Abstract
Background:
Pseudomonas aeruginosa might be converted to coccoid bacteria under antibiotic stress. Bacterial conversion would increase resistance to antibiotics due to changes in cell wall crosslink or decreased metabolic activity. Morphology of P. aeruginosa under stress conditions (presence of antibiotics) can be changed to elongated bacilli, U shape and finally coccoid bacteria. Results of several researches showed that coccoid bacteria are one of the most important aspects of drug resistance. It would be the major reason for treatment failure.
Objectives:
The aim of this study was to determine in vitro morphological and bactericidal effects of amikacin, meropenem and imipenem on P. aeruginosa isolated from clinical specimens.
Materials and Methods:
Eight P. aeruginosa isolates obtained from clinical samples of burned patients and standard strain ATCC 27853 were used in this study. Isolates were identified by biochemical tests and confirmed by PCR method using ITS specific primer. Minimum inhibitory concentrations (MICs) of three antibiotics were determined by E-test method. Bacteria were exposed to antibiotics at different concentrations. Bacterial morphology in different days was examined by specific microscope and viability of isolates was examined by flow cytometry
Results:
All used antibiotics at sub MIC concentration had capability to induce coccoid bacteria. The highest rate of induced coccoid bacteria was 98.2% after 8 days, with contribution of imipenem and meropenem at 2 μg/mL concentration. Amikacin at 4 μg/mL concentration induced lower rate of coccoid bacteria (55.05%). Amikacin had a strong bactericidal effect on coccoid bacteria at 8 μg/mL concentration. Imipenem and meropenem showed very weak bactericidal effect on coccoid bacteria.
Conclusions:
Induction of coccoid form of P. aeruginosa may be one of the important reasons for antibiotic treatment failure; therefore, prescribed dose of antibiotics should be carefully managed to prevent increasing antibiotic resistance and coccoid bacteria induction.
Keywords: Coccoid Bacteria, Drug Resistance, Pseudomonas aeruginosa
1. Background
Pseudomonas aeruginosa, due to production of different virulence factors, is one of the most important causes of many disorders such as bacteremia, burn infection and urinary tract infections (1). Pseudomonas aeruginosa because of limited nutrition requirements and naturally resistance against different antimicrobial agents is widely distributed in the environment. Multidrug-resistant P. aeruginosa (MDR P. aeruginosa) is responsible for life threatening infections, especially in burned patients. Under stress conditions, such as lack of food, pH and temperature changes, oxygen depletion, increased osmotic pressure and antibiotic stress, morphology of P. aeruginosa can be changed to U shape and finally coccoid bacteria (2, 3).
During conversion to coccoid bacteria, cell size is reduced because of shrinkage of cytoplasmic membrane, decreased periplasmic space and condensed DNA (4, 5). Results of several studies showed that presence of bacteria in minimum inhibitory concentration (MIC) and sub MIC of antibiotics affected adherence, hydrophobic cell surface and mobility (6, 7). Bacterial conversion to coccoid bacteria may increase resistance to antibiotics, hydrogen peroxide, osmotic pressure and thermal shock. This resistance can be due to changes in cell wall crosslink or decreased metabolic activity (2, 8-10).
Pseudomonas aeruginosa is a major threat in burn patients, because this bacterium can convert to the coccoid bacteria under stress conditions. When environmental conditions change to favorable, it would return to the bacilli form (11-13). Various genes are involved in conversion of bacilli to coccoid bacteria; one of the major genes are rpos used by P. aeruginosa to survive in these conditions by encoding different sigma factors. Sigma factor encoded by rpoS gene, is a positive regulator of many genes including those involved in the synthesis of exotoxin and antibiotic tolerance. Studies showed that cells with mutations in the rpoS gene have less ability to survive than those without mutation in rpoS (14, 15). Due to increased mortality rate in burned patients infected with P. aeruginosa, we decided to study in vitro morphological effect of routine antibiotics used for patients' treatment in our hospital (amikacin, meropenem and imipenem) on P. aeruginosa.
2. Objectives
The aim of this study was to evaluate in vitro morphological and bactericidal effects of amikacin, meropenem and imipenem on P. aeruginosa isolated from clinical specimen.
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
In our study we used eight P. aeruginosa isolates obtained from clinical samples of burned patients and standard strain ATCC 27853. Primary identification of isolates was performed using conventional biochemical tests as described previously. Then, presumptive identification was confirmed by ITS (16S-23S rDNA internally transcribed spacer)-PCR.
3.2. Antibiotics
The selected antibiotics were amikacin, meropenem and imipenem (MAST, UK) according to highest consumption in burn unit of the hospital.
3.3. Determination of Minimal Inhibitory Concentration (MIC)
MIC for amikacin, meropenem and imipenem were determined by E-test strip (Liofilchem, Italy). McFarland 0.5 standard turbidity samples were prepared and spread on Muller Hinton agar. One E-test strip was placed onto the surface of each cultured plate. Results after an overnight incubation at 37°C were observed and determined according to manufacture instruction. Results are presented in Table 1.
Table 1. MIC of Isolates.
| Antibiotica | Standard Strain | Clinical Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| Imipenem | 1 | 4 | 2 | 1.5 | 2 | 2 | 1.5 | 2 | 2 |
| Meropenem | 1 | 4 | 3 | 2 | 1.5 | 2 | 2 | 2 | 2 |
| Amikacin | 2.5 | 6 | 4 | 4 | 6 | 4 | 4 | 4 | 6 |
aµg⁄mL.
3.4. Induction to Coccoid Form of Bacteria
The bacilli form of fresh colonies of P. aeruginosa was confirmed by Gram staining. Then inoculated into BHI (Brain Heart Infusion) broth (Merck, Germany) and incubated in antibiotic stress overnight at 37°C. A tube without antibiotics was used as a control. We examined samples each 24 hours for 10 day, by Gram staining and observation by camera light microscopy (Digital DP 72-BX 51, Olympus, Japan) at a magnification of x1000. We screened 100 bacteria/field for their morphology and size of cells. When more than 90% of bacterial coccoid were seen, viability of them was evaluated.
3.5. Membrane Integrity and Cell Viability Assay
Bacterial suspensions in BHI broth containing 90% - 100% coccoid bacteria were centrifuged for 10 minutes at 1000 g then washed three times with PBS (phosphate-buffered saline pH8) ( Sigma, USA). Three microliter of propidium iodide (PI) fluorescent stain (Sigma, USA) was added to precipitate and incubated for 30 minutes in the dark. Flow cytometry was performed by FAC Scan Calibur (BD, Becton Dickinson, USA). PI is a fluorescent dye that cannot cross the cell membrane consequently; it is only able to stain dead cells. Forward and side scattered of cells (both in a logarithmic setting) were measured. The red fluorescence of 10/000 bacteria was analyzed with FL2 plotted. The histogram of gated bacteria was detected as the percent of dead and alive cells. A suspension of fresh bacteria and killed bacteria with hypochlorite (5%) were used as control cells.
3.6. Antibiotic Susceptibility Test for Coccoid Bacteria
Amikacin, meropenem and imipenem antibiotics were added to the broth culture containing 100% viable coccoid bacteria at concentrations of 1, 2, 4 and 8 µg/mL for amikacin and 0.25, 0.75, 1.5, 2 and 4 µg/mL for meropenem and imipenem. After 24 hours sensitivity of viable coccoid bacteria to antibiotics was assessed by flow cytometry.
3.7. Statistical Analysis
Analysis of data was performed using SPSS version 20 by one way ANOVA and Post Hoc (LSD) tests. P < 0.05 considered as statistically significant.
4. Results
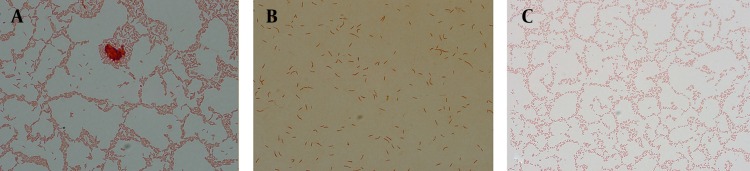
At initiation of experiment (24 hours), 100% of bacteria were bacilli (Figure 1 A). During the time-tested forms of bacilli, U shape and elongated bacilli (mixed morphology) were seen (Figure 1 B). After eight days of incubation, populations of cells were pure coccoid bacteria (100%, Figure 1 C).
Figure 1. Microscopic Images of Gram Stained P. aeruginosa.
Microscopic images under camera light microscopy (Digital DP 72-BX 51, Olympus, Japan) showing pure bacilli form at 24 hours incubation (1A), mixed forms, bacilli, U-shapes and elongated bacilli after exposure to antibiotics (1B) and pure induced coccoid bacteria after 8 days (1C).
A significant result was that all three antibiotics induced coccoid bacteria in MIC and sub MIC. The highest rate of induced coccoid bacteria was 98.2% after 8 days, which contributed to imipenem and meropenem at 2 μg/mL concentration. The rate of induced coccoid bacteria by amikacin was 55.05% at 4 µg⁄mL after eight days (Table 2).
Table 2. Concentration of Antibiotics That Induced and Eradicated Coccoid Bacteria Form in Clinical Isolates and Standard Strain.
| Antibiotica | Clinical Samples | Standard Strain | ||
|---|---|---|---|---|
| Concentration of antibiotics that induced coccoid bacteria | Concentration to antibiotics eradicate coccoid bacteria | Concentration of antibiotics that induced coccoid bacteria | Concentration to antibiotics eradicate coccoid bacteria | |
| Imipenem | 2 | 4 | 0.75 | 1.5 |
| Meropenem | 2 | 4 | 0.75 | 1.5 |
| Amikacin | 4 | 8 | 2 | 4 |
aµg⁄mL.
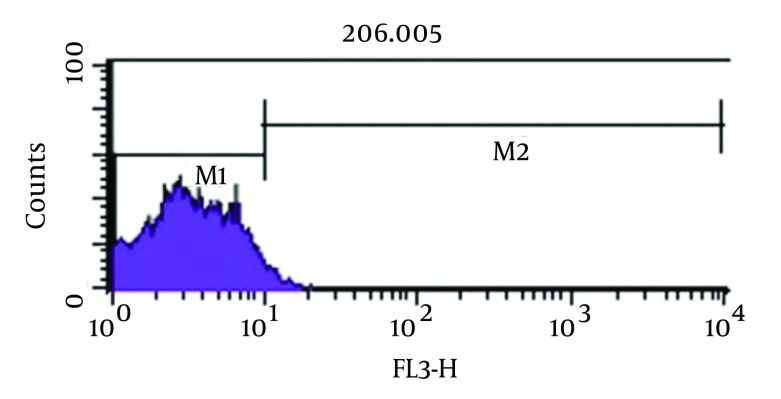
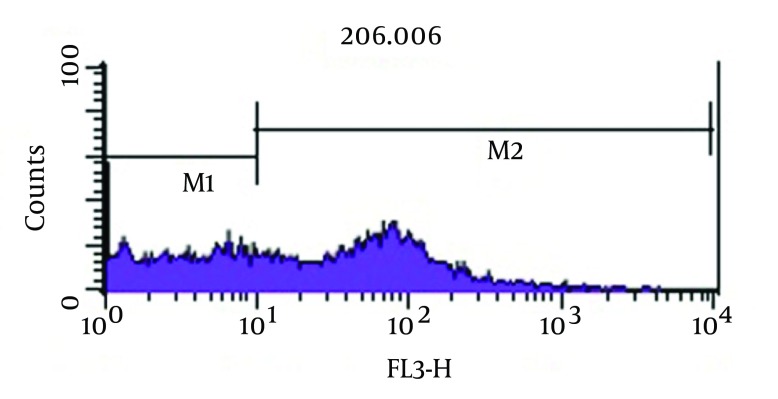
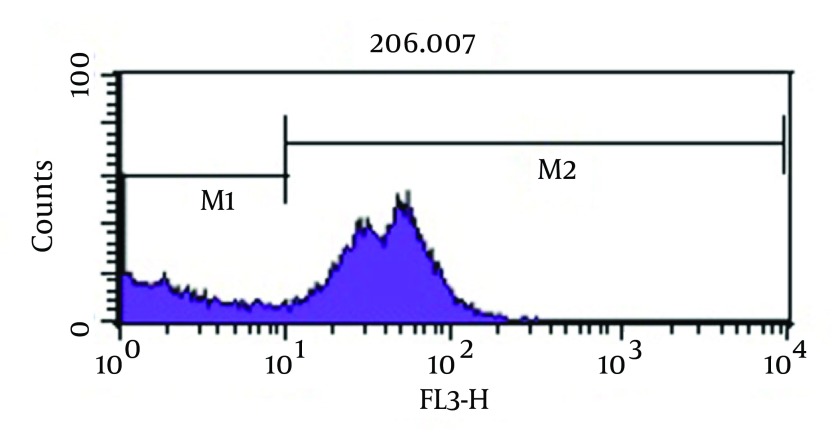
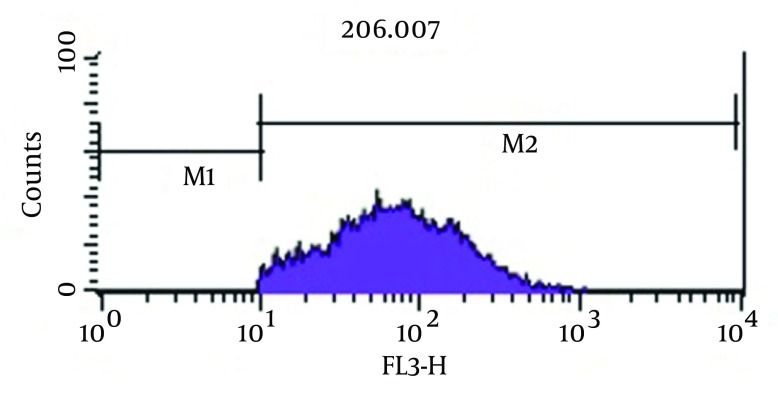
As shown in Figure 3, analysis of viability test by flow cytometry, tubes with imipenem at concentration of 2 μg/mL had significantly the greatest population (98.2%) of viable coccoid bacteria compared with amikacin (P = 0.01). Figure 4 shows that amikacin significantly induced lower rate of viable coccoid bacteria (55.05%).
Figure 3. Flow Cytometry Analysis.
Percentage of Induced Coccoid Bacteria by Imipenem. M1, Percentage of Alive Bacteria; M2, the Percentage of Dead Cells.
Figure 4. Flow Cytometry Analysis.
Percentage of Induced Coccoid Bacteria by Amikacin.
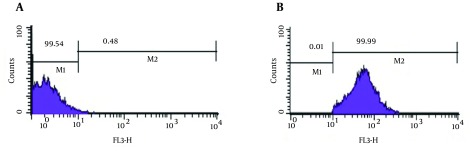
Figure 2. Flow Cytometry Analysis.
Positive Control (Live P. aeruginosa) (2A) and Dead Hypochlorite-Treated P. aeruginosa as Negative Control (2B).
Figure 5 shows bactericidal effect of imipenem at 2 × MIC (4 µg/mL) on viable coccoid bacteria by flow cytometry, in which after 2 days, 59.16% of cells were killed. Results showed (Figure 6) that amikacin at MIC and 2 × MIC (8 µg/mL) concentration had the highest bactericidal effect on viable coccoid bacteria of P. aeruginosa (> 90%). Treatment with MIC and 2 × MIC of meropenem was similar to imipenem result.
Figure 5. Flow Cytometry Analysis.
Bactericidal Effect of Imipenem on Viable Coccoid Bacteria.
Figure 6. Flow Cytometry Analysis.
Bactericidal Effect of Amikacin on Viable Coccoid Bacteria.
5. Discussion
Pseudomonas aeruginosa might be converted to coccoid bacteria under antibiotic stress. Bacterial conversion would increase resistance to antibiotics due to changes in cell wall crosslink or decreased metabolic activity (2, 8-10). It would be the major reason for treatment failure. The purpose of this study was to increase the knowledge about the effect of antibiotics on morphological changes of P. aeruginosa and also the effect of antibiotics on viability of P. aeruginosa coccoid form.
One of the main observed behaviors in environmental stress of Gram negative bacteria is cell size reduction and change in morphology from bacilli to coccoid bacteria (2). Rearrangement of peptidoglycan in cell wall of coccoid bacteria occurs and leads to size reduction (9). In the present study, eight P. aeruginosa isolates obtained from clinical samples of burned patients and standard strain ATCC 27853 were investigated. Isolates were identified by biochemical tests and confirmed by PCR method using ITS specific primer. Bacteria were exposed to antibiotics (amikacin, meropenem and imipenem) at different concentrations and MICs of three antibiotics were determined by E-test method (Table 1). Bacterial morphology in different days was examined by specific microscope and their viability assessed by flow cytometry. We used flow cytometry for bacterial viability assay because it is a fast, easy and reliable method.
Some beta-lactam antibiotics, such as imipenem and meropenem have affinity for penicillin-binding proteins (PBPS) and induce changes in cell morphology, while amikacin is protein synthesis inhibitor (6). The stresses used in this study were antibiotics including amikacin, meropenem and imipenem. These antibiotics were chosen because of routine use for eradication of P. aeruginosa infection in hospital burn unit. Our results showed that all of three antibiotics could induce bacterial coccoid bacteria. As shown in Figure 3, the highest rate of morphological conversion was related to imipenem at concentration of 2 μg/mL (MIC) after 8 days. In this study, we tested susceptibility of viable coccoid bacteria to three antibiotics and showed that amikacin had the highest bactericidal effect on these types of cells; MIC of this antibiotic for coccoid bacteria was 8 µg/mL (Figure 6).
It seems that protein synthesis inhibitor antibiotics such as amikacin have appropriate bactericidal effect on coccoid bacteria. Cefali et al. demonstrated that P. aeruginosa under stress condition begin to change morphology from 9th day, while in our study changes occurred on the 5th day. This difference could be due to differences in characteristics of studied strains, the type of stress and the type of culture medium (2). Faghri et al. performed a similar study on Helicobacter pylori and showed that bacteria changed to coccoid bacteria after three days in exposure to sub MIC of antibiotics. Faghri et al. studied Campylobacter jejuni food stress and found that morphologic changes observed on day 14 (16). This difference could be due to the type of bacteria, differences in rates of bacterial tolerance to stress, their inherent resistance to antibiotics and different type of antibiotics. Besnard et al. studied C. jejuni food stress and showed that morphologic changes were observed on day 14 (17).
The results of this study revealed that prescribed concentration of antibiotics should be carefully ordered to prevent induction of coccoid bacteria that may increase antibiotic resistance. Our results also showed that eradication of coccoid bacteria derived from clinical isolates is possible by higher concentrations of antibiotics (3). There are various methods for confirming viability and integrity of coccoids. We used quick, unpretentious and reliable flow cytometry method for bacterial viability assay. Our study revealed that different antibiotics near to MIC concentration in exposure to P. aeruginosa can induce morphologic transformation from bacilli to coccoid bacteria. It is one of the main reasons for failure in antibiotics treatment. Based on the present study results, susceptibility testing alone may not be sufficient to determine clinical effect of antibiotics on P. aeruginosa infection. Morphological conversion should be investigated by flow cytometry as a fast, easy and reliable method.
Acknowledgments
The authors would like to express their thanks to Imam Musa Kazem hospital laboratory personnel.
Footnotes
Authors’ Contribution:All authors contributed in the research equally.
Financial Disclosure:The vice chancellor of research of Isfahan university of medical sciences financially supported this study.
Funding/Support:This work was financially supported by Isfahan university of medical sciences.
References
- 1.Alikhani MY, Karimi Tabar Z, Mihani F, Kalantar E, Karami P, Sadeghi M, et al. Antimicrobial Resistance Patterns and Prevalence of blaPER-1 and blaVEB-1 Genes Among ESBL-producing Pseudomonas aeruginosa Isolates in West of Iran. Jundishapur J Microbiol. 2014;7(1):e25250. doi: 10.5812/jjm.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cefali E, Patane S, Arena A, Saitta G, Guglielmino S, Cappello S, et al. Morphologic variations in bacteria under stress conditions: near-field optical studies. Scanning. 2002;24(6):274–83. doi: 10.1002/sca.4950240601. [DOI] [PubMed] [Google Scholar]
- 3.Oliver JD. The viable but nonculturable state in bacteria. J Microbiol. 2005;43(Special issue):93–100. [PubMed] [Google Scholar]
- 4.Sachidanandham R, Gin KY, Poh CL. Monitoring of active but non-culturable bacterial cells by flow cytometry. Biotechnol Bioeng. 2005;89(1):24–31. doi: 10.1002/bit.20304. [DOI] [PubMed] [Google Scholar]
- 5.Elabed H, Bakhrouf A, Hamza R, Azaiez M, Gaddour K. Evidence of the adaptive response in Pseudomonas aeruginosa to 14 years of incubation in seawater. Ann Microbiol. 2011;62(4):1385–94. doi: 10.1007/s13213-011-0387-2. [DOI] [Google Scholar]
- 6.Fonseca AP, Sousa JC. Effect of antibiotic-induced morphological changes on surface properties, motility and adhesion of nosocomial Pseudomonas aeruginosa strains under different physiological states. J Appl Microbiol. 2007;103(5):1828–37. doi: 10.1111/j.1365-2672.2007.03422.x. [DOI] [PubMed] [Google Scholar]
- 7.Rowan NJ. Viable but non-culturable forms of food and waterborne bacteria: Quo Vadis? Trends Food Sci Technol. 2004;15(9):462–7. doi: 10.1016/j.tifs.2004.02.009. [DOI] [Google Scholar]
- 8.McDougald D, Rice SA, Weichart D, Kjelleberg S. Nonculturability: adaptation or debilitation? FEMS Microbiol Ecol. 1998;25(1):1–9. doi: 10.1111/j.1574-6941.1998.tb00455.x. [DOI] [Google Scholar]
- 9.Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 2010;34(4):415–25. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 10.Kana BD, Mizrahi V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol Med Microbiol. 2010;58(1):39–50. doi: 10.1111/j.1574-695X.2009.00606.x. [DOI] [PubMed] [Google Scholar]
- 11.Nystrom T. Nonculturable bacteria: programmed survival forms or cells at death's door? Bioessays. 2003;25(3):204–11. doi: 10.1002/bies.10233. [DOI] [PubMed] [Google Scholar]
- 12.Lu X, Liu Q, Wu D, Al-Qadiri HM, Al-Alami NI, Kang DH, et al. Using of infrared spectroscopy to study the survival and injury of Escherichia coli O157:H7, Campylobacter jejuni and Pseudomonas aeruginosa under cold stress in low nutrient media. Food Microbiol. 2011;28(3):537–46. doi: 10.1016/j.fm.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Lleo MM, Pierobon S, Tafi MC, Signoretto C, Canepari P. mRNA detection by reverse transcription-PCR for monitoring viability over time in an Enterococcus faecalis viable but nonculturable population maintained in a laboratory microcosm. Appl Environ Microbiol. 2000;66(10):4564–7. doi: 10.1128/aem.66.10.4564-4567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Japoni A, Anvarinejad M, Farshad S, Giammanco GM, Ranjbar R, Rafaatpour N, et al. Antibiotic Susceptibility Patterns and Molecular Epidemiology of Metallo-β-Lactamase Producing Pseudomonas Aeruginosa Strains Isolated From Burn Patients. Iran Red Crescent Med J. 2014;16(5):e25250. doi: 10.5812/ircmj.10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorgensen F, Bally M, Chapon-Herve V, Michel G, Lazdunski A, Williams P, et al. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology. 1999;145 ( Pt 4):835–44. doi: 10.1099/13500872-145-4-835. [DOI] [PubMed] [Google Scholar]
- 16.Faghri J, Poursina F, Moghim S, Zarkesh Esfahani H, Nasr Esfahani B, Fazeli H, et al. Morphological and Bactericidal Effects of Different Antibiotics on Helicobacter pylori. Jundishapur J Microbiol. 2014;7(1):e25250. doi: 10.5812/jjm.8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Besnard V, Federighi M, Cappelier JM. Evidence of Viable But Non-Culturable state in Listeria monocytogenes by direct viable count and CTC-DAPI double staining. Food Microbiol. 2000;17(6):697–704. doi: 10.1006/fmic.2000.0366. [DOI] [Google Scholar]