Abstract
Bacterial symbionts of fungus-growing ants occupy a highly specialized ecological niche and face the constant existential threat of displacement by another strain of ant-adapted bacteria. As part of a systematic study of the small molecules underlying this fraternal competition, we discovered an analog of the antitumor agent rebeccamycin, a member of the increasingly important indolocarbazole family. While several gene clusters consistent with this molecule's newly reported modification had previously been identified in metagenomic studies, the metabolite itself has been cryptic. The biosynthetic gene cluster for 9-methoxyrebeccamycin is encoded on a plasmid in a manner reminiscent of plasmid-derived peptide antimicrobials that commonly mediate antagonism among closely related Gram-negative bacteria.
Graphical abstract
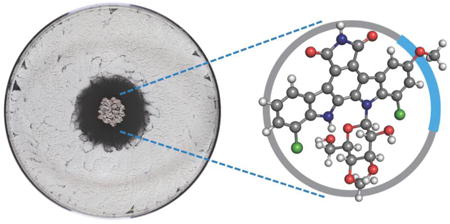
Many, arguably most, of our clinically used antibacterial agents are produced by other bacteria, and this observation has led to the widely held if poorly sourced view that these molecules evolved to mediate bacterial competition in natural environments.1,2 The bacterial symbionts of fungus-growing ants – Actinobacteria in the genus Pseudonocardia – provide a model system in which to define the ecological roles of these bacterially produced small molecules. Fungus-growing ants collect plant material, which they feed to the fungal crops in their gardens, and their bacterial symbionts provide chemical defenses against pathogenic fungi and other microbial competitors.3 The bacterial symbionts are fed by the ants, and their greatest existential threat is displacement by another Pseudonocardia strain. An earlier phenotypic analysis confirmed that Pseudonocardia produce diffusible small molecules, antibiotics, that suppress other Pseudonocardia strains, and in this report we begin to establish the molecular and genetic basis for this competition between these closely related bacteria.4
We began with a small set of ant-associated Pseudonocardia and used an intruder assay to assess competition between them. In this assay, one strain, the ‘resident,’ is placed in the center of a Petri dish and allowed to establish a colony before a second strain, the ‘intruder,’ is introduced around the dish's periphery. The zone of inhibition, if present, represents the ability of the resident to suppress the intruder.4 A few of these assays are shown in Figure 1.
Figure 1.
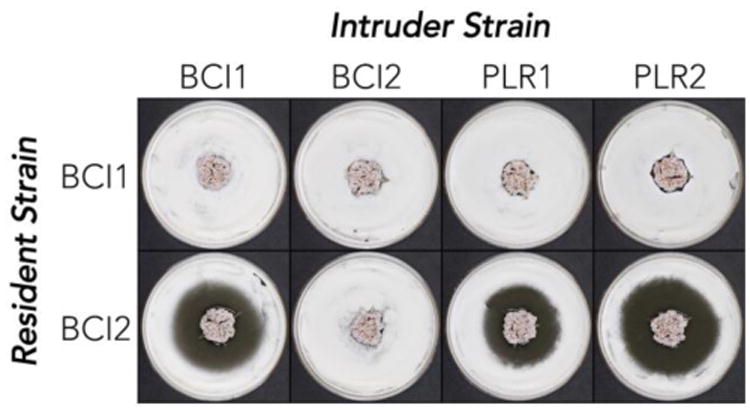
Intruder assay for ant-associated Pseudonocardia strains. The ability of the resident strain (center) to suppress growth of the intruder (periphery) is judged by the size of the zone of inhibition.
We focused on two strains, BCI1 and BCI2, which are both Pseudonocardia sp. collected from Apterostigma dentigerum ant colonies on Barro Colorado Island, Panama. The two ant colonies, which were located near each other on a small island, represent the same ant species and grow identical fungal crops in their gardens. By usual taxonomic standards the BCI1 and BCI2 bacterial isolates are also identical: they share 100% 16S ribosomal DNA sequence identity, and their chromosomal DNA shares ∼98.3% sequence identity across the entire nucleotide sequence (see below). However, as illustrated in Figure 1, their performance in the intruder assay differs dramatically. BCI1 had no measurable defensive activity against any intruder strains while BCI2 produced sizeable inhibitory zones (≥1.4 cm) against all intruder strains except itself (Figure 1).
To investigate the molecular basis for the puzzling dichotomy between the taxonomic and genetic identity and the starkly different assay performance, we tested the organic soluble extracts from bacterial cultures. The BCI1 extract had no activity, while the BCI2 extract potently inhibited a panel of six ant-associated Pseudonocardia (Supplemental Figure S2). We used activity against Pseudonocardia strain PLR1 for activity-guided fractionation, and C18 flash chromatography followed by reverse-phase HPLC led to a single molecule with potent inhibitory activity. High resolution ESI-MS indicated a highly unsaturated, dichlorinated structure with a molecular formula of C28H23Cl2N3O8 ([M+H]+ calc 600.0935, expt 600.0920), which is absent from natural product databases. We noted a distinctive UV-visible spectrum (λmax 207, 244, 296, 317, 401 nm) similar to the dichlorinated indolocarbazole natural product rebeccamycin (1). 1D and 2D NMR revealed the anti-Pseudonocardia compound to be an unreported analog of rebeccamycin bearing a methoxy group, 9-methoxyrebeccamycin (2). All 1H and 13C resonances for the indolocarbazole core and the pendant 4-O-methylglucose of rebeccamycin were evident in this compound's 1D 1H and 13C spectra and could be assigned by COSY and HMBC couplings (Supplemental Figure S6). HMBC couplings indicated that the additional methoxy group in 2 is at position C9 on the glycosylated indole moiety. This placement is corroborated by a characteristic Jmeta of 1.9 Hz between aromatic protons at C8 and C10. The absolute stereochemistry of the sugar was not assigned in this study and is shown as that of rebeccamycin.
Rebeccamycin (1) was first isolated from the Panamanian soil Actinobacterium Lechevalieria aerocolonigenes (formerly Saccharothrix aerocolonigenes) in a screen for antiproliferative agents.5,6 It belongs to the indolocarbazole structural class of tryptophan dimer natural products that are well known in drug discovery efforts.7 The founding member of the indolocarbazoles, first described in 1977, is the kinase inhibitor staurosporine (3).8 Many indolocarbazole natural products are now known, though the maleimide moiety is limited to rebeccamycin, its close analog AT2433-A1 (4),9 and a handful of other structures. Recent bioinformatic analysis of soil metagenomes from the desert southwest of the United States indicated the existence of other rebeccamycin analogs, though the gene products of these biosynthetic gene clusters (BGCs) have not been reported.10 To our knowledge, 9-methoxyrebeccamycin is the only naturally occurring molecule sharing rebeccamycin's dichlorinated indolocarbazole core. Rebeccamycin's antiproliferative activity sparked both a medicinal chemistry effort to advance analogs as chemotherapeutics and biosynthetic engineering efforts to generate novel indolocarbazoles.11,12 Several rebeccamycin analogs have now been in clinical trials for a variety of cancers.11,13 Rebeccamycin's antitumor activity has been ascribed to mammalian topoisomerase I inhibition.14
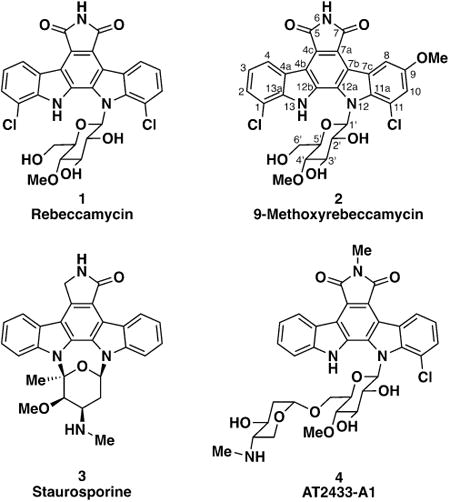
While its antitumor activity has attracted the most attention, rebeccamycin is also a potent antibacterial agent, as is AT2433-A1 (4).6,9 We quantified the antibacterial activity of 9-methoxyrebeccamycin in an agar-based assay. It has potent, nanomolar activity against a small panel of Pseudonocardia (590 nM against BCI1 and 150 nM against PLR1) – roughly two-fold more potent than rebeccamycin (Supplemental Table S3). Not surprisingly, BCI2 is strongly resistant to both 9-methoxyrebeccamycin and rebeccamycin (>300 μM).
PacBio sequencing of BCI1 and BCI2 revealed that each had a circular chromosome and a megaplasmid that were essentially identical in gene content (Supplemental Figure S9). A comparison using in silico DNA/DNA hybridization (DDH) calculations yielded a value of ∼98.3% across the entire chromosomes and ∼96.3% between conserved megaplasmids (pBCI1-1 and pBCI2-1; Supplemental Table S6). In addition to these two conserved replicons, each organism carried a third strain-specific replicon: BCI1 has an unstable linear plasmid (pBCI1-2, 207 kb) while BCI2 harbors a stable circular plasmid (pBCI2-2, 119 kb) with a rebeccamycin-like biosynthetic gene cluster (Figure 2). Niche defense is thus a plasmid-specific trait.
Figure 2.
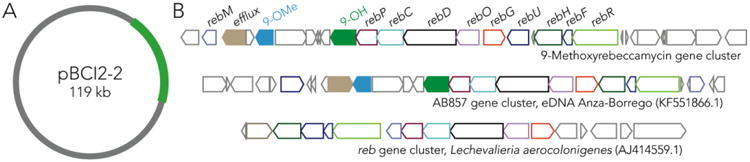
The genetic basis of BCI2 niche defense. (A) BCI2-specific plasmid encoding the biosynthetic gene cluster for 9-methoxyrebeccamycin. (B) The 9-methoxyrebeccamycin biosynthetic gene cluster and two related clusters. The conserved biosynthetic genes are named and color-coded.
The plasmid-borne 9-methoxyrebeccamycin biosynthetic gene cluster (BGC) contains the full suite of genes required for rebeccamycin biosynthesis previously annotated for the rebeccamycin producing strain Lechevalieria aerocolonigenes. The 9-methoxyrebeccamycin cluster additionally encodes enzymes that could install the 9-methoxy group specific to this molecule. These enzymes include a flavin-dependent monooxygenase and a SAM-dependent O-methyltransferase that likely perform the tailoring chemistry (Figure 2B, green and blue shaded arrows). We also compared the 9-methoxyrebeccamycin BGC to other reported rebeccamyin-like gene clusters from eDNA.10 AB857, derived from a California desert soil sample, was the most similar (Figure 2B). This BGC has been predicted to encode a rebeccamycin analog, though the product of this cluster has not been identified.10 All rebeccamycin biosynthesis genes are conserved but interestingly the AB857 BGC also contains the tailoring enzymes that presumably install the 9-methoxy group, which are located on a shared subcluster. Notably, 4/7 rebeccamycin-like gene clusters – the two presented above, AB1533, and NM74710 – encode for the tailoring enzymes for the 9-methoxy group, suggesting that it is a relatively common natural modification to the rebeccamycin scaffold, discovered 30 years ago. Identification of these putative tailoring enzymes could be useful for biosynthetic engineering of rebeccamycin analogs. Knockout of the methyltransferase, for example, would yield a phenolic handle for further elaboration into analogs.
BCI2's high tolerance for 9-methoxyrebeccamycin implies a resistance mechanism on the pBCI2-2 plasmid. The integral membrane transporter RebT, a putative efflux pump in the major facilitator family, has been shown to confer rebeccamycin resistance when heterologously expressed in an otherwise sensitive Actinomycete, Streptomyces albus.15 A distinct member of this transporter family is encoded in both the BCI2 9-methoxyrebeccamycin and eDNA AB857 clusters (Figure 2B, brown shaded arrows) and is the most likely candidate for self-resistance. An uncharacterized putative transporter gene, rebU, is also present in all three clusters and could also contribute to resistance.
A few plasmid-encoded antibiotic BGCs have been reported, although the rapid pace of complete bacterial genome sequencing is likely to reveal many more.16,17 Indeed, we have recently discovered other examples of plasmid-derived BGCs from ant-associated Pseudonocardia isolates.18 Plasmid-encoded niche defense was first reported long ago in E. coli and other Gram-negative bacteria for the colicins and microcins – ribosomally synthesized peptide antimicrobials – which are thought to provide narrow spectrum defense against closely related bacteria.19-21
This analysis of niche defense in an increasingly well studied multilateral symbiosis demonstrates a clear population-level diversity between the two seemingly identical strains we examined. This diversity argues that the fungus-growing ant system will be an even more fruitful source of molecular diversity than was originally thought.22 And the production of a new version of a therapeutically promising molecule that had been identified genetically is an additional argument for further exploration of this system.
Supplementary Material
Acknowledgments
We thank the Duke GCB Genome Sequencing Shared Resource, which provided PacBio sequencing service and the Harvard Medical School Information Technology Department for access to the Orchestra High Performance Computing Cluster. We thank Emily Mevers for measurement of optical rotation. C.S.S. was supported by an Alberta Innovates Health Solutions Fellowship and a Banting Postdoctoral Fellowship; A.C.R. was supported by a Harvard Medical School – Merck Fellowship; The work was also supported by NIH R01 GM086258 (J.C.) and NIH U19 AI09673 (E.B.V., J.C., & C.R.C.).
Footnotes
Supporting Information. Experimental details, supplemental figures, data tables, and spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Sources: The authors declare no competing financial interests.
References
- 1.Davies J. J Ind Microbiol Biotechnol. 2006;33:496. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 2.Davies J. J Antibiot (Tokyo) 2013;66:361. doi: 10.1038/ja.2013.61. [DOI] [PubMed] [Google Scholar]
- 3.Currie CR. Annu Rev Microbiol. 2001;55:357. doi: 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- 4.Poulsen M, Erhardt DP, Molinaro DJ, Lin TL, Currie CR. PLoS ONE. 2007;2:e960. doi: 10.1371/journal.pone.0000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nettleton DE, Doyle TW, Krishnan B, Matsumoto GK, Clardy J. Tetrahedron Lett. 1985;26:4011. [Google Scholar]
- 6.Bush JA, Long BH, Catino JJ, Bradner WT, Tomita K. J Antibiot (Tokyo) 1987;40:668. doi: 10.7164/antibiotics.40.668. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez C, Méndez C, Salas JA. Nat Prod Rep. 2006;23:1007. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- 8.Omura S, Iwai Y, Hirano A, Nakagawa A, Awaya J, Tsuchya H, Takahashi Y, Masuma R. J Antibiot (Tokyo) 1977;30:275. doi: 10.7164/antibiotics.30.275. [DOI] [PubMed] [Google Scholar]
- 9.Matson JA, Claridge C, Bush JA, Titus J, Bradner WT, Doyle TW, Horan AC, Patel M. J Antibiot (Tokyo) 1989;42:1547. doi: 10.7164/antibiotics.42.1547. [DOI] [PubMed] [Google Scholar]
- 10.Chang FY, Ternei MA, Calle PY, Brady SF. J Am Chem Soc. 2013;47:17906. doi: 10.1021/ja408683p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prudhomme M. Eur J Med Chem. 2003;38:123. doi: 10.1016/s0223-5234(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez C, Zhu L, Braña AF, Salas AP, Rohr J, Méndez C, Salas JA. Proc Natl Acad Sci U S A. 2005;102:461. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Her C. Biomolecules. 2015;5:1652. doi: 10.3390/biom5031652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira ER, Belin L, Sancelme M, Prudhomme M, Ollier M, Rapp M, Sevère D, Riou JF, Fabbro D, Meyer T. J Med Chem. 1996;39:4471. doi: 10.1021/jm9603779. [DOI] [PubMed] [Google Scholar]
- 15.Sánchez C, Butovich IA, Braña AF, Rohr J, Méndez C, Salas JA. Chem Biol. 2002;9:519. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- 16.Kinashi H. J Antibiot (Tokyo) 2010;64:19. doi: 10.1038/ja.2010.146. [DOI] [PubMed] [Google Scholar]
- 17.Stinear TP, Mve-Obiang A, Small PLC, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PDR, Davies JK, Lee RE, Adusumilli S, Garnier T, Haydock SF, Leadlay PF, Cole ST. Proc Natl Acad Sci U S A. 2004;101:1345. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sit CS, Ruzzini AC, Van Arnam EB, Ramadhar TR, Currie CR, Clardy J. Proc Natl Acad Sci U S A. 2015;112:13150. doi: 10.1073/pnas.1515348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeWitt W, Helinski DR. J Mol Biol. 1965;13:692. [Google Scholar]
- 20.Duquesne S, Destoumieux-Garzón D, Peduzzi J, Rebuffat S. Nat Prod Rep. 2007;24:708. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 21.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Microbiol Mol Biol Rev. 2007;71:158. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clardy J, Fischbach MA, Currie CR. Curr Biol. 2009;19:R437. doi: 10.1016/j.cub.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


