Abstract
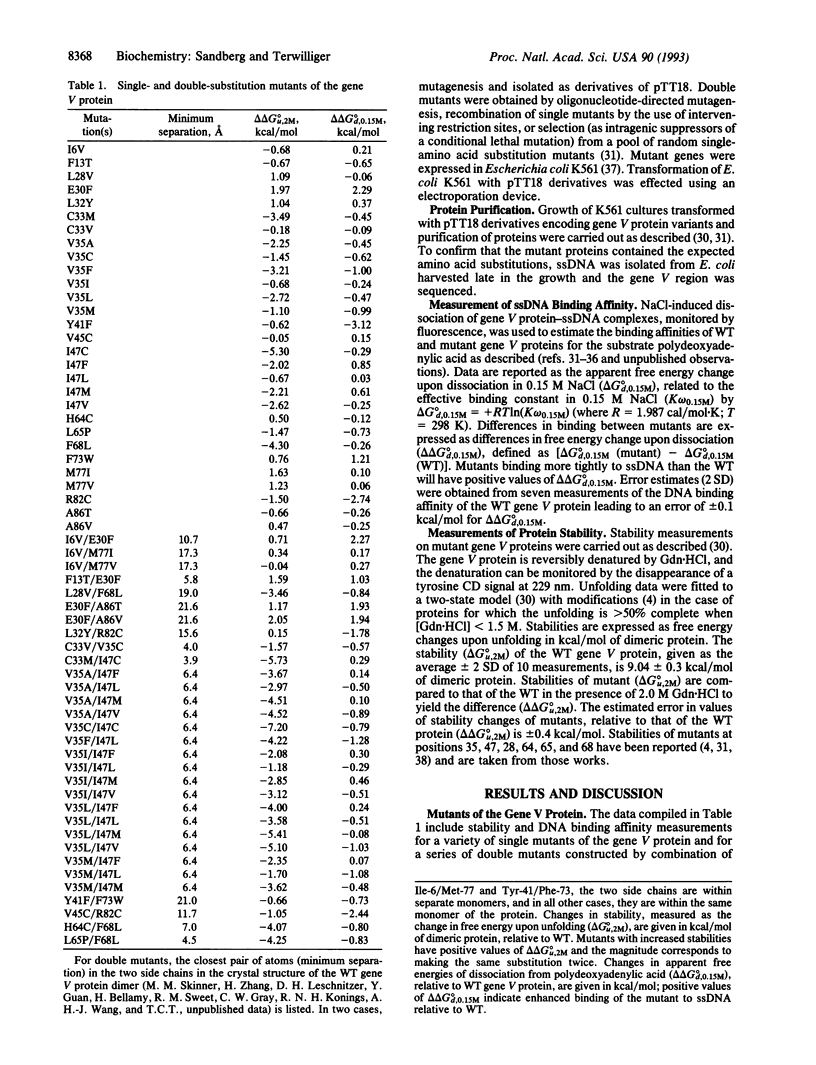
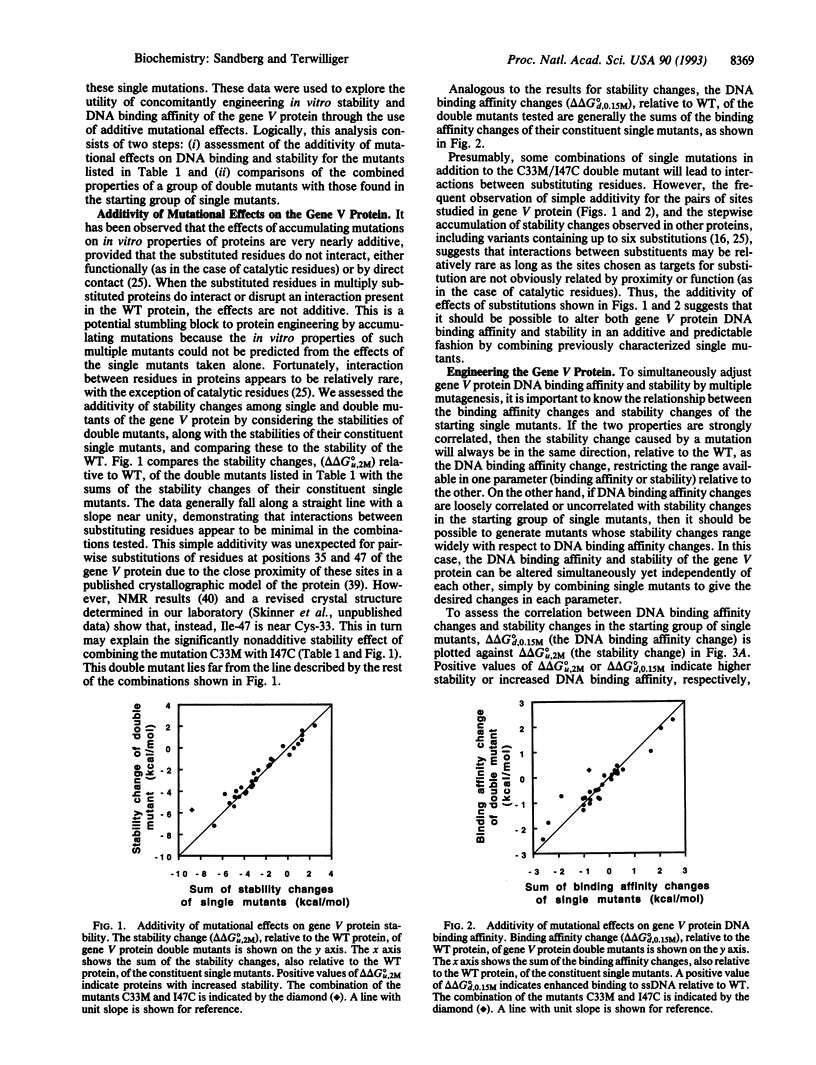
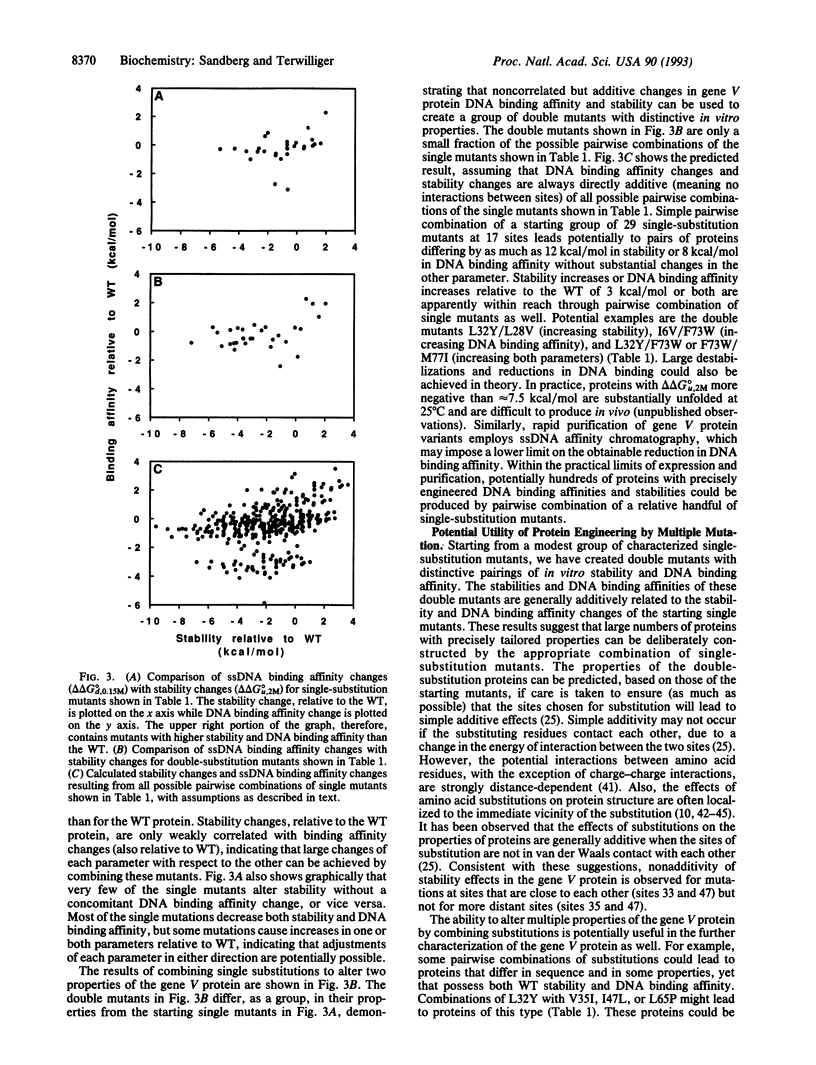
A method for simultaneously engineering multiple properties of a protein, based on the observed additivity of effects of individual mutations, is presented. We show that, for the gene V protein of bacteriophage f1, effects of double mutations on both protein stability and DNA binding affinity are approximately equal to the sums of the effects of the constituent single mutations. This additivity of effects implies that it is possible to deliberately construct mutant proteins optimized for multiple properties by combination of appropriate single mutations chosen from a characterized library.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Sun D. P., Wilson K., Wozniak J. A., Cook S. P., Matthews B. W. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. 1987 Nov 5;330(6143):41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- Alma N. C., Harmsen B. J., de Jong E. A., Ven J., Hilbers C. W. Fluorescence studies of the complex formation between the gene 5 protein of bacteriophage M13 and polynucleotides. J Mol Biol. 1983 Jan 5;163(1):47–62. doi: 10.1016/0022-2836(83)90029-3. [DOI] [PubMed] [Google Scholar]
- Bryan P. N., Rollence M. L., Pantoliano M. W., Wood J., Finzel B. C., Gilliland G. L., Howard A. J., Poulos T. L. Proteases of enhanced stability: characterization of a thermostable variant of subtilisin. Proteins. 1986 Dec;1(4):326–334. doi: 10.1002/prot.340010406. [DOI] [PubMed] [Google Scholar]
- Bulsink H., Harmsen B. J., Hilbers C. W. Specificity of the binding of bacteriophage M13 encoded gene-5 protein to DNA and RNA studied by means of fluorescence titrations. J Biomol Struct Dyn. 1985 Oct;3(2):227–247. doi: 10.1080/07391102.1985.10508413. [DOI] [PubMed] [Google Scholar]
- Carter P., Nilsson B., Burnier J. P., Burdick D., Wells J. A. Engineering subtilisin BPN' for site-specific proteolysis. Proteins. 1989;6(3):240–248. doi: 10.1002/prot.340060306. [DOI] [PubMed] [Google Scholar]
- Carter P., Wells J. A. Dissecting the catalytic triad of a serine protease. Nature. 1988 Apr 7;332(6164):564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- Carter P., Wells J. A. Engineering enzyme specificity by "substrate-assisted catalysis". Science. 1987 Jul 24;237(4813):394–399. doi: 10.1126/science.3299704. [DOI] [PubMed] [Google Scholar]
- Dao-Pin S., Baase W. A., Matthews B. W. A mutant T4 lysozyme (Val 131----Ala) designed to increase thermostability by the reduction of strain within an alpha-helix. Proteins. 1990;7(2):198–204. doi: 10.1002/prot.340070208. [DOI] [PubMed] [Google Scholar]
- Davis N. G., Boeke J. D., Model P. Fine structure of a membrane anchor domain. J Mol Biol. 1985 Jan 5;181(1):111–121. doi: 10.1016/0022-2836(85)90329-8. [DOI] [PubMed] [Google Scholar]
- Folkers P. J., van Duynhoven J. P., Jonker A. J., Harmsen B. J., Konings R. N., Hilbers C. W. Sequence-specific 1H-NMR assignment and secondary structure of the Tyr41----His mutant of the single-stranded DNA binding protein, gene V protein, encoded by the filamentous bacteriophage M13. Eur J Biochem. 1991 Dec 5;202(2):349–360. doi: 10.1111/j.1432-1033.1991.tb16382.x. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Sturtevant J. M., Sauer R. T. Stabilization of lambda repressor against thermal denaturation by site-directed Gly----Ala changes in alpha-helix 3. Proteins. 1986 Sep;1(1):43–46. doi: 10.1002/prot.340010108. [DOI] [PubMed] [Google Scholar]
- Howell E. E., Villafranca J. E., Warren M. S., Oatley S. J., Kraut J. Functional role of aspartic acid-27 in dihydrofolate reductase revealed by mutagenesis. Science. 1986 Mar 7;231(4742):1123–1128. doi: 10.1126/science.3511529. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Shibazaki M., Takagi M. A new way of enhancing the thermostability of proteases. Nature. 1986 Dec 18;324(6098):695–697. doi: 10.1038/324695a0. [DOI] [PubMed] [Google Scholar]
- Karpusas M., Baase W. A., Matsumura M., Matthews B. W. Hydrophobic packing in T4 lysozyme probed by cavity-filling mutants. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8237–8241. doi: 10.1073/pnas.86.21.8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz B. A., Kossiakoff A. The crystallographically determined structures of atypical strained disulfides engineered into subtilisin. J Biol Chem. 1986 Nov 25;261(33):15480–15485. [PubMed] [Google Scholar]
- Kellis J. T., Jr, Nyberg K., Fersht A. R. Energetics of complementary side-chain packing in a protein hydrophobic core. Biochemistry. 1989 May 30;28(11):4914–4922. doi: 10.1021/bi00437a058. [DOI] [PubMed] [Google Scholar]
- Kellis J. T., Jr, Nyberg K., Sali D., Fersht A. R. Contribution of hydrophobic interactions to protein stability. Nature. 1988 Jun 23;333(6175):784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- Liang H., Terwilliger T. C. Reversible denaturation of the gene V protein of bacteriophage f1. Biochemistry. 1991 Mar 19;30(11):2772–2782. doi: 10.1021/bi00225a006. [DOI] [PubMed] [Google Scholar]
- Liao H., McKenzie T., Hageman R. Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc Natl Acad Sci U S A. 1986 Feb;83(3):576–580. doi: 10.1073/pnas.83.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura M., Becktel W. J., Matthews B. W. Hydrophobic stabilization in T4 lysozyme determined directly by multiple substitutions of Ile 3. Nature. 1988 Aug 4;334(6181):406–410. doi: 10.1038/334406a0. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Signor G., Matthews B. W. Substantial increase of protein stability by multiple disulphide bonds. Nature. 1989 Nov 16;342(6247):291–293. doi: 10.1038/342291a0. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Yasumura S., Aiba S. Cumulative effect of intragenic amino-acid replacements on the thermostability of a protein. 1986 Sep 25-Oct 1Nature. 323(6086):356–358. doi: 10.1038/323356a0. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Sauer R. T. Lambda repressor mutations that increase the affinity and specificity of operator binding. Cell. 1985 Sep;42(2):549–558. doi: 10.1016/0092-8674(85)90112-6. [DOI] [PubMed] [Google Scholar]
- Pantoliano M. W., Whitlow M., Wood J. F., Dodd S. W., Hardman K. D., Rollence M. L., Bryan P. N. Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding. Biochemistry. 1989 Sep 5;28(18):7205–7213. doi: 10.1021/bi00444a012. [DOI] [PubMed] [Google Scholar]
- Pretorius H. T., Klein M., Day L. A. Gene V protein of fd bacteriophage. Dimer formation and the role of tyrosyl groups in DNA binding. J Biol Chem. 1975 Dec 25;250(24):9262–9269. [PubMed] [Google Scholar]
- Russell A. J., Fersht A. R. Rational modification of enzyme catalysis by engineering surface charge. Nature. 1987 Aug 6;328(6130):496–500. doi: 10.1038/328496a0. [DOI] [PubMed] [Google Scholar]
- Sandberg W. S., Terwilliger T. C. Energetics of repacking a protein interior. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1706–1710. doi: 10.1073/pnas.88.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg W. S., Terwilliger T. C. Influence of interior packing and hydrophobicity on the stability of a protein. Science. 1989 Jul 7;245(4913):54–57. doi: 10.1126/science.2787053. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Shortle D., Stites W. E., Meeker A. K. Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry. 1990 Sep 4;29(35):8033–8041. doi: 10.1021/bi00487a007. [DOI] [PubMed] [Google Scholar]
- Stearman R. S., Frankel A. D., Freire E., Liu B. S., Pabo C. O. Combining thermostable mutations increases the stability of lambda repressor. Biochemistry. 1988 Sep 20;27(19):7571–7574. doi: 10.1021/bi00419a059. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C. Construction of a synthetic variant of the bacteriophage f1 gene V by assembling oligodeoxynucleotides corresponding to only one strand of DNA. Gene. 1988 Nov 15;71(1):41–47. doi: 10.1016/0378-1119(88)90075-3. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C. Simple and highly efficient site-specific mutagenesis, by ligation of an oligodeoxyribonucleotide into gapped heteroduplex DNA in which the template strand contains deoxyuridine. Gene. 1988 Sep 30;69(2):317–324. doi: 10.1016/0378-1119(88)90441-6. [DOI] [PubMed] [Google Scholar]
- Veiko N. N., Gromova E. S., Shabarova Z. A. Vzaimodeistvie belka gena 5 faga f1 s odno- i dvukhspiral'nymi DNK i polinukleotidami. Mol Biol (Mosk) 1979 Sep-Oct;13(5):1136–1146. [PubMed] [Google Scholar]
- Wells J. A. Additivity of mutational effects in proteins. Biochemistry. 1990 Sep 18;29(37):8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- Wells J. A., Cunningham B. C., Graycar T. P., Estell D. A. Recruitment of substrate-specificity properties from one enzyme into a related one by protein engineering. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5167–5171. doi: 10.1073/pnas.84.15.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A., Powers D. B., Bott R. R., Graycar T. P., Estell D. A. Designing substrate specificity by protein engineering of electrostatic interactions. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1219–1223. doi: 10.1073/pnas.84.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel R., Perry L. J., Baase W. A., Becktel W. J. Disulfide bonds and thermal stability in T4 lysozyme. Proc Natl Acad Sci U S A. 1988 Jan;85(2):401–405. doi: 10.1073/pnas.85.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde J. A., Bolton P. H., Dell'Acqua M., Hibler D. W., Pourmotabbed T., Gerlt J. A. Identification of residues involved in a conformational change accompanying substitutions for glutamate-43 in staphylococcal nuclease. Biochemistry. 1988 May 31;27(11):4127–4132. doi: 10.1021/bi00411a033. [DOI] [PubMed] [Google Scholar]
- Zabin H. B., Terwilliger T. C. Isolation and in vitro characterization of temperature-sensitive mutants of the bacteriophage f1 gene V protein. J Mol Biol. 1991 May 20;219(2):257–275. doi: 10.1016/0022-2836(91)90566-o. [DOI] [PubMed] [Google Scholar]
- Zhang X. J., Baase W. A., Matthews B. W. Toward a simplification of the protein folding problem: a stabilizing polyalanine alpha-helix engineered in T4 lysozyme. Biochemistry. 1991 Feb 26;30(8):2012–2017. doi: 10.1021/bi00222a001. [DOI] [PubMed] [Google Scholar]
- de Jong E. A., Harmsen B. J., Konings R. N., Hilbers C. W. Binding of IKe gene 5 protein to polynucleotides. Fluorescence binding experiments of IKe gene 5 protein and mutual cooperativity of IKe and M13 gene 5 proteins. Biochemistry. 1987 Apr 7;26(7):2039–2046. doi: 10.1021/bi00381a037. [DOI] [PubMed] [Google Scholar]



