Abstract
Background
Dual specificity phosphatases are a class of tumor-associated proteins involved in the negative regulation of the MAP kinase pathway. Downregulation of the dual specificity phosphatase 2 (DUSP2) has been reported in cancer. Epigenetic silencing of tumor suppressor genes by abnormal promoter methylation is a frequent mechanism in oncogenesis. It has been shown that the epigenetic factor CTCF is involved in the regulation of tumor suppressor genes.
Methods
We analyzed the promoter hypermethylation of DUSP2 in human cancer, including primary Merkel cell carcinoma by bisulfite restriction analysis and pyrosequencing. Moreover we analyzed the impact of a DNA methyltransferase inhibitor (5-Aza-dC) and CTCF on the epigenetic regulation of DUSP2 by qRT-PCR, promoter assay, chromatin immuno-precipitation and methylation analysis.
Results
Here we report a significant tumor-specific hypermethylation of DUSP2 in primary Merkel cell carcinoma (p = 0.05). An increase in methylation of DUSP2 was also found in 17 out of 24 (71 %) cancer cell lines, including skin and lung cancer. Treatment of cancer cells with 5-Aza-dC induced DUSP2 expression by its promoter demethylation, Additionally we observed that CTCF induces DUSP2 expression in cell lines that exhibit silencing of DUSP2. This reactivation was accompanied by increased CTCF binding and demethylation of the DUSP2 promoter.
Conclusions
Our data show that aberrant epigenetic inactivation of DUSP2 occurs in carcinogenesis and that CTCF is involved in the epigenetic regulation of DUSP2 expression.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2087-6) contains supplementary material, which is available to authorized users.
Keywords: Cancer, Dual specificity phosphatase 2, Epigenetic, Merkel cell carcinoma, CTCF, DNA methylation
Background
Dual specificity phosphatases (DUSPs) are negative regulators of mitogen-activated protein kinases (MAPK) that regulate proliferative signaling pathways, which are often activated in cancer [1–3]. DUSP2 encodes a dual-specificity phosphatase that inactivates ERK1/2 and p38 MAPK [4, 5]. DUSP2 has also been found to regulate p53- and E2F1-regulated apoptosis [6, 7]. Previously it has been reported that DUSP2 expression is markedly reduced or completely absent in many human cancers [8, 9].
Epigenetic silencing of tumor suppressor genes (TSG) is one of the most relevant molecular alteration that occurs during carcinogenesis [10]. Promoter hypermethylation of TSG occurs in cancer through methylation at the DNA level at C5 of cytosine (5mC), when found as a dinucleotides with guanine. DNA methylation in CpG islands of TSG leads to epigenetic silencing of the according transcript [11, 12]. The ten-eleven-translocation methylcytosine dioxygenases (TET1-3) catalyze the oxidation of 5mC and generate cytosine derivatives including 5-hydroxymethylcytosine (5hmC) [13, 14]. TET proteins are involved in diverse biological processes, as the zygotic epigenetic reprogramming, hematopoiesis and the development of leukemia [15–17]. The frequency of 5hmC suggests that these modified cytosine bases play an important role in epigenetic gene regulation [18]. Aberrant levels of 5hmC have been reported in human cancer [19, 20]. Recently it has been shown that TET proteins bind the CCCTC binding factor (CTCF) [21]. CTCF is associated with altered expression of tumor suppressor genes, such as E-cadherin (CDH1), retinoblastoma 1, RASSF1A, CDKN2A/p16 and TP53 [22–25]. It has also been postulated, that CTCF itself acts as a tumor suppressor [26, 27].
Here we analyzed the epigenetic inactivation and regulation of the dual specificity phosphate 2 (DUSP2) in human cancers. Our data show that DUSP2 is aberrantly methylated in primary Merkel cell cancer and in different human cancer cell lines. Moreover, we observed that 5-Aza-dC and CTCF induce DUSP2 expression by its promoter demethylation.
Methods
Primary tissues and cell lines
The analyzed primary tissues include 22 Merkel cell carcinoma [28, 29], 20 pheochromocytoma [30, 31], six small cell lung cancer [32], 12 breast carcinoma [25, 33] and 12 benign nevus cell nevi [34]. RNA samples from normal tissues (liver, breast, kidney and lung) were obtained from Agilent Technologies (Santa Clara, USA). All patients signed informed consent at initial clinical investigation. The study was approved by local ethic committees (City of Hope Medical Center, Duarte, USA or Martin-Luther University, Halle, Germany). All cell lines were cultured in a humidified atmosphere (37 °C) with 5 % CO2 and 1× Penicillin/Streptomycin in according medium. Cells were transfected with 4 μg of constructs on 3.5 cm plates, using Polyethylenimine or X-tremeGENE HP (Roche Applied Science, Germany). TREx293 cells, that stably express the Tet repressor (LifeTechnologies), were transfected with the expression vector pcDNA4TO-CTCF and selected with Zeocin™ (Invitrogen). CTCF was induced by tetracycline (5 μl/ml of a 1 mg/ml stock) over 48 h.
Methylation analysis
DNA was isolated by phenol-chloroform extraction and then bisulfite treated prior to combined bisulfite restriction analysis (COBRA) and pyrosequencing [35]. 200 ng were subsequently used for semi nested PCR with primer DUSP2BSU2 (GGGATTTGTATTTGAGAAGTTGGGTTTT) and DUSP2BSL2 (CCTCCAACCCCATAACCACC) in a first PCR. For the second PCR DUSP2BSU1 (GTTTTTTTTYGGTGTGTTGGTTTT) and the 5′-biotinylated primer DUSP2BSBIO (CCTCCAACCCCATAACCACC) were used. Products were digested with 0.5 μl TaqI (Fermentas) 1 h at 65 °C and resolved on 2 % TBE agarose gel. Methylation status was quantified utilizing the primer DUSP2BSSeq1 (TTTTGTTTTTTTTTTTAATTTTTTTT) and DUSP2BSSeq2 (GTTTTTTTGTTTTGTTTTTGTATGGTGTT) and PyroMark Q24 (Qiagen). Five CpGs are included in the analyzed region with primer DUSP2BSSeq1 and seven in the region analyzed with primer DUSP2BSSeq2. For in vitro methylation of genomic DNA we used M.SssI methylase (NEB).
Expression analysis
RNA was isolated using the Isol-RNA lysis procedure (5′Prime). 25 μg of breast, kidney, liver and lung RNA of normal human samples were obtained from Agilent Technologies (Santa Clara, CA, USA). RNA was DNase (Fermentas GmbH, St.Leon-Rot, Germany) digested and then reversely transcribed [36]. RT-PCR was performed with primers listed in Additional file 1: Table S1. Quantitative PCR (qRT-PCR) was performed in triplicates with SYBR® Select Master Mix (Life Technologies) using Rotor-Gene 3000 (Corbett Research, Qiagen).
Promoter assay
The DUSP2 promoter was amplified with primers DUSP2BglIIU1: CAGATCTGAGTGGCTTGGGACAGGTCA and DUSP2PromL1: CAGCAGCAGCGTGCGTTCCG from genomic DNA. The 454 bp promoter fragment was cloned into the BglII sites of pRLnull (Promega, Mannheim, Germany) and sequenced. In vitro methylation of the promoter construct was done with M.SssI methylase (NEB, Frankfurt, Germany). HEK293 were transfected with 1 μg of pRL-DUSP2 promoter construct and 0.35 μg of pGL3 control vector (Promega, Mannheim, Germany). Cells were isolated 24 h after transfection and studied using Dual-Luciferase Reporter Assay (Promega, Mannheim, Germany).
Depletion of CTCF by RNAi
Five small interfering RNAs against CTCF: HSS173820 (Stealth siRNA), HSS116456 (Stealth siRNA), HSS116455 (Stealth siRNA), siCTCF1: UCACCCUCCUGAGGAAUCACCUUAA, siCTCF2: GAUGCGCUCUAAGAAAGAA and a control siRNA: CUACGAUGAAGCACUAUUATT were obtained from Invitrogen (Carlsbad, CA, USA) and have been characterized previously [37, 38]. Cells were transfected with a pool of five specific siRNAs against CTCF or a control siRNA according to the manufacturer manual using Lipofectamin RNAiMax from Invitrogen (Carlsbad, CA, USA) on two consecutive days and incubated for a total of 96 h. RNA and protein was isolated.
Chromatin immunoprecipitation (ChIP)
Cells were fixed using 37 % formaldehyde (CalBiochem) with a final concentration of 1 % for 10 min at room temperature. Incubation of 1/7 volume of 1 M glycine for 5 min stopped the fixation process. Cells were washed with PBS and harvested in PBS + 1 mM PMSF. After centrifugation for 2 min at 2000 rpm at 4 °C the supernatant was removed and cells were lysed using 1 ml SDS lysis buffer (0,5 % SDS, 10 mM EDTA, 50 mM Tris HCl pH 8.1) supplemented with protease inhibitors (Complete Mini, Roche) per 107 cells for 10 min on ice. After sonification (400-800 bp) the samples were centrifuged for 10 min at 4 °C and maximum speed. The supernatant was diluted 1:10 with dilution buffer (0,01 % SDS, 1,1 % Triton X-100, 1.2 mM EDTA, 16.7 mM Tris/HCl pH 8.1, 167 mM NaCl) and 1 ml of the dilution was used for each ChIP. 10 % of the chromatin used for one ChIP was preserved as an input sample and stored at -20 °C. The lysate was pre-cleared by rotation at 4 °C for 2 h using 1 ml of the dilution and 20 μl ProteinG Plus/ProteinA Agarose (Calbiochem). After centrifugation at 4 °C for 1 min at 2000 rpm the supernatant was incubated with the corresponding antibody: IgG (46540; Santa Cruz Biotechnologie), Histon H3 (1791; Abcam), α-CTCF-(N2.2, [39]) rotating overnight at 4 °C. Binding of the immune-complexes occurs afterwards by incubation of the chromatin with 20 μl of ProteinG Plus/Protein A agarose for 2 h at 4 °C. After incubation the beads were washed for 5 min rotating at 4 °C one time with low salt buffer (0,05 % SDS, 1 % TritonX100, 2 mM EDTA, 20 mM Tris/HCl pH 8.1, 150 mM NaCl), one time with high salt buffer (0,05 % SDS, 1 % TritonX100, 2 mM EDTA, 20 mM Tris/HCl pH 8.1, 500 mM NaCl), one time with LiCl buffer (0,25 M LiCl, 1 % NP40, 1 % Deoxycholat, 1 mM EDTA, 10 mM Tris/HCl pH 8.1) and two times with TE buffer (10 mM Tris, 1 mM EDTA pH 8.0). Chromatin bound to beads and input material were resuspended in 100 μl TE buffer, 1 μl of 10 mg/ml RNase was added followed by an incubation of 30 min at 37 °C. 5 μl of 10 % SDS, 1 μl of 20 mg/ml Proteinase K was added and incubated for further 2-4 h at 37 °C. The reverse crosslink was performed by incubating the samples over night at 65 °C. DNA was recovered by using QIAquick PCR Purification Kit (Qiagen) and PCR amplification with the following primer: DUSP2CIPU1: TTTGAGGGCCTTTTCCGCTACAAGAG, DUSP2CIPL1: GCCTCCGCTGTTCTTCACCCAGTC, DUSP2CIPU2: GGGTGGGCGCAAAAACGGAGGG, DUSP2CIPL2: CCGGGGCACCATACAAGGGCAGA, DUSP2CIPU3: GGCCACGTCAC CCTCTCAGTGTCTC, DUSP2CIPL3: GCCTCAGCCAAGTTGCCCAGACA. PCR was done with U1/L1 (233 bp), U2/L2 (236 bp) and U3/L3 (120 bp) for positive site, DUSP2 promoter site and negative site, respectively. Quantitative PCR was performed in triplicates with SYBR® Select Master Mix (Life Technologies) using Rotor-Gene 3000 (Corbett Research, Qiagen).
Methylated DNA-immunoprecipitation (MeDIP)
MeDIP was performed according to the protocol of Mohn et al. [40] with antibodies: IgG (46540; Santa Cruz Biotechnologie), 5mC (MAb-081-010; Diagenode) and 5hmC (MAb-31HMC-020; Diagenode). The following primers were used for semi quantitative PCR amplification: DUSP2CIPU2: GGGTGGGCGCAAAAACGGAGGG, DUSP2CIPL2: CCGGGGCACCATACAAGGGCAGA, bACTRTFW: CCTTCCTTCCTGGGCATGGAGTC, bACTRTFW: CGGAGTACTTGCGCTCAGGAGGA.
Western blot
Cell lysates were resolved in SDS-PAGE, immunoblotted, and probed with primary anti-CTCF (N2.2) and anti-GAPDH antibody (GAPDH rabbit polyclonal IgG FL-335, Santa Cruz) [39]. Afterwards an incubation with HRP-coupled secondary antibodies followed. Immunocomplexes were detected by enhanced chemiluminescence reagent (Western Chemiluminescent Immobilon HRP-Substrate, Millipore) according to the manufacturer’s instructions.
Constructs
CTCF and BORIS were generous gifts from Rainer Renkawitz (Justus-Liebig-University, Giessen, Germany) and deletions and mutations were generated with QuikChange Lightning Site-Directed Mutagenesis Kit (Promega, Heidelberg, Germany) with the primers listed in Additional file 2: Table S2.
Statistical evaluation
Statistical analysis was performed using Excel (Microsoft, Redmond, USA) and GraphPad Quick Calcs (GraphPad Software, La Jolla, USA). Data are represented as mean ± standard deviation. Unpaired t-test was used to determine significant differences between groups. For statistical analysis of methylation differences between Merkel cell carcinoma and benign controls a two tailed Fisher exact test was performed. All reported p-values are considered significant for p ≤ 0.05.
Results
Aberrant promoter methylation of DUSP2 in human cancer
Previously it has been shown that expression of the MAPK-specific phosphatase DUSP2 is markedly reduced or completely absent in many human cancers and that its level of expression inversely correlates with cancer malignancy [8]. Here we aimed to dissect the epigenetic mechanisms involved in the aberrant downregulation of DUSP2 in carcinogenesis. DUSP2 is located on 2q11.2 (Fig. 1a) and contains a 1011 bp CpG island in its promoter region (chr2: 96810444-96811454, UCSC genome browser). We analyzed the promoter hypermethylation of DUSP2 in primary tumors including Merkel cell carcinoma (MCC), pheochromocytoma, small cell lung cancer and breast carcinoma by COBRA (Fig. 1 and Additional file 3: Figure S1). In 12 breast carcinoma, 20 pheochromocytoma and six small cell lung cancer samples the DUSP2 promoter was unmethylated (Additional file 3: Figure S1). Interestingly, 10 out of 22 (45 %) Merkel cell carcinoma showed a DUSP2 hypermethylation (Fig. 1b). MCC is a rare but aggressive cutaneous malignancy. In the control tissue (benign nevus cell nevi) only one out of 12 (8 %) analyzed samples exhibited a DUSP2 hypermethylation (NCN12; Fig. 1b). Thus in MCC a significant tumor-specific hypermethylation of DUSP2 was detected (p = 0.05, two tailed Fisher exact test).
Fig. 1.
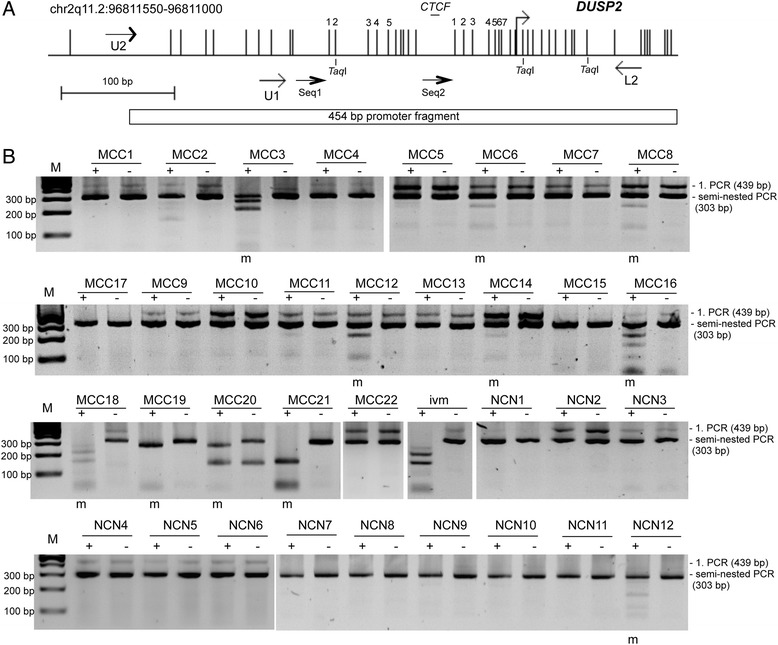
Hypermethylation of DUSP2 in primary Merkel cell carcinoma (MCC). a. Structure of the DUSP2 CpG island promoter on chromosome 2q11.2. Vertical lines indicate CpGs and the transcriptional start site is marked. A CTCF motif sequence (GGCAGAGCA; CTCFBSDB2.0) is marked [47]. Primers used for COBRA and sequencing (Seq1 and Seq2) are depicted by arrows. TaqI restriction sites for COBRA and CpGs analyzed by pyrosequencing are indicated. The 454 bp DUSP2 fragment for the luciferase promoter assay is indicated. b. Methylation of DUSP2 in MCC (m = methylated). For COBRA bisulfite-treated DNA from MCC, benign nevus cell nevi (NCN) and in vitro methylated DNA (ivm) was amplified by semi-nested PCR. First and second PCR products are indicated (439 bp and 303 bp, respectively). Products were digested with TaqI (+) or mock digested (-) and resolved on 2 % agarose gels with a 100 bp marker (M)
To reveal the epigenetic status of DUSP2 in human cancers in more detail, we have analyzed the methylation of its promoter in several human cancer cell lines by COBRA (Fig. 2a) and by pyrosequencing (Fig. 2b). Human fibroblasts (HF53) and the HB2 mammary luminal epithelial cells are unmethylated (methylation level <5 %) (Fig. 2a and b). We detected increased DUSP2 methylation (level ≥5 %) in lung cancer (A427, H322, A549), breast cancer (MCF-7, ZR75-1), melanoma (SKMel13, IGR1, buf1280, SKMel28, MeWo), ovarian cancer (Skov, Ovar, ES2, OAW42), hepatocarcinoma (Hep2G) and sarcoma (LMS6/93, U2OS) cell lines (Fig. 2a and b). Lung cancer cell lines H358 and HTB171, melanoma C8161, ovarian cancer CAOV3, pancreatic cancer PaCa2, thyroid cancer FTC133 and HeLa were rather unmethylated (<5 %). Furthermore the embryonic kidney cell line HEK293 showed an aberrant DUSP2 promoter methylation (29 %). In summary, 17 out of 24 (71 %) human cancer cell lines exhibited increased DUSP2 promoter methylation.
Fig. 2.
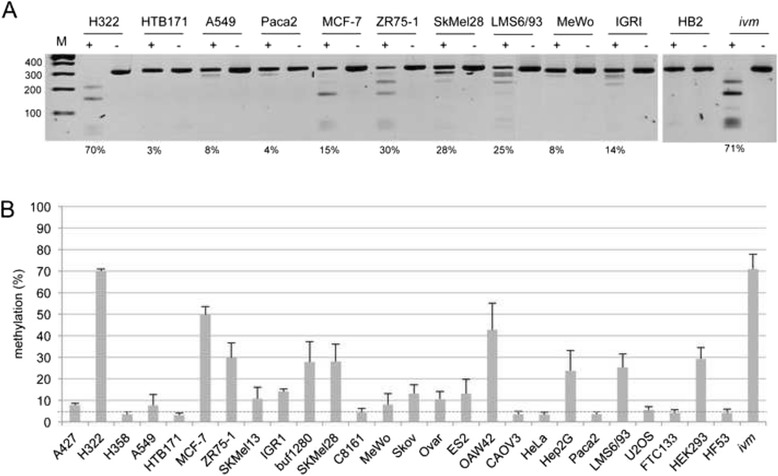
Promoter hypermethylation of DUSP2 in human cancers. a. Combined bisulfite restriction analysis (COBRA) of DUSP2. Bisulfite-treated DNA from the indicated cancer cell lines, normal epithelial breast cells (HB2) and in vitro methylated DNA (ivm) was amplified, digested with TaqI (+) or mock digested (-) and resolved on 2 % agarose gels with a 100 bp marker (M). Methylation levels obtained from pyrosequencing are indicated in percentage. b. Bisulfite pyrosequence analysis of DUSP2 in human cells. The mean methylation levels of five CpGs were analyzed by pyrosequencing (Seq1). The analysis included the results of three independent experiments. The dashed line marks 5 % threshold
Decreased expression of DUSP2 is associated with its promoter hypermethylation
To further analyze the impact of epigenetic regulation of DUSP2 in carcinogenesis we investigated its expression in normal tissues and cancer cell lines (Fig. 3). DUSP2 expression was found in normal breast, kidney, liver, lung tissues and in HEK293 cells (Fig. 3a). In HEK293 cells a genome wide expression array (human ST1.0 S, Affymetrix) detected a 0.02-fold reduced level of DUSP2 compared to beta-actin [41]. We cloned a 454 bp fragment of the DUSP2 promoter in a luciferase reporter system (Fig. 1a), in vitro methylated (ivm) the construct and analyzed its activity (Fig. 3b). Methylation of the DUSP2 promoter construct significantly reduced its expression (Fig. 3b). In cancer cells lines (H322, ZR75-1) and HEK293 cells, that exhibit a methylated promoter, expression of DUSP2 was reduced compared to HeLa and H358 cells, which harbor unmethylated promoter regions (Fig. 3c). H322, ZR75-1 and HEK293 cells were treated with 5-Aza-dC (Aza), a cytidine analogue that inhibits DNA methyltransferases and reactivates epigenetically inactivated TSG [42, 43]. After four days of Aza treatment an induced expression of DUSP2 was found in H322, ZR75-1 and HEK293 cells (Fig. 3c). However, in HeLa and H358 cells, that harbor unmethylated DUSP2 promoters, DUSP2 expression was rather unaffected by Aza (Fig. 3c). For H322 cells the fourfold increased DUSP2 expression after 5 μM Aza treatment was correlated with a significant 1.6-fold demethylation of seven analyzed CpGs at the DUSP2 promoter (Fig. 3d and e). Thus, we observed a methylation dependent silencing of DUSP2 in cancer cell lines, which was reversed by inhibiting DNA methylation.
Fig. 3.
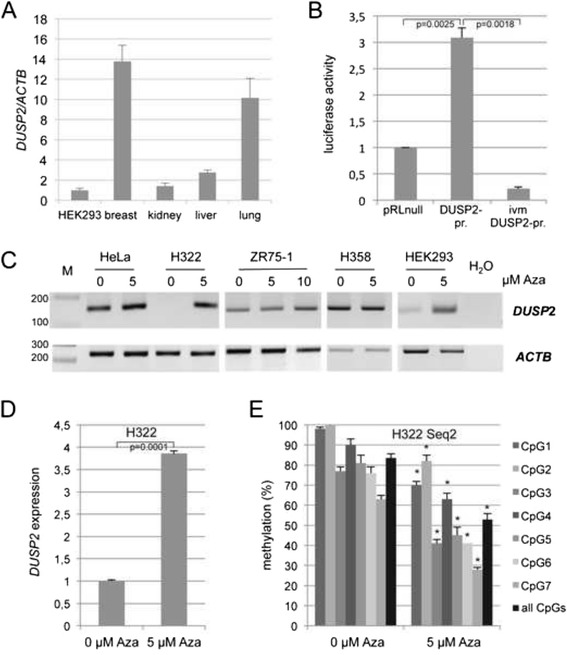
DUSP2 expression and methylation after 5-Aza-2′-deoxycytidine (Aza) treatment. a. Expression of DUSP2 in normal breast, kidney, liver, lung tissues (Agilent Technologies) and HEK293 cells was analyzed by qRT-PCR and normalized to ACTB (HEK293 = 1). b. A DUSP2 promoter fragment (454 bp) was cloned in the pRLnull vector and in vitro methylated (ivm). DUSP2 promoter constructs (DUSP2-pr.) were transfected in HEK293 cells and expression of renilla luciferase was measured and normalized to the expression of the co-transfected firefly plasmid pGL3.1 (pRLnull = 1). The analysis included the results (measurement in triplicates) of three independent experiments and significance is indicated (t-test) c. Expression analysis of DUSP2 in several cell lines after treatment with Aza (0, 5 and 10 μM) for four days. Expression of DUSP2 and ACTB was revealed by semi-quantitative RT-PCR and products (136 bp and 226 bp, respectively) were resolved on a 2 % agarose gel with a 100 bp marker ladder (M). d. Expression of DUSP2 in Aza treated H322 cells analyzed by qRT-PCR and normalized to ACTB. Data of three independent experiments, whereby each PCR was performed in triplicates and significance is indicated (t-test). e. Methylation analysis of DUSP2 after Aza treatment in H322 cells. Methylation of seven CpGs at proximal DUSP2 transcription start site (Seq2, see also Fig. 1) was analyzed by bisulfite pyrosequencing. Data were calculated from three independent experiments and significance is indicated (* = p < 0.01; 0 μM vs. 5 μM Aza)
Regulation of DUSP2 by the epigenetic factor CTCF
It has been shown that the insulator binding protein CTCF is involved in the epigenetic regulation of tumor suppressor genes [22–25, 44, 45]. Wendt and Barksi et al. have reported that silencing of CTCF by RNA interference caused repression of DUSP2 in HeLa cells [38, 46]. Database analysis of CTCF ChipSeq Encode data revealed that CTCF binds at the DUSP2 promoter in HeLa cells (chr2: 96811040-96811190) and a search at the CTCFBSDB2.0 site identified a CTCF motif sequence (GGCAGAGCA chr2: 96811243-96811251) upstream of the DUSP2 transcription start site (Fig. 1a) [47]. To investigate the role of CTCF in the epigenetic regulation of DUSP2, we performed siRNA mediated knock down of CTCF in HEK293, HeLa and HTB171 cells (Fig. 4). In HEK293, a five-fold reduction of CTCF on RNA and protein levels was accomplished by transfection of CTCF-specific siRNA (Fig. 4a and b). This downregulation of CTCF resulted in a 2.5-fold reduction of DUSP2 level (Fig. 4a). Knock down of CTCF in HeLa and HTB171 cells induced a 1.7-fold reduction of DUSP2 (Fig. 4c).
Fig. 4.
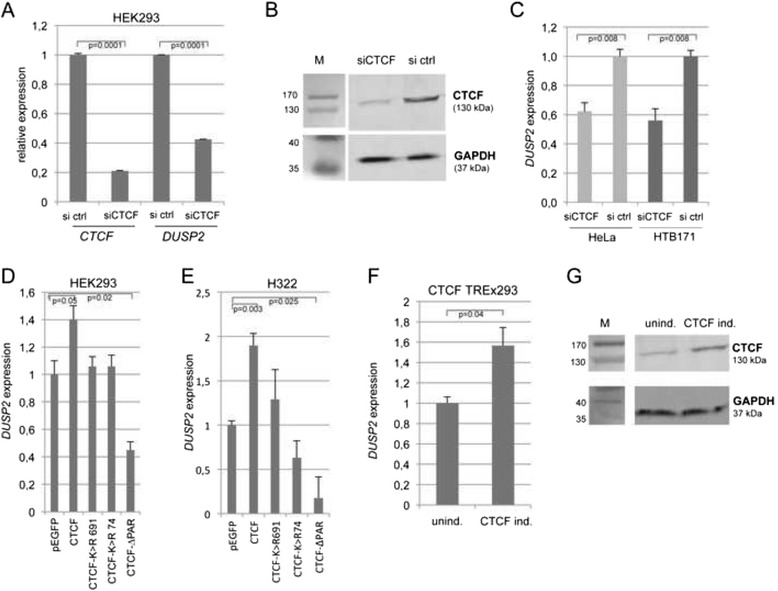
CTCF-dependent expression of DUSP2. a. HEK293 cells were transfected on two consecutive days with a pool of five different siRNAs against hCTCF (siCTCF) or a control siRNA (si ctrl). After 96 h the RNA was isolated and expression of CTCF and DUSP2 was analyzed by RT-PCR and normalized to ACTB (si control = 1). CTCF knockdown was performed 2 times and the qRT-PCR was done in triplicates and significance is indicated (same procedure for C). b. Reduction of CTCF protein after RNA interference was analyzed by western blot. GAPDH expression was utilized as control. c. Reduction of DUSP2 expression after siCTCF transfection in HeLa and HTB171 cells. d. Different CTCF constructs; wildtype, mutated SUMO site K > R at position 74 (CTCF-K > R74) and 691 (CTCF-K > R691), deletion of CTCF PARylation site (CTCF-ΔPAR) and vector control (pEGFP) were transfected in HEK293. After two days DUSP2 expression was analyzed by RT-PCR and normalized to ACTB (normal control = 1). Triplicates were determined and the data of three independent experiment were averaged and significance was calculated (same procedure for e and f) e. Expression of DUSP2 after transfection of different CTCF constructs in H322 cells (for details see d) f. DUSP2 expression was analyzed in CTCF-inducible TREx293 cells, a stable HEK293 cell line (CTCF TREx293) that allows inducible CTCF expression by tetracycline (5 μg/ml). After two days expression of DUSP2 was analyzed by RT-PCR and normalized to ACTB and compared to the uninduced control (unind. = 1). g. Expression of CTCF in TREx293 cells was analyzed by western blot
Next, we tested the effect of CTCF overexpression in cancer cells (Fig. 4 and Additional file 4: Figure S2). In HEK293 and H322 cells that harbor a methylated DUSP2 promoter CTCF transfection resulted in a 1.4- and 2-fold increased expression of DUSP2, respectively (Fig. 4d and e). In HeLa cells that exhibit an unmethylated promoter this CTCF-induced expression of DUSP2 was absent (Additional file 4: Figure S2A). The testis-specific paralogue of CTCF, termed CTCFL or BORIS was unable to induce DUSP2 expression in HEK293 cells (Additional file 4: Figure S2B). Previously, it has been reported that SUMOylation and PARylation of CTCF are involved in its regulatory function [23, 48, 49]. Therefore we generated different CTCF constructs that lack either the N- or C-terminal SUMOylation site (K > R substitution) at position 74 (CTCF-K > R74) and 691 (CTCF-K > R691), respectively or harbor a deletion of its PARylation sites from position 216 to 243 (CTCF-ΔPAR). Expression of the two SUMOylation-site deficient CTCF constructs in HEK293 and H322 cells resulted in a lack of DUSP2 induction compared to wildtype CTCF (Fig. 4d and e). Transfection of the PARylation site deficient CTCF construct (CTCF-ΔPAR) downregulated DUSP2 expression significantly (2.2- and 5.6-fold, respectively) compared to the vector control in HEK293 and H322 cells (Fig. 4d and e). Furthermore, we have generated a stable HEK293 cell line (CTCF TREx293) that allows tetracycline-inducible CTCF expression (Fig. 4f and g). Induction of CTCF in TREx293 cells resulted in 1.6-fold higher DUSP2 expression (Fig. 4f).
Increased CTCF binding at the DUSP2 promoter is associated with reduction in methylation levels and induced DUSP2 expression
To analyze the mechanism of CTCF regulated DUSP2 expression in detail, we analyzed the binding of CTCF at the DUSP2 locus in CTCF TREx293 cells by ChIP (Fig. 5a). Therefore, we utilized CTCF and histone H3 antibody and quantified the precipitation of the DUSP2 promoter, a bona fide CTCF target site at 1 kb downstream (positive site) and negative site 1 kb upstream of the DUSP2 transcriptional start site by qPCR. After CTCF induction we observed a significant 2.3-fold increased binding of CTCF at the DUSP2 promoter. Histone H3 binding and CTCF binding at the negative site were not altered. At the positive site binding of CTCF was significantly increased by 1.6 times after tetracycline-induced CTCF expression (Fig. 5a). Histone H3 levels were reduced after CTCF induction at the positive site (Fig. 5a).
Fig. 5.
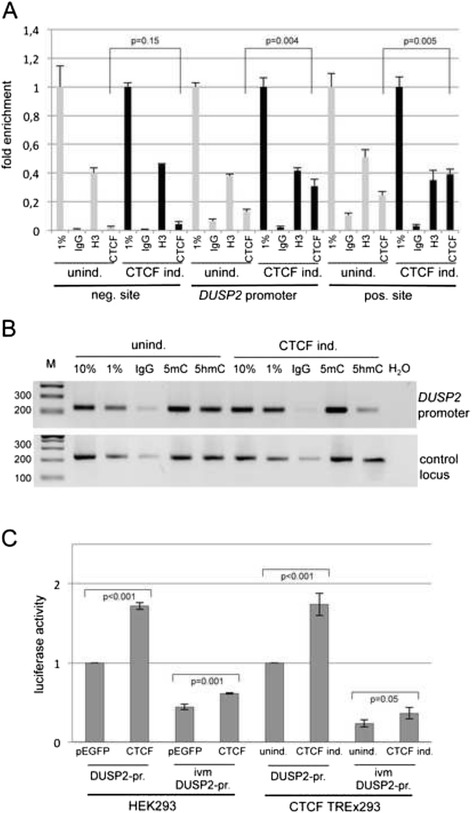
Epigenetic regulation of the DUSP2 promoter by CTCF. a. Binding of CTCF at the DUSP2 promoter analyzed by quantitative ChIP. CTCF expression was induced in CTCF TREx293 cells by tetracycline (5 μg/ml) for 48 h and uninduced cells were used as control. The chromatin was prepared, precipitated with a CTCF-, histone H3- or control IgG-antibodies and amplified with gene specific primers for the DUSP2 promoter, a negative site and a bona fide positive site within the DUSP2 locus. CTCF binding was quantified by qPCR (triplicates from 2 independent experiments) and significance was calculated. Values of the precipitated sample were normalized to 1 % input (=1). b. Methyl-DNA immunoprecipitation (MeDIP) analysis of the DUSP2 promoter. MeDIP with 5mC- and 5hmC-antibodies was done with DNA from uninduced or induced CTCF TREx293 cells. The detection of the 5hmC and 5mC level was performed with semi-quantitative PCR of the DUSP2 promoter and a control locus. PCR products were separated together with a 100 bp marker (M) in 2 % agarose gel. c. Effect of CTCF on the DUSP2 promoter. A DUSP2 promoter fragment (454 bp) was cloned in the pRLnull vector and in vitro methylated (ivm). 2.7 μg DUSP2 promoter constructs (DUSP2-pr.) were transfected in HEK293 or CTCF TREx293 cells and GFP-CTCF or GFP vector (1 μg each) was co-transfected or CTCF was induced for 24 h, respectively. Expression of renilla luciferase was measured and normalized to the expression control vector or to the co-transfected firefly plasmid pGL3.1 (300 ng), respectively. Significance is indicated (t-test)
Subsequently, we analyzed changes in DNA methylation after CTCF induction at the DUSP2 promoter (Fig. 5). Conventional bisulfite sequencing is not able to distinguish between 5-methylcytosine (5mC) or 5-hydroxy-methylcytosine (5hmC) levels. Therefore we precipitated different DNA regions with antibodies that bind 5mC or 5hmC in induced or uninduced CTCF TREx293 cells. Interestingly, after CTCF induction we observed a decrease in 5hmC-precipitated DUSP2 promoter sequences compared to a control locus control (chr5q14.1) (Fig. 5b). This result suggests that CTCF induces dehydroxy-methylation of the DUSP2 promoter. To analyze the effect of CTCF on the DUSP2 promoter, we performed luciferase reporter assays (Fig. 5c). Therefore the DUSP2 promoter construct was in vitro methylated (ivm) and transfected together with CTCF in HEK293 cells. CTCF transfection significantly induced luciferase reporter activity of the unmethylated DUSP2 promoter (1.7-fold) and ivm DUSP2 promoter (1.4-fold) compared to the pEGFP control vector (Fig. 5c). Moreover CTCF induction in CTCF TREx293 cells resulted in a 1.7-fold (unmethylated) or 1.5-fold (ivm) increased DUSP2 promoter activity compared to uninduced cells (Fig. 5c). Taken together these data suggest that CTCF epigenetically activates DUSP2 expression.
Discussion
Previously it has been reported that DUSP2 expression is downregulated in many human cancers [8]. However the mechanism of its silencing was not analyzed in details. Deletion of the DUSP2 locus at 2q11.2 is rather infrequent in cancer. Here we show that the promoter of DUSP2 is hypermethylated in different human cancer cell lines including lung, breast and skin cancers and in HEK293 cells (Figs. 1 and 2). In primary Merkel cell cancer (MCC) we observed a significant tumor specific methylation of DUSP2. MCC is one of the most aggressive cancers of the skin and we have reported frequent hypermethylation of the Ras Association Family Members RASSF1A and RASSF10 in this tumor entity [28, 29]. Hypermethylation of DUSP2 and murine Dusp2 and has been reported in breast cancer cell lines, however methylation in primary human mammary tumors was absent [50], which was also observed in our study (Additional file 3: Figure S1). By inhibiting DNA methyltransferases with the cytidine analogue 5-aza-dC we found that the DUSP2 gene is epigenetically reactivated by its demethylation (Fig. 3). The promoter methylation of DUSP2 in HEK293 consists of 5mC and 5hmC epigenetic marks (Fig. 5). Additionally, we have revealed that CTCF reactivates DUSP2 and this is associated with demethylation of its CpG island promoter (Figs. 4 and 5b). CTCF is a DNA binding factor well known for its multiple functions in gene regulation. Depending on the participating genetic locus it is involved in transcriptional activation [51–53], transcriptional repression [54, 55] or enhancer blocking [56]. Here we show increased binding of CTCF to the DUSP2 promoter (Fig. 5a) and CTCF-dependent induction of the DUSP2 promoter activity (Fig. 5c). Thus it will be interesting to analyze the exact mechanism of CTCF induced DUSP2 expression and promoter dehydroyx-methylation in further details.
DUSP2 encodes a dual-specificity phosphatase that inactivates ERK1/2 and p38 MAPK [4, 5]. DUSP2 has also been found to regulate p53- and E2F1-regulated apoptosis [6, 7]. Dual-specificity phosphatases are negative regulators of the MAPK signal transduction, proliferative pathways that are often activated in cancers [1]. Downregulation of DUSP2 was detected in human acute leukemia coupled with activation of MEK and hyperexpression of ERK [9]. Especially in acute myeloid leukemia, translocation and mutations of TET1 and TET2 gene are frequently observed [57–59]. Moreover it has been reported that CTCF binds TET proteins [21]. Thus it will be interesting to analyze if TET proteins are directly involved in the epigenetic regulation of DUSP2. Here we observed increased binding of CTCF to its target site in the DUSP2 promoter after CTCF induction (Fig. 5a). This binding may alter distinct TET- or DNMT-associated chromatin complexes at the DUSP2 promoter region involved in CTCF-regulated DNA methylation as previously reported [21, 25, 60, 61] and revealed in Fig. 5b. However CTCF-dependent regulation of DUSP2 may also involve its function in chromosome configuration, chromatin insulation or transcriptional regulation [62, 63].
We also observed that the CTCF paralogue CTCFL/BORIS was unable to reactivate DUSP2 expression (Additional file 4: Figure S2). Since the cancer-testis specific BORIS is aberrantly expressed in cancer, an oncogenic role for BORIS has been proposed [64, 65]. It was reported that CTCF, unlike BORIS, cannot bind to methylated binding sites [66]. Therefore, it is interesting to note that the CTCF consensus site in the DUSP2 promoter sequence lacks CpG sites (Fig. 1a) and ChIP data show an enhanced CTCF binding in CTCF induced TREx293 cells at this site (Fig. 5a). It was also postulated that CTCF itself acts as a tumor suppressor [26, 27]. CTCF contains a N-terminal PARylation site [49]. Here we observed that overexpression of CTCF lacking its PARylation site resulted in repression of DUSP2 expression in H322 and HEK293 cells (Fig. 4d and e). This result suggests that the PARylation site of CTCF is important for its activating function. It has been reported that defective CTCF PARylation and dissociation from the molecular chaperone nucleolin occurs in CDKN2A- and CDH1-silenced cells, abrogating its TSG function [23]. Using CTCF mutants, the requirement of PARylation for optimal CTCF function in transcriptional activation of the p19ARF promoter and inhibition of cell proliferation has been demonstrated [49]. In this model CTCF and Poly(ADP-ribose) polymerase 1 form functional complexes [49]. Furthermore, CTCF contains two SUMOylation sites [48]. Overexpresssion of the CTCF construct with mutated SUMOylation sites in the CTCF N-terminus or C-terminus resulted in a lack of DUSP2 reactivation in the lung cancer H322 and HEK293 cells (Fig. 4d and e). SUMOylation of CTCF has been associated with its tumor suppressive function in c-myc expression [48]. There is also a report that CTCF SUMOylation modulates a CTCF domain, which activates transcription and decondenses chromatin [67].
Conclusions
Downregulation of the negative regulator DUSP2 of the oncogenic MAPK signaling pathway has been reported in cancer. In the present study we have investigated the epigenetic regulation of DUSP2 in detail and we show that DUSP2 is epigenetically silenced by promoter methylation in human cancer. Especially in primary Merkel cell carcinoma a tumor-specific hypermethylation of DUSP2 was revealed. Thus it will be interesting to further analyzed primary tumor tissues regarding an aberrant DUSP2 promoter hypermethylation. Moreover our data indicate that the insulator-binding factor CTCF is involved in the epigenetic regulation of DUSP2. Further research will elucidate the exact mechanism of the CTCF-mediated induction of DUSP2.
Acknowledgements
The work was supported by grants (TRR81, UGMLC) from the DFG and Land Hessen to Reinhard Dammann. These organizations had no involvement in the study design, acquisition, analysis, data interpretation, writing of the manuscript and in the decision to submit the manuscript for publication.
Abbreviations
- Aza
5-aza-dC
- COBRA
combined bisulfite restriction analysis
- CTCF
CCCTC binding factor
- DUSP2
dual specificity phosphatase 2
- MCC
Merkel cell carcinoma
Additional files
List of primers for RT-PCR. (DOCX 17 kb)
Primers for site directed mutagenesis of CTCF. (DOCX 47 kb)
Four examples of methylation analysis of DUSP2 in primary pheochromocytomas (Pheo), small cell lung cancer (SCLC) and breast cancer (BrCa). (PDF 2955 kb)
CTCF- and BORIS‐dependent expression of DUSP2 in HeLa and HEK293 cells, respectively A. (PDF 1891 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests. The work was supported by grants (TRR81, LOEWE) from the DFG and Land Hessen to RHD. These organizations had no involvement in the study design, acquisition, analysis, data interpretation, writing of the manuscript and in the decision to submit the manuscript for publication.
Authors’ contributions
RHD has created the study. TH and RHD participated in the design of the study. TH, AMR, APJ and MBS acquired data. TH, MBS, AMR, APJ and RHD controlled analyzed and interpreted data. RHD and TH prepared the manuscript. TH, MBS, AMR, APJ and RHD read, corrected and approved the final manuscript.
Contributor Information
Tanja Haag, Email: Tanjahaag2408@gmail.com.
Antje M. Richter, Email: Antje.M.Richter@gen.bio.uni-giessen.de
Martin B. Schneider, Email: Martin.bernd.schneider@onlinehome.de
Adriana P. Jiménez, Email: Adriana.Jimenez@gen.bio.uni-giessen.de
Reinhard H. Dammann, Phone: +49-641-99-35462, Email: reinhard.dammann@gen.bio.uni-giessen.de
References
- 1.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26(22):3203–13. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 2.Bermudez O, Pages G, Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299(2):C189–202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- 3.Huang CY, Tan TH. DUSPs, to MAP kinases and beyond. Cell Biosci. 2012;2(1):24. doi: 10.1186/2045-3701-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohan PJ, Davis P, Moskaluk CA, Kearns M, Krutzsch H, Siebenlist U, et al. PAC-1: a mitogen-induced nuclear protein tyrosine phosphatase. Science. 1993;259(5102):1763–6. doi: 10.1126/science.7681221. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Q, Muller M, Chen CH, Zeng L, Farooq A, Zhou MM. New insights into the catalytic activation of the MAPK phosphatase PAC-1 induced by its substrate MAPK ERK2 binding. J Mol Biol. 2005;354(4):777–88. doi: 10.1016/j.jmb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Jin YJ, Calaf GM, Huang WL, Yin Y. PAC1 is a direct transcription target of E2F-1 in apoptotic signaling. Oncogene. 2007;26(45):6526–35. doi: 10.1038/sj.onc.1210484. [DOI] [PubMed] [Google Scholar]
- 7.Yin Y, Liu YX, Jin YJ, Hall EJ, Barrett JC. PAC1 phosphatase is a transcription target of p53 in signalling apoptosis and growth suppression. Nature. 2003;422(6931):527–31. doi: 10.1038/nature01519. [DOI] [PubMed] [Google Scholar]
- 8.Lin SC, Chien CW, Lee JC, Yeh YC, Hsu KF, Lai YY, et al. Suppression of dual-specificity phosphatase-2 by hypoxia increases chemoresistance and malignancy in human cancer cells. J Clin Invest. 2011;121(5):1905–16. doi: 10.1172/JCI44362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SC, Hahn JS, Min YH, Yoo NC, Ko YW, Lee WJ. Constitutive activation of extracellular signal-regulated kinase in human acute leukemias: combined role of activation of MEK, hyperexpression of extracellular signal-regulated kinase, and downregulation of a phosphatase, PAC1. Blood. 1999;93(11):3893–9. [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20(3):274–81. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 12.Taberlay PC, Jones PA. DNA methylation and cancer. Prog Drug Res. 2011;67:1–23. doi: 10.1007/978-3-7643-8989-5_1. [DOI] [PubMed] [Google Scholar]
- 13.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25(23):2436–52. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin. 2013;6(1):10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, et al. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22(3):467–77. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2(8):627–37. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150(6):1135–46. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubois-Chevalier J, Oger F, Dehondt H, Firmin FF, Gheeraert C, Staels B, et al. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res. 2014;42(17):10943–59. doi: 10.1093/nar/gku780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soto-Reyes E, Recillas-Targa F. Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines. Oncogene. 2010;29(15):2217–27. doi: 10.1038/onc.2009.509. [DOI] [PubMed] [Google Scholar]
- 23.Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell. 2009;34(3):271–84. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De La Rosa-Velazquez IA, Rincon-Arano H, Benitez-Bribiesca L, Recillas-Targa F. Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res. 2007;67(6):2577–85. doi: 10.1158/0008-5472.CAN-06-2024. [DOI] [PubMed] [Google Scholar]
- 25.Haag T, Herkt CE, Walesch SK, Richter AM, Dammann RH. The apoptosis associated tyrosine kinase gene is frequently hypermethylated in human cancer and is regulated by epigenetic mechanisms. Genes Cancer. 2014;5(9-10):365–74. doi: 10.18632/genesandcancer.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiorentino FP, Giordano A. The tumor suppressor role of CTCF. J Cell Physiol. 2012;227(2):479–92. doi: 10.1002/jcp.22780. [DOI] [PubMed] [Google Scholar]
- 27.Tiffen JC, Bailey CG, Marshall AD, Metierre C, Feng Y, Wang Q, et al. The cancer-testis antigen BORIS phenocopies the tumor suppressor CTCF in normal and neoplastic cells. Int J Cancer. 2013;133(7):1603–13. doi: 10.1002/ijc.28184. [DOI] [PubMed] [Google Scholar]
- 28.Helmbold P, Lahtz C, Enk A, Herrmann-Trost P, Marsch WC, Kutzner H, et al. Frequent occurrence of RASSF1A promoter hypermethylation and merkel cell polyomavirus in merkel cell carcinoma. Mol Carcinog. 2009;48(10):903–9. doi: 10.1002/mc.20540. [DOI] [PubMed] [Google Scholar]
- 29.Richter AM, Haag T, Walesch S, Herrmann-Trost P, Marsch WC, Kutzner H, et al. Aberrant promoter hypermethylation of RASSF family members in merkel cell carcinoma. Cancers. 2013;5(4):1566–76. doi: 10.3390/cancers5041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dammann R, Schagdarsurengin U, Seidel C, Trumpler C, Hoang-Vu C, Gimm O, et al. Frequent promoter methylation of tumor-related genes in sporadic and men2-associated pheochromocytomas. Exp Clin Endocrinol Diabetes. 2005;113(1):1–7. doi: 10.1055/s-2004-830522. [DOI] [PubMed] [Google Scholar]
- 31.Richter AM, Zimmermann T, Haag T, Walesch SK, Dammann RH. Promoter methylation status of Ras-association domain family members in pheochromocytoma. Front Endocrinol. 2015;6:21. doi: 10.3389/fendo.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helmbold P, Lahtz C, Herpel E, Schnabel PA, Dammann RH. Frequent hypermethylation of RASSF1A tumour suppressor gene promoter and presence of Merkel cell polyomavirus in small cell lung cancer. Eur J Cancer. 2009;45(12):2207–11. doi: 10.1016/j.ejca.2009.04.038. [DOI] [PubMed] [Google Scholar]
- 33.Dammann R, Yang G, Pfeifer GP. Hypermethylation of the cpG island of Ras association domain family 1A (RASSF1A), a putative tumor suppressor gene from the 3p21.3 locus, occurs in a large percentage of human breast cancers. Cancer Res. 2001;61(7):3105–9. [PubMed] [Google Scholar]
- 34.Helmbold P, Richter AM, Walesch S, Skorokhod A, Marsch W, Enk A, et al. RASSF10 promoter hypermethylation is frequent in malignant melanoma of the skin but uncommon in nevus cell nevi. J Invest Dermatol. 2012;132(3 Pt 1):687–94. doi: 10.1038/jid.2011.380. [DOI] [PubMed] [Google Scholar]
- 35.Dammann G, Teschler S, Haag T, Altmuller F, Tuczek F, Dammann RH. Increased DNA methylation of neuropsychiatric genes occurs in borderline personality disorder. Epigenetics. 2011;6(12):1454–62. doi: 10.4161/epi.6.12.18363. [DOI] [PubMed] [Google Scholar]
- 36.Richter AM, Walesch SK, Wurl P, Taubert H, Dammann RH. The tumor suppressor RASSF10 is upregulated upon contact inhibition and frequently epigenetically silenced in cancer. Oncogenesis. 2012;1 doi: 10.1038/oncsis.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunther K, Rust M, Leers J, Boettger T, Scharfe M, Jarek M, et al. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic Acids Res. 2013;41(5):3010–21. doi: 10.1093/nar/gkt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 39.Weth O, Paprotka C, Gunther K, Schulte A, Baierl M, Leers J, et al. CTCF induces histone variant incorporation, erases the H3K27me3 histone mark and opens chromatin. Nucleic Acids Res. 2014;42(19):11941–51. doi: 10.1093/nar/gku937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 41.Haag T. Epigenetic analysis of tumor suppressor genes and their regulation by CTCF. Giessen: Justus-Liebig-Universität Giessen; 2014. [Google Scholar]
- 42.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 43.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25(3):315–9. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 44.Filippova GN, Qi CF, Ulmer JE, Moore JM, Ward MD, Hu YJ, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62(1):48–52. [PubMed] [Google Scholar]
- 45.Mendez-Catala CF, Gretton S, Vostrov A, Pugacheva E, Farrar D, Ito Y, et al. A novel mechanism for CTCF in the epigenetic regulation of Bax in breast cancer cells. Neoplasia. 2013;15(8):898–912. doi: 10.1593/neo.121948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Ziebarth JD, Bhattacharya A, Cui Y. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res. 2013;41(Database issue):D188–94. doi: 10.1093/nar/gks1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29(3):714–25. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farrar D, Rai S, Chernukhin I, Jagodic M, Ito Y, Yammine S, et al. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol Cell Biol. 2010;30(5):1199–216. doi: 10.1128/MCB.00827-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demircan B, Dyer LM, Gerace M, Lobenhofer EK, Robertson KD, Brown KD. Comparative epigenomics of human and mouse mammary tumors. Genes Chromosomes Cancer. 2009;48(1):83–97. doi: 10.1002/gcc.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engel N, Thorvaldsen JL, Bartolomei MS. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum Mol Genet. 2006;15(19):2945–54. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 52.Vostrov AA, Quitschke WW. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem. 1997;272(52):33353–9. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- 53.Xu J, Huo D, Chen Y, Nwachukwu C, Collins C, Rowell J, et al. CpG island methylation affects accessibility of the proximal BRCA1 promoter to transcription factors. Breast Cancer Res Treat. 2010;120(3):593–601. doi: 10.1007/s10549-009-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5(12):1743–53. [PubMed] [Google Scholar]
- 55.Renaud S, Loukinov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33(21):6850–60. doi: 10.1093/nar/gki989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98(3):387–96. doi: 10.1016/S0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 57.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–7. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17(3):637–41. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 59.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62(14):4075–80. [PubMed] [Google Scholar]
- 60.Feldmann A, Ivanek R, Murr R, Gaidatzis D, Burger L, Schubeler D. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 2013;9(12) doi: 10.1371/journal.pgen.1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zampieri M, Guastafierro T, Calabrese R, Ciccarone F, Bacalini MG, Reale A, et al. ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem J. 2012;441(2):645–52. doi: 10.1042/BJ20111417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herold M, Bartkuhn M, Renkawitz R. CTCF: insights into insulator function during development. Development. 2012;139(6):1045–57. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 63.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15(4):234–46. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin-Kleiner I. BORIS in human cancers -- a review. Eur J Cancer. 2012;48(6):929–35. doi: 10.1016/j.ejca.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Marshall AD, Bailey CG, Rasko JE. CTCF and BORIS in genome regulation and cancer. Curr Opin Genet Dev. 2014;24:8–15. doi: 10.1016/j.gde.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 66.Nguyen P, Cui HM, Bisht KS, Sun LC, Patel K, Lee RS, et al. CTCFL/BORIS is a methylation-independent DNA-binding protein that preferentially binds to the paternal H19 differentially methylated region. Cancer Res. 2008;68(14):5546–51. doi: 10.1158/0008-5472.CAN-08-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitchen NS, Schoenherr CJ. Sumoylation modulates a domain in CTCF that activates transcription and decondenses chromatin. J Cell Biochem. 2010;111(3):665–75. doi: 10.1002/jcb.22751. [DOI] [PubMed] [Google Scholar]


