ABSTRACT
Skeletal myogenesis in vertebrates is initiated at different sites of skeletal muscle formation during development, by activation of specific control elements of the myogenic regulatory genes. In the mouse embryo, Myf5 is the first myogenic determination gene to be expressed and its spatiotemporal regulation requires multiple enhancer sequences, extending over 120 kb upstream of the Mrf4-Myf5 locus. An enhancer, located at −57/−58 kb from Myf5, is responsible for its activation in myogenic cells derived from the hypaxial domain of the somite, that will form limb muscles. Pax3 and Six1/4 transcription factors are essential activators of this enhancer, acting on a 145-bp core element. Myogenic progenitor cells that will form the future muscle masses of the limbs express the factors necessary for Myf5 activation when they delaminate from the hypaxial dermomyotome and migrate into the forelimb bud, however they do not activate Myf5 and the myogenic programme until they have populated the prospective muscle masses. We show that Msx1 and Meox2 homeodomain-containing transcription factors bind in vitro and in vivo to specific sites in the 145-bp element, and are implicated in fine-tuning activation of Myf5 in the forelimb. Msx1, when bound between Pax and Six sites, prevents the binding of these key activators, thus inhibiting transcription of Myf5 and consequent premature myogenic differentiation. Meox2 is required for Myf5 activation at the onset of myogenesis via direct binding to other homeodomain sites in this sequence. Thus, these homeodomain factors, acting in addition to Pax3 and Six1/4, fine-tune the entry of progenitor cells into myogenesis at early stages of forelimb development.
KEY WORDS: Myf5 transcription, Msx1, Meox2, Mouse embryo, Limb myogenesis
Summary: Homeodomain factors Msx1 and Meox2, acting in addition to Pax3 and Six1/4, fine-tune the entry of progenitor cells into myogenesis at early stages of forelimb development via modulation of limb enhancer gene Myf5.
INTRODUCTION
Skeletal muscles in the trunk and limbs derive from myogenic progenitor cells present in the somites of the vertebrate embryo and their formation depends on myogenic regulatory factors controlling muscle cell determination and differentiation (see Tajbakhsh and Buckingham, 2000). Myf5 is expressed at the onset of myogenesis in the mouse embryo (Ott et al., 1991) when, together with Mrf4, it determines myogenic cell fate (Braun et al., 1992; Kassar-Duchossoy et al., 2004). Thereafter, MyoD is expressed and can direct cells into the myogenic programme when Myf5 and Mrf4 are absent (Braun et al., 1992). The absence of these three myogenic determination factors leads to the absence of skeletal muscles (Kassar-Duchossoy et al., 2004; Rudnicki et al., 1993). Skeletal muscle in the limbs is formed by muscle progenitor cells that delaminate from the hypaxial dermomyotome of the somites and migrate into the limb field. These cells express the paired/homeodomain transcription factor Pax3 and in its absence they fail to migrate and subsequently undergo apoptosis (see Buckingham and Relaix, 2007). Migration in response to the ligand HGF depends on the c-met receptor (Bladt et al., 1995), and on CXCR4, the receptor for the ligand SDF, which like HGF, is expressed by mesenchymal cells in the limb bud (Vasyutina et al., 2005). The c-met gene is a target for Pax3 (Epstein et al., 1996) and CXCR4 is genetically downstream of Lbx1 which is also expressed in Pax3-positive migratory cells. In the absence of Lbx1, the ventral muscle mass fails to form and these myogenic progenitors remain in the vicinity of the somite where they can adopt other cell fates (see Buckingham, 2001; Schäfer and Braun, 1999). Six homeodomain factors, like Pax3, are important upstream regulators of myogenesis (see Buckingham and Relaix, 2007) and Six1/4 are also expressed in myogenic progenitors that migrate to the limbs. In the absence of Six1/4, limb muscles do not form correctly and Pax3 expression in the hypaxial somite is compromised (Grifone et al., 2005).
Transcriptional regulation of the myogenic determination genes has been extensively studied. Myf5 and Mrf4 are closely linked on mouse chromosome 10 and their transcriptional regulatory elements extend over a region of at least 120 kb, 5′ to and within the Mrf4-Myf5 locus. A number of enhancers have been characterised which direct different aspects of the complex spatiotemporal regulation of Myf5 in the embryo (see Buckingham and Vincent, 2009; Carvajal and Rigby, 2010; Daubas and Buckingham, 2013; Moncaut et al., 2012; Ribas et al., 2011).
A regulatory region required for Myf5 transcription in the limb and in the more mature hypaxial somite is located at −48/−58 kb 5′ of Myf5 (Hadchouel et al., 2000). Within this region, complete expression in the developing limbs requires the concerted activity of at least three sub-regions, with the main limb enhancer located at −57/−58 kb (Hadchouel et al., 2000, 2003). Within this enhancer a 145-bp core sequence contains an essential Pax3 paired domain binding site (Bajard et al., 2006) and an adjacent Six1/4 binding site, required for complete activity (Giordani et al., 2007). Mutation in a homeodomain X-vent-type site, between the Pax and Six sites, negatively affects enhancer activity (Buchberger et al., 2007). Pax3 and Six1/4 are expressed in myogenic progenitor cells that migrate into the limb buds prior to Myf5 activation. Within the developing limb, a proportion of these cells will proliferate, with subsequent expression of Pax7, and do not immediately enter the myogenic programme (see Buckingham and Relaix, 2007). Once myogenic progenitor cells have populated the prospective limb muscle masses, Sonic Hedgehog (Shh) drives myogenesis specifically within the ventral muscle mass, to enhance Myf5 transcription through essential Gli-binding sites located immediately 3′ of the core 145-bp element in the −57/−58 kb enhancer (Anderson et al., 2012). However during the delamination/migration of the myogenic progenitor cells, it is not clear what other regulatory factors interact with the 145-bp sequence to prevent premature activation of Myf5. When this sequence is present in multiple copies in a reporter transgene, premature activation is observed, suggesting saturation of potential repressor mechanisms (Bajard et al., 2006). Two other homeodomain factors, Meox2 and Msx1, are also expressed in migratory limb myogenic progenitor cells. In Meox2 mutants, Myf5 activation in the limb buds is delayed and there are later muscle defects, attributed to secondary effects of Meox2 in connective tissue (Mankoo et al., 1999). Meox2 binds in vitro to the Xvent2 sequence, but when a BAC transgene encompassing the entire Mrf4/Myf5 locus and regulatory regions was placed in a Meox2 mutant background no effects on Myf5-nLacZ expression were detected (Buchberger et al., 2007). Msx1 is expressed in mesenchymal cells throughout the early forelimb bud, and also in the myogenic progenitor cells that migrate from the mouse somite to the forelimbs (Houzelstein et al., 1999). In contrast, Msx2 has a distinct expression pattern but its expression in myogenic cells has not been reported. Msx1 is known to inhibit myogenic differentiation (Song et al., 1992) and MyoD activation (Woloshin et al., 1995) when over-expressed in cultured muscle cells and has been shown to recruit the repressive polycomb complex (Wang et al., 2011). ChIP Seq experiments in an ex vivo over-expression context, show Msx1 binding to the MyoD core enhancer and to the −57/−58 kb Myf5 regulatory region. Over-expression of Msx1 in the chick limb bud prevented MyoD activation and resulted in reduced skeletal muscle formation, with a proposed mechanism through direct repression by Msx1 binding to the MyoD core enhancer and also by Msx1/Pax3 complex formation that inhibits Pax3 binding to its targets (Bendall et al., 1999). We therefore decided to investigate more closely the potential role of Msx1 and Meox2 homeodomain factors in modulating the activity of the 145-bp core element of the Myf5 limb enhancer in myogenic progenitor cells that migrate to the forelimb, at the onset of myogenesis in the mouse embryo.
RESULTS
Meox2 and Msx homeoproteins bind in vitro to the 145-bp Myf5 enhancer
We first examined the 145-bp Myf5 regulatory element for homeodomain consensus binding (HBox) sequences (Noyes et al., 2008), in addition to the binding sites for Pax3 (Bajard et al., 2006) and Six1/4 (Giordani et al., 2007) which are known to be functionally important. Three binding sites, HBox1, 2 and 3 were identified (Fig. 1A), where HBox2 is part of the previously described Xvent2 site (Buchberger et al., 2007) and HBox1 and 3 are located 5′ and 3′ respectively of the central Pax3-HBox2-Six1/4 domain. Electrophoretic mobility shift assays (EMSA) show that Meox2 protein interacts with all three HBox sequences (Fig. 1B). Specificity of interaction is shown by the presence of supershifted bands with anti-Meox2 antibodies (Fig. 1B, lanes 2) and by competition with an excess of unlabelled probe, but not with a probe in which the HBox sequence is mutated (see Fig. 1B, lanes 3, 4 for HBox2 example). Meox2 binding is abolished when the HBox sites are mutated (results not shown). Similar experiments with Msx1 and Msx2 proteins show that they bind HBox2 (Fig. 1C, lanes 2 and 4), but not HBox1 and 3 (results not shown). In the absence of reliable Msx antibodies, recombinant proteins with haemaglutinin (C-terminal-HA) or Flag (C-terminal-Flag) tags for Msx1 or Msx2 respectively were used. Antibodies against HA or Flag substantially reduced binding (Fig. 1C, lanes 3 and 5). In this case a supershift is not seen, presumably because the protein/DNA complex is disrupted. In conclusion, Meox2 protein can bind in vitro to HBox1, 2 and 3, whereas Msx1/2 binds only to HBox2 of the 145-bp Myf5 regulatory sequence.
Fig. 1.
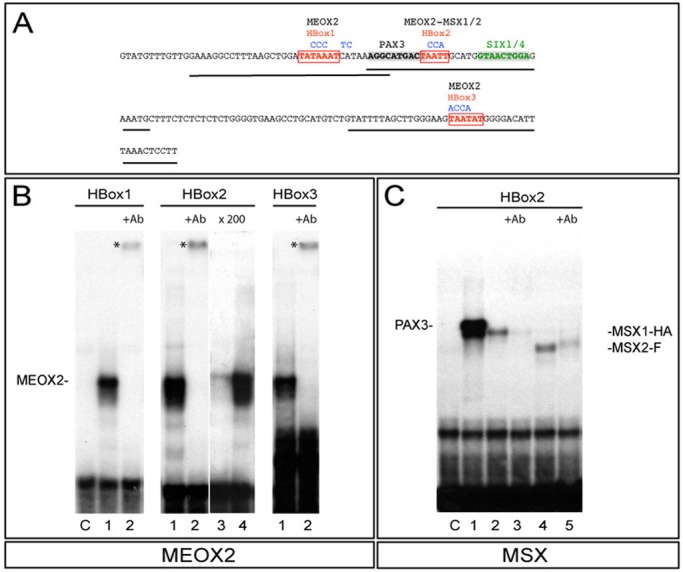
EMSA experiments showing that the 145-bp Myf5 element contains HBox sites to which Meox2 and Msx1/2 proteins bind in vitro. (A) The DNA sequence of the 145-bp Myf5 enhancer. HBox1-3 binding sites are in red. Blue letters above the sequence show nucleotide changes in mutated oligos used in EMSA. HBox1-3 DNA probes used in gel shifts are indicated by solid lines under the sequence. Pax3 (black) and Six1/4 (green) binding sites (Bajard et al., 2006; Giordani et al., 2007) are indicated in bold. (B) EMSA experiments were performed using 3 different DNA probes containing putative HBox1, 2 or 3 binding sites. In vitro synthesized Meox2 protein was added to the probes (lanes 1-2) and polyclonal anti-Meox2 antibodies were subsequently added (lanes 2). Each probe binds Meox2 protein and the complex is supershifted when antibodies are added (asterisks). Competition with unlabelled probe is shown for HBox2 (lanes 3, 4). A 200 molar excess of unlabelled probe competes the binding (lane 3), whereas unlabelled probe with a mutated binding site does not compete (lane 4). A control with crude reticulocyte lysate is shown in lane C. (C) Example of an EMSA experiment showing that Msx1 and 2 proteins can bind to the HBox2 probe. Binding occurs when Msx1-Cterm-HA (Msx1-HA) or Msx2-Cterm-Flag (Msx2-F) proteins are present (lanes 2 and 4) and specificity is demonstrated by the reduction of the shifted bands when antibodies, anti-HA (lane 3) or anti-Flag (lane 5), are added. Controls are shown with crude reticulocyte lysate (lane C) or with Pax3 protein as a positive control (lane 1).
Meox2 or Msx1 do not bind in vitro simultaneously with Pax3 or Six proteins to the HBox2 region
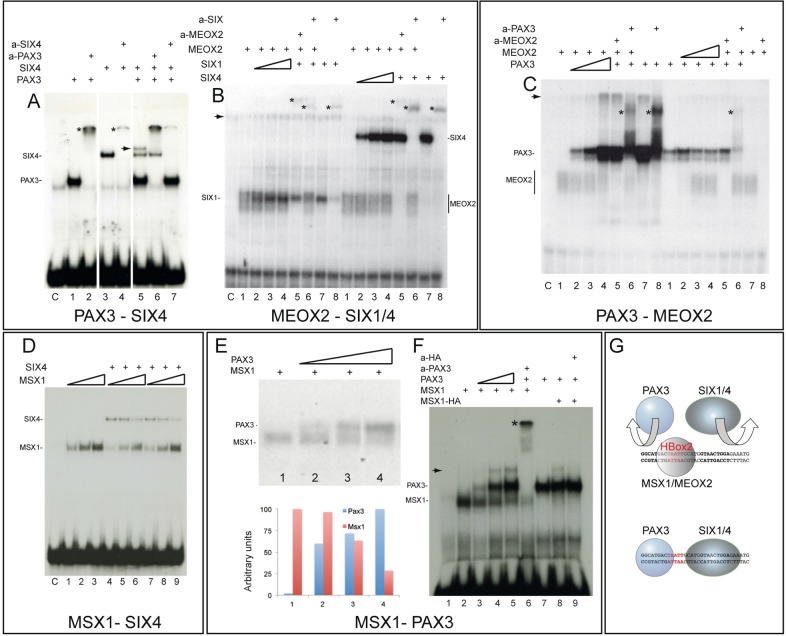
The close proximity of Pax3 and Six1/4 binding sites to the HBox2 site, which binds Meox2 and Msx proteins, raises the question of whether these proteins can bind simultaneously. We used limiting molar amounts of labelled DNA probe to test this in gel shift assays. Binding of Pax3 and Six4 together is demonstrated by the appearance of a supplementary slower migrating band (Fig. 2A, lane 5) which is disrupted when either anti-Pax3 or anti-Six4 antibodies are added (Fig. 2A, lanes 6, 7). A similar result was obtained with Pax3 and Six1 proteins (not shown). This indicates that Pax3 and Six proteins can bind together on the same Pax3/HBox2/Six DNA sequence. In contrast, when constant amounts of Meox2 are added, with increasing amounts of either Six1 or Six4, we found no evidence for co-binding on HBox2 and Six binding sites since no supplementary slower migrating band was detected (Fig. 2B). When constant amounts of Meox2 are added with increasing amounts of Pax3, or constant amounts of Pax3 with increasing amounts of Meox2, no supplementary band was detectable and we conclude that Pax3 and Meox2 cannot bind simultaneously on the same DNA sequence (Fig. 2C). Similar experiments with Msx1 and Six4 proteins (Fig. 2D) also indicated that Msx1 cannot bind together with Six4. In these experiments, competition for DNA binding was observed. When increasing amounts of Msx1 are added, Six4 binding is disrupted (Fig. 2D, lanes 4-6). This also occurs if Msx1 is added subsequently, after Six4 binding to the oligo (lanes 7-9). Competition is also observed between Pax3 and Msx1 and is illustrated in Fig. 2E, where increasing amounts of Pax3, relative to a constant amount of Msx1, led to a diminution of Msx1 binding. Again no additional slower migrating band was observed, even on long exposures of the autoradiogram (result not shown). In order to test whether steric hindrance is the cause of this competition, we engineered an elongated version of the probe where a mutated HBox2* sequence, which does not bind Msx but does not interfere with Pax3 binding (see Fig. 4), is introduced on both sides of the bona fide HBox2 site, increasing the distance between the Pax3 and Six1/4 sites. In this case, we can detect an additional slower migrating band, indicating that Pax3 and Msx1 are bound to the same elongated DNA sequence (Fig. 2F). We therefore propose that homeodomain protein binding to the HBox2 site interferes with binding of Pax3, which is essential for the function of the 145-bp Myf5 regulatory sequence (Bajard et al., 2006). Interference with Six binding will also have a negative impact on the activity of the limb enhancer (Giordani et al., 2007). This may be important in preventing premature activity of the 145-bp sequence in migrating myogenic progenitors which contain Pax3 and Six factors, but in which Myf5 is not yet transcribed (Fig. 2G).
Fig. 2.
EMSA experiments with combinations of proteins to test co-binding or competition on the HBox2 probe. The labelled HBox2 probe, which contains Pax3 and Six binding sites was used in limiting molar amounts, compared to the proteins added. (A) Pax3 and Six4 proteins can bind together to the same DNA sequence. Pax3 binding (lane 1) is supershifted with anti-Pax3 antibodies (lane 2, asterisk). Six4 binding (lane 3) is supershifted when anti-Six4 antibodies are present (lane 4, asterisk). When both proteins are present, an additional slower migrating band is detected (lane 5, arrow) and the intensity of the band corresponding to Six4 binding alone is reduced, compared to lane 3. When anti-Pax3 (lane 6) or anti-Six4 (lane7) antibodies are added, this band, which is therefore due to co-binding of Pax3 and Six4 on the same oligo, is disrupted and the bands due to Pax3 or Six4 binding alone are supershifted. Similar results were obtained with Pax3 and Six1, with disruption of the Pax3/Six1 complex with anti-Pax3 antibodies (results not shown). Lane C is the control with crude lysate. (B) Meox2 and Six1/4 co-binding is not detectable. EMSA was performed with constant amounts of Meox2 and increasing amounts of either Six1 (left side of panel) or Six4 (right side of panel). Presence of anti-Meox2 (lanes 5) or anti-Six1 (left, lanes 6,8) or Six4 (right, lanes 6,8) antibodies are indicated above the panel. No additional slower migrating band, suggesting co-binding of Meox2 and Six proteins, is detected. A weak band (indicated by an arrow) was detected in most samples, including the control with crude lysate (lane C). Asterisks indicate the position of bands supershifted by antibodies. (C) Meox2 and Pax3 co-binding is not detectable. In the presence of constant amounts of Meox2 (left side of panel), no Pax3 (lane 1) or increasing amounts of Pax3 were added (lanes 2-4). No supplementary band is detected when both proteins are present. Controls are shown with Pax3 alone (lane 7) or with Pax3 and anti-Pax3 antibody (lane 8) or lysate alone (C). In the presence of constant amounts of Pax3 (right side of panel), no Meox2 (lane 1) or increasing amounts of Meox2 were added (lanes 2-4). No additional slower migrating band is detected when both proteins are present in the binding reaction. A weak band (arrow) was detected in most samples, including those without Meox2 and appears to represent a Pax3 complex with this lysate. Addition of anti-Meox2 (lanes 5, 8) or anti-Pax3 (lane 6) antibodies disrupt the bandshifts and, in the case of Pax3, generate a supershift (asterisk). Controls are shown with Meox2 alone (lane 7) or with Meox2 and anti-Meox2 antibody (lane 8). (D) Msx1 and Six4 co-binding is not detectable. Increasing amounts of Msx1 alone (lanes 1-3) or with constant amounts of Six4 added at the same time (lanes 4-6) do not produce an additional slower migrating band when both proteins are present. Six4 binding is reduced when higher amounts of Msx1 are added (lanes 4-6). This displacement occurs even if Six4 protein is added before Msx1 (lanes 7-9). (E) Msx1 binding is displaced by increasing amounts of Pax3. When constant amounts of Msx1 without (lane 1) or with 2.5, 5, or 10 fold amounts of Pax3 (lanes 1-4) are added, reduction of Msx1 binding is seen as Pax3 levels increase, under gel conditions which facilitate distinction of Pax3 and Msx1 binding with lower levels of Pax3. This is confirmed by the histogram showing a scan of this autoradiograph with ImageJ 1.47v. (F) Msx1 and Pax3 can bind together when the spacing between the sites is increased. One copy of a mutated HBox2* site, which does not bind Msx1 (see Fig. 4), was intercalated on either side of the bona fide HBox2 site, thus increasing the distance between Pax3 and Six binding sites. In the presence of constant amounts of Msx1 (lanes 2-6) and increasing amounts of Pax3 (lanes 3-5), a supplementary slower migrating complex appears (arrow). When anti-Pax3 antibody is added, this band disappears and Pax3 complexes are supershifted (asterisk, lane 6). This band does not appear if Pax3 is added alone (lane 7) but is detected when Pax3 and Msx1-HA proteins are added together or sequentially (lane 8) and disappears if anti-HA antibodies are added (lane 9). Note that bands corresponding to Pax3 or Msx1-HA complexes alone migrate at almost the same positions as shown in Fig. 1 (G) The drawing resumes the competition between homeoproteins such as Msx1 and Meox2 and Pax3/Six proteins for in vitro binding to the oligonucleotide probe in the EMSA experiments presented. The binding of a homeoprotein at the HBox2 site (red) prevents the binding of Pax3 or Six1/4.
Fig. 4.
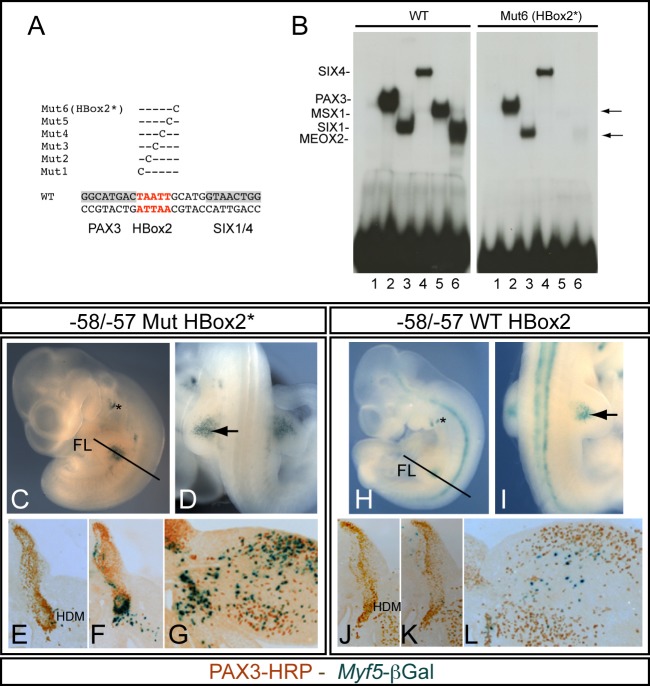
Mutation of HBox2, without affecting Pax3 and Six1/4 binding, results in premature activation of the −58/−57 kb Myf5 limb enhancer. (A). Single mutation scanning in the HBox2 binding site (red) region by successive replacements of nucleotides by a cytosine (C) (Mut1 to 6). Pax3 and Six1/4 binding sites are shown as shadowed boxes. (B) EMSA experiments were performed with the wild-type (WT) sequence or the mutated probe corresponding to Mut6 in order to test the binding of Pax3, Six1/4, Msx1 and Meox2 in vitro synthesized proteins. Compared with the wild-type sequence (WT), mutation 6 (Mut6-HBox2*) does not affect the binding of Pax3 (lanes 2), Six 1 (lanes 3) and Six4 (lanes 4), but compromises the binding of Msx1 (lanes 5) and Meox2 (lanes 6) (arrows). Controls with crude lysate are shown in lanes 1. (C-L) Examples of X-gal stained transient transgenic embryos at E10.5 (35/36 somites), obtained with a −58/−57baMyf5nLacZ transgene in which the 145-bp sequence within the −58/−57 region contains a mutated HBox2* sequence (C-G) or a wild-type HBox2 (H-L). Whole mount X-Gal stained embryos are shown in C and H, with close-ups of the forelimb region in D and I, where the arrow points to X-gal staining. FL, forelimb bud, asterisk shows branchial arch expression – this and variable neural tube expression are due to sequences in the Myf5 proximal promoter region used (baMyf5). (E-G,J-L) Serial cryostat sections at the forelimb level where the plane of section is shown by a line in C for E,F, and in H for J,K. Sections including the hypaxial somite and proximal forelimb are shown in G and L. These X-Gal stained sections (Myf5-β-Gal) were treated with anti-Pax3 antibodies, revealed by horse radish peroxidase (PAX3-HRP) to label Pax3-positive myogenic progenitors (HDM, hypaxial dermomyotome).
Meox2 and Msx1 bind the 145-bp Myf5 sequence in vivo
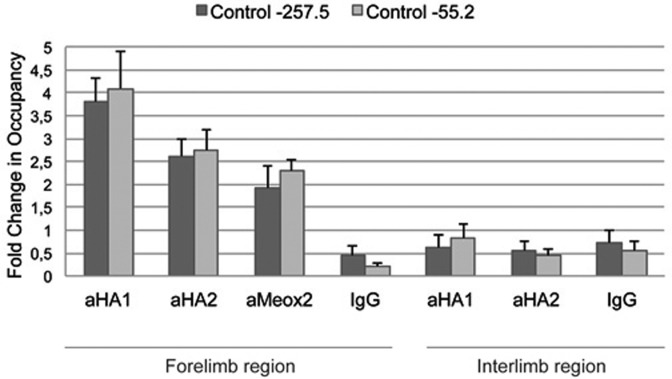
We next investigated Msx1 and Meox2 binding in vivo to the 145-bp Myf5 regulatory sequence by chromatin immunoprecipitation (ChIP) experiments, using sonicated chromatin prepared from the thoracic region of the trunk including the forelimbs of embryonic day (E)10-E10.5 mouse embryos. An anti-Meox2 antibody was used on wild-type embryos, and anti-HA antibodies were used to immunoprecipitate HA-tagged Msx1 protein prepared from Msx1Tag/Tag embryos. The Tag does not interfere with Msx1 function (Duval et al., 2014). ChIP results are presented as a ratio of binding to the 145-bp element at −57.5 kb compared to that obtained with two negative control regions at −275.5 kb and −55.2 kb upstream of the Myf5 locus, previously used as control for ChIP (Bajard et al., 2006; Giordani et al., 2007). Msx1 binding is not observed with chromatin from interlimb regions excluding limb buds, of E10.5 embryos (Fig. 3). These experiments therefore show in vivo binding of Msx1 and Meox2 to the 145-bp sequence at forelimb level.
Fig. 3.
Msx1 and Meox2 proteins interact in vivo with the 145-bp Myf5 regulatory sequence. ChIP experiments were carried out with two different anti-HA antibodies (aHA1-2), to immunoprecipitate chromatin prepared from the thoracic region of the trunk including forelimb buds (forelimb region), of E10-E10.5 Msx1Tag/Tag embryos, or with anti-Meox2 antibodies to immunoprecipitate chromatin prepared from the equivalent region of wild-type embryos at E10.5. Histograms represent the fold change in occupancy of the 145-bp Myf5 element, versus two different negative control regions located respectively at −257.5 kb (black) and −55.2 kb (grey) upstream of the Myf5 gene transcription start site (Bajard et al., 2006; Giordani et al., 2007). A control using IgGs from non-immune serum is shown for experiments with wild-type chromatin. Results on the right part of the figure were obtained with ChIP on extracts from interlimb regions, excluding limb buds, of Msx1Tag/Tag embryos at E10.5, using anti-HA antibodies or control non immune IgGs. Biological replicates of these ChIP experiments were carried out with three different preparations of chromatin. Error bars indicate s.e.m.
Functional importance of the HBox2 site for the correct timing of Myf5 activation in forelimb buds
Pax3, Six1/4, Meox2 and Msx1 are all expressed in myogenic progenitor cells of the forelimb (Houzelstein et al., 1999; Mankoo et al., 1999; Relaix and Buckingham, 1999) but Myf5 is not activated until these cells have migrated from the hypaxial somite into the forelimb bud, at the 35 somite stage (E10.5) (Tajbakhsh and Buckingham, 1994). Since both Meox2 and Msx1 proteins bind to HBox2, we investigated the importance of this binding site in regulating the activation of Myf5. We screened to find a mutation in or close to HBox2 that prevents Msx1 and Meox2 binding without disrupting the essential binding of Pax3 and Six1/4. Single nucleotide mutations were introduced (Fig. 4A) and the resulting DNA sequences tested for Pax3, Six1/4, Msx1 and Meox2 binding by EMSA. Mutations 2 and 3 abolished both Pax3 and Msx1 binding. Mutation 1 perturbed Msx1 binding and gave additional bands with the reticulocyte lysate. Mutations 4 and 5 both abolished Msx1 binding. In these mutations, Pax3 and/or Six1 binding was also affected (results not shown). This effect on Pax3 and Six1/4 binding probably explains the loss of Myf5 expression after mutating the Xvent2 site, reported by Buchberger et al. (2007). However we found that mutation 6 (named Mut6-HBox2*) does not perturb Pax3 or Six binding whereas the interaction with both Msx1 (lane 5) and Meox2 (lane 6) was severely compromised (Fig. 4B). The effect of this HBox2* mutation was then tested in vivo by transient transgenesis. For these experiments the −58/−57 kb region containing the 145-bp sequence, which gives robust expression, was placed in front of the Myf5 proximal promoter region and the nLacZ reporter. Embryos were collected at E10.5. The HBox2* mutation resulted in premature activation of the transgene, with excessive β-galactosidase labelling in the forelimb bud (Fig. 4C,D) compared to that obtained with the transgene containing a wild-type Hbox2 sequence (Fig. 4H,I). Labelling in the branchial arches and variable labelling in the neural tube is due to sequences present in the proximal promoter region (Carvajal et al., 2001) and was observed with both constructs (Table S2). On serial sections, X-gal staining shows labelled cells within and adjacent to the hypaxial dermomyotome (Fig. 4E,F), whereas such cells are very rare in controls (Fig. 4J,K). A magnified view of a section shows an accumulation of β-galactosidase positive cells adjacent to the somite in the proximal forelimb bud (Fig. 4G), whereas normally only a few dispersed β-galactosidase positive cells are present within the limb (Fig. 4L). Furthermore, with the HBox2* mutated transgene, many more β-galactosidase-positive cells are present within the limb. This indicates that premature activation of the transgene also extends to progenitors within the forelimb, where only a proportion of Pax3-positive myogenic progenitors normally activate Myf5 and enter myogenesis, while the rest provides a reserve cell population for future muscle growth (Buckingham and Relaix, 2007). We therefore conclude that the HBox2* mutation, which compromises homeobox protein binding, leads to precocious and ectopic activation of the Myf5 enhancer in myogenic progenitors of the forelimb bud in vivo.
Role of Msx proteins in the early repression of Myf5 in the hypaxial somite and forelimb bud
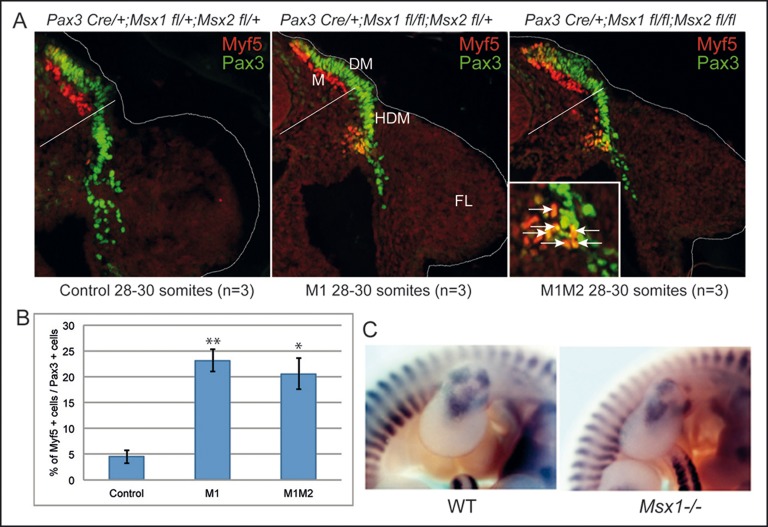
To test the potential role of the Msx1 protein in the delay of Myf5 gene activation in myogenic progenitor cells, we examined embryos in which Msx conditional alleles had been inactivated specifically in myogenic progenitor cells by the use of a Pax3Cre allele, at E9.75 (28-30 somites) when they have just begun to delaminate from the hypaxial dermomyotome and enter the forelimb bud. This is the time window when Msx1 is maximally expressed in the hypaxial dermomyotome at forelimb level (Houzelstein et al., 1999). The efficiency of Cre recombinase to recombine Msx1 floxed alleles in Pax3 expressing cells was assessed by Msx1 in situ hybridization on cryosections of Pax3Cre/+Msx1fl/+ and Pax3Cre/+Msx1fl/fl embryos at E10.5. The control in situ signal was too faint in the hypaxial somite, but it was clear that Msx1 transcripts were absent in Pax3Cre/+Msx1fl/fl embryos, in Pax3 expressing cells of the dorsal neural tube, a region where Msx1 is also expressed (Liem et al., 1995), whereas Msx1 transcripts are present in the non-myogenic distal mesenchyme of the forelimb (Fig. S1). In control embryos at this stage (Fig. 5A, left panel) only rare Myf5-positive (+) cells could be detected in the hypaxial dermomyotome and in adjacent delaminating cells. In contrast, in embryos where both Msx1 alleles have been recombined, Myf5 expression was observed within the Pax3-expressing population of cells in the hypaxial dermomyotome and in adjacent delaminating cells (Fig. 5A, middle panel). Pax3 expressing cells in the hypaxial region were counted as well as those co-expressing Myf5. 23.2% of Pax3+ cells are also Myf5+ when Msx1 alleles are inactivated, compared to 4.5% in control embryos. Inactivation of both Msx1 and Msx2 alleles gave a similar percentage of Myf5+ cells/Pax3+ cells (20.6%) (Fig. 5A, right panel; Fig. 5B). We also examined sections at forelimb bud level at E10.25 (34-36 somites). Pax3+ cells co-expressing Myf5 are clearly detected in the dorsal-most part of the forelimb bud, and counting of these cells showed a similar tendency between control versus Msx1 mutant embryos (9.5% of Myf5+ cells/Pax3+ cells, vs 11.8% - results not shown). At later stages, differences in the forelimbs between mutant and wild type were less evident, as shown by Myf5 in situ hybridisation for dorsal muscle masses at E11.5 (Fig. 5C). By this stage, as previously shown by Houzelstein et al. (1999), domains of Myf5 expression in the forelimb are adjacent to those where Msx1-nLacZ is expressed, Myf5 being expressed in future ventral and dorsal muscle masses and Msx1 in the distal mesenchyme. We conclude that lack of the Msx1 protein in vivo leads to premature onset of Myf5 expression in early Pax3-positive myogenic progenitors as they delaminate from the hypaxial dermomyotome, but does not affect Myf5 transcription in those cells that continue to migrate into the developing forelimb bud and does not affect later myogenesis.
Fig. 5.
Msx1 Cre-mediated inactivation in Pax3-positive cells leads to premature expression of Myf5 in the hypaxial somite. (A) Immunochemistry on transverse cryosections at forelimb level of Pax3Cre/+;Msx1fl/+;Msx2fl/+(control, left), Pax3Cre/+;Msx1fl/fl;Msx2fl/+ (M1, middle) and Pax3Cre/+;Msx1fl/fl;Msx2fl/fl (M1M2, right) embryos at E9.75 (28-30 somites), showing merged images after immunostaining with anti-Pax3 (green) and anti-Myf5 antibodies (red). Contrary to control embryos, double positive Myf5+/Pax3+ cells are found in the region of the hypaxial dermomyotome (HDM), indicated by white arrows in the higher magnification shown for the M1M2 embryo. White lines in merged images represent the upper limit (corresponding to the ventral part of the neighbouring neural tube) below which Myf5+ cells in the Pax3+ population were counted. The contour of the embryos is marked by a white line. FL, forelimb; DM, dermomyotome; M, myotome. (B) Histogram representing the percentage of Myf5+ cells among the Pax3+ cells counted in cryosections equivalent to those shown in (A), with 870-960 Pax3+ cells counted from three embryos for each genotype. Error bars indicate s.e.m. 0.01<*P<0.05; 0.001<**P<0.01; versus control. (C) Comparison of wild-type Msx1+/+ and Msx1nLacZ/nLacZ (Msx1−/−) mutant embryos hybridized with an anti-sense Myf5 riboprobe. Whole mount lateral enlarged views at the forelimb level of embryos at E11.5 are shown.
Meox2 is necessary for the early activation of the 145-bp Myf5 regulatory element
Since the Meox2 protein can bind in vitro to the three HBox sites of the 145-bp sequence and because Meox2 is also present in limb myogenic progenitors in vivo, we examined the regulation of early Myf5 expression by this factor. We crossed Myf5nLacZ/+ mice (Tajbakhsh et al., 1996) with the Meox2+/− mouse line (Mankoo et al., 1999) and examined expression between E10.5 and E12.5, when one or two alleles of Meox2 are inactivated. There was a clear delay in nLacZ expression in forelimb buds of the Meox2 homozygous mutant versus heterozygous embryos at 36-39 somite stages (Fig. 6A-C compared with D-F). Our observations correlate with the decrease in Myf5 transcripts reported at E10.5 in forelimb buds of Meox2−/− mutant embryos (Mankoo et al., 1999). We then crossed a transgenic 145-baMyf5nLacZ mouse line, with the Meox2+/− line and observed the same delay in nLacZ expression in forelimbs of Meox2−/− mutants at E10.5 (Fig. 6G-L), indicating that the onset of Myf5 expression in the forelimbs depends on activation of the 145-bp Myf5 sequence by Meox2 in Pax3-positive myogenic progenitor cells, which are also clearly present in the mutant (Fig. S2). This delay does not persist at later stages of development, when 145-baMyf5-nLacZ expression in forelimb buds is observed in Meox2−/− embryos at E12.5 (Fig. S3), consistent with previous observations (Buchberger et al., 2007). At E12.5, expression of the 145-baMyf5nLacZ transgene in the hindlimb was delayed in the mutant (Fig. S3). This demonstrates that the mechanism of Myf5 activation by Meox2 is common to fore- and hindlimbs. In order to determine whether this is a direct effect, we examined transgene function when HBox1, 2 and 3, that bind Meox2, are mutated, using the HBox2* mutation which does not disrupt the binding of Pax3 and Six1/4. These mutations in the 145-bp sequence were tested in the context of the −58/−57 kb Myf5 enhancer. Compared with the non-mutated -58/-57baMyf5nLacZ transgene (Fig. 6M), the nLacZ reporter was not expressed in forelimb buds at E10.5 when all 3 HBox sites were mutated (Fig. 6N,O). These results demonstrate the role of Meox2 in the direct activation of Myf5 through the 145-bp sequence in limb buds at the onset of myogenesis.
Fig. 6.
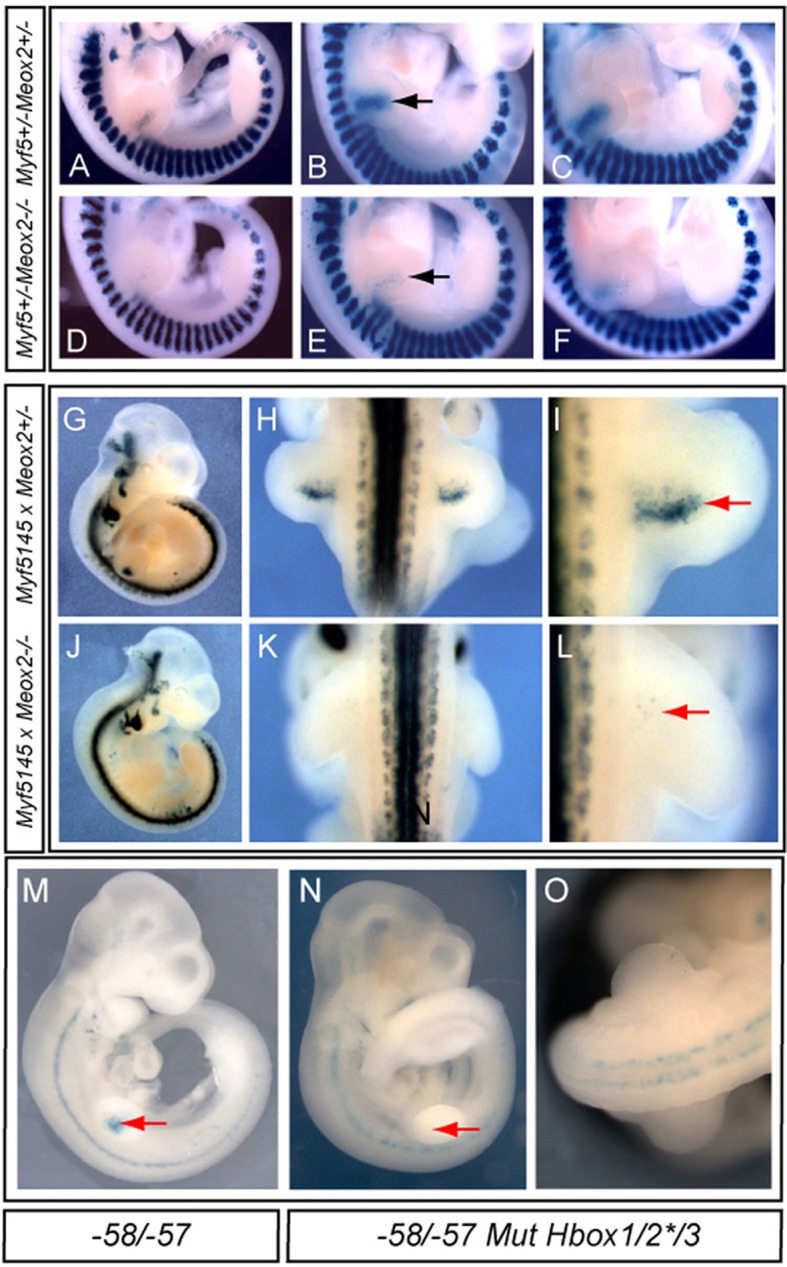
Meox2 inactivation induces a delay in Myf5 expression in forelimb buds, which operates via HBox sequences in the 145-bp Myf5 element. (A-F) Lateral views showing X-Gal staining at 36 (A,D), 37 (B,E) and 39 (C,F) somite stages of Meox2+/−Myf5+/nLacZ (Myf5+/−A-C) or Meox2−/−Myf5+/nLacZ (Myf5+/− D-F) embryos. Activation of the Myf5nLacZ allele in myogenic progenitor cells in the forelimb bud is delayed in the absence of Meox2 (arrows in B,E). (G-L) X-Gal stained Meox2+/− (G-I) and Meox2−/− (J-L) embryos (E10.5) obtained after crossing with a transgenic 145-baMyf5nlacZ line, with magnifications of the dorsal aspect of the forelimbs shown in H,I,K,L. There is a striking reduction in β-galactosidase positive cells in the mutant at this stage (red arrows). (M-O) X-Gal stained transient transgenic embryos at E10.5, with a −58/−57baMyf5nlacZ transgene (−58/−57) (M) or with the same transgene in which HBoxes 1-3 sequences in the 145-bp sequence have been mutated (−58/−57 Mut HBox1/2*/3) (N,O). An enlargement at the forelimb level is shown in O. When all 3 HBox sequences are mutated transgene expression in the forelimb (red arrow) is absent at E10.5. Expression in the branchial arches and neural tube is due to sequences in the proximal promoter region of Myf5 (baMyf5), present in the transgene.
DISCUSSION
The Myf5 myogenic determination gene is not activated in limb muscle progenitor cells when they delaminate from the somite and migrate to the limb bud, despite expression of Pax3 and Six1/4 which can activate the Myf5 limb regulatory element. In this context, we show that direct binding of Msx1 and Meox2 controls the precise onset of Myf5 activation in the forelimb bud in vivo. Msx1 prevents precocious activation by competing Pax3 and Six1/4 binding while Meox2 is required to initiate expression once cells have reached the forelimb buds.
We have characterised three homeodomain-binding sites present in this 145-bp sequence, located within an enhancer at −58/−57 kb from the Myf5 gene. We show that all three HBox sites bind Meox2 in vitro and that Meox2 is bound to the 145-bp sequence in vivo in preparations from the trunk region, including forelimbs, of E10.5 embryos. At this stage, mutation of all three HBox sites results in a loss of activity of the limb enhancer in −57/−58baMyf5nLacZ transgenic embryos. Pax3 and Meox2 proteins are co-expressed in cells that leave the hypaxial dermomyotome and migrate into the limb (Mankoo et al., 1999). These authors also reported delayed activation of Myf5 transcription in the early forelimb bud in the absence of Meox2. We now confirm this with the β-galactosidase reporter from a Myf5nLacZ allele and conclude from our transgenic experiments that the HBox sequences in the 145-bp element are implicated in this delay in –57/–58baMyf5nLacZ transgenic embryos. This delay is not due to interference with essential Pax3 or Six1/4 binding to adjacent sites, since the HBox2* mutation used permits Pax3 and Six1/4 binding in vitro and this mutation alone results in premature activation of the transgene. Pax3 and Meox2 proteins have been shown to interact in vitro with each other (Stamataki et al., 2001), however in EMSA experiments we saw no indication of co-binding of Meox2 and Pax3 or Six proteins to the Hbox2 region. In Meox2 mutants, there is a reduction in Pax3 transcripts (Mankoo et al., 1999) and a lower level of Pax3 may also impact the −145-bp enhancer, however from our transgenic experiments we conclude that Meox2 directly affects Myf5 transcription. In the absence of Meox2, the delay in activation of the transgene corresponds to a four somite interval at E10.5. The low level of expression of the 145-Myf5nLacZ transgene in forelimb buds of Meox2−/− E10.5 embryos is not due to a lack of myogenic progenitor cells because these are clearly detected at a similar stage using Pax3 whole mount in situ hybridization (Fig. S2). Buchberger et al. (2007), who did not detect a difference in Meox2 mutants, probably missed the delay we observe at E10.5 by examining E11.5-E13.5 embryos. We found that the delay of Myf5 activation in Meox2 mutants, occurring via the 145-bp element, is compensated in forelimbs by E12.5 and indeed no difference in Myosin Heavy Chain expression was observed in Meox2 mutant limbs (B.S.M., unpublished). At E12.5, the delay is still observed in hindlimbs, which suggests that the mechanism of early Myf5 activation by Meox2 is similar in fore- and hindlimbs. Since mutation of Hbox1, Hbox2 and Hbox3 sites prevents the correct onset of transgene expression, while the HBox2* mutation alone leads to premature activation of the transgene, we conclude that HBox1 and HBox3 are the sites implicated in activation by Meox2, acting together with Pax3 and Six1/4 on their respective binding sites. This suggests that these factors, and especially Pax3, which is critical for activation, compete favourably over Meox2 binding to HBox2 at the onset of myogenesis, thus avoiding the steric hindrance seen when this site is occupied by Msx1. Our results provide the first demonstration that Meox2 directly activates a skeletal muscle determination gene, fine-tuning the onset of myogenesis in the limb. Subsequently Meox2 is not required in this context and other factors may intervene, such as Mef1 or NFat which also have binding sites in the 145-bp sequence (Buchberger et al., 2007).
Msx1, which is also expressed in myogenic progenitors that migrate to the forelimb (Houzelstein et al., 1999), binds in vitro to HBox2, but not to HBoxes 1 and 3. The HBox2 sequence is identical to the consensus site for Msx1 binding to DNA characterized in vitro (Catron et al., 1993; Hovde et al., 2001). ChIP experiments confirm that Msx1 binds in vivo to the 145-bp sequence, presumably at HBox2, at a stage when myogenic progenitor cells delaminate from the hypaxial dermomyotome and enter the forelimb bud from the somite, prior to Myf5 activation. Msx1 is only expressed in the hypaxial dermomyotome at this axial level, as confirmed by ChIP experiments on interlimb somites which show no occupancy by Msx1. Transplantation of early forelimb level somites from Msx1nlacZ/+ embryos into chick embryos at the same axial level showed that β-galactosidase-positive cells subsequently left the somites and entered the limb bud (Houzelstein et al., 1999), leading us to conclude that we are dealing with limb muscle progenitors. We saw no evidence for formation of an inactivating complex between Pax3 and Msx1, in the 145-bp Myf5 context, as reported previously for the MyoD enhancer (Bendall et al., 1999). Instead we show that Msx1 binding to HBox2 competes with binding of Pax3 or Six1/4, respectively to sites 5′ and 3′of HBox2, due to steric hindrance since this is alleviated by increasing the distance between these sites. Thus Msx1 binding to HBox2 prevents binding of essential activators and also probably exerts a direct repressive effect by recruiting Polycomb as shown for MyoD (Wang et al., 2011) and/or by recruitment of methyltransferase G9a and repressive H3K9me2 as shown for this Myf5 regulatory region in C2 muscle cells overexpressing Msx1 (Wang and Abate-Shen, 2012). In transgenic analysis, when HBox2 is mutated, premature activation of Myf5 in the hypaxial somite region and in myogenic progenitor cells in the forelimb bud is observed, consistent with a role for Msx1 in repressing Myf5 activation via this site. Expression of the Msx1lacZ reporter allele in the Pax3-positive myogenic progenitors, is rapidly lost when they reach the proximal region of the limb where myogenic cells accumulate (Houzelstein et al., 1999). Thus we propose down-regulation of Msx1 releases repression and permits Pax3 and Six1/4 binding. This repression by Msx1 on Myf5 and also on MyoD (Bendall et al., 1999), will effectively prevent the premature onset of myogenesis. When we examine the phenotype of conditional Msx1flox/flox embryos where the floxed alleles are inactivated by Cre recombinase in Pax3-expressing cells (Pax3Cre/+), we do not detect a major difference in Myf5 activation in these cells in the developing forelimb, but we see significantly more Myf5-positive cells in the hypaxial dermomyotome as well as immediately adjacent to this part of the somite at forelimb bud level, at the onset of delamination and migration. Similar results were obtained with Msx1flox/flox; Msx2flox/flox embryos, indicating that Msx1 is primarily responsible and indeed Msx2 expression has not been reported in somites (Bensoussan et al., 2008). This Msx1 mutant phenotype is transitory, most evident at the 28-30 somite stage. By E11.5, we did not detect an increase in Myf5-positive cells in the forelimb. In contrast, an increase in Myf5 transcripts had been reported in the forelimbs of Msx1 mutant embryos at this stage (Wang et al., 2011), however, since the in situ signal was generally higher, it is not clear whether the control and mutant embryos were strictly comparable. Since the effect that we observe is transitory and limb muscle defects have not been reported in the absence of Msx1 (Houzelstein et al., 1997; Satokata and Maas, 1994), it is probable that other factors also intervene to repress premature Myf5 activation. Up-regulation of the Myf5 transgene in myogenic progenitors in the forelimb, as well as the hypaxial somite region, when Hbox2 is mutated suggests that another homeodomain binding factor may be involved. Lbx1 is a potential candidate, however we did not observe binding of Lbx1 in EMSA experiments (results not shown). Furthermore, although binding might have exerted steric hindrance, Lbx1 appears to be an activator in the myogenic context (Vasyutina et al., 2005). Other repressive mechanisms may also operate, to prevent premature transcription of Myf5. In addition to the 145-bp element within the −58/−57 kb sequence used to test the functional effect of mutating HBox2, a second conserved region contains a Smad binding site, which, when mutated, led to ectopic Myf5 activation in the vicinity of the somite (Buchberger et al., 2007). Bmp4 in lateral mesoderm adjacent to the hypaxial somite has been shown to prevent premature activation of MyoD in the chick embryo (Pourquié et al., 1996) and it is therefore likely that BMP signalling acting through the Smad site also re-inforces repression of Myf5 transcription during the migration of myogenic progenitor cells to the limb. In these cells, MyoD transcription has also been shown to be regulated by transcriptional repression exerted by the bHLH-PAS transcription factor Sim2, acting on the MyoD core enhancer (Havis et al., 2012). Repression of Myf5 was not observed when Sim2 was overexpressed under conditions where MyoD was downregulated in chick and Xenopus embryos (Havis et al., 2012). This suggests that the two myogenic determination genes are repressed by a combination of different mechanisms, to prevent premature entry into the myogenic programme.
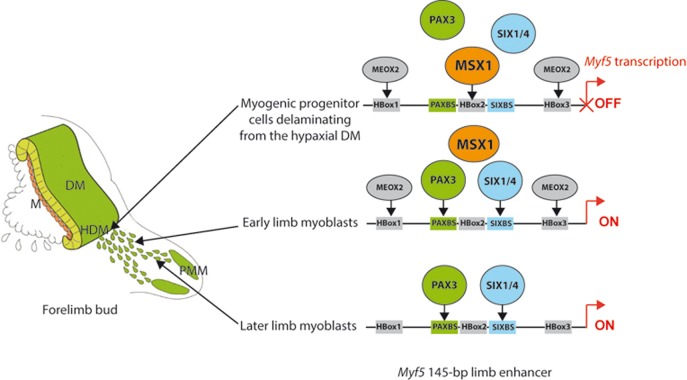
In conclusion, fine-tuning of the onset of Myf5 activation in myogenic progenitor cells that migrate from the somite to the forelimb depends on the binding of homeodomain factors that inhibit or activate transcription via the 145-bp regulatory sequence at −57.5 kb. We propose that inhibition is normally exerted by binding of Msx1 that interferes with the binding of Pax3 and Six1/4 and may also act as a transcriptional repressor. This prevents premature activation of this myogenic determination gene in Pax3-positive progenitor cells which would compromise the correct localisation of skeletal muscles and also the maintenance of myogenic progenitors required for subsequent muscle growth. In addition to Pax3 and Six1/4 activation of the 145-bp limb element, the initiation of Myf5 transcription also depends on Meox2 that binds to additional homeodomain sites. A model recapitulating the onset of Myf5 gene activation during forelimb development is shown in Fig. 7. This example, for a myogenic determination gene, illustrates the importance of transcriptional fine-tuning to ensure the precise spatiotemporal expression of key regulatory factors during development.
Fig. 7.
Model for activation of Myf5 transcription via the 145-bp core element, during early forelimb bud development. Binding of Msx1 interferes with binding of Pax3 and Six1/4 to the enhancer, and this prevents premature activation of Myf5 expression in myogenic progenitors in the hypaxial somite. Binding of Pax3 and Six1/4 in conjunction with Meox2 binding to flanking homeobox sites in the enhancer is required for normal activation of Myf5 expression. Maintenance of Myf5 expression in the myoblasts once they have migrated into the limb is dependent on Pax3 and Six1/4 but is independent of Meox2 activity. Abbreviations: DM, dermomyotome; M, myotome; HDM, hypaxial dermomyotome; PMM, pre-muscle masses.
MATERIALS AND METHODS
Plasmid constructions used for transgenesis
All plasmid constructs used for transgenesis are derived from plasmid pbaMyf5-nLacZ (Hadchouel et al., 2000), p−58/−57 and p−145-baMyf5nLacZ (Bajard et al., 2006; Hadchouel et al., 2003) constructs. Mutagenesis of homeodomain binding sites (HBox) was first performed by PCR amplification with the Expand High Fidelity PCR System (Roche), using as a template a plasmid in which the −58/−57 fragment had been subcloned into the pGEMT-Easy vector (Promega) and two primers, forward and reverse, complementary to their 5′ extremities as described in Daubas et al. (2009). Mutations were successively carried out in HBox2, 3 and 1 using primers as described in Table S1. Then a HBox2* mutation was introduced into the −58/−57 fragment, mutated in HBoxes 1, 2 and 3 using a primer HBox2* (see Table S1) and the QuickChange Multi Site-Directed Mutagenesis kit (Stratagene). Mutations were checked by DNA sequencing (GATC Biotech) and the mutated fragments were isolated from pGEMT-Easy plasmids (Promega) and recloned into the pbaMyf5-nLacZ vector.
Mouse lines, transgenesis and embryo analysis
The Meox2+/− and Msx1nLacZ:+ mouse lines have been described in Mankoo et al. (1999) and Houzelstein et al. (1997). The Msx1Tag/Tag mouse line was produced by inserting HA Tag in-frame in front of the Msx1 stop codon followed by an IRES-nls mCherry cassette. After ES cell selection and blastocyst injection, F1 mice born from germline chimaeras were bred with FLPe transgenic mice to remove the neomycin cassette. Heterozygous Msx1Tag/+ and homozygous Msx1Tag/Tag mice were viable and fertile, indicating that the Msx1Tag allele is functional (Duval et al., 2014). Immunodetection of the HA Tag in E10.5 Msx1Tag/+ embryos showed correct expression. Conditional Msx1 (a kind gift of Dr Robert Maxson, Los Angeles, CA, USA) and Msx2 floxed alleles are respectively described in Fu et al. (2007) and Bensoussan et al. (2008). The Pax3Cre/+ mouse line is described in Engleka et al. (2005). Transgenic mice were obtained on a C57BL/6JxSJL genetic background. Experiments were performed in accordance with the European Community guidelines (2010/63/UE) and with French national regulations for the care and use of laboratory animals. All transgenesis experiments were carried out by injecting vectorless plasmid inserts and performed by the Centre d'Ingénierie Génétique Murine of the Pasteur Institute. The −58/−57baMyf5nLacZ transgenic mouse line was as described previously (Hadchouel et al., 2003). Hetezygous transgenic males were crossed with non-transgenic females (C57BL/6JxSJL F1). Embryos were staged taking E0.5 as the day of the vaginal plug and somites were counted for more precise staging. Most functional studies were carried out by transient transgenesis. Embryos were screened for transgene expression by X-Gal staining as described in Tajbakhsh et al. (1996). The number of transgenic embryos analysed in each experiment is indicated in Table S2.
Electrophoretic mobility shift assays
A 1044 bp SphI-XhoI fragment corresponding to mouse Meox-2 cDNA was blunted and cloned into the EcoRV site of the pcDNA3 plasmid (Invitrogen). The Pax3 expression plasmid was a gift from F. Relaix (Paris Est-Créteil University, Créteil) and consisted of a full length mouse Pax3 cDNA cloned into a pcDNA3.1 vector (Invitrogen). Six1 and Six4 expression vectors were gifts from P. Maire (Institut Cochin, Paris). Full length Six1 and Six4 cDNAs were cloned into the pCR3 CMV T7 expression vector (Invitrogen), as described in Spitz et al. (1998). Plasmid constructions for recombinant Msx1-C-HA and Msx2-C-Flag proteins were designed by N.D. Fragments corresponding to full length coding regions of Msx1-C-HA and Msx2-C-Flag were subcloned into pcDNA3.1+ (Invitrogen) and pCMV-TnT (Promega) vectors, respectively. Expression plasmid DNAs were used to synthesize proteins in vitro with the TnT T7 Quick Coupled Transcription/Translation System (Promega). Electrophoretic mobility shift assays (EMSA) were performed as described in Bajard et al. (2006). P32-γATP labelled oligos, containing HBox1, 2 or 3 are listed in Table S1, as well as the elongated core enhancer probe, LongPaxHBox2Six (containing spacers between the HBox2 site and Pax3/Six binding sites). Except when specified otherwise, proteins to be tested together were added simultaneously. One microliter of either anti-HA (Roche Applied Science), anti-FLAG M2 (Sigma), rabbit polyclonal anti-Meox2 (B. Mankoo), mouse monoclonal anti-Pax3-C (DSHB) or rabbit anti-Six4 (Sigma) were added to EMSA binding reactions for supershift assays.
Immunocytochemistry
Immunodetection was performed on X-Gal stained 20 µm cryostat sections, using monoclonal mouse anti-Pax3-C antibodies (DSHB) and the Vector M.O.M (Mouse on Mouse) Peroxidase kit (Vector Laboratories). Sections of control and Msx mutants shown in Fig. 5A were 10 µm thick. Monoclonal mouse anti-Pax3-C antibodies (1:250, DSHB), rabbit polyclonal anti-Myf5-C20 (1:250, Santa Cruz), Alexa Fluor 488 Anti-Mouse IgGs and Alexa Fluor 546 Anti-Rabbit IgGs (1:500, Molecular Probes) were employed for immunofluorescence experiments. Nuclear staining was with Hoechst solution.
In situ hybridisation
Whole-mount in situ hybridization on mouse embryos was performed as described in Daubas et al. (2000) using digoxigenin-labelled antisense Myf5 (Ott et al., 1991) or Pax3 (kindly provided by Dr P. Gruss, Max Planck Institute, Gottingen) riboprobes. Automated in situ hybridization on cryostat embryo sections was performed with an InsituPro VSi apparatus (Intavis Bioanalytical Instruments) using a digoxigenin-labelled antisense Msx1 riboprobe (Lyons et al., 1992).
ChIP-qPCR
The trunk at forelimb level, including forelimbs, or interlimb regions were dissected from E10-10.5 mouse embryos and the protocol used for chromatin immunoprecipitation (ChIP) and qPCR analysis was as described in Daubas and Buckingham (2013). The following antibodies were used: rat monoclonal anti-HA (high affinity, clone 3F10, Roche – aHA1), rabbit polyclonal anti-HA (ChIP grade, Abcam – aHA2), rabbit polyclonal anti-Meox2 (B.S.M.), rabbit polyclonal anti-histone H3 (tri-methyl K4) (ChIP grade, Abcam) and IgG from rabbit serum (Sigma). qPCR primer sequences are listed in Table S1. All analyses were carried out in 96-well plates using a StepOnePlus PCR machine (Applied Biosystems) and the FastStart Universal SYBR Green Master (Rox) (Roche). qPCR reactions were performed in triplicate with each of 3 different preparations of chromatin prepared either from forelimb or interlimb regions of pooled embryos. 5 µl of DNA solution were used per reaction, corresponding to 1 or 0.1 µl of immunoprecipitated DNA or Input DNA, respectively. Standard curves of all primers were performed to check for efficient amplification (above 90%). Melting curves were also performed to verify production of single DNA species with each primer pair. Relative levels of expression in each assay were obtained through the ΔΔCt method (Livak and Schmittgen, 2001). Fold changes in occupancy, compared to negative control regions, are equal to 2−ΔΔCt.
Acknowledgements
We thank Drs P. Maire (Institut Cochin, Paris) and F. Relaix (Paris Est-Créteil University, Créteil) for Six1, Six4 and Pax3 expression vectors and Dr R. Maxson (University of Southern California, Los Angeles) for the Msx1flox mouse strain. Drs J. F Ouimette and C. Rougeulle are acknowledged for their advice on ChIP experiments. C. Bodin provided tissue cryostat sectioning. Dr P. Maire is acknowledged for his constant support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
P.D. and M.B. designed the experiments. P.D., N.D. and B.S.M. performed the experiments. P.D. and M.B. wrote the paper, with contributions from L.B. and B.R.
Funding
This work was supported by the Pasteur Institute and the Centre national de la recherche scientifique and by grants from the Association Française contre les Myopathies (AFM), and European Union programmes: EuroSyStem [Grant no. 200720], Optistem [Grant no. 223098 and MYORES [Grant no. 511978].
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/suppl/doi:10.1242/bio.014068/-/DC1
References
- Anderson C., Williams V. C., Moyon B., Daubas P., Tajbakhsh S., Buckingham M. E., Shiroishi T., Hughes S. M. and Borycki A.-G. (2012). Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity. Genes Dev. 26, 2103-2117. 10.1101/gad.187807.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajard L., Relaix F., Lagha M., Rocancourt D., Daubas P. and Buckingham M. E. (2006). A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 20, 2450-2464. 10.1101/gad.382806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall A. J., Ding J., Hu G., Shen M. M. and Abate-Shen C. (1999). Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development 126, 4965-4976. [DOI] [PubMed] [Google Scholar]
- Bensoussan V., Lallemand Y., Moreau J., Cloment C. S., Langa F. and Robert B. (2008). Generation of an Msx2-GFP conditional null allele. Genesis 46, 276-282. 10.1002/dvg.20390 [DOI] [PubMed] [Google Scholar]
- Bladt F., Riethmacher D., Isenmann S., Aguzzi A. and Birchmeier C. (1995). Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376, 768-771. 10.1038/376768a0 [DOI] [PubMed] [Google Scholar]
- Braun T., Rudnicki M. A., Arnold H.-H. and Jaenisch R. (1992). Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell 71, 369-382. 10.1016/0092-8674(92)90507-9 [DOI] [PubMed] [Google Scholar]
- Buchberger A., Freitag D. and Arnold H.-H. (2007). A homeo-paired domain-binding motif directs Myf5 expression in progenitor cells of limb muscle. Development 134, 1171-1180. 10.1242/dev.02798 [DOI] [PubMed] [Google Scholar]
- Buckingham M. (2001). Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11, 440-448. 10.1016/S0959-437X(00)00215-X [DOI] [PubMed] [Google Scholar]
- Buckingham M. and Relaix F. (2007). The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell Dev. Biol. 23, 645-673. 10.1146/annurev.cellbio.23.090506.123438 [DOI] [PubMed] [Google Scholar]
- Buckingham M. and Vincent S. D. (2009). Distinct and dynamic myogenic populations in the vertebrate embryo. Curr. Opin. Genet. Dev. 19, 444-453. 10.1016/j.gde.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Carvajal J. J. and Rigby P. W. J. (2010). Regulation of gene expression in vertebrate skeletal muscle. Exp. Cell Res. 316, 3014-3018. 10.1016/j.yexcr.2010.07.005 [DOI] [PubMed] [Google Scholar]
- Carvajal J. J., Cox D., Summerbell D. and Rigby P. W. (2001). A BAC transgenic analysis of the Mrf4/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development 128, 1857-1868. [DOI] [PubMed] [Google Scholar]
- Catron K. M., Iler N. and Abate C. (1993). Nucleotides flanking a conserved TAAT core dictate the DNA binding specificity of three murine homeodomain proteins. Mol. Cell. Biol. 13, 2354-2365. 10.1128/MCB.13.4.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubas P. and Buckingham M. E. (2013). Direct molecular regulation of the myogenic determination gene Myf5 by Pax3, with modulation by Six1/4 factors, is exemplified by the -111kb-Myf5 enhancer. Dev. Biol. 376, 236-244. 10.1016/j.ydbio.2013.01.028 [DOI] [PubMed] [Google Scholar]
- Daubas P., Tajbakhsh S., Hadchouel J., Primig M. and Buckingham M. (2000). Myf5 is a novel early axonal marker in the mouse brain and is subjected to post-transcriptional regulation in neurons. Development 127, 319-331. [DOI] [PubMed] [Google Scholar]
- Daubas P., Crist C. G., Bajard L., Relaix F., Pecnard E., Rocancourt D. and Buckingham M. (2009). The regulatory mechanisms that underlie inappropriate transcription of the myogenic determination gene Myf5 in the central nervous system. Dev. Biol. 327, 71-82. 10.1016/j.ydbio.2008.11.031 [DOI] [PubMed] [Google Scholar]
- Duval N., Daubas P., Bourcier de Carbon C., St Cloment C., Tinevez J.-Y., Lopes M., Ribes V. and Robert B. (2014). Msx1 and Msx2 act as essential activators of Atoh1 expression in the murine spinal cord. Development 141, 1726-1736. 10.1242/dev.099002 [DOI] [PubMed] [Google Scholar]
- Engleka K. A., Gitler A. D., Zhang M., Zhou D. D., High F. A. and Epstein J. A. (2005). Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 280, 396-406. 10.1016/j.ydbio.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Epstein J. A., Shapiro D. N., Cheng J., Lam P. Y. and Maas R. L. (1996). Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc. Natl. Acad. Sci. USA 93, 4213-4218. 10.1073/pnas.93.9.4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Ishii M., Gu Y. and Maxson R. (2007). Conditional alleles of Msx1 and Msx2. Genesis 45, 477-481. 10.1002/dvg.20316 [DOI] [PubMed] [Google Scholar]
- Giordani J., Bajard L., Demignon J., Daubas P., Buckingham M. and Maire P. (2007). Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc. Natl. Acad. Sci. USA 104, 11310-11315. 10.1073/pnas.0611299104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R., Demignon J., Houbron C., Souil E., Niro C., Seller M. J., Hamard G. and Maire P. (2005). Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 132, 2235-2249. 10.1242/dev.01773 [DOI] [PubMed] [Google Scholar]
- Hadchouel J., Tajbakhsh S., Primig M., Chang T. H., Daubas P., Rocancourt D. and Buckingham M. (2000). Modular long-range regulation of Myf5 reveals unexpected heterogeneity between skeletal muscles in the mouse embryo. Development 127, 4455-4467. [DOI] [PubMed] [Google Scholar]
- Hadchouel J., Carvajal J. J., Daubas P., Bajard L., Chang T., Rocancourt D., Cox D., Summerbell D., Tajbakhsh S., Rigby P. W. J. et al. (2003). Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development 130, 3415-3426. 10.1242/dev.00552 [DOI] [PubMed] [Google Scholar]
- Havis E., Coumailleau P., Bonnet A., Bismuth K., Bonnin M.-A., Johnson R., Fan C.-M., Relaix F., Shi D.-L. and Duprez D. (2012). Sim2 prevents entry into the myogenic program by repressing MyoD transcription during limb embryonic myogenesis. Development 139, 1910-1920. 10.1242/dev.072561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzelstein D., Cohen A., Buckingham M. E. and Robert B. (1997). Insertional mutation of the mouse Msx1 homeobox gene by an nlacZ reporter gene. Mech. Dev. 65, 123-133. 10.1016/S0925-4773(97)00065-8 [DOI] [PubMed] [Google Scholar]
- Houzelstein D., Auda-Boucher G., Chéraud Y., Rouaud T., Blanc I., Tajbakhsh S., Buckingham M. E., Fontaine-Pérus J. and Robert B. (1999). The homeobox gene Msx1 is expressed in a subset of somites, and in muscle progenitor cells migrating into the forelimb. Development 126, 2689-2701. [DOI] [PubMed] [Google Scholar]
- Hovde S., Abate-Shen C. and Geiger J. H. (2001). Crystal structure of the Msx-1 homeodomain/DNA complex†,‡. Biochemistry 40, 12013-12021. 10.1021/bi0108148 [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V. and Tajbakhsh S. (2004). Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431, 466-471. 10.1038/nature02876 [DOI] [PubMed] [Google Scholar]
- Liem K. F., Tremml G., Roelink H. and Jessell T. M. (1995). Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell 82, 969-979. 10.1016/0092-8674(95)90276-7 [DOI] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lyons G. E., Houzelstein D., Sassoon D., Robert B. and Buckingham M. E. (1992). Multiple sites of Hox-7 expression during mouse embryogenesis: comparison with retinoic acid receptor mRNA localization. Mol. Reprod. Dev. 32, 303-314. 10.1002/mrd.1080320402 [DOI] [PubMed] [Google Scholar]
- Mankoo B. S., Collins N. S., Ashby P., Grigorieva E., Pevny L. H., Candia A., Wright C. V. E., Rigby P. W. J. and Pachnis V. (1999). Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature 400, 69-73. 10.1038/21892 [DOI] [PubMed] [Google Scholar]
- Moncaut N., Cross J. W., Siligan C., Keith A., Taylor K., Rigby P. W. J. and Carvajal J. J. (2012). Musculin and TCF21 coordinate the maintenance of myogenic regulatory factor expression levels during mouse craniofacial development. Development 139, 958-967. 10.1242/dev.068015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes M. B., Christensen R. G., Wakabayashi A., Stormo G. D., Brodsky M. H. and Wolfe S. A. (2008). Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133, 1277-1289. 10.1016/j.cell.2008.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M. O., Bober E., Lyons G., Arnold H. and Buckingham M. (1991). Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development 111, 1097-1107. [DOI] [PubMed] [Google Scholar]
- Pourquié O., Fan C.-M., Coltey M., Hirsinger E., Watanabe Y., Bréant C., Francis-West P., Brickell P., Tessier-Lavigne M. and Le Douarin N. M. (1996). Lateral and axial signals involved in avian somite patterning: a role for BMP4. Cell 84, 461-471. 10.1016/S0092-8674(00)81291-X [DOI] [PubMed] [Google Scholar]
- Relaix F. and Buckingham M. (1999). From insect eye to vertebrate muscle: redeployment of a regulatory network. Genes Dev. 13, 3171-3178. 10.1101/gad.13.24.3171 [DOI] [PubMed] [Google Scholar]
- Ribas R., Moncaut N., Siligan C., Taylor K., Cross J. W., Rigby P. W. J. and Carvajal J. J. (2011). Members of the TEAD family of transcription factors regulate the expression of Myf5 in ventral somitic compartments. Dev. Biol. 355, 372-380. 10.1016/j.ydbio.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki M. A., Schnegelsberg P. N. J., Stead R. H., Braun T., Arnold H.-H. and Jaenisch R. (1993). MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351-1359. 10.1016/0092-8674(93)90621-V [DOI] [PubMed] [Google Scholar]
- Satokata I. and Maas R. (1994). Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 6, 348-356. 10.1038/ng0494-348 [DOI] [PubMed] [Google Scholar]
- Schäfer K. and Braun T. (1999). Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nat. Genet. 23, 213-216. 10.1038/13843 [DOI] [PubMed] [Google Scholar]
- Song K., Wang Y. and Sassoon D. (1992). Expression of Hox-7.1 in myoblasts inhibits terminal differentiation and induces cell transformation. Nature 360, 477-481. 10.1038/360477a0 [DOI] [PubMed] [Google Scholar]
- Spitz F., Demignon J., Porteu A., Kahn A., Concordet J.-P., Daegelen D. and Maire P. (1998). Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl. Acad. Sci. USA 95, 14220-14225. 10.1073/pnas.95.24.14220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamataki D., Kastrinaki M.-C., Mankoo B. S., Pachnis V. and Karagogeos D. (2001). Homeodomain proteins Mox1 and Mox2 associate with Pax1 and Pax3 transcription factors. FEBS Lett. 499, 274-278. 10.1016/S0014-5793(01)02556-X [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S. and Buckingham M. E. (1994). Mouse limb muscle is determined in the absence of the earliest myogenic factor myf-5. Proc. Natl. Acad. Sci. USA 91, 747-751. 10.1073/pnas.91.2.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S. and Buckingham M. (2000). The birth of muscle progenitor cells in the mouse: spatiotemporal considerations. Curr. Top. Dev. Biol. 48, 225-268. 10.1016/S0070-2153(08)60758-9 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Bober E., Babinet C., Pournin S., Arnold H. and Buckingham M. (1996). Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev. Dyn. 206, 291-300. [DOI] [PubMed] [Google Scholar]
- Vasyutina E., Stebler J., Brand-Saberi B., Schulz S., Raz E. and Birchmeier C. (2005). CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 19, 2187-2198. 10.1101/gad.346205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. and Abate-Shen C. (2012). The Msx1 homeoprotein recruits G9a methyltransferase to repressed target genes in myoblast cells. PLoS ONE 7, e37647 10.1371/journal.pone.0037647 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang J., Kumar R. M., Biggs V. J., Lee H., Chen Y., Kagey M. H., Young R. A. and Abate-Shen C. (2011). The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev. Cell 21, 575-588. 10.1016/j.devcel.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshin P., Song K., Degnin C., Killary A. M., Goldhamer D. J., Sassoon D. and Thayer M. J. (1995). MSX1 inhibits MyoD expression in fibroblast×10T½ cell hybrids. Cell 82, 611-620. 10.1016/0092-8674(95)90033-0 [DOI] [PubMed] [Google Scholar]