criteria of the International Study Group for BD36. This study obtained the approval of the Local Ethics Research Council. before the collection of blood, all the investigated individuals had signed the informed consent.
Ethical considerations
Before the collection of blood, all the investigated individuals had signed the informed consent. The investigation protocols obtained the approval of the Clinical Research Ethics Committee of the Zhongshan Ophthalmic Center of Sun Yat-sen University and the First Affiliated Hospital of Chongqing Medical University. All experiments were conducted in accordance with the approved guidelines and regulations. This study was conducted according to the tenets of the Declaration of Helsinki.
SNP selection
We selected nineteen SNPs of NLR family genes including NOD1//(rs2075818, rs2907748, rs2907749), NOD2//(rs8057431, rs3135499), NLRP1//(rs6502867, rs878329, rs12150220, rs8079034), NLRP3//(rs10754558, rs10925019, rs4925648, rs3806265, rs2027432) and CIITA//(rs12932187, rs1107438, rs8048002, rs6498122, rs4774) on the basis of three standards : 1. According to previous reports, the relevant SNP had been proved to be associated with an autoimmune or auto-inflammatory disease. 2. Allele or genotype data existed in the National Center of Biotechnology Information (NCBI) single nucleotide polymorphisms (dbSNP) database in the Chinese Han population. 3. All SNPs had to have a minor allele frequency (MAF) that was larger than 0.05 and an R2 threshold of 0.80; The R2 value reflects the degree to which captured alleles are in linkage disequilibrium (LD) with the tag SNP. Genotyping data for each of the 5 NLRP genes (Chinese Han population) was downloaded from the HapMap Web site (http://hapmap.ncbi.nlm.nih.gov/index.html.en) .
DNA extraction and genotyping
Genomic DNA was extracted from the blood of patients and healthy individuals using the QIAmp DNA Blood Mini Kit (Qiagen Inc., Valencia, CA, USA), all the samples were stored at −80 °C until used. All SNPs except NLRP3//rs10925019 were genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). All the primers were designed using primer software 5.0 (Premier Biosoft International, Palo Alto, CA, USA). Primers and restriction enzymes are shown in Table 3. NLRP3//rs10925019 (TaqMan assay ID: C_26646027_10) genotyping was performed on the Applied Biosystems 7500 Real-Time PCR system using TaqMan® SNP assay (Applied Biosystems, CA, USA). The analysis was performed by TaqMan Genotyper Software. To verify the accuracy of genotyping, direct sequencing was carried out (Beijing Biomed Co. Ltd. China) using randomly selected samples (10% of all samples). The genotyping success rate was above 95%.
Table 3. Primers and restriction enzymes used for RFLP analysis of the NOD1, NOD2, NLRP1, NLRP3 and CIITA genes.
| Gene | SNP number | Primers | Restriction enzyme |
|---|---|---|---|
| NOD1 | rs2075818 | GCAATCGGGAACTTCTGGTCACT | HaeIII |
| GGGGCAGGCACACACAATCTC | |||
| rs2907748 | AAGGCTCTCCAGCTATGCAGAT | PvuIII | |
| GTGGGCTCCTCTACAGGCA | |||
| rs2907749 | CCCCCACACACACAGCAGGTT | BccI | |
| GCTGGAGGCTGACTGTGTGTGAC | |||
| NOD2 | rs8057431 | CTGACTGAGGCAGCGGGAGTTTA | DraI |
| CAGGAGACCAAGGCAGGAAGACC | |||
| rs3135499 | CGGCCTCTCACAAAAGACCGGAT | BamHI | |
| GGAATGGCCTGGATGGATGAGT | |||
| NLRP1 | rs8079034 | CGCAGACAAAGGTCCTTAGGTA | BstEII |
| AACTTGAAGGGAAGTCTAGCAGT | |||
| rs12150220 | GCTTGGAGACTCATGGTCTG | BseRI | |
| CCCTCTACTTCAACATGGTTTTCA | |||
| rs6502867 | GGACAGAATTAAGACTGATAA | ApaLI | |
| TTATCAGTCTTAATTCTGTCC | |||
| rs878329 | CCGGGCTGCATCAACCTTCT | Bsp1286I | |
| GCCCCAACCACCAACATGAGAC | |||
| NLRP3 | rs10754558 | CAGGACAATGACAGCATCGGGTGTTGAT | MboI |
| GCTGCCATAAAATTTCAACATAA | |||
| rs4925648 | TTCCTGGTTCTAAACCCCTCTG | PstI | |
| CACAGGCTAGGCACTCACT | |||
| rs3806265 | TTGGCAGGTGGACAGCAGCA | PvuII | |
| GACCCCAAACATCCCCCAAATCA | |||
| rs2027432 | CACCATACACCTTTTTTCTCGGGC | BstEII | |
| GGGCCTCCATTTTCTCATCTGTG | |||
| CIITA | rs12932187 | TGCCCCTGAAGAAGTCGTTT | TaqI |
| CTTAAGGCTGCACCCAACCAC | |||
| rs1107438 | ACAGTCATCATCTTCCCCATTTTAC | HinfI | |
| GCCCTCAGTTATTGTTTTCAGAGAT | |||
| rs8048002 | CACCATACACCTTTTTTCTCGGGC | HaeIII | |
| GGGCCTCCATTTTCTCATCTGTG | |||
| rs6498122 | GTCCCTCAGTTTTGCTCCTATC | Csp6I | |
| GTCCTCTCCCTCAATAATATGGT | |||
| rs4774 | TCCCCTGCCATTGCTTGA | Hin1I | |
| AACCTCGGAGCAGCTTCTTCT |
Real-time PCR
In this study, peripheral blood mononuclear cells (PBMCs) were obtained from healthy controls by Ficoll-Hypaque density-gradient centrifugation. Cells were stimulated with or without anti-CD3 (0.5ug/ml) and anti-CD28 antibodies (0.1ug/ml, eBioscience, San Diego, CA, USA) to analog antigen presentation or lipopolysaccharide (LPS, 5ug/ml, Fluka, Buchs, Switzerland) to analog an inflammatory signal for 72 hours at a density of 1 × 106 cells/ml. RNA was acquired from the cultured cells by TRIzol (Invitrogen), after reserve transcription (transcription kit, Takara Biotechnology Co. Ltd., Dalian, China.), mRNA expression of NOD1 gene (forward: 5′ TTGACCACCCTGAGTCTTGC 3′, reserve: 5′ TCATTTTGGGTCAGCCACAG 3′) and CIITA gene (forward: 5′ TGAGGCTGTGTGCTTCTGAG 3′, reserve: 5′ ACACTGTGAGCTGCCTTGG 3′) was measured by using real-time PCR equipment with a commercial dye kit (Applied Biosystems), β-Actin was selected as the internal reference gene and its expression was detected by the following primers: forward 5′-GGATGCAGAAGGAGATCACTG -3′ and reverse 5′-CGATCCACACGGAGTACTT-3′. Data were normalized to mRNA beta-actin and expression levels were calculated by the 2−△△ method.
Cytokine Measurements
The human Duoset enzyme-linked immunosorbent assay (ELISA) development kit (R&D System, Minneapolis, MN) was used to measure the concentration of IL-6, IL-8, IL-10, IL-1β, TNF-α and MCP-1 in cell culture supernatant in accordance with the manufacturers’ instructions.
Statistical analysis
Differences in alleles and genotypes of all SNP variations were evaluated by the Fisher’s exact test or X2 test using SPSS (version 17.0; SPSS Inc, Chicago, IL). The p values were corrected with the Bonferroni correction method and a Pc <0.05 was considered to be significant. The X2 test was used to determine the Hardy-Weinberg equilibrium (HWE). The independent samples t test or nonparametric Mann-Whitney U test was used to compare CIITA, NOD1 and cytokine (IL-6, IL-8, IL-10, TNF-α, IL-1βand MCP-1) expression levels among three genotype groups.
Additional Information
How to cite this article: Li, L. et al. Genetic Variations of NLR family genes in Behcet's Disease. Sci. Rep. 6, 20098; doi: 10.1038/srep20098 (2016).
Supplementary Material
Acknowledgments
The authors would like to thank all donors enrolled in the present study. This work was supported by Natural Science Foundation Major International (Regional) Joint Research Project (81320108009), Key Project of Natural Science Foundation (81130019), National Natural Science Foundation Project (31370893, 81200678), Basic Research program of Chongqing (cstc2013jcyjC10001), Fundamental and Advanced Research Program of Chongqing (cstc2015jcyjA10022), Science and Technology Project of Chongqing Municipal Education Commission (KJ1500236), Scientific Research Program of Science and Technology Commission of Yuzhong District of Chongqing (20150102),Chongqing Key Laboratory of Ophthalmology (CSTC, 2008CA5003), National Key Clinical Specialties Construction Program of China, Key Project of Health Bureau of Chongqing (2012-1-003), Research fund for Traditional Chinese Medicine of Chongqing Health and Family Planning Commission (ZY201401013).
Footnotes
Author Contributions L.L. and P.Y. conceived the idea and designed the experiments. L.L., H.Y. and Y.J. performed the experiments, L.L., H.Y., L.B. and B.D. analyzed the data. L.L., H.Y., A.K. and P.Y. wrote the manuscript. L.L. prepared figures. All authors reviewed the manuscript.
References
- Yang P. et al. Clinical features of chinese patients with Behcet’s disease. Ophthalmology 115, 312–318 e314 (2008). [DOI] [PubMed] [Google Scholar]
- Ideguchi H. et al. Behcet disease: evolution of clinical manifestations. Medicine (Baltimore) 90, 125–132 (2011). [DOI] [PubMed] [Google Scholar]
- Yu H., Liu Y., Bai L., Kijlstra A. & Yang P. Predisposition to Behcet’s disease and VKH syndrome by genetic variants of miR-182. J Mol Med (Berl) 92, 961–967 (2014). [DOI] [PubMed] [Google Scholar]
- Fang J. et al. Association of TLR2 gene polymorphisms with ocular Behcet’s disease in a Chinese Han population. Invest Ophthalmol Vis Sci 54, 8384–8392 (2013). [DOI] [PubMed] [Google Scholar]
- Hu K., Hou S., Jiang Z., Kijlstra A. & Yang P. JAK2 and STAT3 polymorphisms in a Han Chinese population with Behcet’s disease. Invest Ophthalmol Vis Sci 53, 538–541 (2012). [DOI] [PubMed] [Google Scholar]
- Hou S. et al. Identification of a susceptibility locus in STAT4 for Behcet’s disease in Han Chinese in a genome-wide association study. Arthritis Rheum 64, 4104–4113 (2012). [DOI] [PubMed] [Google Scholar]
- Chen F. et al. CD40 gene polymorphisms confer risk of Behcet’s disease but not of Vogt-Koyanagi-Harada syndrome in a Han Chinese population. Rheumatology (Oxford) 51, 47–51 (2012). [DOI] [PubMed] [Google Scholar]
- Hou S., Kijlstra A. & Yang P. Molecular Genetic Advances in Uveitis. Prog Mol Biol Transl Sci 134, 283–298 (2015). [DOI] [PubMed] [Google Scholar]
- Yu H. et al. FAS Gene Copy Numbers are Associated with Susceptibility to Behcet Disease and VKH Syndrome in Han Chinese. Hum Mutat 36, 1064–1069 (2015). [DOI] [PubMed] [Google Scholar]
- Carneiro L. A., Travassos L. H. & Girardin S. E. Nod-like receptors in innate immunity and inflammatory diseases. Ann Med 39, 581–593 (2007). [DOI] [PubMed] [Google Scholar]
- Hugot J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603 (2001). [DOI] [PubMed] [Google Scholar]
- van Heel D. A. et al. Inflammatory bowel disease susceptibility loci defined by genome scan meta-analysis of 1952 affected relative pairs. Hum Mol Genet 13, 763–770 (2004). [DOI] [PubMed] [Google Scholar]
- Huebner C. et al. Nucleotide-binding oligomerization domain containing 1 (NOD1) haplotypes and single nucleotide polymorphisms modify susceptibility to inflammatory bowel diseases in a New Zealand caucasian population: a case-control study. BMC Res Notes 2, 52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini L. et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20, 319–325 (2004). [DOI] [PubMed] [Google Scholar]
- Hoffman H. M., Mueller J. L., Broide D. H., Wanderer A. A. & Kolodner R. D. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet 29, 301–305 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church L. D., Cook G. P. & McDermott M. F. Primer: inflammasomes and interleukin 1beta in inflammatory disorders. Nat Clin Pract Rheumatol 4, 34–42 (2008). [DOI] [PubMed] [Google Scholar]
- Sui J. et al. NLRP1 gene polymorphism influences gene transcription and is a risk factor for rheumatoid arthritis in han chinese. Arthritis Rheum 64, 647–654 (2012). [DOI] [PubMed] [Google Scholar]
- Magitta N. F. et al. A coding polymorphism in NALP1 confers risk for autoimmune Addison’s disease and type 1 diabetes. Genes Immun 10, 120–124 (2009). [DOI] [PubMed] [Google Scholar]
- Taieb A. NALP1 and the inflammasomes: challenging our perception of vitiligo and vitiligo-related autoimmune disorders. Pigment Cell Res 20, 260–262 (2007). [DOI] [PubMed] [Google Scholar]
- Martinez A. et al. Role of the MHC2TA gene in autoimmune diseases. Ann Rheum Dis 66, 325–329 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanberg M. et al. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat Genet 37, 486–494 (2005). [DOI] [PubMed] [Google Scholar]
- Belguendouz H. et al. Cytokines Modulate the “Immune-Metabolism” Interactions during Behcet Disease: Effect on Arginine Metabolism. Int J Inflam 2015, 241738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. Y., Chen S. L., Shen N. & Lu Y. Cytokines and Behcet’s disease. Autoimmun Rev 11, 699–704 (2012). [DOI] [PubMed] [Google Scholar]
- Devaiah B. N. & Singer D. S. CIITA and Its Dual Roles in MHC Gene Transcription. Front Immunol 4, 476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J. P. & Trowsdale J. Genetic control of MHC class II expression. Cell 109 Suppl, S21–33 (2002). [DOI] [PubMed] [Google Scholar]
- Steimle V., Otten L. A., Zufferey M. & Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). 1993. J Immunol 178, 6677–6688 (2007). [PubMed] [Google Scholar]
- Bronson P. G. et al. CIITA variation in the presence of HLA-DRB1*1501 increases risk for multiple sclerosis. Hum Mol Genet 19, 2331–2340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. et al. CIITA rs4774 and rs6498122 polymorphisms are associated with oral lichen planus in Chinese people: a case-control study. Eur J Oral Sci 121, 69–75 (2013). [DOI] [PubMed] [Google Scholar]
- Ronninger M. et al. Interaction analysis between HLA-DRB1 shared epitope alleles and MHC class II transactivator CIITA gene with regard to risk of rheumatoid arthritis. PLoS One 7, e32861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J. S. et al. Genetic association analysis of CIITA variations with nasal polyp pathogenesis in asthmatic patients. Mol Med Rep 7, 927–934 (2013). [DOI] [PubMed] [Google Scholar]
- Chang C. H., Guerder S., Hong S. C., van Ewijk W. & Flavell R. A. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity 4, 167–178 (1996). [DOI] [PubMed] [Google Scholar]
- Saraiva M. & O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol 10, 170–181 (2010). [DOI] [PubMed] [Google Scholar]
- Ahn J. K., Yu H. G., Chung H. & Park Y. G. Intraocular cytokine environment in active Behcet uveitis. Am J Ophthalmol 142, 429–434 (2006). [DOI] [PubMed] [Google Scholar]
- Day T. G. et al. Autoinflammatory genes and susceptibility to psoriatic juvenile idiopathic arthritis. Arthritis Rheum 58, 2142–2146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med 356, 1216–1225 (2007). [DOI] [PubMed] [Google Scholar]
- Varela Aguilar J. M. & Sanchez Roman J. [Diagnosis of Behcet’s disease. Which criteria should be used?]. An Med Interna 7, 165 (1990). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


