Abstract
Myeloid differentiation primary response gene 88 (MYD88), a universal adapter protein, plays an important role in activating the nuclear factor-κB (NF-κB) and regulating the expression of proinflammatory genes like tumor necrosis factor (TNF) and interleukin-1 (IL-1), which were highly involved in Salmonella Pullorum infection. To detect the relationship between polymorphisms of the MyD88 gene and Salmonella Pullorum disease, we screened the coding region (CDS) of the MYD88 gene by DNA pool construction and sequencing based on case-control study. Eight single nucleotide polymorphisms (SNPs) in the sequenced fragment (5 exons), 7 known loci and one novel mutation named G4810372T (SNP8), were found in the fifth exon. In addition, we found 7 nonsynonymous substitutions. The allele frequency of only one SNP, g.4810191C > T (SNP1), was significantly different (P < 0.05) between case and control groups. The genotype frequencies of SNP1 (g.4810191C > T) and SNP3 (g.4810257G > T) were of significant difference between the case and the control groups (P < 0.05). Collectively, SNPs of the MyD88 gene were significantly associated with susceptibility to Salmonella Pullorum infection, which can be used as a disease-resistant marker in chicken. These results provided a theoretical basis for future research on chicken breeding by marker-assisted selection.
1. Introduction
Salmonella Pullorum is a typical bacterial disease that has threated the modern poultry industry over the past years. Chicken becomes the carrier in the spread of Salmonella Pullorum and may cause economic losses worldwide through mortality, morbidity, and reductions in egg production [1]. Vaccination, antibiotics, and other drugs are the major preventive measure, but antibiotics will cause resistance to pathogens and antibiotic residues in poultry products and other issues. Generations of purifying elimination has been applied in domestic chicken populations, which would largely decrease genetic resistance of Salmonella Pullorum. In vertebrates, two types of immunity are developed to protect the host from infections: innate and adaptive [2]. The innate immune system is the first line of defense against microbial pathogens and of key importance early in bacterial infections. During infection, host inflammatory reaction is mediated by the recognition of pattern-recognition receptors (PRRs), which was activated by pathogen-associated molecular patterns (PAMPs) [3]. The TLR family is a major class of PRRs, which is critical for induction of the immune response to the given microbial challenge [4]. After recognized PAMPs, the TLR signaling pathway can be segregated into two specific ways: the MyD88-dependent pathway and the MyD88 independent pathway [5, 6]. The first pathway results in the activation of the nuclear factor-κB (NF-κB), and the expression of proinflammatory genes like tumor necrosis factor (TNF) and interleukin-1 (IL-1) [7, 8]. The second one upregulates interferon 3 (IRF3-) mediated expression of type I interferons (IFN) and IFN-inducible genes. The MyD88-dependent pathway is prominent for all TLRs except TLR3 [9]. As a universal adaptor protein, MyD88 plays an important role in activating the innate immune system [10]. The typical structure composed of amino terminal death domain of MyD88 recruits the downstream immune molecules, binding interactions with Interleukin-1 receptor-associated kinases (IRAKs) [11]. Some study on the relationship between TLR4 signaling pathway including related molecule and the susceptibility to disease showed that the signal pathways have played a major role in the infection [12, 13]. The expression of TLR4 and immune related genes, such as Gal 1, Gal 2, IL-8, IL-18, and IFN-γ, established different degree of correlation against salmonella in hens [14].
In large yellow croaker, MyD88 was reported to play a crucial role in defensing against pathogenic infection for its extremely differential expression between spleen and muscle [15, 16]. The lack of MyD88 protein in mice may result in susceptibility to leishmania major infection [17]. Though some studies broadly reveal the function of MyD88 in TLR signaling pathways [18], the relationship between polymorphisms of the MyD88 gene and Pullorum infection in chicken has not been reported. In view of the importance of the MyD88 gene, it is necessary to analyze its sequence variations and to study whether its polymorphisms are associated with intersubject differences against salmonella. In this study, we selected chickens infected and uninfected with Salmonella Pullorum to conduct an association analysis with MyD88, aiming to provide a theoretical reference for poultry marker-assisted selection.
2. Materials and Methods
2.1. Pullorum Detection and Sample Collection
Based on case-control design, the method of whole blood glass plate agglutination (SN/T 1222-2003, AQSIQ) was used to test infection with Salmonella Pullorum. Whole blood sample (20 μL) was mixed with Pullorum reagent (20 μL) on a glass slide that was kept undisturbed for 3 min at room temperature (25 to 30°C). With the naked eye, agglutinations (clumping of RBCs) were read in infected samples. The level of infection have divided into “+++,” “++,” and “+” for 100%, 75%, and 50% aggregation, while no agglutination was read in uninfected ones (“−”). Grouped chickens were marked with “+++,” “++,” and “+” into the case.
Following common laying hen immunization program, chicken were vaccinated timely of immunization. 4,334 Er-lang mountainous hens have been tested at the time of 300 days of age, among which 128 infected subjects were collected as the cases and 163 uninfected subjects were collected as controls. The protocol was approved by the Committee on the Care and Use of Laboratory Animals of the State-Level Animal Experimental Teaching Demonstration Center of Sichuan Agricultural University. Blood samples were stored at −20°C.
2.2. DNA Extraction and DNA Pool Construction
The genomic DNA of all the samples was extracted by using the standard phenol/chloroform method. DNA samples were diluted or enriched at the level of (100 ± 3) ng/μL with Nano Drop (ND-2000, Thermo Scientific) for homogeneity of DNA pool. A case DNA pool and a control one were composed of 30 samples that each contains 2 μL DNA selected at random, respectively.
2.3. Primer Design
Using the Primer Premier 5 software (Premier BioSoft, Palo Alto, CA, USA), five pairs of primers were designed to cover the entire coding region (CDS) of the MyD88 gene (Table 1) from Gallus gallus (GenBank accession number NM_001030962).
Table 1.
The primer sequences to amplify the CDS of MyD88 gene in chicken.
| Name | Amplicon size (bp) | Sequence (5′-3′) | Production (bp) | Anneal temperature (°C) | Region |
|---|---|---|---|---|---|
| P1 | 19 | F: GGCTCCTTCCAACCCAAAC | 793 | 59.0 | exon1 |
| 21 | R: AGACCGATCTCACCTCACCAC | ||||
|
| |||||
| P2 | 22 | F: CAAGAAGGCACTGGGTAAACTC | 483 | 54.6 | exon2 |
| 21 | R: GAATAGGCAACGGGAAGAATG | ||||
|
| |||||
| P3 | 20 | F: TCTGTCAAAGGCTGGGGAAG | 297 | 54.0 | exon3 |
| 19 | R: ACACTGAGCTGCCCCAAGC | ||||
|
| |||||
| P4 | 24 | F: GTTCCTGCTCAACCACAACTAAAG | 443 | 52.5 | exon4 |
| 22 | R: GGGTTCTGGTTCAGTAGGCATC | ||||
|
| |||||
| P5 | 23 | F: TAGAAGCAGGATGTGAGTGTGGC | 501 | 59.6 | exon5 |
| 25 | R: GCAGTGACTCAGTCTTTAAGCGAAT | ||||
2.4. PCR Amplification and Sequencing
Polymerase chain reaction (PCR) amplification was performed in a volume of 25 μL reaction mixture containing 50–100 ng of DNA, 0.3 μM of each forward and reverse primer, and 15 μL of 2x Taq PCR MasterMix (Tiangen Biotech Co., China). The procedure was carried out with one cycle of denaturalization at 94°C for 5 min, 35 cycles of 94°C for 40 s, appropriate annealing temperature (Table 1) for 30 s, and 72°C for 40 s, followed by a final extension cycle at 72°C for 8 min. All PCR products of DNA pool and individual were directly sequenced by the Shenzhen BGI Biotechnology Company (Beijing, China).
2.5. Statistical Analysis
The sequencing electrophoretogram was read by Chromas software. Sequence variations, composition, and variable sites were identified using DNAstar software (DNAstar Inc., Madison, WI, USA). Hardy-Weinberg equilibrium, pairwise linkage disequilibrium (D′), and association analysis were conducted by Haploview software (version 3.32, http://www.broad.mit.edu/mpg/haploview/). Complementary analysis, such as the standard Chi-squared test of genotype frequency, was performed by use of R software (version 3.0.2, The R Foundation for Statistical Computing).
3. Result
3.1. Sequence Variations in CDS Region of the MyD88 Gene
No mutation was observed in MYD88 CDS region except for the exon5. A total of 8 SNPs were detected in exon5, including 7 known SNPs (http://www.ncbi.nlm.nih.gov/projects/SNP/) and a novel mutation locating at 4810372 bp of the chicken genome named G4810372T (SNP 8). 7 SNPs (SNP1, SNP2, SNP3, SNP4, SNP5, SNP6, and SNP8) were nonsynonymous variants leading to amino acids changes, while the allele change of SNP7 leads to a synonymous mutation (Table 2).
Table 2.
Change of alleles and amino acids in the CDS region of MyD88 gene.
| Markers | ID | Position | Obs HET | Expt HET | Allele change | Amino acids change | Mutation |
|---|---|---|---|---|---|---|---|
| SNP1 | rs317890917 | 4810191 | 0.457 | 0.412 | C>T | 307 Leucine > phenylalanine | Nonsynonymous |
| SNP2 | rs14131328 | 4810253 | 0.258 | 0.349 | G>C | 327 Glutamine > Histidine | Nonsynonymous |
| SNP3 | rs14131329 | 4810257 | 0.405 | 0.419 | G>T | 329 Valine > Leucine | Nonsynonymous |
| SNP4 | rs14131330 | 4810266 | 0.481 | 0.481 | T>C | 332 Cysteine > Arginine | Nonsynonymous |
| SNP5 | rs14131331 | 4810276 | 0.436 | 0.483 | G>A | 335 Arginine > Histidine | Nonsynonymous |
| SNP6 | rs14131332 | 4810293 | 0.471 | 0.477 | G>C | 341 Glycine > Arginine | Nonsynonymous |
| SNP7 | rs14131333 | 4810352 | 0.474 | 0.483 | A>G | 360 Leucine > Leucine | Synonymous |
| SNP8 | G4810372T | 4810372 | 0.072 | 0.082 | G>T | 367 Serine > Isoleucine | Nonsynonymous |
Obs HET is observed heterozygosity; Expt HET is expected heterozygosity.
3.2. The Hardy-Weinberg Equilibrium
The Hardy-Weinberg equilibrium tests of the 8 SNPs in the case and control group were shown in Tables 2 and 3. The observed heterozygosity of all SNPs was at a general level as expected. Most of SNPs fit the assumption of the Hardy-Weinberg equilibrium except SNP2 (P < 0.05) that was removed from the analysis. The minor allele frequencies (MAF) of all the mutations were more than 0.01.
Table 3.
Allele frequency of mutation loci in the MyD88 gene.
| Markers | Alleles | χ 2, P value | OR | 95% CI | HWE (P) | MAF | |
|---|---|---|---|---|---|---|---|
| SNP1 (rs317890917) | T | C |
χ
2 = 4.604 P = 0.0319 |
0.6752 | 0.4712–0.9674 | 0.0873 | 0.29 |
| Cases | 170 (0.664) | 86 (0.336) | |||||
| Controls | 243 (0.745) | 83 (0.255) | |||||
|
| |||||||
| SNP3 (rs14131329) | T | G |
χ
2 = 2.384 P = 0.1226 |
0.7555 | 0.5290–1.079 | 0.6509 | 0.299 |
| Cases | 171 (0.668) | 85 (0.332) | |||||
| Controls | 237 (0.727) | 89 (0.273) | |||||
|
| |||||||
| SNP4 (rs14131330) | T | C |
χ
2 = 0.2487 P = 0.6180 |
1.0888 | 0.7794–1.521 | 1.0 | 0.402 |
| Cases | 156 (0.609) | 100 (0.391) | |||||
| Controls | 192 (0.589) | 134 (0.411) | |||||
|
| |||||||
| SNP5 (rs14131331) | G | A |
χ
2 = 0.3047 P = 0.5810 |
1.098 | 0.7869–1.534 | 0.122 | 0.407 |
| Cases | 155 (0.605) | 101 (0.395) | |||||
| Controls | 190 (0.583) | 136 (0.417) | |||||
|
| |||||||
| SNP6 (rs14131332) | G | C |
χ
2 = 0.0155 P = 0.9009 |
1.022 | 0.7306–1.4282 | 0.8877 | 0.393 |
| Cases | 156 (0.609) | 100 (0.391) | |||||
| Controls | 197 (0.604) | 129 (0.396) | |||||
|
| |||||||
| SNP7 (rs14131333) | G | A |
χ
2 = 0.3166 P = 0.5736 |
0.9089 | 0.6517–1.2677 | 0.8152 | 0.409 |
| Cases | 148 (0.578) | 108 (0.422) | |||||
| Controls | 196 (0.601) | 130 (0.399) | |||||
|
| |||||||
| SNP8 (G4810372T) | T | G |
χ
2 = 0.1708 P = 0.6794 |
0.8445 | 0.3786–1.884 | 0.1766 | 0.043 |
| Cases | 244 (0.953) | 12 (0.047) | |||||
| Controls | 313 (0.960) | 13 (0.040) | |||||
CI, confidence interval; OR, odds ratio; HWE (P), P value of the Hardy-Weinberg equilibrium test; and MAF, minimum allele frequency.
3.3. Allele and Genotype Frequency of the Mutated Loci
The results of the allele and genotype frequency of the 8 SNPs in the case and control groups were shown in Tables 3 and 4. The Chi-squared test was used to compare the allele frequencies in the MyD88 gene between the case and control groups. The data showed that only SNP1 (χ 2 = 4.604, P = 0.0319) was significantly correlated with Salmonella Pullorum at the allelic level. At the genotype level, two SNPs, SNP1 (χ 2 = 7.924, P = 0.019, OR = 0.6752, and 95% CI = 0.4712–0.9674) and SNP3 (χ 2 = 8.353, P = 0.015, OR = 0.7555, and 95% CI = 0.5290–1.079), showed significant correlations with Salmonella Pullorum in the chicken population.
Table 4.
Genotype frequency of mutation loci in the MyD88 gene.
| Markers | Genotypes | χ 2, P value | ||
|---|---|---|---|---|
| SNP1 (rs317890917) | TT | TC | CC |
χ
2 = 7.924 P = 0.019 |
| Cases | 50 (0.391) | 70 (0.547) | 8 (0.072) | |
| Controls | 90 (0.552) | 63 (0.387) | 10 (0.061) | |
|
| ||||
| SNP3 (rs14131329) | TT | TG | GG |
χ
2 = 8.353 P = 0.015 |
| Cases | 54 (0.422) | 63 (0.492) | 11 (0.086) | |
| Controls | 92 (0.564) | 53 (0.325) | 18 (0.110) | |
|
| ||||
| SNP4 (rs14131330) | TT | TC | CC |
χ
2 = 3.2 P = 0.202 |
| Cases | 44 (0.344) | 68 (0.531) | 16 (0.125) | |
| Controls | 60 (0.368) | 72 (0.442) | 31 (0.190) | |
|
| ||||
| SNP5 (rs14131331) | GG | GA | AA |
χ
2 = 2.174 P = 0.337 |
| Cases | 47 (0.367) | 61 (0.477) | 20 (0.156) | |
| Controls | 62 (0.380) | 66 (0.405) | 35 (0.215) | |
|
| ||||
| SNP6 (rs14131332) | GG | GC | CC |
χ
2 = 2.805 P = 0.246 |
| Cases | 45 (0.352) | 66 (0.516) | 17 (0.133) | |
| Controls | 64 (0.393) | 69 (0.423) | 30 (0.184) | |
|
| ||||
| SNP7 (rs14131333) | GG | GA | AA |
χ
2 = 2.999 P = 0.223 |
| Cases | 40 (0.313) | 68 (0.531) | 20 (0.156) | |
| Controls | 63 (0.387) | 70 (0.429) | 30 (0.184) | |
|
| ||||
| SNP8 (G4810372T) | TT | TG | GG |
χ
2 = 2.843 P = 0.241 |
| Cases | 118 (0.922) | 8 (0.063) | 2 (0.016) | |
| Controls | 150 (0.920) | 13 (0.080) | 0 (0) | |
3.4. Association between Haplotypes and Susceptibility to Pullorum
Figure 1 revealed that the degree of the linkage disequilibrium (LD) indicates the correlation between polymorphic variants at different positions in the MyD88 gene. With 2 blocks in dark red in the D′ plot, it was clear that the SNPs of block 1 (SNP1, SNP3, and SNP4) and block 2 (SNP5, SNP6, SNP7, and SNP8) are of high D′, respectively. Nevertheless, SNP6 and SNP8 were in equilibrium and independent of one another in block 2. Haplotype analysis showed that the haplotype groups CTT (χ 2 = 4.604, P = 0.0319) and TTC (χ 2 = 11.643, P = 6.0E − 4) in block 1 of the MyD88 gene correlated significantly with resistance to Salmonella Pullorum infection (Figure 1, Table 5). Haploid type distribution of block 2, composed by SNP5, SNP6, SNP7, and SNP8, was not significantly different (P > 0.05) in the case and control groups. Haploid types in block 2 had no relationship with disease resistance.
Figure 1.
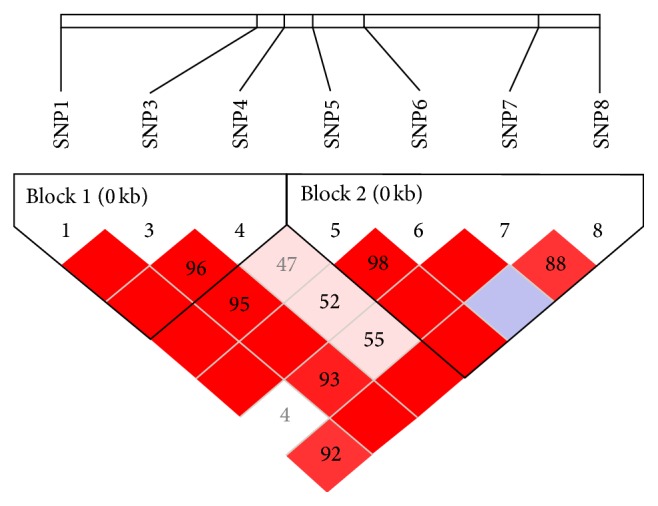
LD value within each diamond represents the correlation between pairs of SNPs (measured as D′) in the CDS of the MyD88 gene. The diamond without a number means complete LD (D′ = 1). Darker red of the diamonds indicates higher D′, while white indicates lower D′.
Table 5.
The haplotype analysis of 8 MyD88 SNPs.
| Haplotype groups | Frequency (cases) | Frequency (control) | χ 2 | P value | |
|---|---|---|---|---|---|
| Block 1 | TTT | 0.273 | 0.325 | 1.818 | 0.1776 |
| TGC | 0.332 | 0.263 | 3.222 | 0.0727 | |
| CTT | 0.336 | 0.255 | 4.604 | 0.0319 | |
| TTC | 0.059 | 0.148 | 11.643 | 6.0E − 4 | |
|
| |||||
| Block 2 | ACGT | 0.383 | 0.396 | 0.1 | 0.7515 |
| GGAT | 0.380 | 0.359 | 0.272 | 0.6023 | |
| GGGT | 0.171 | 0.184 | 0.17 | 0.6803 | |
| GGAG | 0.042 | 0.040 | 0.016 | 0.8979 | |
| AGGT | 0.012 | 0.022 | 0.805 | 0.3695 | |
4. Discussion
Infectious disease has an effect on the food safety and spreads broadly in chicken, especially in commercial lines. In the past years, the contradiction between antibiotics abuse and food safety has increasingly grown [19, 20]. In addition, selection based heavily on production performance could disadvantageously affect individual immunity leaving chickens less resistive to pathogenic bacteria [21]. Recently, the intersubject differences in immunity that associated with missense mutations in innate immune genes, such as major histocompatibility complex (MHC) [22, 23] and myxovirus resistance gene (Mx) [24], have significantly increased in chicken. Consequently, studying the basic knowledge of mutations in innate immune genes has an important significance for chicken disease-resistant breeding.
In previous study, the use of SNP of innate immune genes, such as natural resistance associated macrophage protein 1 (Nramp1) [25], TLR4 [26], CD28, and MD-2 [27], leads to enhancement of Salmonella Pullorum resistance in chicken. Yet, in another study, SNP797T/C genotype in the first intron of the MyD88 gene in swine was of no significant correlation with resistance to Salmonella Pullorum infection [28]. Susceptibility to Salmonella Pullorum infection showed the differences across species [29]. Through the analysis, the polymorphisms of the CDS area in MyD88 gene and the correlation with resistance to salmonella suggested that MyD88 gene may be one of the major Salmonella Pullorum resistant genes in innate immune system.
The Hardy-Weinberg equilibrium is influenced by many factors, including selection, the rate of recombination, the rate of mutation, genetic drift, the system of mating, population structure, and genetic linkage. Due to the selection and foreign blood imported in the chicken population, the alleles and genotypes of SNP2 loci were unstable and SNP2 was removed from statistic analysis. We screened out a newly identified SNP site named G4810372T (SNP 8), and the follow-up studies may be needed to test the functional significance for the newly identified SNP site [30].
In genetics, a missense mutation, a type of nonsynonymous substitution, results in truncation of the resulting protein and protein nonfunctional. According to the Human Gene Mutation Database (HGMD), missense mutation of CRYBB2 leads to congenital cataract in a family of Croatian origin [31]. In the CDS of MyD88, there are 376 amino acid residues [32], 7 of which have changed for they have underwent nonsynonymous substitutions. The homology of DNA sequence (1122/1130) was presented higher than the amino acid sequence homology (369/376). In other words, the MyD88 gene polymorphism was of higher performance on the amino acid levels. All the mutations locating only in extron 5 declared that MyD88 gene was highly conserved. In analysis by the SMART software (http://smart.embl-heidelberg.de/), the 7 missense mutations locate out of the typical structure of MyD88, the N-terminal death domain, C-terminal toll-interleukin-1 receptor (TIR) domain, and intermediate domain. Therefore, the 7 substitutions of amino acid caused by polymorphisms of the MyD88 gene may have the potential of resistance to bacteria and pathogens.
To reveal the relationship between polymorphisms of the MyD88 gene and disease resistance, we conducted a Mendelian population-based case-control study in chicken [33]. The OR and 95% confidence interval are used to determine the resistance or susceptibility to Pullorum infection. The OR value less than 1 means resistance effect, while value greater than 1 means susceptibility effect [34]. At the same time, the P values determine the significance of its association. SNP1 alleles distribution, of the lowest OR value (OR = 0.6752), made significant differences in case and control samples (P < 0.05). The genotypes distribution of SNP3 and SNP1 revealed significant differences in case and control samples (P < 0.05). Results indicated that polymorphisms of the MyD88 gene and susceptibility to Pullorum infection have significant correlation.
In the study of the multiple loci in linkage disequilibrium (LD) and the correlation of disease, the correlation of haploid types is more effective than a single locus analysis [35]. Many phenotypic traits are often the result of the interaction between multiple loci, especially in a haploid type block, and caused by interaction among a set of mutations in a certain area of the chromosome [36, 37]. In the correlation analysis of haploid type, the first thing is to determine the strength of LD (D′) and type in the group. LD exists widely in the family of chicken population in this study. The results showed that the SNPs among block 1 and block 2 groups were in strong linkage disequilibrium state (r 2 > 0.33, |D′ | > 0.8). Containing the analysis results of SNP1 loci, difference between haploid type composed by SNP1, SNP3, and SNP4 loci and resistance to disease was very significant. The result showed that the analysis of haploid types had better statistical effect (P < 0.01). Effects between SNPs loci in haploid type set canceled each other out on account of interactive effects of genes, leading to no significant difference between SNP5, SNP6, SNP7, and SNP8 loci haploid type and disease resistance.
In conclusion, after comparison and analysis of the genetic variation of MyD88 gene, we found that the novel mutation G4810372T may have an effect on the individual immune. But further functional studies are necessary to evaluate the molecular mechanism caused by polymorphisms of the MyD88 gene. The correlation analysis of polymorphisms of the MyD88 gene and susceptibility of Salmonella Pullorum in chicken showed that, in each mutation, alleles in SNP1 locus and genotypes of SNP1 and SNP3 had a significant effect against salmonella. What is more, the advantaged haploid type (TTC) combined by SNP1, SNP3, and SNP4 loci played a very significant role in genetic resistance to Salmonella Pullorum infection. Polymorphisms of the MyD88 gene or advantaged haploid type in a particular area had a certain positive effect against susceptibility to Pullorum infection. From the above, the MyD88 gene can be used as a candidate gene for follow-up study, which could provide a theoretical reference for poultry marker-assisted selection.
Acknowledgment
Research was supported by the Open Fund of Farm Animal Genetic Resources Exploration and Innovation Key Laboratory of Sichuan Province (Grant no. 2011NZ0099-6).
Conflict of Interests
All authors have not declared conflict of interests.
References
- 1.Shivaprasad H. L. Fowl typhoid. In: Beard C. W., McNulty M. S., editors. Diseases of Poultry: World Trade and Public Health Implications Scientific and Technical Review. Vol. 19. Paris, France: Office International des Epizooties; 2000. pp. 405–424. [Google Scholar]
- 2.Iwasaki A., Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327(5963):291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liniger M., Summerfield A., Zimmer G., McCullough K. C., Ruggli N. Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. Journal of Virology. 2012;86(2):705–717. doi: 10.1128/jvi.00742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Akira S., Takeda K. Toll-like receptor signalling. Nature Reviews Immunology. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 6.Takeda K., Kaisho T., Akira S. Toll-like receptors. Annual Review of Immunology. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 7.Vasselon T., Detmers P. A. Toll receptors: a central element in innate immune responses. Infection and Immunity. 2002;70(3):1033–1041. doi: 10.1128/iai.70.3.1033-1041.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D., Ding A. MyD88-mediated stabilization of interferon-γ-induced cytokine and chemokine mRNA. Nature Immunology. 2006;7(4):375–381. doi: 10.1038/ni1308. [DOI] [PubMed] [Google Scholar]
- 9.Naiki Y., Michelsen K. S., Zhang W., Chen S., Doherty T. M., Arditi M. Transforming growth factor-β differentially inhibits MyD88-dependent, but not TRAM- and TRIF-dependent, lipopolysaccharide-induced TLR4 signaling. The Journal of Biological Chemistry. 2005;280(7):5491–5495. doi: 10.1074/jbc.c400503200. [DOI] [PubMed] [Google Scholar]
- 10.Walker W. E., Nasr I. W., Camirand G., Tesar B. M., Booth C. J., Goldstein D. R. Absence of innate MyD88 signaling promotes inducible allograft acceptance. The Journal of Immunology. 2006;177(8):5307–5316. doi: 10.4049/jimmunol.177.8.5307. [DOI] [PubMed] [Google Scholar]
- 11.Cao Z., Henzel W. J., Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science. 1996;271(5252):1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 12.Moynagh P. N. TLR signalling and activation of IRFs: revisiting old friends from the NF-κB pathway. Trends in Immunology. 2005;26(9):469–476. doi: 10.1016/j.it.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Medzhitov R., Janeway C., Jr. The Toll receptor family and microbial recognition. Trends in Microbiology. 2000;8(10):452–456. doi: 10.1016/s0966-842x(00)01845-x. [DOI] [PubMed] [Google Scholar]
- 14.Sadeyen J.-R., Trotereau J., Protais J., et al. Salmonella carrier-state in hens: study of host resistance by a gene expression approach. Microbes and Infection. 2006;8(5):1308–1314. doi: 10.1016/j.micinf.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Yao C.-L., Kong P., Wang Z.-Y., et al. Molecular cloning and expression of MyD88 in large yellow croaker, Pseudosciaena crocea . Fish & Shellfish Immunology. 2009;26(2):249–255. doi: 10.1016/j.fsi.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Qiu L., Song L., Yu Y., Zhao J., Wang L., Zhang Q. Identification and expression of TRAF6 (TNF receptor-associated factor 6) gene in Zhikong Scallop Chlamys farreri . Fish and Shellfish Immunology. 2009;26(3):359–367. doi: 10.1016/j.fsi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Muraille E., De Trez C., Brait M., De Baetselier P., Leo O., Carlier Y. Genetically resistant mice lacking MyD88-adapter protein display a high susceptibility to Leishmania major infection associated with a polarized Th2 response. The Journal of Immunology. 2003;170(8):4237–4241. doi: 10.4049/jimmunol.170.8.4237. [DOI] [PubMed] [Google Scholar]
- 18.Barton G. M., Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300(5625):1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 19.An H., Zhang D. H., Chen X. L., et al. Isolation and identification of Salmonella pullorum . Agricultural Science & Technology. 2012;13:661–663. [Google Scholar]
- 20.Livant E. J., Avendano S., McLeod S., et al. MX1 exon 13 polymorphisms in broiler breeder chickens and associations with commercial traits. Animal Genetics. 2007;38(2):177–179. doi: 10.1111/j.1365-2052.2007.01577.x. [DOI] [PubMed] [Google Scholar]
- 21.Swaggerty C. L., Pevzner I. Y., He H., et al. Selection of broilers with improved innate immune responsiveness to reduce on-farm infection by foodborne pathogens. Foodborne Pathogens and Disease. 2009;6(7):777–783. doi: 10.1089/fpd.2009.0307. [DOI] [PubMed] [Google Scholar]
- 22.Chaves L. D., Faile G. M., Krueth S. B., Hendrickson J. A., Reed K. M. Haplotype variation, recombination, and gene conversion within the turkey MHC-B locus. Immunogenetics. 2010;62(7):465–477. doi: 10.1007/s00251-010-0451-2. [DOI] [PubMed] [Google Scholar]
- 23.Eimes J. A., Bollmer J. L., Dunn P. O., Whittingham L. A., Wimpee C. Mhc class II diversity and balancing selection in greater prairie-chickens. Genetica. 2010;138(2):265–271. doi: 10.1007/s10709-009-9417-4. [DOI] [PubMed] [Google Scholar]
- 24.Haller O., Staeheli P., Kochs G. Protective role of interferon-induced Mx GTPases against influenza viruses. Revue Scientifique et Technique. 2009;28(1):219–231. doi: 10.1128/JVI.01718-06. [DOI] [PubMed] [Google Scholar]
- 25.Beaumont C., Protais J., Pitel F., et al. Effect of two candidate genes on the Salmonella carrier state in fowl. Poultry Science. 2003;82(5):721–726. doi: 10.1093/ps/82.5.721. [DOI] [PubMed] [Google Scholar]
- 26.Li P., Xia P., Wen J., et al. Up-regulation of the MyD88-dependent pathway of TLR signaling in spleen and caecum of young chickens infected with Salmonella serovar Pullorum. Veterinary Microbiology. 2010;143(2–4):346–351. doi: 10.1016/j.vetmic.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Malek M., Hasenstein J. R., Lamont S. J. Analysis of chicken TLR4, CD28, MIF, MD-2, and LITAF genes in a Salmonella enteritidis resource population. Poultry Science. 2004;83(4):544–549. doi: 10.1093/ps/83.4.544. [DOI] [PubMed] [Google Scholar]
- 28.Li X., Liu H., Yang S., et al. Characterization analysis and polymorphism detection of the porcine Myd88 gene. Genetics and Molecular Biology. 2009;32(2):295–300. doi: 10.1590/S1415-47572009000200015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langothe H., Palmer C. N. A., Morris A. D., et al. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57(11):3129–3135. doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariani P., Barrow P. A., Cheng H. H., Groenen M. A., Negrini R., Bumstead N. Localization to chicken chromosome 5 of a novel locus determining salmonellosis resistance. Immunogenetics. 2001;53(9):786–791. doi: 10.1007/s00251-001-0387-7. [DOI] [PubMed] [Google Scholar]
- 31.Weisschuh N., Aisenbrey S., Wissinger B., Riess A. Identification of a novel CRYBB2 missense mutation causing congenital autosomal dominant cataract. Molecular Vision. 2012;18:174–180. [PMC free article] [PubMed] [Google Scholar]
- 32.Schlesselman J. J., Schneiderman M. A. Case control studies: design, conduct, analysis. Journal of Occupational and Environmental Medicine. 1982;24:p. 879. [Google Scholar]
- 33.Wheaton S., Lambourne M. D., Sarson A. J., Brisbin J. T., Mayameei A., Sharif S. Molecular cloning and expression analysis of chicken MyD88 and TRIF genes. DNA Sequence. 2007;18(6):480–486. doi: 10.1080/10425170701295856. [DOI] [PubMed] [Google Scholar]
- 34.Durrant C., Zondervan K. T., Cardon L. R., Hunt S., Deloukas P., Morris A. P. Linkage disequilibrium mapping via cladistic analysis of single-nucleotide polymorphism haplotypes. The American Journal of Human Genetics. 2004;75(1):35–43. doi: 10.1086/422174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin E. R., Lai E. H., Gilbert J. R., et al. SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in alzheimer disease. The American Journal of Human Genetics. 2000;67(2):383–394. doi: 10.1086/303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson G. C. L., Esposito L., Barratt B. J., et al. Haplotype tagging for the identification of common disease genes. Nature Genetics. 2001;29(2):233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Zhao L. P., Dudoit S. A fine-scale linkage-disequilibrium measure based on length of haplotype sharing. The American Journal of Human Genetics. 2006;78(4):615–628. doi: 10.1086/502632. [DOI] [PMC free article] [PubMed] [Google Scholar]


