Abstract
The Hippo signaling network is a key regulator of cell fate. In the recent years, it was shown that its implication in cancer goes well beyond the sole role of YAP transcriptional activity and its regulation by the canonical MST/LATS kinase cascade. Here we show that the motin family member AMOTL1 is an important effector of Hippo signaling in breast cancer. AMOTL1 connects Hippo signaling to tumor cell aggressiveness. We show that both canonical and noncanonical Hippo signaling modulates AMOTL1 levels. The tumor suppressor Merlin triggers AMOTL1 proteasomal degradation mediated by the NEDD family of ubiquitin ligases through direct interaction. In parallel, YAP stimulates AMOTL1 expression. The loss of Merlin expression and the induction of Yap activity that are frequently observed in breast cancers thus result in elevated AMOTL1 levels. AMOTL1 expression is sufficient to trigger tumor cell migration and stimulates proliferation by activating c-Src. In a large cohort of human breast tumors, we show that AMOTL1 protein levels are upregulated during cancer progression and that, importantly, the expression of AMOTL1 in lymph node metastasis appears predictive of the risk of relapse. Hence we uncover an important mechanism by which Hippo signaling promotes breast cancer progression by modulating the expression of AMOTL1.
Introduction
The Hippo signaling pathway controls organ size primarily through the inhibition of cell proliferation and the stimulation of apoptosis. The canonical Hippo pathway is composed of a cascade of kinases (MST1/2 and LATS1/2) leading to the phosphorylation and inhibition of two transcriptional cofactors, YAP and TAZ. In recent years, the implication of this pathway in cancer development has been progressively documented, focusing largely on the role of YAP and TAZ [38].
An impressive number of new components of the pathway were progressively identified that connect Hippo signaling to various environmental cues such as mechanical or metabolic stress, cell density, or adhesion. Among them, the motin family of proteins (AMOT, AMOTL1, and AMOTL2) was recognized as bona fide component of the Hippo pathway [1].
AMOT was originally identified as a receptor for the antiangiogenic factor angiostatin and a regulator of endothelial cell motility [2], [3]. Conservation of sequence, structure and interactors between motins suggests redundancy in the family. Indeed, all three motins were found to interact with actin and to regulate cellular polarity, cell adhesion and [4], [5]. But the mechanisms regulating these functions are still largely unknown.
The motins contribute to Hippo signaling in different ways. They bind to YAP and, depending on the experimental setting, either inhibit or promote its activity [6], [7]. They also bind to LATS kinases, acting both as regulators and substrate [8]. Furthermore, AMOT was shown to interact with the upstream Hippo regulator Merlin, resulting in the modulation of the activity of the Rac1/MAPK pathway [9]. Thus, the emerging picture is that the motins act as molecular nodes allowing cross talk between major signaling pathways involved in cell proliferation, migration, or polarity.
Few studies have addressed a possible involvement of motins in cancer. AMOT was found to be highly expressed in blood vessels of Kaposi’s sarcoma [2]. High mRNA levels of AMOT are associated with a poor clinical outcome in breast cancer [10]. Recently, AMOT expression was linked to venous invasion and poor prognosis and was proposed to represent a potential prognostic marker in clear cell renal carcinoma [11]. However, the regulation and the role of the motin family members during cancer development and progression remain largely unexplored.
In this report, we investigated the role of the motins in breast cancer. We show that AMOTL1 expression is linked to breast cancer aggressiveness and its expression in lymph node metastasis is predictive of disease relapse. In mice, AMOTL1 expression in tumor xenografts stimulates their growth. In vitro, we show that AMOTL1 promotes the migration of tumor cells but also stimulates tumor growth by activating c-Src. Finally, we demonstrate that AMOTL1 activity is tightly regulated at different levels in breast cancer cells. Indeed, YAP/TAZ stimulates AMOTL1 expression, whereas NF2/Merlin induces the phosphorylation of the protein leading to its degradation mediated by the NEDD family of ubiquitin ligase. Thus, our results show that AMOTL1 expression is regulated by both Yap-mediated canonical and NF2/Merlin driven noncanonical Hippo signaling during breast cancer progression.
Materials and Methods
Human Sample Analysis
Analyses of human samples were performed in accordance with the French Bioethics Law 2004-800 and the French National Institute of Cancer Ethics Charter and after approval by the Institut Curie review board and ethics committee that waived the need for written informed consent from the participants. Women were informed of the research use of their tissues and did not declare any opposition for such research. Data were analyzed anonymously.
Breast Cancer Tissue Microarray (TMA)
Samples of primary breast tumors were surgically removed before any radiation, hormonal, or chemotherapy treatment at Institut Curie from 2005 to 2006. Our series of invasive breast tumors comprised all TNBC and HER2 tumors available and equal number of consecutively treated Luminal A and Luminal B tumors from the same period following the same criteria. Based on clinicopathological examination, these tumors were classified as invasive ductal carcinoma (IDC) and ductal carcinoma in situ (DCIS). Inclusion of DCIS tumors in the TMA was performed using the same criteria as for IDCs and were classified into the subtypes TNBC, HER2, and Luminal A and B [31], [32], [33], [34]. Breast molecular subtypes were defined as follows: Luminal A + B according to [35]: Luminal A: estrogen-receptor (ER) ≥ 10%, progesterone-receptor (PR) ≥ 20%, Ki-67 < 14%; Luminal B: ER ≥ 10%, PR < 20%, Ki-67 ≥ 14%; ER − PR − HER2 +: ER < 10%, PR < 10%, HER2 2 + amplified or 3 +; ER − PR − HER2 − (triple-negative breast cancers): ER < 10%, PR < 10%, HER2 0/1 + or 2 + nonamplified according to the American Society of Clinical Oncology guidelines [36]. The second cohort of 126 invasive breast tumors has been described elsewhere [37].
Gene Expression Analysis
Raw .cel files from two primary breast tumor gene expression data sets were downloaded from NCBI GEO (GSE21653) and ArrayExpress (E-MTAB-365) repositories and normalized with RMA algorithm before selection of the optimal probeset for each gene using Jetset method [39]. The AMOTL1 signature was defined as follow: for both breast tumor gene expression data set, we selected AMOTL1 correlated genes with a Spearman's rho coefficient above 0.3 and a P value below .05 after a Bonferroni correction. Functional analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery. The same strategy was used for the analysis of ovarian and colon cancer cohorts presented in the supplementary data section.
Cell Lines
MDA-MB-468, HEK293, Hela, and MCF10A cells were obtained from the ATCC. BC52 cell line was developed at the Laboratory of Preclinical Investigation in Curie Institute.
Cells and Tumor Extracts
Cells and tumors were lysed in 50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 4% SDS, 1 mM EDTA, protease (P8340), and phosphatase (P5726) inhibitors (Sigma-Aldrich).
Antibodies
For Western blots
The following were used: Merlin (sc-332) and NEDD4 (sc-25508) from Santa Cruz, CA; phospho-histone H3 ser10 (H0412), AMOTL1 (HPA001196), and actin (A2228) from Sigma-Aldrich; YAP (#4912), p27 (#2552), AIP4 (#12117), and cleaved caspase 3 (#9661) from Cell Signaling Technology (Ozyme, France); E-cadherin (610182) from BD Biosciences; phospho-Src Y418 (44-660G) and phospho-FAK Y397 (44-624G) from Invitrogen; AMOTL1 p-S262 Covalab (pab0956-P, Lyon, France); and Tead2 Covalab (pab0961-P, Lyon, France).
For immunofluorescence
The following were used: Merlin (sc-332, Santa Cruz), AMOTL1 (HPA001196), and anti Flag M2 (F3165) from Sigma-Aldrich. Images were acquired using a Leica DM 6000B epifluorescence microscope and a 40 × or a 63 × oil immersion objective.
For aggregate cryosections
The following were used: AMOTL1 (HPA001196, Sigma Aldrich), phospho-histone H3 (H0412, Sigma-Aldrich), and cleaved caspase 3 (#9661, Cell Signaling Technology).
For immunohistochemistry (IHC) on animal tissues
The following were used: AMOTL1 (HPA001196, Sigma Aldrich) at 1/100, YAP (sc-15407, Santa cruz) at 1/100, E-cadherin (#610182, BD Biosciences) at 1/100, and cleaved caspase 3 (#9661, Cell Signaling Technology) at 1/50.
siRNAs and shRNAs
Transfection of siRNA and shRNA was performed using lipofectamine RNAimax solution (Invitrogen). The following were used: human AMOTL1 on-target plus smartpool siRNA (#L-017595-01-0005) and nontargeting control siRNAs (D-001810-03), Dharmacon (Thermo Scientific, France); pLKO-shRNA lentiviral constructs targeting human Merlin (TRCN0000018338, called sh #1 and TRCN0000039974, called sh #2), AMOTL1 (TRCN0000130193), and nontargeting control shRNA (SHC002V), Sigma, France.
Animal Studies
All mice studies were conducted following the guidelines of the American Association of Laboratory Animal Care under an approved protocol. Eight-week-old female nu/nu mice (Charles River, France) were inoculated with 2 × 106 BC52 tet on AL1 cells in the neck mammary fat pad. AMOTL1 expression was obtained by addition of 2 mg/ml of doxycycline to the drinking water supplemented with 10% sucrose. The tumors were measured twice per week.
IHC Analysis of Breast Cancer TMA
TMA consisted of replicate 1 mm–diameter tumor cores selected from the most representative tumor areas (high tumor cell density) of each tumor sample and a matched tissue core from adjacent nontumoral (normal) breast epithelium. Three-micrometer paraffin sections were deparaffinized before heat-induced epitope retrieval for 20 minutes in EnVision FLEX Target Retrieval Solution (pH 6.1). Then, endogenous activity was blocked with a 3% hydrogen peroxide solution. Staining with anti-AMOTL1 antibody (Covalab, Lyon, France), pab0883-P, 1:100, was done using a Dako Autostainer plus stainer.
IHC of Mouse Tissues
Tissues were fixed using alcohol, formalin, and acetic acid and embedded in paraffin. Four-micrometer–thick tissue sections were prepared according to conventional procedures.
Cell Growth in Matrigel
Ninety-six–well cell culture plates were coated with 50 μl of 1:3 Matrigel (BD Biosciences) in DMEMoverlaid by a 1:1 mix of 5 × 103 cells and Matrigel. After 30 minutes, this gel was overlaid by 100 μl of medium.
Migration and Invasion Assay
A total of 5 × 105 cells were added to the top chamber with 8.0-μm pore membranes (for migration evaluation) or Matrigel-coated 8.0-μm pore membranes (for invasion evaluation) (BD Biosciences, France). After 16 hours, the cells that had migrated to the other side of the insert were stained with phalloidin-rhodamine and counted.
AMOTL1 Half-Life Evaluation and Proteasome Inhibition by MG132
BC52 tet on cells were transiently transfected with pRetroXAL1 or PRetroXAL1S262A and either pBabe-Merlin or empty pBabe vector for 24 hours; then the cells were split into 6-well plates, and doxycycline at 1 μg/ml was added in the medium. After 24 hours, the doxycycline was washed out, cells were rinsed in PBS three times, and protein extraction was performed at the indicated time points. AMOTL1 half-life was determined as the time needed to get 50% decrease of the initial amount of AMOTL1 (time = 0 hour). For the proteasome activity inhibition experiment, BC52 cells expressing shRNA control or shRNA against AMOTL1 were treated overnight by 5 μM MG132 before protein extraction.
Anti-AMOTL1 p-S262 Antibody
A 15-mer peptide corresponding to human AMOTL1 sequence and containing phosphorylated S262 was coupled to keyhole lympet hemocyanin. Rabbit immunization and purification against phosphor and nonphospho peptides were done by Covalab (pab0956-P, Lyon, France).
Results
Expression of Motins in Human Breast Tumors
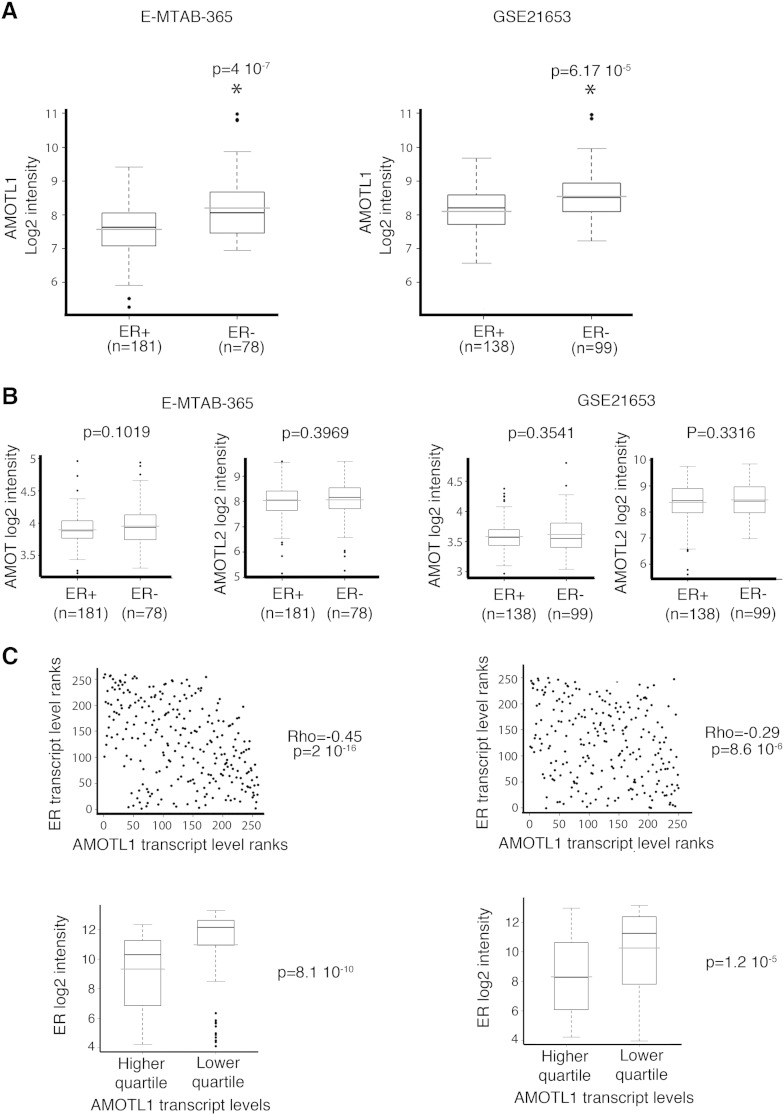
To evaluate the pattern of expression of the motins in breast tumors, their transcript levels were analyzed in two independent published gene expression data sets (E-MTAB-365 and GSE21653). No pattern of expression specific to Luminal A, Luminal B, Her2-enriched, and basal-like subtypes was observed (not shown). However, in both data sets, AMOTL1 expression was upregulated in tumors with ER-negative status (Figure 1A). This was not the case for AMOT or AMOTL2 (Figure 1B), suggesting that AMOTL1 expression might be more specifically associated to this more invasive tumor type. In addition, for both cohorts, AMOTL1 transcript levels were found to be significantly higher in breast tumors expressing low levels of ER transcripts. Finally, we also observed that AMOTL1 and ER transcripts displayed an inverted correlation (Figure 1C).
Figure 1.
AMOTL1 expression is higher in ER − breast tumors. (A) AMOTL1 transcripts levels in ER − and ER + breast tumors in E-MTAB-365 and GSE21653 data sets (Mann-Whitney test). (B) AMOT and AMOTL2 transcripts levels in relation to the ER status in both data sets (Mann-Whitney test). (C) AMOTL1 and ER transcript levels are negatively correlated in both data sets (Spearman test, top). (Bottom) The ER transcript levels are higher in the quartile with the highest AMOTL1 expression (Mann-Whitney test).
Angiomotin-like protein 1 (AMOTL1) Upregulation in Invasive Tumors and Local Metastasis
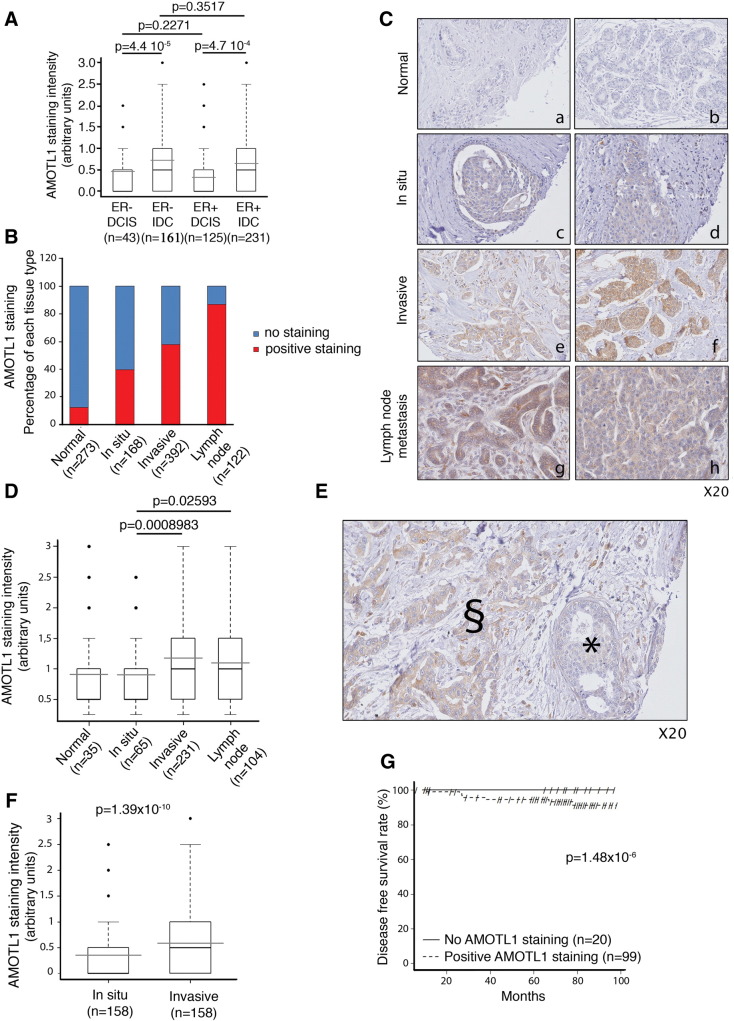
We next evaluated AMOTL1 protein levels in a human breast cancer TMA composed of 392 IDCs associated with noninvasive DCISs (n = 168), invaded lymph nodes (local metastasis, n = 122), and normal tissues (n = 273) (see Supplementary Table S1 for a description of the tumors). AMOTL1 staining intensity was not linked to the ER status. However, it was stronger in IDC than in DCIS both in ER + and ER − tumors, suggesting that AMOTL1 expression might be linked to invasiveness (Figure 2A). Overall, AMOTL1 was detected in about 13% of normal tissues (compared with 80% of positive endothelial cells, Supplementary Figure S1), 40% of DCISs, 58% of IDC tissues, and more than 87% of lymph node metastases (Figure 2, B and C). In general, when detected, AMOTL1 staining intensity was significantly higher in invasive lesions (IDC and lymph node metastasis) than in benign ones (DCIS) or in normal tissues (Figure 2D). Increase of AMOTL1 protein levels was also significant between DCIS (mean intensity = 0.36) and IDC (mean intensity = 0.59) from the same tumor (Figure 2, E and F). Finally, we observed that only patients presenting a detectable AMOTL1 staining in the invaded lymph nodes suffered of relapse during the follow-up period of 100 months as opposed to none of the AMOTL1-negative group (Figure 2G). Despite an important difference in the size of the groups, log-rank test showed that the difference of clinical outcome was significant. Altogether, our results show that AMOTL1 expression is linked to breast cancer progression and the risk of relapse.
Figure 2.
AMOTL1 protein expression is upregulated during breast tumor progression. (A) AMOTL1 staining intensity is higher in IDC compared with to DCIS of the ER same status (Mann-Whitney test). (B) The proportions of normal breast tissue, DCIS, IDC, and lymph node tumors with detectable AMOTL1 by IHC are presented. AMOTL1 detection is more frequent with tumor progression. (C) Examples of AMOTL1 expression in breast normal epithelium (a, b, see also Supplementary Figure S1), DCIS (c, d), IDC (e, f), and in lymph node metastasis (g, h). The staining intensity appears higher in invasive lesions. (D) The quantification of AMOTL1 staining intensity in normal breast epithelium cells, DCIS, IDC, and lymph node tumor cells confirms that the expression is higher in invasive lesions (Mann-Whitney test). (E) Staining of AMOTL1 shows that its expression is higher in IDC than in DCIS from the same biopsy. ⁎DCIS area; §IDC area. (F) The quantification of the staining intensity of AMOTL1 in IDC and DCIS of the same biopsy confirms higher expression in the invasive lesions (Mann-Whitney test). (G) A Kaplan-Meyers analysis shows that the risk of relapse is significantly higher in patients with AMOTL1 positive staining of the lymph node metastasis.
AMOTL1 protein expression is upregulated during breast tumor progression. (A) AMOTL1 staining intensity is higher in IDC compared with to DCIS of the ER same status (Mann-Whitney test). (B) The proportions of normal breast tissue, DCIS, IDC, and lymph node tumors with detectable AMOTL1 by IHC are presented. AMOTL1 detection is more frequent with tumor progression. (C) Examples of AMOTL1 expression in breast normal epithelium (a, b, see also Supplementary Figure S1), DCIS (c, d), IDC (e, f), and in lymph node metastasis (g, h). The staining intensity appears higher in invasive lesions. (D) The quantification of AMOTL1 staining intensity in normal breast epithelium cells, DCIS, IDC, and lymph node tumor cells confirms that the expression is higher in invasive lesions (Mann-Whitney test). (E) Staining of AMOTL1 shows that its expression is higher in IDC than in DCIS from the same biopsy. ⁎DCIS area; §IDC area. (F) The quantification of the staining intensity of AMOTL1 in IDC and DCIS of the same biopsy confirms higher expression in the invasive lesions (Mann-Whitney test). (G) A Kaplan-Meyers analysis shows that the risk of relapse is significantly higher in patients with AMOTL1 positive staining of the lymph node metastasis.
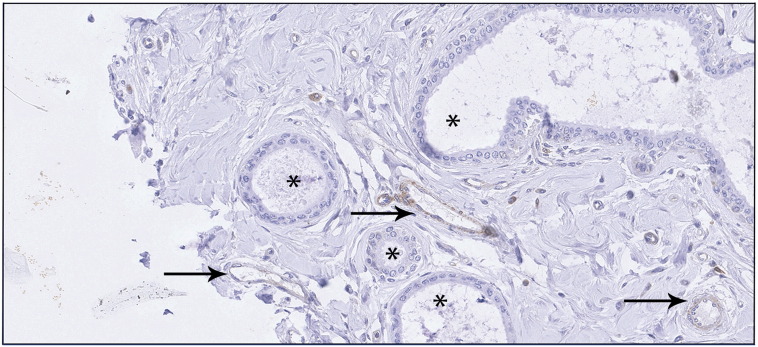
Supplementary Figure S1.
IHC showing staining of AMOTL1 in normal mammary epithelial cells (*) and in endothelial cells (black arrows).
AMOTL1 Levels Are Regulated by Merlin and Yap in Breast Cancer Cells
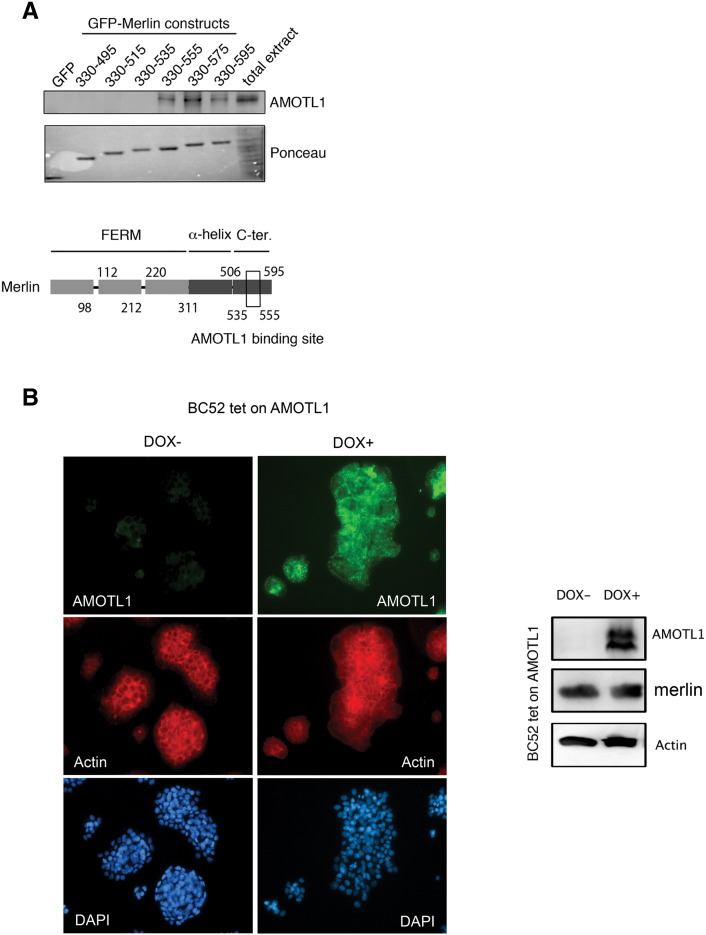
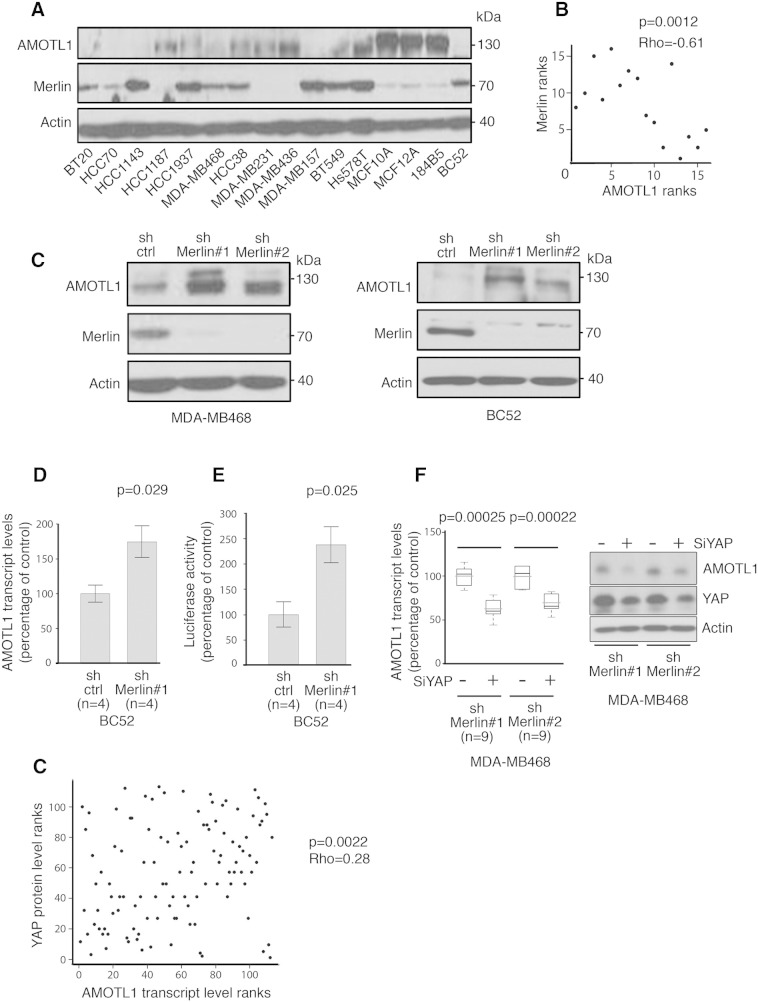
Little is known about the mechanisms that control motins protein levels. AMOTL1, like the other motins, interacts with C-terminal domain of the NF2/Merlin tumor suppressor [9] (Supplementary Figure S2A), suggesting possible mutual regulation. Remarkably, when we evaluated AMOTL1 and Merlin expression by Western blot in a set of 16 breast cancer cell lines (see Supplementary Table S2 for a description), the two proteins presented an obvious opposite expression profile (Figure 3, A and B). When Merlin expression was inhibited with shRNA in MDA-MB-468 and BC52 cell lines, AMOTL1 levels increased (Figure 3C). In contrast, the overexpression of AMOTL1 in the doxycycline inducible BC52 tet on AMOTL1 cell line (Supplementary Figure S2B) or its downregulation by siRNA (not shown) had no effect on Merlin levels. These results demonstrate that AMOTL1 protein levels in breast tumor cells, and possibly in breast cancer, are negatively regulated by Merlin.
Supplementary Figure S2.
(A, upper panel) Identification of AMOTL1 binding site to Merlin by GFP-Trap with GFP-Merlin truncated constructs and GFP alone as a negative control. (Lower panel) Scheme of Merlin protein showing AMOTL1 binding site between amino acids 535 and 575 of the C-terminal domain. (B) BC52 cells expressing AMOTL1 under the doxycycline inducible promoter were generated, and the protein induction was confirmed by immunofluorescence staining (immunofluorescence at left). The induction of AMOTL1 expression in BC52 tet on AMOTL1 cells had no impact on endogenous Merlin levels (Western blot at right).
Figure 3.
AMOTL1 expression is negatively regulated by Merlin. (A) The expression levels of AMOTL1 and Merlin proteins in a set of 16 breast cancer cell lines display a clear opposite pattern. (B) The levels of Merlin and AMOTL1 quantified by Western blot are negatively correlated (Spearman's test). (C) The level of AMOTL1 increases when Merlin protein expression is inhibited in BC52 and MDA-MB-468 using shRNA (sh Merlin #1 and #2 and control shRNA, sh ctrl). (D) The inhibition of Merlin expression in BC52 cells (sh Merlin #1) leads to the upregulation of AMOTL1 mRNA levels measured by real-time PCR (mean ± SD) (Mann-Whitney test). (E) The transcriptional activity of YAP/TAZ was measured in BC52 cells expressing shRNA control or shRNA against Merlin using the YAP/TAD-responsive reporter 8xGTIIC-lux (mean ± SD) (Mann-Whitney test). (F) Real-time PCR analysis of the AMOTL1 transcript levels upon siRNA-mediated YAP inhibition in BC52 cells where NF2 is downregulated by specific shRNA (sh Merlin #1 and #2, control shRNA: sh ctrl) (left panel) (Mann-Whitney test). Immunoblot analysis of AMOTL1 and YAP protein level upon siRNA-mediated YAP inhibition in BC52 cells stably expressing shRNA control (sh ctrl) of NF2 (sh Merlin #1 and #2) (right panel). (G) Correlation between AMOTL1 transcript levels and YAP protein levels (measured by RPPA) in a set of 126 human breast tumors (Spearman test).
We then measured AMOTL1 mRNA expression by real-time PCR in BC52 cells upon inhibition of Merlin expression and found a near two-fold increase (Figure 3D). Merlin is not a transcription factor but inhibits the activity of the TEAD cotranscription factors YAP/TAZ. When Merlin expression was abolished in BC52 cells, we observed more than two-fold increase of YAP/TAZ transcriptional activity measured with a synthetic YAP/TAZ-responsive luciferase reporter (Figure 3E). Furthermore, in MDA-MB468 cells depleted for Merlin, the extinction of YAP expression by siRNA resulted in a decrease of AMOTL1 mRNA and protein levels (Figure 3F). Finally, in a set of 126 invasive breast tumors, distinct from the cohort used for IHC, we observed a significant positive correlation between YAP protein levels and AMOTL1 mRNA expression (P = .0022) (Figure 3G). Altogether, our results show that AMOTL1 expression is regulated by Hippo signaling mediated by Merlin and Yap in breast cancer cells and possibly in tumors and that Merlin likely acts upstream of Yap in this context.
Merlin Triggers AMOTL1 Degradation through Direct Interaction
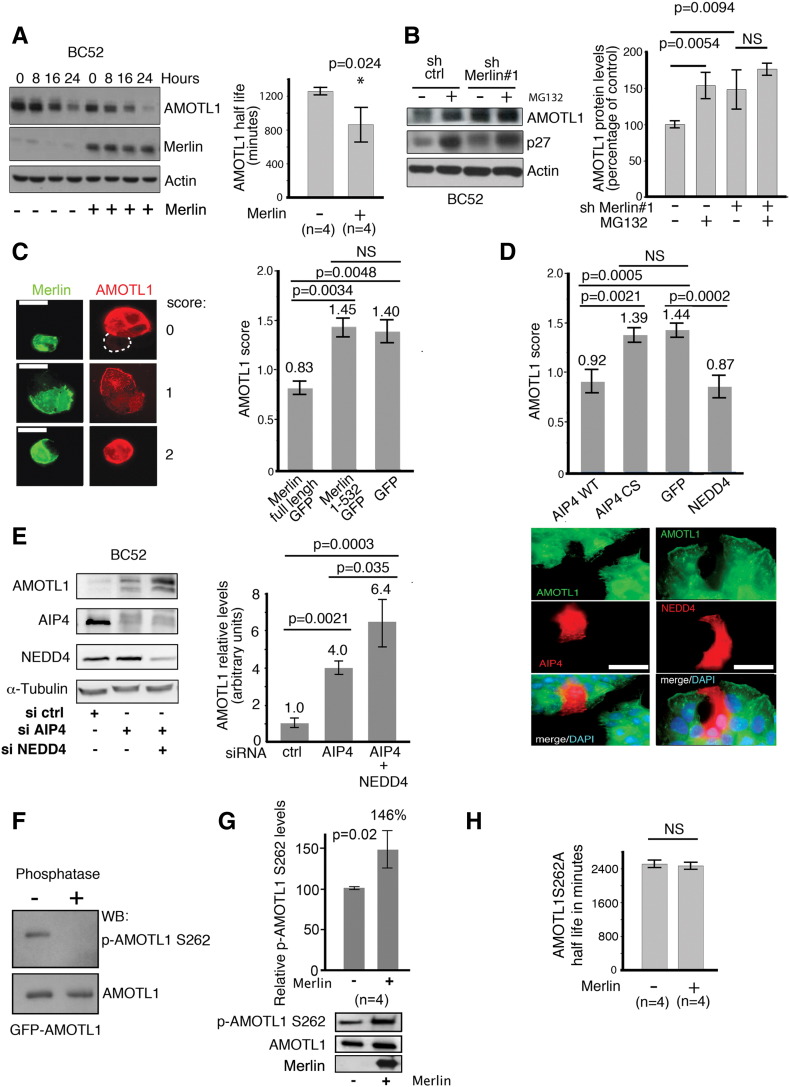
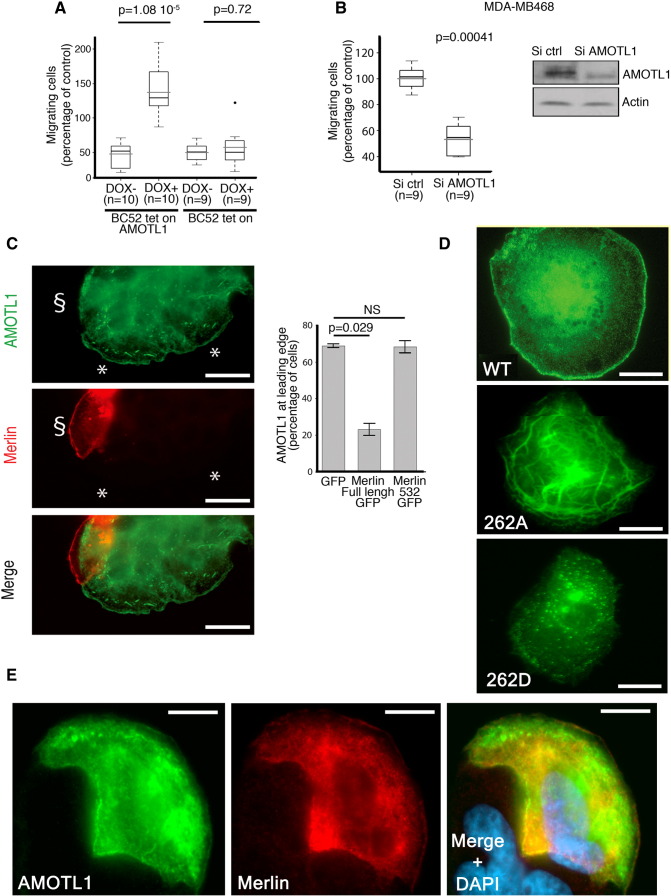
The interaction between the two proteins also suggests that Merlin may regulate AMOTL1 stability directly. AMOTL1 expression was transiently induced with a short pulse of doxycycline in BC52 cells that express it under an inducible promoter. AMOTL1 decay was then followed over time. When Merlin was coexpressed, the quantification of four independent experiments showed that the AMOTL1 half-life was 30% shorter (Figure 4A). Furthermore, when Merlin expression was inhibited by shRNA, the level of AMOTL1 did not significantly increase if cells were already exposed to MG132, suggesting that the proteasome was involved in Merlin downregulation of AMOTL1 (Figure 4B). By immunofluorescence, we also observed that the expression of GFP-Merlin was leading to a decrease of overexpressed AMOTL1 signal in inducible BC52 tet on AMOTL1, whereas a mutant of Merlin (1-532) unable to bind to AMOTL1 as well as the GFP alone had no effect (Figure 4C). Altogether, these results show that AMOTL1 degradation by the proteasome requires direct interaction with Merlin. The ubiquitin ligases NEDD4 and AIP4 were shown to regulate the stability of the motins [12], [13]. In BC52, we observed that AIP4 and NEDD4 expression resulted in a loss of AMOTL1 expression compared with the control, and a mutant AIP4 devoid of ubiquitin ligase activity (AIP4 CS) had no impact on AMOTL1 (Figure 4, D, upper and lower panels). In contrast, the inhibition of AIP4 and NEDD4 expression using siRNA resulted in higher endogenous AMOTL1 levels (Figure 4E). AIP4 was shown to preferentially bind to motins phosphorylated on a conserved serine in position 262 in human AMOTL1 [12]. Using an antibody specific of AMOTL1 phosphorylated on serine 262 (Figure 4F), we showed that Merlin induces an increase of about 50% of S262 phosphorylation when cotransfected with AMOTL1 in 293 T cells (Figure 4G). Finally, using the same strategy as in Figure 4A, we also observed that Merlin expression did not modify the half-life of the S262A mutant of AMOTL1 that cannot be phosphorylated (Figure 4H). Altogether, our results show that Merlin induces the phosphorylation of AMOTL1 on S262, leading to its degradation, likely by increasing the binding of NEDD family of ubiquitin ligases.
Figure 4.
Merlin induces phosphorylation and degradation of AMOTL1 mediated by NEDD ubiquitin ligases. (A) AMOTL1 half-life is reduced upon Merlin expression in BC52 tet on AMOTL1. (Left) AMOTL1 levels following doxycycline withdrawal when cotransfected with Merlin vector (+) or control vector (−). (Right) Quantification of three independent experiments (Mann-Whitney test). (B) When Merlin expression is inhibited by shRNA, overnight MG132 treatment of BC52 does not modify AMOTL1 protein levels (sh Merlin#1 vs sh ctrl). p27 was used as a positive control of MG132 activity (representative Western blot at left). The average of four independent experiments is presented on the right graph (Kruskal-Wallis test). (C) AMOTL1 signal (in red) drops in BC52 tet on AMOTL1 cells expressing GFP-Merlin (left). Numbers refer to the intensity score (0: no signal, 1: intermediate, and 2: strong). Scale bar is 50 μm. The quantification of AMOTL1 intensity score upon expression of Merlin, a defective AMOTL1-binding Merlin mutant (1-532), or GFP alone is presented on the right graph. AMOTL1 staining was scored in 150 transfected cells (right, Kruskall Wallis test). (D) The same approach was used for AIP4, an ubiquitin ligase activity dead mutant of AIP4 (CS) or GFP-NEDD4 (n = 150 cells). GFP was used as a negative control (quantification on top with Kruskal-Wallis test, immunofluorescence at the bottom). Scale bar is 50 μm. (E) The level of endogenous AMOTL1 increases in BC52 cells when the expression of AIP4 and NEDD4 is inhibited with siRNA (right). [Left, quantification (n = 3) Kruskal-Wallis test]. (F) The specificity of the anti-phospho-AMOTL1 S262 was tested on immunoprecipitated GFP-AMOTL1 treated (+) or not (−) with alkaline phosphatase (top). (G) Merlin induces the phosphorylation of GFP-AMOTL1 in 293 HEK cells. The level of phosphorylation of AMOTL1 was quantified by Western blot on immunoprecipitated GFP-AMOTL1 and normalized from the amount of GFP-AMOTL1 (n = 4, Mann-Whitney test). (H) When tested using the same strategy as in panel A, Merlin expression showed no significant impact on the half-life of AmotL1S262A mutant (n = 4, Mann-Whitney test). For A, B, C, E, G, and H, values are provided as mean ± SEM.
AMOTL1 Expression Is Linked to Proliferation, Migration, and EMT
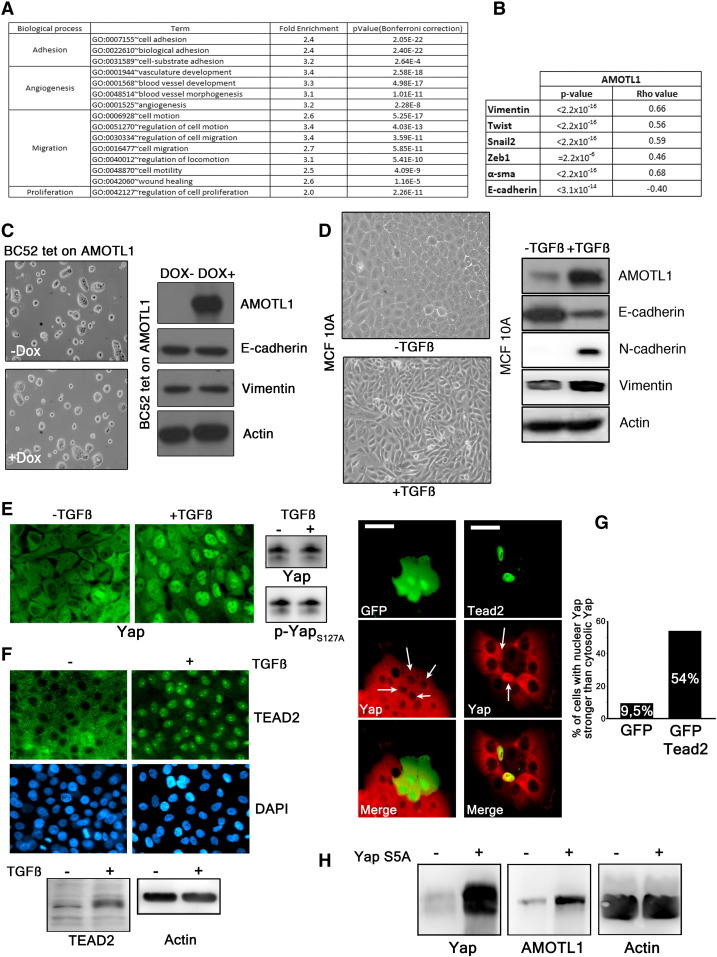
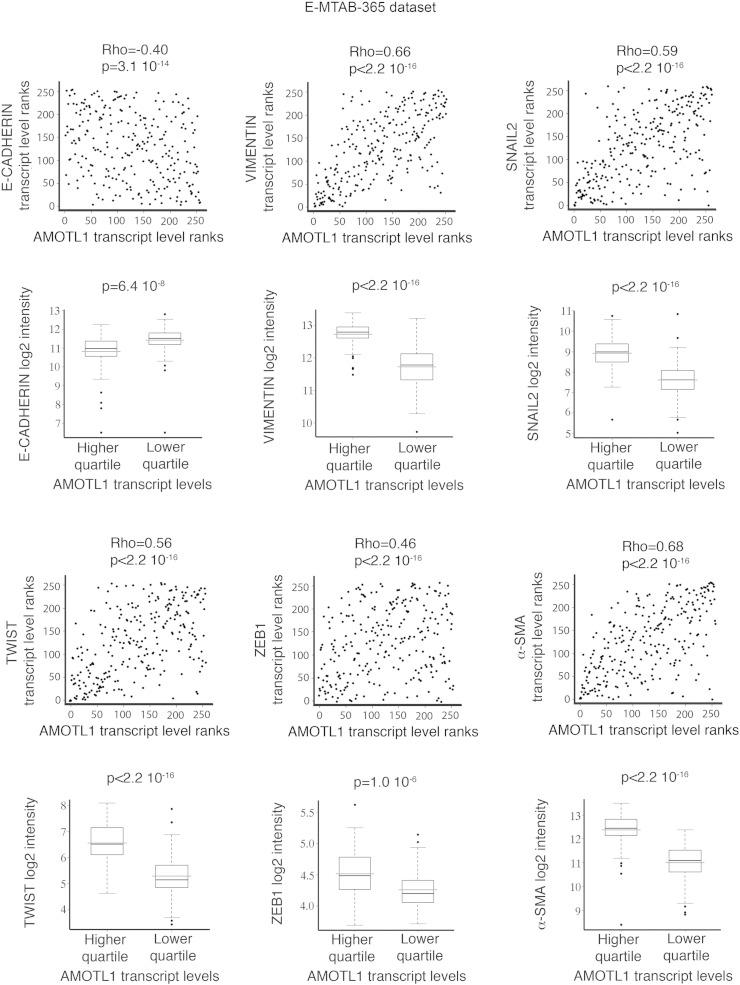
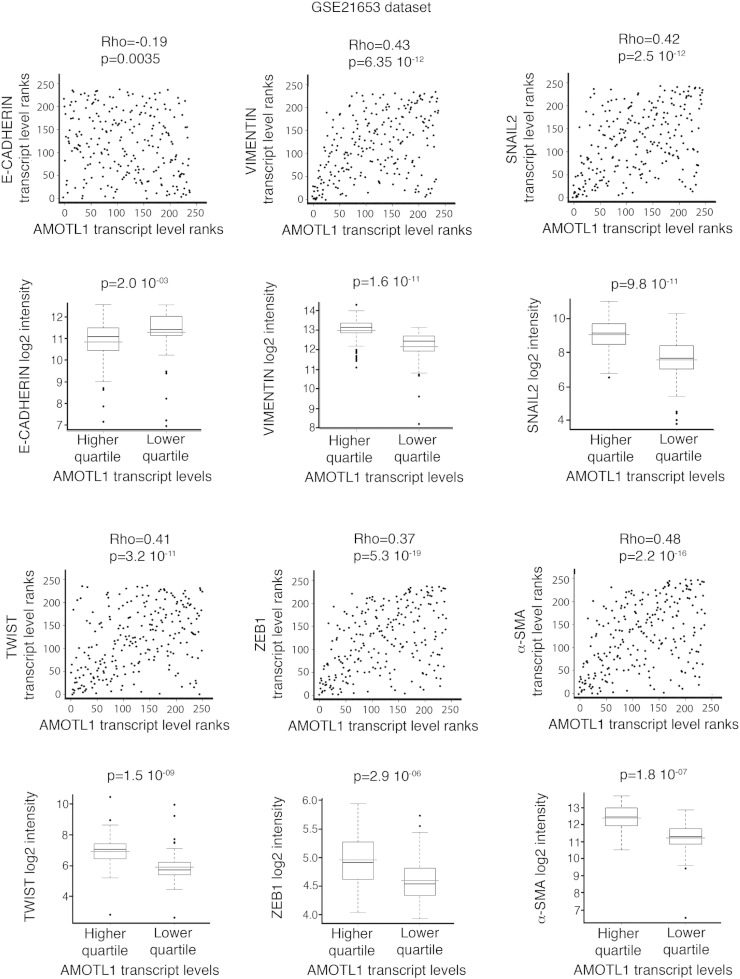
Taking advantage of the two gene expression data sets previously described (E-MTAB-365 and GSE21653), we defined an AMOTL1 transcriptional signature corresponding to the genes with the highest positive correlation to AMOTL1 transcript levels (Supplementary Table S3 and experimental procedures). With this signature, we performed an unsupervised analysis and pathway enrichment study using all Gene Ontology (GO) terms. The main GO terms associated with AMOTL1 signature (with P values smaller than 10− 3 after Bonferroni correction) corresponded to adhesion, angiogenesis, migration, and proliferation (Figure 5A and Supplementary Table S4) for both E-MTAB-365 and GSE21653 data sets. The angiogenesis signature was expected from AMOTL1 role in angiogenesis and endothelial cell regulation [4], [10]. We also confirmed the involvement of AMOTL1 in proliferation and migration. An AMOTL1 signature was also produced from a set of colon carcinoma (GSE64857) and ovarian tumor (GSE63885). They appeared different from the breast cancer signature, suggesting a different role for AMOTL1 depending on the tumor type, although some processes, such as angiogenesis, were conserved (Supplementary Table S5). Strikingly, in breast tumors, the list of genes whose expression strongly correlated with AMOTL1 (P < 10- 6) included EMT markers such as Vimentin, Twist, Zeb1, Snail2, and α-smooth muscle actin (α-sma) in addition to a strong negative correlation (P < 10− 13) with E-cadherin (Figure 5B and Supplementary Figure S3., Supplementary Figure S4.). EMT is recognized as an important mechanism for carcinoma progression. AMOTL1 overexpression in BC52 however did not lead to noticeable changes in cell morphology or in EMT markers’ expression (Figure 5C). But when we stimulated MCF10A with TGFβ, a classical model of EMT induction, we observed a strong increase in AMOTL1 expression together with Vimentin and N-cadherin and a reduction of E-cadherin levels (Figure 5D). In the light of our results on the regulation of AMOTL1 expression, we tested if its induction by TGFβ might require Yap and Merlin. TGFβ treatment of MCF10A or BC52 had no impact on Merlin levels or phosphorylation (not shown). However, we observed a marked nuclear accumulation of Yap in the nuclei (Figure 5E). This was not linked to a change in Yap expression or phosphorylation on S127 residue (Figure 5E), arguing for a Hippo-independent mechanism. However, we observed an increase of Tead2 expression by Western blot and its accumulation in the nuclei (Figure 5F). We then tested if Tead2 could recruit Yap to the nucleus by overexpressing GFP-Tead2 in MCF10A and evaluating the localization of endogenous Yap. Indeed, GFP-Tead2 overexpression induced a very obvious accumulation of Yap in the nuclei of MCF10A (Figure 5G). Finally, when a constitutively active Yap5SA mutant was stably expressed in MCF10A, it led to elevated levels of AMOTL1 protein (Figure 5H). Together, our results suggest that, upon TGFβ stimulation, Tead2 accumulates and recruits Yap into the nucleus where together they stimulate AMOTL1 expression. Thus, AMOTL1 expression is part of an EMT program in breast tumor cells and likely contributes to it. However, its expression alone is not sufficient to trigger EMT.
Figure 5.
AMOTL1 is associated with proliferation, migration, and EMT in human breast tumors. (A) List of the GO terms associated to the AMOTL1 signature in data set E-MTAB-365. (B) Summary of the correlations found between AMOTL1 and EMT markers’ expression for the same data set (Spearman test, E-MTAB-365) (see also Supplementary Figure S3). (C) BC52 tet on AMOTL1 cells induced for 48 hours with doxycycline show no EMT-like morphological changes (left). The overexpression of AMOTL1 in BC52 cells does not induce the expression of the EMT markers (right). (D) EMT was induced by exposing MCF10A to 10 ng/ml of TGFβ for 24 hours (phase contrast at right) leading to the downregulation of E-cadherin and increased AMOTL1, N-cadherin, and Vimentin levels (left). (E) Three days of TGFβ treatment induces Yap nuclear translocation in MCF10A (left). However, Western blot analysis of the cells shows no impact on the levels and phosphorylation of Yap on S127 (right). (F) Three days of TGFβ at 10 ng/ml induces Tead2 nuclear accumulation in MCF10A (top). Western blot shows that the treatment induces Tead2 expression (bottom). (G) GFP expression does not affect Yap nuclear localization, whereas GFP-Tead2 promotes Yap nuclear accumulation (left). Scale bar is 25 μm. The quantification of the experiment is presented on the right. More than 100 cells were counted for each condition. (H) The expression of constitutively nuclear Yap mutant (Yap 5SA) in MCF10A induces the expression of AMOTL1.
AMOTL1 is associated with proliferation, migration, and EMT in human breast tumors. (A) List of the GO terms associated to the AMOTL1 signature in data set E-MTAB-365. (B) Summary of the correlations found between AMOTL1 and EMT markers’ expression for the same data set (Spearman test, E-MTAB-365) (see also Supplementary Figure S3). (C) BC52 tet on AMOTL1 cells induced for 48 hours with doxycycline show no EMT-like morphological changes (left). The overexpression of AMOTL1 in BC52 cells does not induce the expression of the EMT markers (right). (D) EMT was induced by exposing MCF10A to 10 ng/ml of TGFβ for 24 hours (phase contrast at right) leading to the downregulation of E-cadherin and increased AMOTL1, N-cadherin, and Vimentin levels (left). (E) Three days of TGFβ treatment induces Yap nuclear translocation in MCF10A (left). However, Western blot analysis of the cells shows no impact on the levels and phosphorylation of Yap on S127 (right). (F) Three days of TGFβ at 10 ng/ml induces Tead2 nuclear accumulation in MCF10A (top). Western blot shows that the treatment induces Tead2 expression (bottom). (G) GFP expression does not affect Yap nuclear localization, whereas GFP-Tead2 promotes Yap nuclear accumulation (left). Scale bar is 25 μm. The quantification of the experiment is presented on the right. More than 100 cells were counted for each condition. (H) The expression of constitutively nuclear Yap mutant (Yap 5SA) in MCF10A induces the expression of AMOTL1.
Supplementary Figure S3.
Scatter plot representation of the correlations between AMOTL1 and EMT-related markers’ transcript levels in E-MTAB-365 data set using Spearman test. Markers are E-cadherin, Vimentin, Snail2, Twist, Zeb1, and α-sma. In addition, for each EMT-related transcript, the level of expression is compared between the highest quartile and the lowest quartile of AMOTL1 expression (Mann-Whitney test).
Supplementary Figure S4.
Scatter plot representation of the correlations between AMOTL1 and EMT-related markers’ transcript levels in GSE21653 data set using Spearman test. Markers are E-cadherin, Vimentin, Snail2, Twist, Zeb1, and α-sma. In addition, for each EMT-related transcript, the level of expression is compared between the highest quartile and the lowest quartile of AMOTL1 expression (Mann-Whitney test).
AMOTL1 Expression Stimulates Migration of Breast Cancer Cells
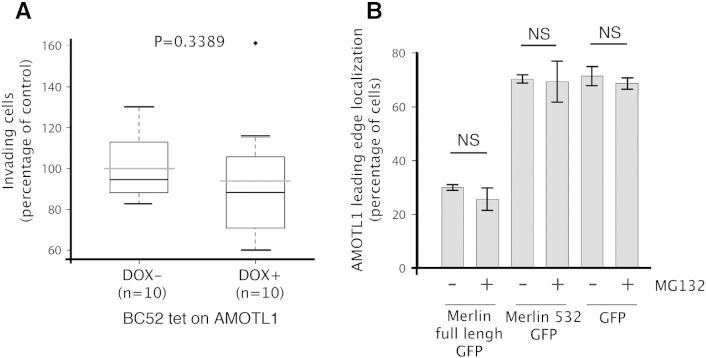
Cells undergoing EMT acquire a more motile phenotype. Hence, we investigated the role of AMOTL1 in breast cancer cell migration. When AMOTL1 expression was induced in BC52 cells, it stimulated migration measured by Transwell assay (Figure 6A). Cell invasion of a Matrigel layer was also increased. However, when the effect of the induction of migration was substracted, it appeared that AMOTL1 had no significant impact on invasion (Supplementary Figure S5A). Conversely, when AMOTL1 expression was inhibited using siRNA in MDA-MB-468 cells, migration was reduced two-folds compared with control (Figure 6B). Taken together, our results confirm that AMOTL1 is a strong inducer of breast cancer cell migration, in turn potentiating tumor invasion.
Figure 6.
AMOTL1 induces cell migration and is negatively regulated by Merlin at the lamellipodia. (A) AMOTL1 induction (+ DOX) in BC52 tet on AMOTL1 triggers migration in Transwell assay (the number of independent experiments is indicated). The intrinsic effect of doxycycline on migration was tested on the parental BC52 tet on cell line and was not significant (Mann-Whitney test). (B) AMOTL1 depletion by siRNA reduces MDA-MB468 cell migration in Transwell assay (Mann-Whitney test). (C) AMOTL1 (green) is delocalized away from the lamellipodia in cells from a colony of BC52 tet on AMOTL1 transfected with a plasmid expressing Merlin (red) (left images and see also Supplementary Figure S5B). Scale bar is 25 μm. The presence of AMOTL1 at the leading edge of BC52 tet on AMOTL1 cells upon GFP, GFP-Merlin, or GFP-Merlin 1-532 expression was measured (right graph) (Kruskal-Wallis test). Values are provided as mean ± SD. (D) The phospho-mimetic mutant of AMOTL1 (262D) does not localize at the leading edge of BC52 cells and shows a punctuated distribution, in contrast to wild-type (WT). The S262A mutant AMOTL1 is more obviously associated to filamentous actin structures. Scale bar is 10 μm. (E) Merlin expression does not delocalize AMOTL1 S262A from the leading edge of BC52 cells. Scale bar is 10 μm.
AMOTL1 induces cell migration and is negatively regulated by Merlin at the lamellipodia. (A) AMOTL1 induction (+ DOX) in BC52 tet on AMOTL1 triggers migration in Transwell assay (the number of independent experiments is indicated). The intrinsic effect of doxycycline on migration was tested on the parental BC52 tet on cell line and was not significant (Mann-Whitney test). (B) AMOTL1 depletion by siRNA reduces MDA-MB468 cell migration in Transwell assay (Mann-Whitney test). (C) AMOTL1 (green) is delocalized away from the lamellipodia in cells from a colony of BC52 tet on AMOTL1 transfected with a plasmid expressing Merlin (red) (left images and see also Supplementary Figure S5B). Scale bar is 25 μm. The presence of AMOTL1 at the leading edge of BC52 tet on AMOTL1 cells upon GFP, GFP-Merlin, or GFP-Merlin 1-532 expression was measured (right graph) (Kruskal-Wallis test). Values are provided as mean ± SD. (D) The phospho-mimetic mutant of AMOTL1 (262D) does not localize at the leading edge of BC52 cells and shows a punctuated distribution, in contrast to wild-type (WT). The S262A mutant AMOTL1 is more obviously associated to filamentous actin structures. Scale bar is 10 μm. (E) Merlin expression does not delocalize AMOTL1 S262A from the leading edge of BC52 cells. Scale bar is 10 μm.
Supplementary Figure S5.
(A) The impact of doxycycline-mediated induction of AmotL1 expression on BC52 tet on AL1 cells invasion was evaluated using Transwell assay and corrected from the intrinsic effect of doxycycline on invasion that was measured on BC52 tet on cells (Mann-Whitney test). (B) The presence of AMOTL1 at the leading edge of BC52 tet on AL1 was measured in cells expressing GFP-Merlin, GFP-Merlin (1-532), and GFP alone in the presence or absence of MG132 treatment (mean ± SD). Inhibition of the proteasome has no effect on AMOTL1 delocalization by Merlin.
Merlin Antagonizes AMOTL1 Function during Cell Migration
When expressed in BC52 cells, AMOTL1 clearly localized to the lamellipodia (Figure 6C). We also observed Merlin presence in this structure. Very interestingly, upon GFP-Merlin expression, AMOTL1 no longer remained at the leading edge (Figure 6, C, left). A mutant of Merlin defective for AMOTL1 binding had no such effect, indicating that the interaction with Merlin is necessary for AMOTL1 delocalization (Figure 6, C, right). MG132 treatment did not prevent AMOTL1 removal from lamellipodia, showing that local degradation by Merlin is the cause (Supplementary Figure S5B). It has been shown that phosphorylation of AMOT on the site equivalent to S262 of AMOTL1 prevents the interaction with F-actin. When expressed in BC52, the S262D phospho-mimicking AMOTL1 mutant did not localize to the lamellipodia but presented a vesicular distribution (Figure 6D). The S262A mutant was strongly associated to actin filaments but could localize to the lamellipodia although not as clearly as the WT AMOTL1 (Figure 6D). Importantly, AMOTL1 262D was found to colocalize at the leading edge of the BC52 cells with Merlin and was not efficiently displaced (Figure 6E). Hence, based on our results, we propose that Merlin induction of S262 phosphorylation releases AMOTL1 from actin, leading to its removal from the lamellipodia.
AMOTL1 Promotes Proliferation of Breast Tumor Cells through Src Activation
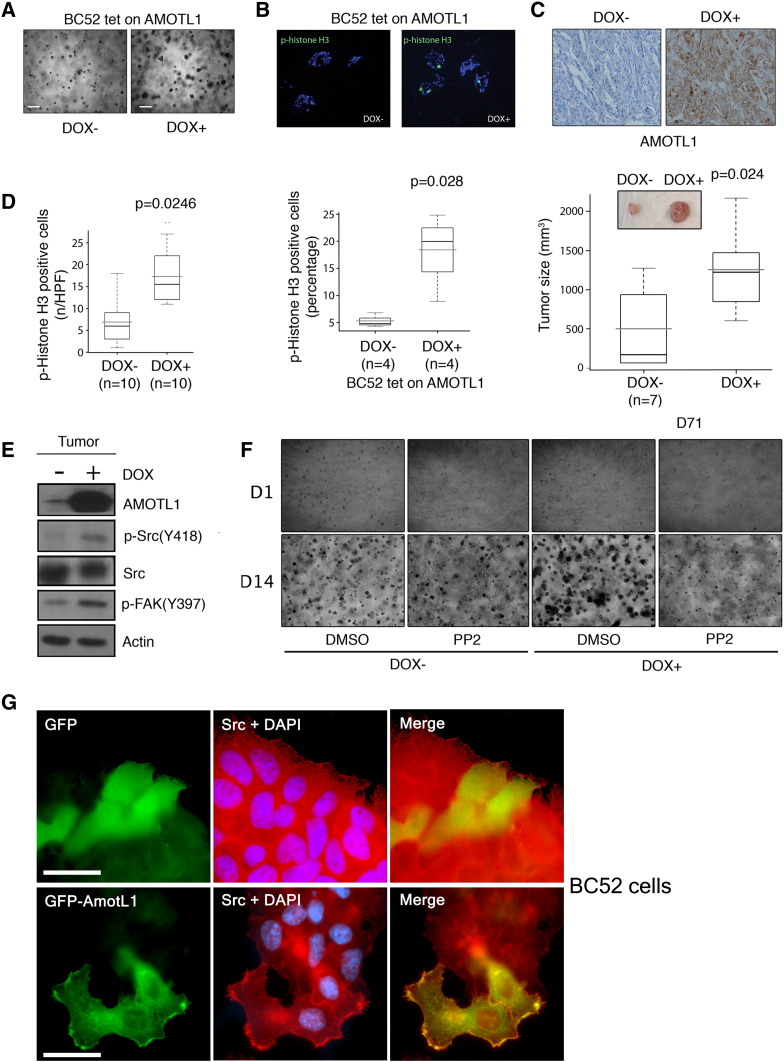
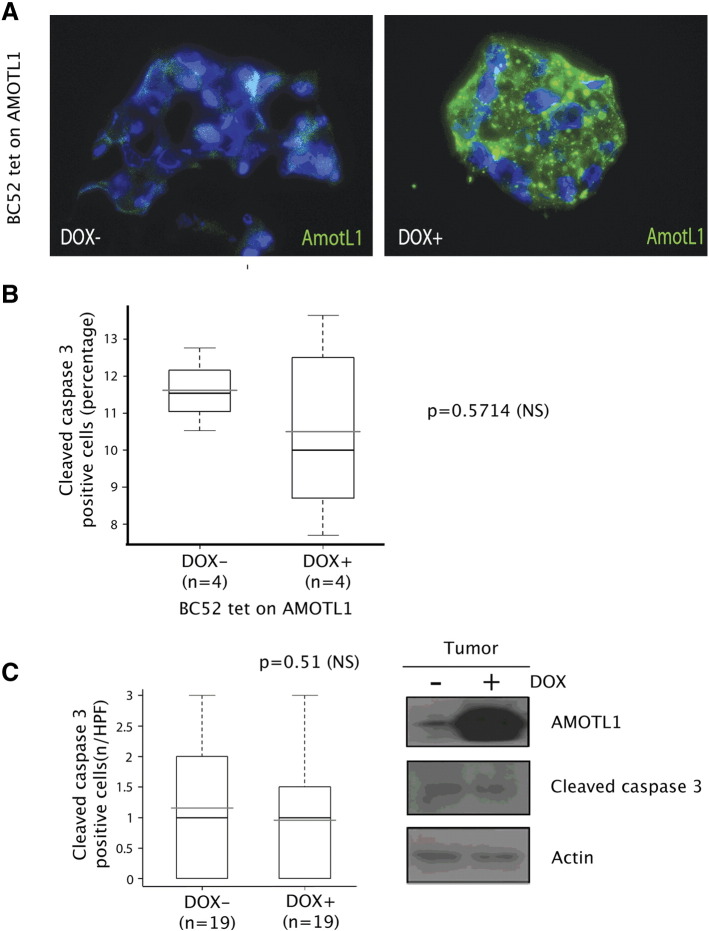
AMOTL1 impact on breast cancer cell proliferation was tested in 3D Matrigel matrix. BC52 tet on AMOTL1 cells grew into bigger aggregates in the presence of doxycycline (Figure 7A) and presented an increased number of p-histone H3-positive cells (Figure 7B). However, there was no modification of apoptosis as measured by cleaved caspase 3 staining (Supplementary Figure S6, A and B). When BC52 tet on AMOTL1 were injected in their fat pad, nude mice that were fed with doxycycline in the drinking water grew bigger tumors compared with control, with a mean volume increase of 3.4-fold (372 vs 1260 mm3) after 71 days (Figure 7C). Tumors overexpressing AMOTL1 expressed higher levels of p-Histone H3 (Figure 7D) but no increase in apoptosis (Supplementary Figure S6C). We next investigated which mitogenic signaling might be activated by AMOTL1 overexpression in mouse tumors. We observed no difference in PAK phosphorylation (not shown) or YAP nuclear localization (Supplementary Figure S6D), two pathways regulated by the motins. Interestingly however, we found that the levels of phosphorylated Src were higher in tumors overexpressing AMOTL1 (Figure 7E). Consistently, the phosphorylation of a target of Src, FAK, was upregulated (Figure 7E). Finally, BC52 tet on AMOTL1 cells were grown as aggregates in 3D and treated with the Src inhibitor PP2. Whereas no obvious effect of PP2 was observed in the absence of doxycycline, the growth of cell aggregates overexpressing AMOTL1 was severely inhibited (Figure 7F). Similarly to what was described for AMOTL2, we observed that, in BC52 cells, AMOTL1 colocalizes with endogenous Src and appears to recruit it to lamellipodia (Figure 7G). Overall, our results show that AMOTL1 promotes proliferation of breast cancer cells in vitro and in vivo by stimulating Src activity, possibly by localizing it to specific domains where it can be activated.
Figure 7.
AMOTL1 regulates proliferation of BC52 cells in vitro and in vivo through Src activation. (A) AMOTL1 induction by doxycycline for 14 days in BC52 leads to bigger aggregates in Matrigel (scale bar: 100 μm). (B) Phospho-histone H3 expression measured on cryosections (top) and quantified (bottom) is higher upon AMOTL1 induction (+ DOX) (Mann-Whitney test). (C) AMOTL1 expression is induced in tumor from mice treated (DOX +) with doxycycline (IHC, upper pictures). The mean tumor size is bigger after 71 days in doxycycline-treated mice (Mann-Whitney test). The insert shows representative tumors. (D) Quantification of phospho-histone H3 staining performed on DOX − and DOX + tumor cryosections (left, Mann-Whitney test). Phospho-histone H3 expression is induced in mouse tumor upon induction of AMOTL1 expression (Western blot, right). (E) AMOTL1, Src, phospho-Src (Y418), and phospho-FAK (Y397) show increased levels in doxycycline-treated mouse tumor (DOX +) compared with untreated (DOX −) (Western blot of a representative tumor). (F) BC52 tet on AMOTL1 aggregates in Matrigel were treated by DMSO as a control or by Src inhibitor PP2 (100 nM) during 14 days. Src inhibition predominantly inhibits the growth of BC52 aggregates that overexpress AMOTL1 (DOX +). (G) When expressed in BC52 cells, GFP-AMOTL1 colocalizes with endogenous Src and appears to recruit it to lamellipodia. GFP is used as a negative control. Scale bar is 25 μm.
Supplementary Figure S6.
(A) Staining of AMOTL1 expression in cryosections of BC52 tet on AL1 cell aggregates in Matrigel, with (DOX +) or without (DOX −) doxycycline induction. (B) The expression of cleaved caspase 3 was evaluated by immunofluorescence staining performed on aggregate cryosections of BC52 tet on AL1 cell aggregates in Matrigel, with (DOX +) or without (DOX −) doxycycline induction. The percentage of cleaved caspase-3 positive is presented. No significant difference was observed (Mann-Whitney test). (C) The expression of cleaved caspase-3 was evaluated by IHC staining performed on tumor sections of mice fed with (DOX +) or without (DOX −) doxycycline in the drinking water. The number of positive caspase-3 cells per high-power field is presented on the left. The right panel shows immunoblot analysis of cleaved caspase 3 levels in tumor samples of mice fed with (DOX +) or without (DOX −) doxycycline. No significant difference was observed (Mann-Whitney test).
Discussion
Evidences of motins’ involvement in cancer remain sparse. In breast tumors, we observe that, in contrast to AMOT and AMOTL2, AMOTL1 mRNA is differently expressed between the ER − and ER + subgroups and that AMOTL1 and estrogen receptor mRNA are negatively correlated. A link between AMOTL1 expression and the ER status was previously reported [10], supporting the notion that AMOTL1 plays a role in this group of tumors. Surprisingly, no difference in staining intensity linked to the ER status was observed. AMOTL1 protein expression however increases during tumor progression, with the highest levels in lymph node metastasis. The apparent discrepancy between AMOTL1 transcripts and protein levels in relation to the ER status could be explained as follows: AMOTL1 protein levels are higher in the more invasive IDC lesion, irrespective of the ER status. However, the more aggressive ER − tumors contain a higher proportion of invasive lesions. AMOTL1 transcripts levels are measured from biopsies that contain variable proportions of DCIS and IDC, and their levels likely reflect the enrichment in IDC lesions in ER − tumors.
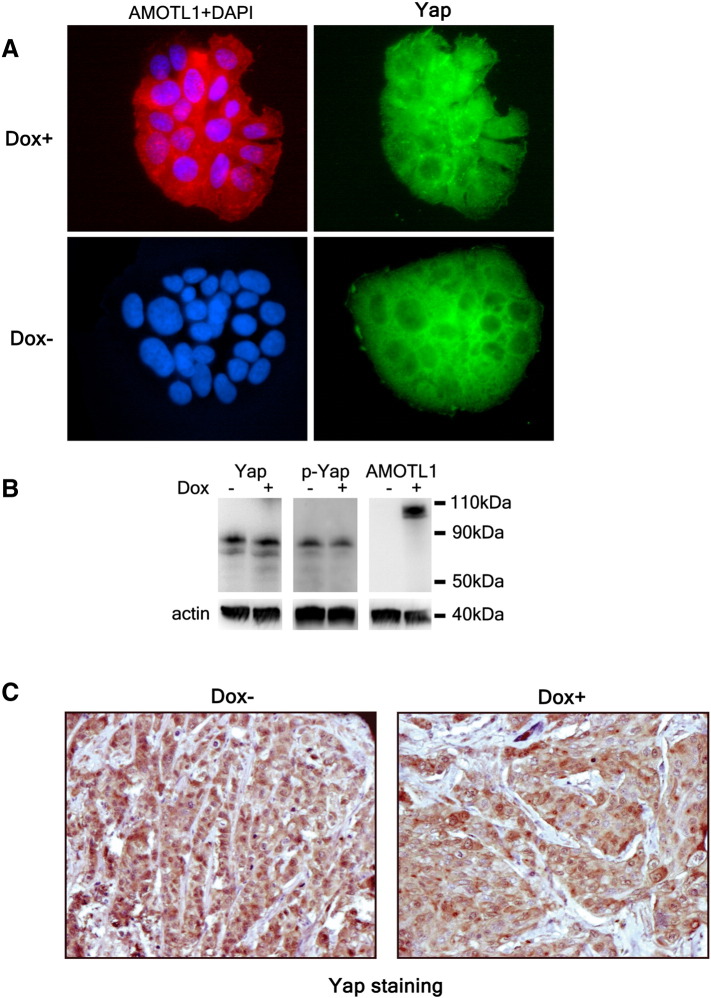
Gene ontology analysis of human tumor data clearly link AMOTL1 expression to EMT. AMOTL1 gene expression levels correlate with several markers of EMT. Its protein expression increases in response to EMT induced by TGFβ in MCF10A cells. We did not observe morphological changes induced by siRNA-mediated downregulation of AMOTL1 in MCF10A (not shown). This is consistent with a previous study [17] and in contrast with the induction of EMT by AMOTL2 knock down in this cell type. AMOTL2 and L1 were shown to bind and sequester Yap in the cytoplasm. However, other studies also demonstrated a positive regulation of Yap by the motins [6]. We observed that the overexpression of AMOTL1 in BC52 cells and xenograft tumors (Supplementary Figure S7) or MCF10A (not shown) had little impact on endogenous Yap nuclear localization. Upon TGFβ stimulation, we saw a clear accumulation of Yap in the nuclei of MCF10A that AMOTL1 elevated levels could not prevent. Our results tend to demonstrate that AMOTL1 expression is a consequence of the activation of Yap in response to TGFβ and does not modulate Yap significantly. The expression of AMOTL1 alone does not trigger EMT in the tumor cells. Therefore, in breast tumors, our results show that AMOTL1 likely participates in EMT, an important mechanism for the transition from benign to invasive tumor. Cells from aggressive tumors are more motile and tend to proliferate faster. Our GO analysis again shows that AMOTL1 is associated to the regulation of proliferation and migration. Indeed, we show that AMOTL1 is a potent inducer of breast tumor cell growth in 3D cultures and in xenograft tumor model. It does not display any growth inhibition activity that was reported for motins in various cellular models [14], [7]. The mechanisms appear distinct from those proposed for AMOT [9], [15]. We did not observe an induction Rac and cdc42 activity by AMOTL1, assessed by the pull down of the activated form of the GTPases (not shown). Contrarily to AMOT [16], AMOTL1 had no impact on p-ERK levels in cells in culture or in tumors (not shown). As mentioned previously, AMOTL1 expression did not lead to the changes in YAP regulation that were reported in previous studies [6], [7], [17], [18] such as YAP cytoplasmic sequestration or change in its phosphorylation state (Supplementary Figure S7, A and B). Our work shows that inducing Yap activity is unlikely to be the major means by which AMOTL1 controls tumor growth. This observation is corroborated by a previous study showing that AMOTL1 expression does not modify the mRNA level of CTGF and Cyr61, bona fide targets of YAP [18]. We also identified genes that were differentially expressed between two xenograft tumors where AMOTL1 expression was stimulated by doxycycline and two control tumors (no doxycycline). The results are presented in Supplementary Table 6 and show no Yap target genes in the list. In contrast, our results show that AMOTL1 stimulates breast cancer cell proliferation by inducing Src activity. Xenografts tumors overexpressing AMOTL1 show elevated levels of active Src, and 3D cultures of BC52 cells become sensitive to PP2 Src inhibitor only when AMOTL1 is overexpressed. The mechanisms of this regulation appear to involve the relocalization of Src in specific compartment such as the lamillipodia, leading to its local activation similarly to what was shown for AMOTL2 [19].
Supplementary Figure S7.
(A) The induction by doxycycline of AMOTL1 expression in BC52 tet on AMOTL1 for 48 hours (Dox +, in red) has no visible impact on endogenous Yap (green) distribution when compared with uninduced cell (Dox −). Yap is essentially excluded from the nuclei of the cells in colonies. (B) Extracts were prepared under the same growth conditions as in A. The induction of AMOTL1 expression shows no noticeable impact on endogenous Yap expression or phosphorylation on S127. (C): IHC staining of YAP in BC52 tumors of mice fed with (DOX +) or without (DOX −) doxycycline in the drinking water. Nuclear and cytoplasmic expression of YAP appears essentially unaffected by AMOTL1 overexpression.
AMOTL1 promotes endothelial cell migration, and we observed that it stimulates breast tumor cells’ migration too. Again, the absence of activation of Rac and Cdc42 by AMOTL1 argues against their involvement in the process. However, we cannot exclude that AMOTL1 may regulate Rac activity locally similarly to the regulation of RhoA activity by AMOT [20]. Indeed, AMOTL1 clearly localizes in the lamellipodia, suggesting an active role in the dynamics of this structure essential for migration. Merlin was previously shown to inhibit migration in various models [21], [22], [23]. More recently, it was shown that Merlin coordinates collective cell migration in response to mechanical forces via the modulation of Rac activity. This function apparently also involves angiomotin [24]. Our results suggest that Merlin delocalization of AMOTL1 from the lamellipodia and subsequent degradation could result in the localized inactivation of Rac and participate in this process. An alternate possibility is that degradation of AMOTL1 prevents local activation of Src, thus impairing its role in cell migration. Delocalization and degradation of AMOTL1 are a consequence of AMOTL1 S262 phosphorylation induced by Merlin. Indeed, this event is sufficient to remove AMOTL1 from the lamellipodia by preventing the interaction with F-actin similarly to what was reported for AMOT [8]. In addition, phosphorylation was shown to facilitate the binding of the NEDD ubiquitin ligase [14] to AMOT and AMOTL1. But conflicting studies reported either degradation [13] or stabilization of the proteins [12] as a consequence. We show that, in BC52 cells, AIP4 and NEDD4 trigger AMOTL1 degradation. S262 is a target for LATS1/2 kinases that bind to motins. Interestingly, Merlin binds and activates LATS1/2 at the plasma membrane [25]. It is thus possible that Merlin locally stimulates the activity of LATS1/2 to phosphorylate AMOTL1.
Importantly, a remarkable study showed that Merlin expression levels are downregulated during breast cancer progression and are almost absent in metastasis, in a striking opposite pattern to the levels of AMOTL1 that we observed in our cohort [26]. This observation, together with our results, supports the idea that AMOTL1 acts as a promoter of breast cancer progression, more specifically under circumstances where the expression of Merlin is degraded. This happens following Merlin phosphorylation, a consequence of AKT activation, which is frequently observed in breast cancer. However, we also showed that AMOTL1 expression is stimulated by Yap in tumor cells and its mRNA levels correlate with Yap protein levels in invasive breast tumors. In conclusion, our work demonstrates that AMOTL1 is involved in breast cancer progression and that it is regulated by Hippo members Yap and Merlin either independently or synergistically. Besides breast cancer, our results show that AMOTL1 could participate in the aggressive phenotype of other cancers where Merlin expression is occasionally lost such as glioma [27] melanoma [28], mesothelioma [29], or renal cell carcinoma [30] or where signaling leading to Yap or Taz activation expression is induced [30].
Conflict of Interest
The authors declare no conflict of interest.
The following are the supplementary data related to this article.
Supplementary Table S1. Clinicopathological characteristics corresponding to the human breast tumors used to prepare the tissue microarray.
Supplementary Table S2. Characteristics of all the breast cancer cell lines used.
Supplementary Table S3. List of the genes with the highest positive correlation to AMOTL1 transcript levels from which was created an AMOTL1 transcriptional signature used to perform an unsupervised analysis and pathway enrichment study using all Gene Ontology (GO) terms.
Supplementary Table S4. Results from the unsupervised analysis and pathway enrichment study using AMOTL1 transcriptional signature for all Gene Ontology (GO) terms. Dataset E-MTAB-365 and GSE21653.
Supplementary Table S5. Results from the unsupervised analysis and pathway enrichment study using AMOTL1 transcriptional signature for all Gene Ontology (GO) terms. Dataset GSE64857 (colon carcinoma) and GSE63885 (ovarian cancer).
Supplementary Table S6. List of the differentially expressed genes between xenografted tumors that overexpressed (Dox+) or not (Dox-) AMOTL1. Cut-off was set at 2 fold for upregulated genes and at 0.8 fold for downregulated genes. Tumor RNAs were extracted with the miniRNeasy kit (Qiagen) and analysed on Affimetryx U133 plus 2 array.
Acknowledgements
We wish to thank Laura Brulle and Sylvie Robine for help in mouse work; Maria Fernandes for AMOTL1 vector; Stefano Piccolo for the 8xGTIIC-Luc vector; and Celine Jacquet, Fatima Dekmous, and Houria Benamghar for their help. We thank the Programme Incitatif Collaboratif Breast Invasion Migration for the preparation and analysis of TMA. We also thank Sophie Richon and Virginie Maire for providing breast cancer cell lines. This work was supported by Institut National du Cancer, Association pour la Recherche sur le Cancer, Ligue contre le Cancer, Association Neurofibromatose et Recklinghausen,Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Institut Curie for D. L. and ANR 11-bsv8-0010-02 INCA-6521 and ARC PGA120140200831 for A. G.
Footnotes
This work was supported by Institut National du Cancer, Association pour la Recherche sur le Cancer, Ligue contre le Cancer, Association Neurofibromatose et Recklinghausen,Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, and the Institut Curie for D. L. and ANR11-bsv8-0010-02 INCA-6521 and ARCPGA120140200831 for A. G.
References
- 1.Bratt A, Wilson WJ, Troyanovsky B, Aase K, Kessler R, Van Meir EG, Holmgren L. Angiomotin belongs to a novel protein family with conserved coiled-coil and PDZ binding domains. Gene. 2002;298:69–77. doi: 10.1016/s0378-1119(02)00928-9. [DOI] [PubMed] [Google Scholar]
- 2.Troyanovsky B, Levchenko T, Månsson G, Matvijenko O, Holmgren L. Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J Cell Biol. 2001;152:1247–1254. doi: 10.1083/jcb.152.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratt A, Birot O, Sinha I, Veitonmäki N, Aase K, Ernkvist M, Holmgren L. Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem. 2005;280:34859–34869. doi: 10.1074/jbc.M503915200. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Y, Vertuani S, Nyström S, Audebert S, Meijer I, Tegnebratt T, Borg JP, Uhlén P, Majumdar A, Holmgren L. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ Res. 2009;105:260–270. doi: 10.1161/CIRCRESAHA.109.195156. [DOI] [PubMed] [Google Scholar]
- 5.Gagné V, Moreau J, Plourde M, Lapointe M, Lord M, Gagnon E, Fernandes MJ. Human angiomotin-like 1 associates with an angiomotin protein complex through its coiled-coil domain and induces the remodeling of the actin cytoskeleton. Cell Motil Cytoskeleton. 2009;66:754–768. doi: 10.1002/cm.20405. [DOI] [PubMed] [Google Scholar]
- 6.Yi C, Shen Z, Stemmer-Rachamimov A, Dawany N, Troutman S, Showe LC, Liu Q, Shimono A, Sudol M, Holmgren L. The p130 isoform of angiomotin is required for Yap-mediated hepatic epithelial cell proliferation and tumorigenesis. Sci Signal. 2013;6:ra77. doi: 10.1126/scisignal.2004060. [ http://dx.doi.org/10.1126/scisignal.2004060] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B, Li L, Lu Q, Wang LH, Liu C-Y, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai X, She P, Chi F, Feng Y, Liu H, Jin D, Zhao Y, Guo X, Jiang D, Guan KL. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem. 2013;288:34041–34051. doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, Christian N, Persson NL, Shimono A, Speicher DW, Marmorstein R. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang WG, Watkins G, Douglas-Jones A, Holmgren L, Mansel RE. Angiomotin and angiomotin like proteins, their expression and correlation with angiogenesis and clinical outcome in human breast cancer. BMC Cancer. 2006;6:6–16. doi: 10.1186/1471-2407-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Liu P, Tian M, Li Y, Chen W, Li X. Proteomic identification of angiomotin by ProteomeLab PF-2D and correlation with clinical outcome in human clear cell renal cell carcinoma. Int J Oncol. 2013;42:2078–2086. doi: 10.3892/ijo.2013.1889. [DOI] [PubMed] [Google Scholar]
- 12.Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD, Sun Y, Hudmon A, Wells CD. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci U S A. 2013;110:17368–17373. doi: 10.1073/pnas.1308236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, An J, Zhang P, Xu C, Gao K, Wu D, Wang D, Yu H, Liu JO, Yu L. The Nedd4-like ubiquitin E3 ligases target angiomotin/p130 to ubiquitin-dependent degradation. Biochem J. 2012;444:279–289. doi: 10.1042/BJ20111983. [DOI] [PubMed] [Google Scholar]
- 14.Oka T, Schmitt AP, Sudol M. Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene. 2012;31:128–134. doi: 10.1038/onc.2011.216. [DOI] [PubMed] [Google Scholar]
- 15.Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, Kulkarni S, Gish G, Virag C, Lim C. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125:535–548. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 16.Ranahan WP, Han Z, Smith-Kinnaman W, Nabinger SC, Heller B, Herbert B-S, Chan R, Wells CD. The adaptor protein AMOT promotes the proliferation of mammary epithelial cells via the prolonged activation of the extracellular signal-regulated kinases. Cancer Res. 2011;71:2203–2211. doi: 10.1158/0008-5472.CAN-10-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. J Biol Chem. 2011;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Li Z, Xu P, Huang L, Tong J, Huang H, Meng A. Angiomotin-like2 gene (amotl2) is required for migration and proliferation of endothelial cells during angiogenesis. J Biol Chem. 2011;286:41095–41104. doi: 10.1074/jbc.M111.296806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernkvist M, Luna Persson N, Audebert S, Lecine P, Sinha I, Liu M, Schlueter M, Horowitz A, Aase K, Weide T. The Amot/Patj/Syx signaling complex spatially controls RhoA GTPase activity in migrating endothelial cells. Blood. 2009;113:244–253. doi: 10.1182/blood-2008-04-153874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Gutmann DH, Haipek CA, Martinsen BJ, Bronner-Fraser M, Krull CE. Characterization of chicken Nf2/merlin indicates regulatory roles in cell proliferation and migration. Dev Dyn. 2004;229:541–554. doi: 10.1002/dvdy.20002. [DOI] [PubMed] [Google Scholar]
- 22.Poulikakos PI, Xiao GH, Gallagher R, Jablonski S, Jhanwar SC, Testa JR. Re-expression of the tumor suppressor NF2/merlin inhibits invasiveness in mesothelioma cells and negatively regulates FAK. Oncogene. 2006;25:5960–5968. doi: 10.1038/sj.onc.1209587. [DOI] [PubMed] [Google Scholar]
- 23.Hartmann M, Parra LM, Ruschel A, Schubert S, Li Y, Morrison H, Herrlich A, Herrlich P. Tumor suppressor NF2 blocks cellular migration by inhibiting ectodomain cleavage of CD44. Mol Cancer Res. 2015;13(5):879–890. doi: 10.1158/1541-7786.MCR-15-0020-T. [DOI] [PubMed] [Google Scholar]
- 24.Das T, Safferling K, Rausch S, Grabe N, Boehm H, Spatz JP. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat Cell Biol. 2015;17:276–287. doi: 10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- 25.Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrow KA, Das S, Metge BJ, Ye K, Mulekar MS, Tucker JA, Samant RS, Shevde LA. Loss of tumor suppressor Merlin in advanced breast cancer is due to post-translational regulation. J Biol Chem. 2011;286:40376–40385. doi: 10.1074/jbc.M111.250035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau Y-KI, Murray LB, Houshmandi SS, Xu Y, Gutmann DH, Yu Q. Merlin is a potent inhibitor of glioma growth. Cancer Res. 2008;68:5733–5742. doi: 10.1158/0008-5472.CAN-08-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray LB, YuQ Lau Y-KI. Merlin is a negative regulator of human melanoma growth. PLoS One. 2012;7:e43295. doi: 10.1371/journal.pone.0043295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianchi AB, Mitsunaga SI, Cheng JQ, Klein WM, Jhanwar SC, Seizinger B, Kley N, Klein-Szanto AJ, Testa JR. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc Natl Acad Sci U S A. 1995;92:10854–10858. doi: 10.1073/pnas.92.24.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moroishi T, Hansen CG, Guan KL. The emerging roles of Yap and Taz in cancer. Nat Rev Cancer. 2015;15(2):73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami H, Mizuno T, Taniguchi T, Fujii M, Ishiguro F, Fukui T, Demidenko E, Korc M, Shi W, Preis M. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 2011;71:873–883. doi: 10.1158/0008-5472.CAN-10-2164. [DOI] [PubMed] [Google Scholar]
- 32.Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, Perou CM, Mohsin SK, O'Connell P, Tsimelzon A, Medina D. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 33.Hannemann J, Velds A, Halfwerk JBG, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res. 2006;8:R61. doi: 10.1186/bcr1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent-Salomon A, Lucchesi C, Gruel N, Raynal V, Pierron G, Goudefroye R, Reyal F, Radvanyi F, Salmon R, Thiery JP. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14:1956–1965. doi: 10.1158/1078-0432.CCR-07-1465. [DOI] [PubMed] [Google Scholar]
- 35.Yu K, Lee CH, Tan PH, Tan P. Conservation of breast cancer molecular subtypes and transcriptional patterns of tumor progression across distinct ethnic populations. Clin Cancer Res. 2004;10:5508–5517. doi: 10.1158/1078-0432.CCR-04-0085. [DOI] [PubMed] [Google Scholar]
- 36.Prat A, Cheang MCU, Martín M, Parker JS, Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen TO. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maire V, Baldeyron C, Richardson M, Tesson B, Vincent-Salomon A, Gravier E, Marty-Prouvost B, De Koning L, Rigaill G, Dumont A. TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS One. 2013;8:e63712. doi: 10.1371/journal.pone.0063712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Clinicopathological characteristics corresponding to the human breast tumors used to prepare the tissue microarray.
Supplementary Table S2. Characteristics of all the breast cancer cell lines used.
Supplementary Table S3. List of the genes with the highest positive correlation to AMOTL1 transcript levels from which was created an AMOTL1 transcriptional signature used to perform an unsupervised analysis and pathway enrichment study using all Gene Ontology (GO) terms.
Supplementary Table S4. Results from the unsupervised analysis and pathway enrichment study using AMOTL1 transcriptional signature for all Gene Ontology (GO) terms. Dataset E-MTAB-365 and GSE21653.
Supplementary Table S5. Results from the unsupervised analysis and pathway enrichment study using AMOTL1 transcriptional signature for all Gene Ontology (GO) terms. Dataset GSE64857 (colon carcinoma) and GSE63885 (ovarian cancer).
Supplementary Table S6. List of the differentially expressed genes between xenografted tumors that overexpressed (Dox+) or not (Dox-) AMOTL1. Cut-off was set at 2 fold for upregulated genes and at 0.8 fold for downregulated genes. Tumor RNAs were extracted with the miniRNeasy kit (Qiagen) and analysed on Affimetryx U133 plus 2 array.