Abstract
Objectives
Cancer cachexia affects the majority of tumor patients and significantly contributes to high mortality rates in these subjects. Despite its clinical importance, the identity of tumor-borne signals and their impact on specific peripheral organ systems, particularly the heart, remain mostly unknown.
Methods and results
By combining differential colon cancer cell secretome profiling with large-scale cardiomyocyte phenotyping, we identified a signature panel of seven “cachexokines”, including Bridging integrator 1, Syntaxin 7, Multiple inositol-polyphosphate phosphatase 1, Glucosidase alpha acid, Chemokine ligand 2, Adamts like 4, and Ataxin-10, which were both sufficient and necessary to trigger cardiac atrophy and aberrant fatty acid metabolism in cardiomyocytes. As a prototypical example, engineered secretion of Ataxin-10 from non-cachexia-inducing cells was sufficient to induce cachexia phenotypes in cardiomyocytes, correlating with elevated Ataxin-10 serum levels in murine and human cancer cachexia models.
Conclusions
As Ataxin-10 serum levels were also found to be elevated in human cachectic cancer patients, the identification of Ataxin-10 as part of a cachexokine cocktail now provides a rational approach towards personalized predictive, diagnostic and therapeutic measures in cancer cachexia.
Keywords: Cancer cachexia, Ataxin-10, Cardiac dysfunction, Fatty acid metabolism
Highlights
-
•
Cancer cachexia induces remodeling of the heart.
-
•
Tumor-borne secreted factors mediate cardiomyocyte atrophy.
-
•
Selective cachexokines, including Bin1, Stx7, Minpp1, Gaa, Ccl2, Adamtsl4, and Atxn10 provoke aberrant cardiac FA metabolism.
-
•
Ataxin-10 levels are elevated under cachectic conditions in mice and tumor patients.
1. Introduction
Cancer-induced cachexia describes a multi-factorial disease condition characterized by massive loss of adipose tissue and skeletal muscle mass and is believed to be responsible for up to 30% of cancer-related deaths in humans [1]. However, the exact causes of death and in particular the contribution of cardiac dysfunction to deaths of these patients is unclear [2]. Due to the phenotypic heterogeneity of cancer cachexia, which depends on tumor type, size and mass [3], and the mostly unknown etiology at the molecular level, cachexia still represents an immediate unmet medical need as effective and routine therapeutic measures are still lacking to date [4].
Interestingly, the clinical severity of cancer cachexia hardly correlates with tumor mass [5], indicating that the tumor controls peripheral energy balance in critical host tissues via distinct signaling mediators rather than acting as a direct “energy sink” [6]. Indeed, classical experiments demonstrated the para-biotic transfer of cachexia in rats, indicating that circulating factors, rather than local tumor effects, act as causative factors in energy wasting [7].
Since the discovery of tumor necrosis factor (TNF) alpha as the wasting-associated “cachectin” [8], numerous cytokines have been discussed as pro-cachectic mediators, most notably Interleukin (IL) 6 [9]. In addition, zinc-alpha-2-glycoprotein (ZAG), also known as LMF (lipid mobilizing factor), has been identified as an adipokine with direct lipolytic action in white adipose tissue during murine and human cancer cachexia [10]. However, attempts to overcome cachectic energy wasting by therapeutically targeting these pathways remained rather disappointing and/or not feasible [11]. For example, administration of anti-TNF alpha antibodies [12] did not promote weight stabilization in human clinical trials, overall suggesting that the cancer cachectic phenotype results either from the combinatorial action of various factors or from yet to be discovered, individually acting, tumor-borne mediators. Indeed, technological limitations in both appropriate read-out systems as well as specific tumor secretome detection methods have thus far prevented the rational identification of tumor-borne signals and hampered major progress in prognostic/diagnostic/therapeutic options in cancer cachexia.
Whereas the vast majority of previous studies have focused on the loss of skeletal muscle and adipose tissue as phenotypic features of cancer cachexia, recent studies suggest that other organs, i.e. the liver [13], [14] and the heart [15], also play a prominent role in systemic cachexia. Indeed, cardiac failure is responsible for a substantial proportion of cancer cachexia-induced deaths [16], and classical clinical studies by Burch and colleagues already documented the presence of atrophic hearts and impairment in cardiac function in cachectic tumor patients [17]. In this respect, cancer-induced cachexia in mice was found to disrupt myocardial structure and re-activate the fetal gene program [18], [19], correlating with hypo-innervation in the left ventricle [20], overall suggesting that cardiac dysfunction may represent an understudied component of systemic cancer cachexia.
Here we develop and apply cellular high throughput cardiomyocyte phenotyping to test for the cachexia-inducing capacities of a newly defined set of tumor-borne signaling mediators. These cachexokines were found to be both necessary and sufficient to trigger cardiac atrophy and metabolic dysfunction in cardiomyocytes. As a prototype, Ataxin-10 was found to be sufficient to cause cachectic cardiac phenotypes and to signal the existence of cachexia by its serum levels in murine and human model systems as well as in pancreatic cancer patients.
2. Methods
2.1. Chemicals
All chemicals were purchased from Sigma Aldrich (Munich, Germany) unless stated otherwise. Cell culture media and supplements were from Life Technologies (Darmstadt, Germany) unless stated otherwise. Cell culture plastic ware was obtained from BD (Heidelberg, Germany) unless stated otherwise.
2.2. Animal experiments and treatments
Ectopic mouse model: 9–10 week old male Balb/c, C57BL6/N and Fox Chase SCID mice were obtained from Charles River Laboratories (CRL, Brussels, BE). Mice were injected subcutaneously into the right flank with 1.5 × 106 C26 colon carcinoma cells, 5 × 105 MC38 colon carcinoma cells and 5 × 106 SW480 cells, respectively. C26 mouse experiment: Mice were monitored for 21 days after tumor cell implantation and a cohort of mice (6 control and 10 tumor-bearing mice) was sacrificed every week after Pressure-Volume loop measurement. Echocardiography and Pressure-Volume loop (PV loop) measurement were performed weekly to assess the cardiac performance. Body composition was determined by Echo Magnetic Resonance Imaging (EchoMRI; Echo Medical Systems, Houston, TX, USA) analysis once a week. Body weight and food intake were determined in regular intervals. MC38 mouse experiment: Mice were monitored for 30 days after tumor cell inoculation. Cardiac function was assessed by weekly echocardiography. Body composition was also measured once a week. Body weight and food intake were determined in regular intervals. SW480 mouse experiment: Mice were monitored until the onset of cachexia (15–21 days after tumor cell inoculation). Body weight was measured at regular intervals.
Orthotopic mouse model (PDAC): 10 week old male C57BL6/J mice were obtained from Charles River Laboratories (CRL, Brussels, BE). Anesthetized mice treated with 5 mg/kg Carprofen as analgesia were injected with 3000 cells (PDAC cell line #8024 or #8025; kind gift from Dieter Saur, TU Munich, Germany) into the pancreas. Mice were monitored until the onset of cachexia (24–26 days after tumor cell inoculation). Body composition was determined once a week. Body weight was measured in regular intervals.
Genetic mouse model (APC delta 580 mice): APC delta 580 mice on a C57BL6/N background were originally purchased from the National Cancer Institute (NCI) at Frederick (Frederick, MD, USA) and bred at the Animal Facility of the German Cancer Research Center (DKFZ; Heidelberg, Germany). Heterozygous APC delta 580 male mice and age-matched wild-type C57BL/6N mice with an age of 14 weeks were monitored until the onset of cachexia (at 4–6 months of age). Cardiac performance which was measured by echocardiography and body weight were determined in regular intervals.
Diabetic mice: 9–11 week old female ob/ob and db/db mice were obtained from Charles River Laboratories (CRL, Brussels, BE). After 1 week, mice were sacrificed in random fed state.
A detailed description of general animal handling, hemodynamic measurement techniques and histology can be found in the supplemental material. The animal care and all experimental protocols were reviewed and approved by local authorities (Regierungspräsidium Karlsruhe, Germany; G23/08; G12/06; G73/11; G178/13).
2.3. Cell culture
Cell lines were regularly tested for mycoplasma contamination by the company Multiplexion using Multiplex Cell Contamination Test (McCT) [21].
2.4. Preparation of conditioned supernatants (SN)
C26, MC38 (kind gift from C. Bourquin, Clinical Pharmacology, Munich, Germany) and HEK293A cells (ATCC, Manassas, VA, USA) were plated onto 15 cm plates and after reaching a confluency of ∼50% culture medium (DMEM high glucose, 10% FBS, 1% Pen/Strep) was changed. After 48 h, media were collected and the remaining cells were pelleted by centrifugation. For the treatment of cardiomyocytes, conditioned SN was 3:1 diluted in fresh culture medium. A detailed description of used transfection procedures can be found in the supplemental material.
2.5. Primary neonatal rat cardiomyocytes
Hearts were isolated from male and female Wistar rats at the age of 1–2 days. Heart slices were digested in 0.1% pancreatin. To reduce the amount of non-cardiomyocyte like epithelial cells and fibroblasts, cells were pre-plated on 15 cm plates. Afterward, cells were harvested, counted and finally plated on cell culture plates that were coated with 0.1% gelatin (BD, Heidelberg, Germany). Cells were cultured in DMEM high glucose supplemented with 10% FBS and 1% Pen/Strep. After 3 days of plating cells were treated with conditioned SN for 24 h. A detailed description of the high throughput analysis of cardiomyocyte size and immunofluorescence staining can be found in the supplemental material.
2.6. Primary adult mouse cardiomyocytes
Hearts from 9 to 10 week old anesthetized Balb/c male mice were excised and mounted on a Langendorff perfusion apparatus. A retrograde perfusion with nominally Ca2+-free Tyrode's solution (pH 7.42) followed by a perfusion with digestion buffer (Tyrode's solution containing 4 mg/ml liberase 1 (Roche, Mannheim, Germany), trypsin 0.6% and 0.125 mM CaCl2) was performed. After digestion, ventricular tissue was cut, dispersed, and filtered, and cells were pelleted. Cell pellet underwent a Ca2+ reintroduction (0.1, 0.2, 0.4 and 0.8 mM). Afterward, cell pellet was resolved in plating medium (MEM/HBSS medium containing 2 mM glutamine, 1% Pen/Strep, 10% FBS, 10 mM BDM and 2 mM Na-ATP), and cardiomyocytes were plated at 50,000 rod-shaped myocytes/well on laminin-coated 6-well plates. After 1 h, plating medium was removed and cardiomyocytes were treated with conditioned SN for 48 h. A detailed description of engineered heart tissue (EHT) and immunofluorescence staining can be found in the supplemental material.
2.7. Quantitative secretome analysis
C26 as well as MC38 cells were grown on two 10 cm cell culture dishes until they reached 60–70% confluency. Cells were cultured in SILAC medium (DMEM (non-GMP formulation w/o met, arg and lys), 10% FBS (dialyzed), 4 mM l-Glutamine, 100 mg/l Primocin) for 30 min (4 ml per dish). Subsequently, cells were incubated for 24 h in SILAC medium supplemented with 0.1 mM l-azidohomoalanine (AHA) and either 84 μg/ml [13C6, 15N4] l-arginine and 146 μg/ml [13C6, 15N2] l-lysine (heavy label) or 84 μg/ml [13C6] l-arginine and 146 μg/ml [4,4,5,5-D4] l-lysine (intermediate label). Experiments were performed as independent biological duplicates with reversed SILAC labels. Media were collected, centrifuged, supplemented with protease inhibitor (Roche, Mannheim, Germany) and frozen at −80 °C. Enrichment of newly synthesized proteins, sample preparation for mass spectrometry, liquid chromatography tandem mass spectrometry (LC/MS), data analysis and statistics were performed as previously described [22].
2.8. Extracellular flux analysis
Neonatal rat cardiomyocytes were seeded at 5 × 104 per well onto an XF96 PS microplate (Seahorse Bioscience, Copenhagen, DK), which was coated with 0.1% gelatin. Cardiomyocytes were exposed to conditioned SN 24 h prior to the analysis. Then medium was changed to the respective assay medium. Assays were performed according to manufacturers' instructions (Seahorse Bioscience, Copenhagen, DK). Mitochondrial Stress test: Assay compounds were used at the following final concentrations: 2 μM oligomycin, 0.3 μM FCCP, 1 μM rotenone and 1 μM antimycin A. Palmitate Uptake Assay: 10 mM sodium palmitate (Sigma, #P9767) was coupled to fatty-acid free BSA (Sigma, #A8806). Both BSA-palmitate conjugate solution and beta-oxidation inhibitor etomoxir were added at a final concentration of 1 mM. Mitochondrial function assessed by oxygen consumption rate (OCR) was normalized to cell density (absorbance) determined by Sulforhodamine B staining.
2.9. Quantitative Taqman RT-PCR
Total RNA was extracted from frozen organ samples or cardiomyocytes using Qiazol reagent and RNeasy kit (Qiagen, Hilden, Germany). cDNA was prepared by reverse transcription using M-MuLV enzyme and Oligo dt primer (Thermo Scientific, Schwerte, Germany). cDNAs were amplified using assay-on-demand kits and a StepOne Real-time PCR system (Life Technologies, Darmstadt, Germany). RNA expression data were quantified according to the delta CT method as described [23] and normalized to RNA levels of TATA-box binding protein (TBP).
2.10. Gene expression profiling
RNA isolation was performed as described before. cDNA and cRNA synthesis and hybridization to arrays of type Mouse Genome 430 2.0 from Affymetrix were performed according to the manufacturer's recommendations. A Custom CDF Version 14 with Entrez based gene definitions was used to annotate the arrays. The Raw fluorescence intensity values were normalized applying quantile normalization. Differential gene expression was analyzed based on loglinear mixed model ANOVA (Hsieh W.P., 2003; Roy J., 2007), using a commercial software package SAS® JMP7 Genomics (version 4) from SAS® (SAS® Institute, Cary, NC, USA). A false positive rate of a = 0.05 with false discovery rate (FDR) correction was taken as the level of significance. The overrepresentation analysis (ORA) is a microarray data analysis that uses predefined gene sets to identify a significant overrepresentation of genes in data sets [24]. Pathways belonging to various cell functions were obtained from public external databases (KEGG, http://www.genome.jp/kegg/). A Fisher's exact test was performed to detect the significantly regulated pathways. Gene Set Enrichment Analysis (GSEA) was used to determine whether defined lists (or sets) of genes exhibit a statistically significant bias in their distribution within a ranked gene list (http://www.broadinstitute.org/gsea/; [24]).
2.11. Enzyme-linked immunosorbent assay (ELISA)
Levels of human and murine Ataxin-10 were determined in conditioned SNs and sera using pre-coated Sandwich-ELISA (Uscn Life Science, Wuhan, CA). Assays were performed following the manufacturer's instructions. Conditioned SNs and human sera were measured undiluted and mouse serum was diluted 1:10/1:20.
2.12. Human subjects
Human serum samples and clinical data of patients with pathologically confirmed pancreatic ductal adenocarcinoma were derived from the prospective biobank and database of the European Pancreas Center (EPC), Department of General Surgery, Heidelberg University Hospital and were collected as approved by the local ethics committee.
Ataxin-10 (Atxn10) serum levels were determined in pre-surgery serum samples by using pre-coated Sandwich-ELISA (Uscn Life Science, Wuhan, CA, USA). Atxn10 serum levels were compared between control groups of patients with pancreatic ductal adenocarcinoma (PDAC) without cachexia (defined according to consensus classification from 2011 [25]), which did or did not receive neoadjuvant radio- and/or chemotherapy (referred to as neoTx) and groups of PDAC patients with cachexia, which did or did not receive neoTx. Clinico-pathological data include tumor characteristics, therapy and weight loss and are provided in Table 3.
2.13. Statistical analysis
For each experiment, means ± SEM were determined. Statistical analysis was performed with GraphPad Prism (Version 5.0, GraphPad Software Inc., CA, USA). Statistical analyses were performed using student's t-test (Welch's test), 1-way ANOVA or 2-way ANOVA where appropriate. Correlation was determined using Spearman correlation coefficient. p < 0.05 was considered statistically significant.
3. Results
3.1. Distinct colon cancer cell lines exert differential impacts on cardiac dysfunction
Cardiac dysfunction has been described as a severe complication in the cachectic state as induced by different cancer entities in both experimental animals and humans, including tumors of the colon, the lung, and the pancreas [15], [17].
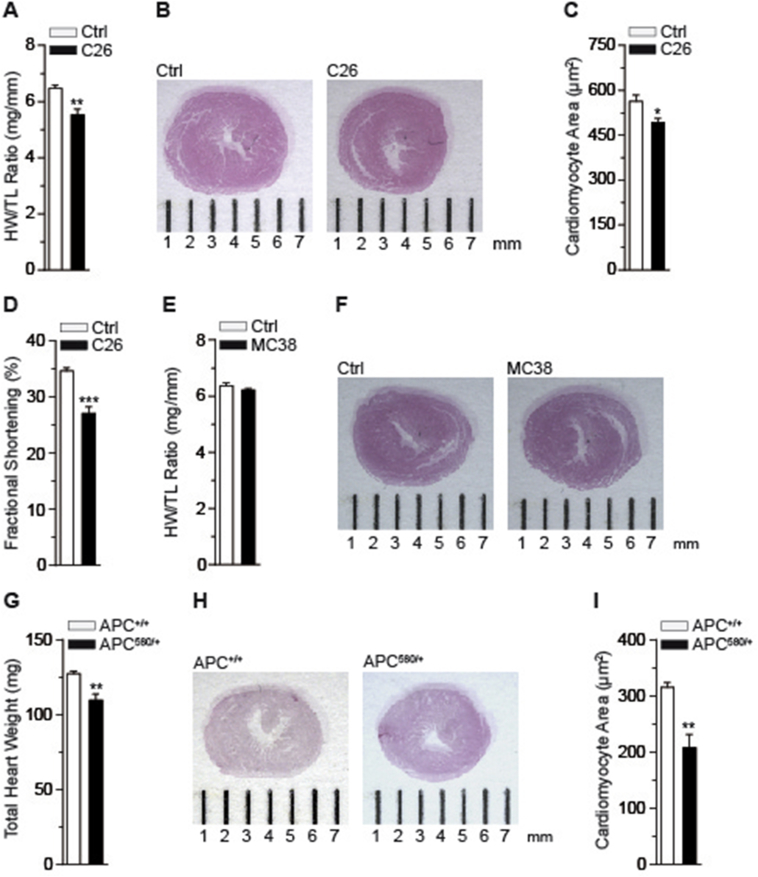
To initially delineate the cachexia-inducing properties of distinct tumor cell models in vivo, we employed comparative metabolic phenotyping in mice using the well-established cachexia-inducing colon (C) 26 and non-cachexia-inducing murine colon (MC) 38 adenocarcinoma transplantation models [26], [27]. Three weeks after cell transplantation, subcutaneous implantation of C26 cells into wild-type mice promoted a roughly 10% loss of body weight (Figure S1A), which was associated with loss of skeletal muscle and adipose tissue mass and decreased food intake as reported previously (Figure S1B–D) [14]. Importantly, C26 tumors triggered a significant loss of heart weight (Figure 1A) with a gross reduction in heart size (Figure 1B) and cardiomyocyte diameter (Figure 1C) as compared with healthy control littermates. Notably, while no cardiac fibrosis was evident in cachectic animals (Figure S1E–H), the observed atrophy was associated with impairment of cardiac function assessed by echocardiography (Figure 1D) and of other hemodynamic parameters as determined by pressure-volume (PV) loop measurements (Figure S2A–I).
Figure 1.
Cancer cachexia induces remodeling of the heart (A) Heart weight (HW)/tibia length (TL) ratio for control (Ctrl) and C26 bearing mice (C26) (n = 6 animals in Ctrl group, n = 10 animals in C26 group). (B) Representative overview images of H&E stained heart cross-sections of a control (Ctrl) and a C26 bearing mouse (C26). (C) Quantification of cardiomyocyte cross-sectional areas for control (Ctrl) and C26 bearing mice (C26) (n ≥ 100 cardiomyocytes per heart; n = 5 hearts per group). (D) Fractional shortening (FS) for control (Ctrl) and C26 bearing mice (C26) determined by using Vevo 2100 (n = 6 animals in Ctrl group, n = 10 animals in C26 group). (A)–(D) Mice were sacrificed on day 21 post PBS (Ctrl) and C26 cell injection. (E) Heart weight (HW)/tibia length (TL) ratio for control (Ctrl) and MC38 carrying mice (MC38) (n = 8 animals in Ctrl group, n = 10 animals in MC38 group). (F) Representative overview images of H&E stained heart cross-sections of a control (Ctrl) and a MC38 bearing mouse (MC38). (G) Total heart weight (HW) of wild-type (APC+/+) and APC delta 580 (APC580/+) mice (n = 6 animals in APC+/+ group, n = 8 animals in APC580/+ group). (H) Representative overview images of H&E stained heart cross-sections of a wild-type (APC+/+) and an APC delta 580 (APC580/+) mouse. (I) Quantification of cardiomyocyte cross-sectional areas of wild-type (APC+/+) and APC delta 580 (APC580/+) mice (n ≥ 100 cardiomyocytes per heart; n = 4 hearts per group). (A), (C), (D), (E), (G) and (I) Data are means ± SEM. *indicates significance using Student's t-test with Welch correction, *p < 0.05, **p < 0.01, ***p < 0.001.
In contrast, transplantation of MC38 colon cancer cells and subsequent tumor formation in wild-type animals had no effect on body weight and fat and skeletal muscle mass as compared to non-tumor-bearing littermates (Figure S3A–C). Also, MC38 tumor formation did not affect heart weight and size (Figure 1E,F; Figure S3D) nor cardiac function in these animals (Figure S3E), indicating that the MC38 adenocarcinoma intrinsically lacks pro-cachectic properties and functionality as compared with the C26 colon cancer model in vivo.
To verify our findings in the C26 model in an independent genetic model of colon cancer-induced cachexia, we employed animals carrying the adenomatous polyposis coli (APC) mutation, the most common human mutation associated with colorectal cancer [28]. APC mutant animals developed numerous intestinal lesions at 5–6 month of age [29]. Lesion appearance triggered a significant reduction in body weight, skeletal muscle and WAT mass and anemia (Figure S3F–I), thereby mimicking the observed cachectic features in the C26 transplantation model. Importantly, APC mutant cachectic animals also displayed reduced heart weight and size as well as impaired cardiac function with no signs of fibrosis (Figure 1G–I; Figure S3J–L), thus validating cardiac atrophy and dysfunction in an independent experimental setting of colon cancer.
3.2. Cardiomyocyte atrophy is induced through tumor-borne secreted factors in a cell-autonomous manner
Our results thus far suggested that cardiac atrophy represents an important read-out parameter in systemic cancer cachexia with differential responsiveness to distinct colon cancer cell types.
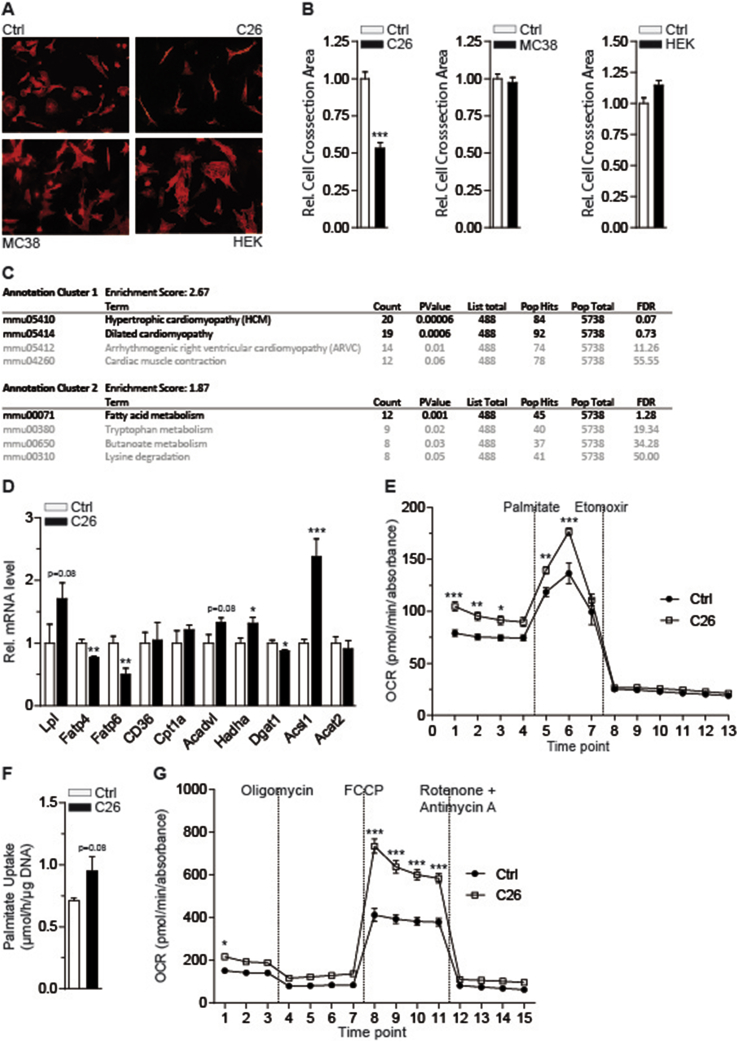
To test whether the cachexia-inducing effect of C26 colon cancer cells is mediated by tumor-borne factors in a cell-autonomous fashion and to exclude host-derived, secondary signals, we developed an in vitro cardiomyocyte phenotyping system. To this end, primary cardiomyocytes were isolated from either neonatal rats or adult wild-type mice, plated onto multi-well cell culture plates, and exposed to conditioned supernatants (SN) from colon cancer and non-tumor control cell lines. Consistent with the cardiac phenotypes in vivo (Figure 1), exposure of primary cardiomyocytes to C26 conditioned SN triggered a substantial reduction of cardiomyocyte size in both neonatal rat and adult mouse cardiomyocytes, respectively (Figure 2A,B; Figure S4A,B). In contrast, neither MC38 nor HEK293A non-tumor SN caused cardiomyocyte atrophy under these conditions (Figure 2A,B), suggesting that C26 colon cancer cells specifically release pro-cachectic mediators that can cause cardiac atrophy in a cell-autonomous manner. In support of this notion, exposure of cardiomyocytes to serum of C26-carrying cachectic mice provoked cellular atrophy in vitro, the strength of which was correlating with the progression of cachexia in the donor mice (Figure S4C).
Figure 2.
Tumor-borne secreted factors mediate cardiomyocyte atrophy and alter cardiomyocyte fatty acid metabolism. (A) Immunofluorescence staining of α-actinin in neonatal rat cardiomyocytes cultured in standard medium (Ctrl) or conditioned medium from C26, MC38 and HEK293A cells for 24 h, respectively (Magnification: 20×). (B) Relative cross-sectional areas of neonatal rat cardiomyocytes of the experiment shown in (A) (n ≥ 100 cardiomyocytes per condition). (C) Top hits (black) of KEGG pathway analysis of genes differentially regulated in primary adult mouse cardiomyocytes after treatment with conditioned medium from C26 cells for 48 h (n = 3 cell culture wells per group). (D) Relative mRNA level of lipoprotein lipase (Lpl), fatty acid transporter 4 (Fatp4), fatty acid transporter 6 (Fatp6), fatty acid translocase (FAT/CD36), carnitine palmitoyltransferase 1a (Cpt1a), acyl-CoA dehydrogenase, very long chain (Acadvl), hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (Hadha), diglyceride acyltransferase 1 (Dgat1), acyl-CoA synthetase long-chain family member 1 (Acsl1) and acetyl-CoA acetyltransferase 2 (Acat2) in neonatal rat cardiomyocytes cultured in standard medium (Ctrl) or conditioned medium from C26 cells (n = 4 cell culture wells per group). (E) Palmitate-driven oxidation of neonatal rat cardiomyocytes cultured in standard medium (Ctrl) or C26 conditioned medium for 24 h (n = 9 cell culture wells for Ctrl group, n = 15 cell culture wells for C26 group). Mitochondrial function is expressed by oxygen consumption rate (OCR; means ± SEM). 1 mM BSA-conjugated palmitate and 1 mM etomoxir were added at the indicated time points. (F) Uptake of 3H labeled palmitate by neonatal rat cardiomyocytes cultured in standard medium (Ctrl) or C26 conditioned medium (C26) for 24 h (n = 6 cell culture wells per group). (G) MitoStress testing of neonatal rat cardiomyocytes cultured in standard medium (Ctrl) or C26 conditioned medium (C26) for 24 h (n = 10 cell culture wells for Ctrl group, n = 15 cell culture wells for C26 group). Mitochondrial function is expressed by oxygen consumption rate (OCR; means ± SEM). 2 μM oligomycin, 0.3 μM FCCP and 1 μM rotenone together with 1 μM antimycin A were added at the indicated time points. (B), (D) and (F) Data are means ± SEM. *indicates significance using Student's t-test with Welch correction, *p < 0.05, **p < 0.01, ***p < 0.001. (E) and (G) Data are means ± SEM. *indicates significance using 2-way ANOVA, Bonferroni post-test, *p < 0.05, **p < 0.01, ***p < 0.001.
To further define the molecular-metabolic properties of cachectic cardiomyocytes, we performed comparative, high throughput transcriptome analysis in hearts from C26 tumor-carrying animals as compared with non-tumor bearing littermates and C26 SN-exposed primary mouse cardiomyocytes as compared with control SN. Remarkably, fatty acid (FA) metabolism appeared amongst the most significantly regulated gene pathways in atrophic cardiomyocytes (Figure 2C), and several genes involved in FA transport, oxidation and storage were found to be dysregulated in response to C26 SN in primary neonatal rat cardiomyocytes as demonstrated by altered mRNA levels (Figure 2D), thus confirming the in vivo data (Figure S4D,E).
To explore the impact of C26 SN on cardiac FA metabolism also at the functional level, we performed metabolic flux analyses in primary neonatal rat cardiomyocytes using Seahorse technology. Consistent with a reduced lipid storage capacity in both C26 SN-exposed isolated neonatal rat cardiomyocytes (Figure S5A) and hearts from C26 transplanted animals (Figure S5B), palmitate-driven FA oxidation in cardiomyocytes was significantly elevated in response to C26 SN as compared with controls (Figure 2E), correlating with a tendency to enhance cellular FA uptake under these conditions (Figure 2F). Noteworthy, mitochondrial stress testing revealed an increased maximal respiratory capacity in cardiomyocytes exposed to C26 SN (Figure 2G).
These findings supported the hypothesis that atrophic cardiomyocytes switch to an energy-wasting mode in response to pro-cachectic cancer cell-derived mediators. To delineate the functional consequences of combined atrophic and metabolic changes in response to tumor cell SN, we employed engineered heart tissue (EHT) [30] in combination with enriched tumor cell SN. In congruence with a functional impairment in contractile heart function in vivo (Figure 1D; Figure S2), exposure of EHT to cachexia-inducing C26 SN diminished the contractile properties of the EHT (Figure S5C,D), thereby further substantiating the conclusion that cachexia-inducing colon cancer cells can directly impair cardiac function through secreted factors, i.e. cachexokines.
3.3. A signature of seven secreted proteins acts in a combined manner to provoke cardiac atrophy and aberrant FA oxidation
The previous experiments indicated that cachectic cardiomyocytes are characterized by two distinct features, namely reduced cell size and aberrant FA oxidation, both triggered through tumor-borne factors intrinsic to C26 but not to MC38 colon cancer cells. Indeed, heat inactivation prevented the atrophy induction by C26 SN, and withdrawal of C26 conditioned medium recovered the atrophic phenotype in cardiomyocytes (Figure S5E,F), overall suggesting that the cellular atrophy is largely conferred through protein mediators. Given the clinical importance of potential cachexokines for preventive, diagnostic and therapeutic applications, these findings prompted us to employ unbiased differential secretome analysis of C26 and MC38 tumor cells.
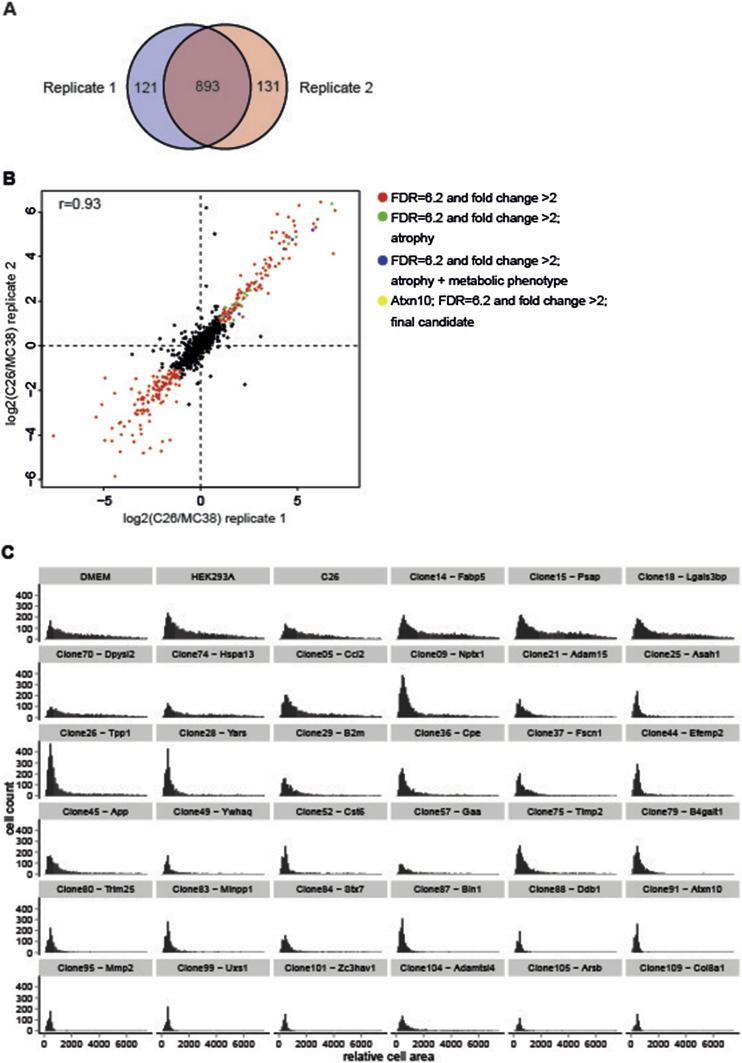
To this end, we combined click chemistry and pulsed stable isotope amino acid labeling in both C26 and MC38 cells to selectively enrich and quantify secreted proteins from both cell lines. The combination of these two labeling approaches allows the identification of secreted proteins irrespective of the complexity of the background proteins, and thus the use of optimal growth conditions (i.e. the presence of serum) [22]. Mass-spectrometry-based identification of C26 and MC38 secretomes and differential secretome mapping established a total of 893 reproducibly secreted proteins from either C26 or MC38 cells, respectively (Figure 3A). Out of these factors, 129 proteins were found to be at least 2-fold more abundant in the C26 secretome as compared with MC38, while 142 proteins were at least 2-fold more abundant in the MC38 secretome (Figure 3B; Table 1).
Figure 3.
Quantification of pro-cachectic factors and analysis of their atrophy-inducing potential. (A) and (B) Quantification of secreted proteins comparing cachexia-inducing C26 cells and non-cachexia-inducing MC38 cells. (A) Number and overlap of proteins quantified in the two biological replicates. (B) Pearson correlation between the two biological replicates is 0.93. 271 of 893 proteins quantified in both replicates show significant differences between the two cell lines (red). 28 of the 129 proteins stronger secreted by C26 cells can induce atrophy (green, blue and yellow). 7 of these additionally induce a metabolic phenotype (blue and yellow). (C) Histogram of the 384-well high throughput screening showing the areas of neonatal rat cardiomyocytes cultured in standard medium (Ctrl), under non atrophy-inducing conditions (conditioned medium from untreated HEK293A cells and HEK293A cells + cDNA clone: 14 (Fabp5), 15 (Psap), 18 (Lgals3bp), 70 (Dpysl2) or 74 (Hspa13)) and under atrophy-inducing conditions (conditioned medium from C26 cells and HEK293A cells + cDNA Clone: 5 (Ccl2), 9 (Nptx1), 21 (Adam15), 25 (Asah1), 26 (Tpp1), 28 (Yars), 29 (B2m), 36 (Cpe), 37 (Fscn1), 44 (Efemp2), 45 (App), 49 (Ywhaq), 52 (Cst6), 57 (Gaa), 75 (Timp2), 79 (B4galt1), 80 (Trim25), 83 (Minpp1), 84 (Stx7), 87 (Bin1), 88 (Ddb1), 91 (Atxn10), 95 (Mmp2), 99 (Uxs1), 101 (Zc3hav1), 104 (Adamtsl4), 105 (Arsb) or 109 (Col8a1)). (n = 6–9 cell culture wells per condition).
We hypothesized that amongst the 129 secreted proteins with relative enrichment in cachexia-inducing C26 tumors, individual or a combination of secreted proteins would be able to impose the cardiac cachectic phenotype described above (Figure 2).
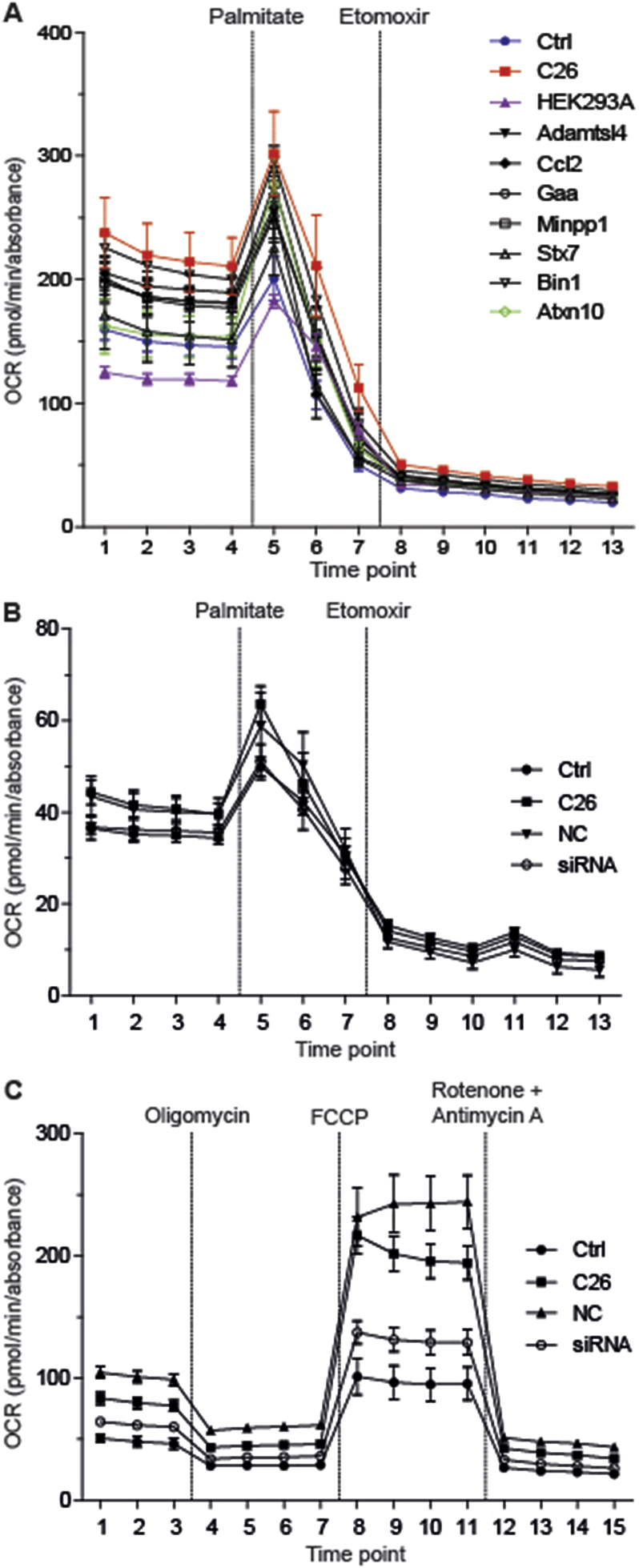
To directly test the pro-cachectic potential of identified candidates in an unbiased fashion, we obtained cDNA clones and over-expressed 109 candidates (Table 2) in non-cancerous HEK293A cells (Figure S5G). Candidate-enriched SN (Figure S5H) from these cells was collected and assayed for its atrophy-inducing potential on primary neonatal rat cardiomyocytes using a high throughput, automated 384-well screening format. In congruence with our hypothesis, SN from 28 out of 109 transfected cell clones indeed caused cardiomyocyte atrophy in repeated screening rounds (Figure 3C), and their impact on cardiomyocyte size could be validated by subsequent small-scale in vitro phenotyping as described above (Figure S5I). Next, the selected C26-derived secreted proteins were further filtered for their potential impact on cardiac FA oxidation as determined by metabolic flux analysis in primary neonatal rat cardiomyocytes. As shown in Figure 4A, enriched SN from 7 out of 28 bona fide atrophy-inducing mediators (Bridging integrator 1 (Bin1), Syntaxin 7 (Stx7), Multiple inositol-polyphosphate phosphatase 1 (Minpp1), Glucosidase alpha acid (Gaa), Chemokine ligand 2 (Ccl2), Adamts like 4 (Adamtsl4) and Ataxin-10 (Atxn10) – enhanced palmitate-driven FA oxidation, thereby mimicking the dual effect of full C26 SN in terms of cardiomyocyte size reduction and FA metabolism (Figure 4A; Figure S6A).
Figure 4.
Selective cachexokines provoke aberrant fatty acid metabolism in cardiomyocytes. (A) Palmitate-driven oxidation of neonatal rat cardiomyocytes cultured in standard medium (Ctrl), conditioned medium from C26 cells or HEK293A cells that were either untreated (HEK293A) or over-expressed one of the selective “cachexokines” (Adamts like 4 (Adamtsl4), Chemokine ligand 2 (Ccl2), Glucosidase alpha acid (Gaa), Multiple inositol-polyphosphate phosphatase 1 (Minpp1), Syntaxin 7 (Stx7), Bridging integrator 1 (Bin1) and Ataxin-10 (Atxn10)) for 24 h (n = 5 cell culture wells for Ctrl, C26, HEK293A, Gaa and Stx7 group, n = 6 cell culture wells for Adamtsl4, Ccl2, Minpp1, Bin1 and Atxn10 group). Mitochondrial function is expressed by oxygen consumption rate (OCR; means ± SEM). 1 mM BSA-conjugated palmitate and 1 mM etomoxir were added at the indicated time points. (B) Palmitate-driven oxidation of neonatal rat cardiomyocytes cultured in standard medium (Ctrl), conditioned medium from untreated C26 cells or C26 cells that were treated with either negative control (NC) siRNA or siRNAs directed against all 7 remaining candidates (siRNA) for 24 h (n = 7 cell culture wells for NC and siRNA group, n = 9 cell culture wells for Ctrl and C26 group). (C) MitoStress testing of neonatal rat cardiomyocytes cultured as described for (B) (n = 10 cell culture wells per group). Mitochondrial function is expressed by oxygen consumption rate (OCR; means ± SEM). 2 μM oligomycin, 0.3 μM FCCP and 1 μM rotenone together with 1 μM antimycin A were added at the indicated time points.
These experiments thus identified a signature panel of 7 cachexokines that were sufficient as individual factors to drive cardiomyocyte dysfunction in response to colon cancer growth independent from secondary host responses.
To next determine whether individual cachexokines were not only sufficient but also necessary to promote a cardiac cachectic phenotype, we employed siRNA-based knockdown strategies to silence the expression of the identified cachexokine signature panel in C26 colon cancer cells. Remarkably, SN from C26 cells depleted in individual cachexokines (Figure S6B) still promoted cellular atrophy of primary neonatal rat cardiomyocytes as compared with SN from control siRNA-transfected or parental C26 cells. In addition, individual cachexokine deficiency in C26 colon cancer cells did not improve the fatty acid metabolism phenotype, promoting the conclusion that individual cachexokines although sufficient to trigger a cardiac cancer cachectic phenotype may normally be required to act in a combinatorial fashion to confer a full cachectic response. Indeed, simultaneous ablation of all seven cachexokines in C26 colon cancer cells (Figure S6C) was able to prevent the SN-induced dysmetabolic phenotype in isolated cardiomyocytes as compared with the parental or control siRNA-transfected C26 SN (Figure 4B,C), underlining the notion that the seven cachexokines act in a combinatorial manner to specifically target cardiomyocyte function and metabolism during cancer cachexia.
3.4. Ataxin-10 correlates with weight loss in murine cachexia models and marks cachexia in cancer patients
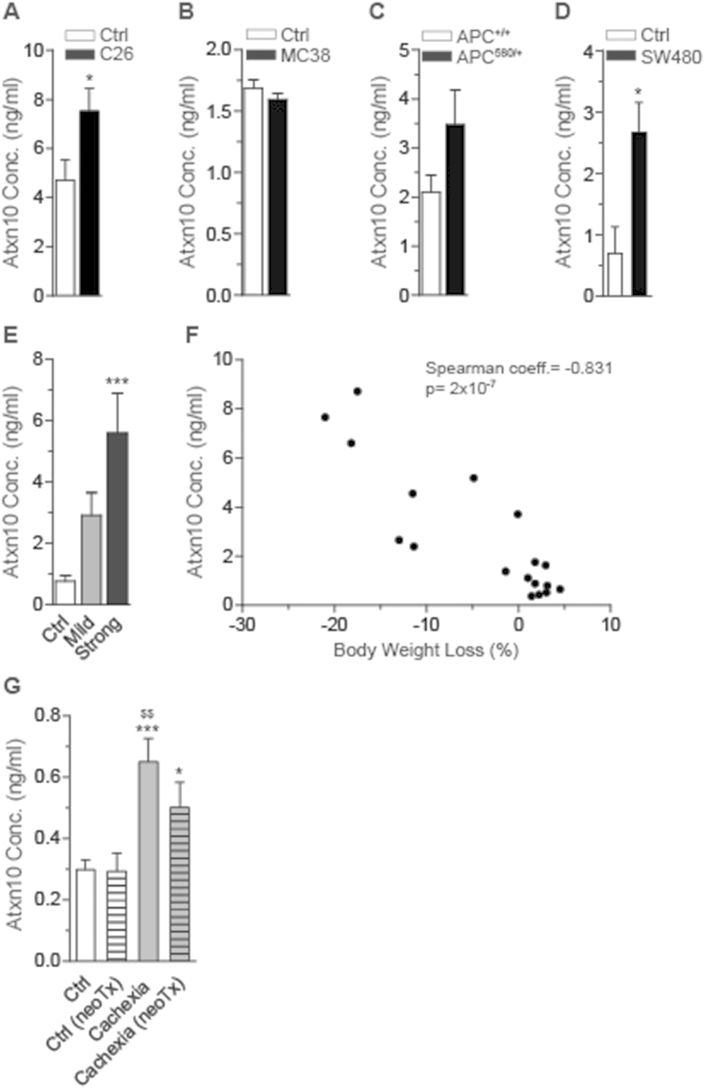
In order to provide proof-of-concept and to test for a potential significance of a prototype cachexokine to serve as diagnostic and/or prognostic marker, we screened the serum levels of Ataxin-10, which had initially been shown to display the most robust differential secretion between C26 and MC38 colon cancer cells (Figure 3B; Table 1; Figure S7A,B) in a variety of cancer cachexia mouse models. Consistent with the pro-cachectic properties of Ataxin-10-enriched SN in cellular co-culture studies, serum levels of Ataxin-10 were elevated in C26-carrying mice and APC mutant mice but not MC38 carrying mice (Figure 5A–C), correlating with the cachectic and non-cachectic phenotype, respectively. Importantly, Ataxin-10 serum levels were also induced 4-fold in an additional independent cachexia model using human SW480 colon cancer cells in a SCID mouse background (Figure 5D). Intriguingly, tumor-induced cachexia mediated by orthotopic implantation of pancreatic cancer cells into C57BL6 wild-type mice (Figure S7C–E) led to an up to 6-fold elevation of circulating Ataxin-10 levels, which tightly correlated with the degree of body wasting (Figure 5E,F), indicating that the elevation of Ataxin-10 serum levels may serve as a prognostic/diagnostic biomarker for both murine as well as human colon cancer. Interestingly, Ataxin-10 levels were also significantly elevated in two mouse models of obesity and type 2 diabetes (Figure S7F,G), suggesting that this marker might also be indicative of cardiac dysfunction under distinct metabolic conditions that resemble the cancer cachectic phenotype [31].
Figure 5.
Serum Ataxin-10 levels are elevated under cachectic conditions. (A)–(D) Ataxin-10 (Atxn10) serum levels in different mouse models: (A) C26 (n = 6 animals in Ctrl group, n = 9 animals in C26 group), (B) MC38 (n = 3 animals in Ctrl group, n = 4 animals in MC38 group), (C) APC delta 580 (n = 5 animals in APC+/+ group, n = 6 animals in APC580/+ group) and (D) SW480 (n = 3 animals in Ctrl group, n = 6 animals in SW480 group). (E) Ataxin-10 (Atxn10) serum levels in wild-type mice (Ctrl) and mice with mild (mean body weight loss: 2.5%) and strong (mean body weight loss: 17%) cachectic phenotype that underwent orthotopic implantation of pancreatic cancer cells (n = 7 animals in Ctrl group, n = 6 animals in mild cachectic group, n = 5 animals in strong cachectic group). (F) Spearman correlation of Ataxin-10 (Atxn10) serum level and body weight loss (%) in the pancreatic orthotopic implantation mouse model (Coefficient (coeff.) = −0.831), p-value: 2 × 10−7). (G) Ataxin-10 (Atxn10) serum levels in pancreatic ductal adenocarcinoma patients (PDAC) without cachexia and neoadjuvant therapy (neoTx) (Ctrl), PDAC patients without cachexia but neoTx (Ctrl (neoTx)), PDAC patients with cachexia but without neoTx (Cachexia) and PDAC patients with cachexia and neoTx (Cachexia (neoTx)) (n = 24 patients in Ctrl group, n = 8 patients in Ctrl (neoTx) group, n = 10 patients in Cachexia group, n = 10 patients in Cachexia (neoTx) group). (A)–(D) Data are means ± SEM. *indicates significance using Student's t-test with Welch correction, *p < 0.05. (E) Data are means ± SEM. *indicates significance using 1-way ANOVA, Bonferroni post-test, compared to Ctrl group, ***p < 0.001. (G) Data are means ± SEM. *indicates significance using 1-way ANOVA, Tukey's multiple comparison test, compared to Ctrl group, *p < 0.05, ***p < 0.001. $indicates significance using 1-way ANOVA, Tukey's multiple comparison test, compared to Ctrl (neoTx) group, $$p < 0.01.
To finally explore the validity of this hypothesis in humans, we determined Ataxin-10 serum levels in a cohort of cachectic and non-cachectic pancreatic cancer patients (Table 3). Of note, Ataxin-10 levels were significantly elevated in serum from cachectic patients that were untreated or received neoadjuvant radio- and/or chemotherapy (RTx, CTx) as compared to weight stable subjects that were either treated with radio- and/or chemotherapy or untreated (Figure 5G), overall demonstrating that Ataxin-10 represents a prototypical and conserved member of the cardiac-directed cachexokine signature that imposes cardiac atrophy, contractile dysfunction and aberrant lipid metabolism in cancer cachexia and may serve as a prognostic factor in human cancer cachectic patients.
4. Discussion
Cancer cachexia represents a severe clinical condition for which no effective diagnostic, preventive or therapeutic measures are available to date [6]. While it has become clear that metabolic disturbances play a major role in the development of cachexia [16], and already classical experiments suggested the presence of circulating pro-cachectic factors [7], the rational identification of tumor-borne cachexokines has been hampered by technological obstacles in the past decades [6], including limitations in reliable detection methods in body fluids and the absence of relevant phenotypic read-out systems.
Our studies now provide a first unbiased and functional screening setup for the discovery of bona fide cachexokines with both sufficient and necessary cachexogenic properties. In this context, the established cardiomyocyte phenotyping system can potentially set the framework for an in vitro diagnostic tool to predict and/or diagnose pro-cachectic mediators in patient samples in a large-scale format.
Of note, the cardiomyocyte setting can be anticipated to monitor only a subset of heart-specific “cachexokines”. Indeed, while the aim of our current study was to specifically screen for heart-specific cachexia signaling cues, the overall cachexokine profile from C26 cells may represent a valuable starting point to interrogate the functionality of additional factors in other tissue compartments.
In addition, it will be interesting to determine the interplay between our newly identified mediators and previously described pharmacological attempts to ameliorate tumor-induced cardiac cachexia [2], including the beta-blocker bisoprolol and the aldosterone antagonist spironolactone. Given the obvious complexity of cachexia-triggering signaling pathways, it will be interesting to determine potential points of convergence amongst these divergent pathways to pinpoint critical intracellular nodes in the cachectic cardiomyocyte.
Of note, impaired heart function is often associated with fibrosis, which results in stiffness [32], and a number of other studies reported an increase in cardiac fibrosis in the tumor-bearing situation [2], [15], [18]. However, in our study, neither induction of fibrosis nor a rise in appropriate markers was observed. Also, trichrome staining did not reveal an increase in collagen deposition in all animal models investigated. We speculate that the reasons for these discrepancies probably lie in the use of distinct C26 sub-clones and/or different degrees of cachexia at the time of sacrifice. In addition, recent studies have highlighted the importance of the immediate tumor environment (e.g. subcutaneous vs. intraperitoneal implantation sites) for the cachexia-inducing properties of a given tumor [33], which might also explain seemingly contradictory observations with respect to a fibrotic phenotype. However, we did observe a change in the type of collagens, i.e. downregulation of collagen III and simultaneous upregulation of collagen I. As a higher collagen I to III ratio leads to reduced tissue function due to enhanced stiffness [34], it is tempting to speculate that the imbalance of the two collagen types promotes the observed cardiac dysfunction in our setting.
With respect to heart function, our studies indicate that alterations in fatty acid and/or lipid metabolism represent distinct features of the cachectic heart. In line with previous studies [35], it is tempting to speculate that inefficient mitochondrial FA oxidation and an increased uncoupling potential in cardiomyocytes, as previously described in cachectic skeletal muscle [36], contributes to an energy deficient state and ultimately cellular atrophy as observed by us and others [15], [18]. In any case, it will be mandatory to put cachexia-associated alterations in FA oxidation into perspective with additional mechanisms contributing to cardiac dysfunction in cachexia, most notably including the induction of autophagy [15], hypo-innervation [20], and altered mTOR signaling [37]. However, while the molecular mechanisms of cachexokine-induced FA metabolic dysfunction and its relative contribution to a cachectic heart phenotype remain to be defined in future studies, it is evident that “metabolocentric” therapeutic approaches clearly have to be considered more extensively to overcome cardiac dysfunction and eventually mortality in cancer cachectic patients.
At the systemic level, most cachectic phenotypes have been ascribed to the combinatorial action of both tumor- and host-derived mediators, most notably including members of the cytokine family [38]. Our results now raise the intriguing possibility that particular tumor-borne cachexokine signatures indeed exist that are sufficient and necessary to provoke a full-blown cachectic phenotype, independent from secondary host responses. Considering that the combined but not the single knockdown of 7 potential cachexokines was sufficient to rescue the metabolic phenotype and that the combined knockdown failed to prevent the cardiac atrophy, it can be hypothesized that the overall cachectic phenotype is induced by combinatorial effects of several tumor secreted factors, influencing very distinct aspects of the overall pathology. Most clinical interventions have focused on single pro-cachectic factors known for decades, and treatments with long-term beneficial effects still do not exist. Our studies support the notion that the development of new therapeutic targets could be based on tumor-specific secretome profiles. In this respect, it is tempting to speculate about the degree of secretome overlaps between different tumor entities. Indeed, while Ataxin-10 represents a common feature of both colon and pancreatic tumor secretomes, future experimental studies will need to define the secretomes of other tumor entities to establish common or distinguishing signatures. We hypothesize that tumor-specific secretome profiling and the identification of cachectic pathways driven by specific cachexokine signatures will increase the chance to find therapeutic modalities in cancer cachectic patients, as previously exemplified by antagonism against the high affinity activin type 2 receptor ActRIIB in skeletal muscle atrophy [39], [40]. These considerations should of course take into account other systemic parameters including the impact of chemotherapy as a confounding factor in many cancer cachectic patients [41].
Given the relevance of our findings also for the human setting (Figure 5), our study may pave the way for as yet unprecedented diagnostic, preventive, and therapeutic approaches in cancer cachexia, potentially using the Ataxin-10 marker as a valid starting point. Noteworthy, as all tumor-induced pathophysiological features of the heart were also described for patients with heart failure [42], one should consider adding the assessment of cardiac function to the list of diagnostic criteria for cancer cachexia. The presented data show that reduction in cardiac function occurred stepwise and already began in the pre-cachectic state which could be used for the prognostic classification of tumor patients in the future.
Acknowledgments
We thank Yvonne Feuchter, Hermann-Josef Gröne, Maria Muciek, Vera Kosova and Tobias Schafmeier for helpful discussions and experimental support.
This work was supported by grants from the Heidelberg University Frontier Excellence Program to J.B. and S.H., the DZHK (Deutsches Zentrum für Herz-Kreislauf-Forschung – German Centre for Cardiovascular Research) and by the BMBF (German Ministry of Education and Research) to M.B., T.E., H.A.K., J.B. and S.H, the European Commission (FP7-Health-2010; MEDIA-261409) to J.B. and the Helmholtz Cross Program Topic “Metabolic Dysfunction” to S.H. The Biobank of the European Pancreas Center was supported by grants from the Heidelberger Stiftung Chirurgie and BMBF (NGFNplus-01GS08114).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2015.11.004.
Contributor Information
Johannes Backs, Email: johannes.backs@med.uni-heidelberg.de.
Stephan Herzig, Email: stephan.herzig@helmholtz-muenchen.de.
Conflict of interest
None declared.
Author contributions
M.S., E.B.-T., M.R., C.U.O., L.H.L., R.B., H.C.V., M.B., D.S., C.S., N.G., K.E., M.N.H., T.W., K.M.-D., J.K., and M.B.D. performed experiments. T.H. and O.S. supervised and coordinated patient recruitment. T.E. and H.A.K. supervised and coordinated special techniques. J.B. and S.H. designed and directed research and wrote the manuscript.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Raw data of secretome analysis of cachexia-inducing C26 cells compared to non-cachexia-inducing MC38 cells (related to Figure 3).
Annotation of all used ready-to-use cDNA overexpression plasmids. All ORF clones are based on the human gene sequence and do not include any tag. cDNA clones were obtained from Source BioScience (Nottingham, UK, USA).
Clinico-pathological data of pancreatic ductal adenocarcinoma (PDAC) patients. Ataxin-10 (Atxn10) serum levels were determined in pre-surgery serum samples of these patients by using Sandwich-ELISA (Fig. 5G). BMI: Body mass index, BW: Body weight, neoTX: neoadjuvant therapy (radio- and/or chemotherapy), pT-, pN-, pM-stage: pathologic (p) tumor staging according to Tumor-Node-Metastasis (TNM) classification, R-status: tumor staging according to residual tumor (R) classification.
References
- 1.Fearon K., Arends J., Baracos V. Understanding the mechanisms and treatment options in cancer cachexia. Nature Reviews Clinical Oncology. 2013;10:90–99. doi: 10.1038/nrclinonc.2012.209. [DOI] [PubMed] [Google Scholar]
- 2.Springer J., Tschirner A., Haghikia A., von Haehling S., Lal H., Grzesiak A. Prevention of liver cancer cachexia-induced cardiac wasting and heart failure. European Heart Journal. 2014;35:932–941. doi: 10.1093/eurheartj/eht302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewys W.D., Begg C., Lavin P.T., Band P.R., Bennett J.M., Bertino J.R. Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative Oncology Group. American Journal of Medicine. 1980;69:491–497. doi: 10.1016/s0149-2918(05)80001-3. [DOI] [PubMed] [Google Scholar]
- 4.Tisdale M.J. Cachexia in cancer patients. Nature Reviews Cancer. 2002;2:862–871. doi: 10.1038/nrc927. [DOI] [PubMed] [Google Scholar]
- 5.Martignoni M.E., Kunze P., Hildebrandt W., Kunzli B., Berberat P., Giese T. Role of mononuclear cells and inflammatory cytokines in pancreatic cancer-related cachexia. Clinical Cancer Research. 2005;11:5802–5808. doi: 10.1158/1078-0432.CCR-05-0185. [DOI] [PubMed] [Google Scholar]
- 6.Fearon K.C., Glass D.J., Guttridge D.C. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metabolism. 2012;16:153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Norton J.A., Moley J.F., Green M.V., Carson R.E., Morrison S.D. Parabiotic transfer of cancer anorexia/cachexia in male rats. Cancer Research. 1985;45:5547–5552. [PubMed] [Google Scholar]
- 8.Beutler B., Cerami A. Cachectin and tumour necrosis factor as two sides of the same biological coin. Nature. 1986;320:584–588. doi: 10.1038/320584a0. [DOI] [PubMed] [Google Scholar]
- 9.Strassmann G., Fong M., Kenney J.S., Jacob C.O. Evidence for the involvement of interleukin 6 in experimental cancer cachexia. The Journal of Clinical Investigation. 1992;89:1681–1684. doi: 10.1172/JCI115767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirai K., Hussey H.J., Barber M.D., Price S.A., Tisdale M.J. Biological evaluation of a lipid-mobilizing factor isolated from the urine of cancer patients. Cancer Research. 1998;58:2359–2365. [PubMed] [Google Scholar]
- 11.Mantovani G., Madeddu C. Cancer cachexia: medical management. Support Care Cancer. 2009;18:1–9. doi: 10.1007/s00520-009-0722-3. [DOI] [PubMed] [Google Scholar]
- 12.Wiedenmann B., Malfertheiner P., Friess H., Ritch P., Arseneau J., Mantovani G. A multicenter, phase II study of infliximab plus gemcitabine in pancreatic cancer cachexia. The Journal of Supportive Oncology. 2008;6:18–25. [PubMed] [Google Scholar]
- 13.Berriel Diaz M., Krones-Herzig A., Metzger D., Ziegler A., Vegiopoulos A., Klingenspor M. Nuclear receptor cofactor receptor interacting protein 140 controls hepatic triglyceride metabolism during wasting in mice. Hepatology. 2008;48:782–791. doi: 10.1002/hep.22383. [DOI] [PubMed] [Google Scholar]
- 14.Jones A., Friedrich K., Rohm M., Schafer M., Algire C., Kulozik P. TSC22D4 is a molecular output of hepatic wasting metabolism. EMBO Molecular Medicine. 2013;5:294–308. doi: 10.1002/emmm.201201869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosper P.F., Leinwand L.A. Cancer causes cardiac atrophy and autophagy in a sexually dimorphic manner. Cancer Research. 2011;71:1710–1720. doi: 10.1158/0008-5472.CAN-10-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tisdale M.J. Are tumoral factors responsible for host tissue wasting in cancer cachexia? Future Oncology. 2010;6:503–513. doi: 10.2217/fon.10.20. [DOI] [PubMed] [Google Scholar]
- 17.Burch G.E., Phillips J.H., Ansari A. The cachetic heart. A clinico-pathologic, electrocardiographic and roentgenographic entity. Diseases of the Chest. 1968;54:403–409. doi: 10.1378/chest.54.5.403. [DOI] [PubMed] [Google Scholar]
- 18.Tian M., Nishijima Y., Asp M.L., Stout M.B., Reiser P.J., Belury M.A. Cardiac alterations in cancer-induced cachexia in mice. International Journal of Oncology. 2010;37:347–353. doi: 10.3892/ijo_00000683. [DOI] [PubMed] [Google Scholar]
- 19.Tian M., Asp M.L., Nishijima Y., Belury M.A. Evidence for cardiac atrophic remodeling in cancer-induced cachexia in mice. International Journal of Oncology. 2011;39:1321–1326. doi: 10.3892/ijo.2011.1150. [DOI] [PubMed] [Google Scholar]
- 20.Muhlfeld C., Das S.K., Heinzel F.R., Schmidt A., Post H., Schauer S. Cancer induces cardiomyocyte remodeling and hypoinnervation in the left ventricle of the mouse heart. PloS One. 2011;6:e20424. doi: 10.1371/journal.pone.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt M., Pawlita M. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Research. 2009;37:e119. doi: 10.1093/nar/gkp581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichelbaum K., Winter M., Diaz M.B., Herzig S., Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nature Biotechnology. 2012;30:984–990. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fearon K., Strasser F., Anker S.D., Bosaeus I., Bruera E., Fainsinger R.L. Definition and classification of cancer cachexia: an international consensus. Lancet Oncology. 2011;12:489–495. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka Y., Eda H., Tanaka T., Udagawa T., Ishikawa T., Horii I. Experimental cancer cachexia induced by transplantable colon 26 adenocarcinoma in mice. Cancer Research. 1990;50:2290–2295. [PubMed] [Google Scholar]
- 27.Rosenberg S.A., Spiess P., Lafreniere R. A new approach to the adoptive immunotherapy of cancer with tumor-infiltrating lymphocytes. Science. 1986;233:1318–1321. doi: 10.1126/science.3489291. [DOI] [PubMed] [Google Scholar]
- 28.Labianca R., Merelli B. Screening and diagnosis for colorectal cancer: present and future. Tumori. 2010;96:889–901. [PubMed] [Google Scholar]
- 29.Kuraguchi M., Wang X.P., Bronson R.T., Rothenberg R., Ohene-Baah N.Y., Lund J.J. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genetics. 2006;2:e146. doi: 10.1371/journal.pgen.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eschenhagen T., Fink C., Remmers U., Scholz H., Wattchow J., Weil J. Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: a new heart muscle model system. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 1997;11:683–694. doi: 10.1096/fasebj.11.8.9240969. [DOI] [PubMed] [Google Scholar]
- 31.Lopaschuk G.D., Ussher J.R., Folmes C.D., Jaswal J.S., Stanley W.C. Myocardial fatty acid metabolism in health and disease. Physiological Reviews. 2010;90:207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- 32.Conrad C.H., Brooks W.W., Hayes J.A., Sen S., Robinson K.G., Bing O.H. Myocardial fibrosis and stiffness with hypertrophy and heart failure in the spontaneously hypertensive rat. Circulation. 1995;91:161–170. doi: 10.1161/01.cir.91.1.161. [DOI] [PubMed] [Google Scholar]
- 33.Matsuyama T., Ishikawa T., Okayama T., Oka K., Adachi S., Mizushima K. Tumor inoculation site affects the development of cancer cachexia and muscle wasting. International Journal of Cancer. Journal International du Cancer. 2015;137:2558–2565. doi: 10.1002/ijc.29620. [DOI] [PubMed] [Google Scholar]
- 34.Lapiere C.M., Nusgens B., Pierard G.E. Interaction between collagen type I and type III in conditioning bundles organization. Connective Tissue Research. 1977;5:21–29. doi: 10.3109/03008207709152608. [DOI] [PubMed] [Google Scholar]
- 35.Busquets S., Almendro V., Barreiro E., Figueras M., Argiles J.M., Lopez-Soriano F.J. Activation of UCPs gene expression in skeletal muscle can be independent on both circulating fatty acids and food intake. Involvement of ROS in a model of mouse cancer cachexia. FEBS Letters. 2005;579:717–722. doi: 10.1016/j.febslet.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 36.Tzika A.A., Fontes-Oliveira C.C., Shestov A.A., Constantinou C., Psychogios N., Righi V. Skeletal muscle mitochondrial uncoupling in a murine cancer cachexia model. International Journal of Oncology. 2013;43:886–894. doi: 10.3892/ijo.2013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manne N.D., Lima M., Enos R.T., Wehner P., Carson J.A., Blough E. Altered cardiac muscle mTOR regulation during the progression of cancer cachexia in the ApcMin/+ mouse. International Journal of Oncology. 2013;42:2134–2140. doi: 10.3892/ijo.2013.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Argiles J.M., Busquets S., Toledo M., Lopez-Soriano F.J. The role of cytokines in cancer cachexia. Current Opinion in Supportive and Palliative Care. 2009;3:263–268. doi: 10.1097/SPC.0b013e3283311d09. [DOI] [PubMed] [Google Scholar]
- 39.Zhou X., Wang J.L., Lu J., Song Y., Kwak K.S., Jiao Q. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell. 2010;142:531–543. doi: 10.1016/j.cell.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Lokireddy S., Wijesoma I.W., Bonala S., Wei M., Sze S.K., McFarlane C. Myostatin is a novel tumoral factor that induces cancer cachexia. The Biochemical Journal. 2012;446:23–36. doi: 10.1042/BJ20112024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Cramer L., Hildebrandt B., Kung T., Wichmann K., Springer J., Doehner W. Cardiovascular function and predictors of exercise capacity in patients with colorectal cancer. Journal of the American College of Cardiology. 2014;64:1310–1319. doi: 10.1016/j.jacc.2014.07.948. [DOI] [PubMed] [Google Scholar]
- 42.Evangelista L.S., Moser D.K., Westlake C., Pike N., Ter-Galstanyan A., Dracup K. Correlates of fatigue in patients with heart failure. Progress in Cardiovascular Nursing. 2008;23:12–17. doi: 10.1111/j.1751-7117.2008.07275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data of secretome analysis of cachexia-inducing C26 cells compared to non-cachexia-inducing MC38 cells (related to Figure 3).
Annotation of all used ready-to-use cDNA overexpression plasmids. All ORF clones are based on the human gene sequence and do not include any tag. cDNA clones were obtained from Source BioScience (Nottingham, UK, USA).
Clinico-pathological data of pancreatic ductal adenocarcinoma (PDAC) patients. Ataxin-10 (Atxn10) serum levels were determined in pre-surgery serum samples of these patients by using Sandwich-ELISA (Fig. 5G). BMI: Body mass index, BW: Body weight, neoTX: neoadjuvant therapy (radio- and/or chemotherapy), pT-, pN-, pM-stage: pathologic (p) tumor staging according to Tumor-Node-Metastasis (TNM) classification, R-status: tumor staging according to residual tumor (R) classification.