Abstract
Acinetobacter baumannii (A. baumannii) is an important nosocomial pathogen in healthcare institutions. β-Lactamase-mediated resistance is the most common mechanism for carbapenem resistance in A. baumannii. The aim of this study was to determine the antibiotic resistance pattern, to detect OXA encoding genes, class A, bla PER-1, and to detect the presence of ISAba1. A total of 124 A. baumannii isolates were collected from hospitalized patients in a teaching hospital in Kashan, Iran. The susceptibility of isolates to different antibiotics was determined by disk-diffusion method. PCR was used to detect bla PER-1, bla OXA-23, bla OXA-24, bla OXA-51, bla OXA-58, and ISAba1 genes. All isolates were resistant to ceftazidime, ceftriaxone, and cefotaxime. All of the isolates revealed susceptibility to polymyxin B and colistin. Ninety-six percent of the isolates were extensive drug resistance (XDR), 5.6% extended spectrum beta-lactamase (ESBL), and 54.8% metallo-beta-lactamase (MBL). All isolates were positive for bla OXA-51 and ISAba1. bla OXA-23, bla OXA-24, and bla OXA-58 were found in 79.8%, 25%, and 3.2%, respectively. The frequency rate of bla PER-1 gene was 52.4%. Multidrug resistant A. baumannii isolates are increasing in our setting and extensively limit therapeutic options. The high rate presence of class D carbapenemase-encoding genes, mainly bla OXA-23 carbapenemases, is worrying and alarming as an emerging threat in our hospital.
1. Introduction
Multidrug resistant A. baumannii is a main cause of hospital acquired infections and recognized to cause a wide spectrum of life-threatening diseases. This organism is resistant to almost all frequently accessible antibiotics which limits treatment options [1]. Emergence of resistance during medication for A. baumannii infections may occur and yields increasing rates of morbidity, mortality, and therapeutic costs [2]. A. baumannii is capable of accumulating multiple antibiotic resistance genes, leading to development of multidrug resistant or extensively drug resistant strains. Acquired resistance characteristics occur as consequences from mutation or acquisition of exogenous resistance determinants and can be mediated by different mechanisms, including beta-lactamases, alterations in cell-wall channels, and efflux pumps. The most worrying clinical resistance mechanism has been acquisition of serine and metallo-beta-lactamases, which present resistance to carbapenems [3]. Therefore, this study was performed to identify the prevalence of beta-lactamases producing multidrug resistant A. baumannii and the molecular characterization of prevalent genes among them, with the purpose of improving the therapeutic options and decreasing the morbidity and mortality in Beheshti Hospital in Kashan, Iran.
2. Materials and Methods
2.1. Sample Collection
From June 2013 to November 2014, a descriptive study was conducted with various clinical samples collected from a tertiary care hospital in Kashan, Iran. A total of 124 A. baumannii isolates were collected from clinical samples including tracheal tube 63 (50.8%), blood, 28 (22.6%), sputum, 10 (8.1%), pleural fluid, 7 (5.6%), urine, 7 (5.6%), cerebrospinal fluids, 4 (3.2%), urine catheter, 3 (2.4%), and wounds, 2 (1.6%), from hospitalized patients. The Ethics Committee of Kashan University of Medical Sciences approved the study protocol.
Bacterial Isolates. A. baumannii isolates were identified by using MICROGEN GNA+B (Microgen Bioproducts Co., UK).
2.2. Determination of Antibiotic Resistance Patterns of A. baumannii
Antibiotic sensitivity test of the isolates was determined using the Kirby-Bauer disk-diffusion breakpoint assay on Mueller-Hinton agar (Merck, Germany); and the cultures were incubated for 24 h at 37°C. Bacteria were classified as susceptible, intermediate, or resistant to antibiotics in accordance with the current Clinical Laboratory Standard Institute recommendations [4]. Piperacillin (100 μg), ampicillin-sulbactam (10/10 μg), piperacillin-tazobactam (100/10 μg), cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cefepime (30 μg), imipenem (10 μg), meropenem (10 μg), amikacin (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), tetracycline (30 μg), trimethoprim-sulfamethoxazole (1.25/23.75 mg), colistin (10 μg), and polymyxin B (300 IU) disks were purchased from MAST, Merseyside, UK. Escherichia coli ATCC 25922 were used as the quality control strain in every susceptibility test.
MDR, XDR, and PDR Definitions. Multidrug resistant (MDR) was applied when the isolate is nonsusceptible to at least one antibacterial agent in ≥3 of the following classes of antibiotics including quinolones, broad-spectrum cephalosporins, beta-lactamaseinhibitor/beta-lactams, aminoglycosides, tetracyclines, trimethoprim-sulfamethoxazole, and carbapenems. Extensive drug resistance (XDR) is defined as bacterial isolates remaining susceptible to only one or two categories. Pan-drug resistant (PDR) is defined as bacterial isolates with nonsusceptibility to all agents in all of the antimicrobial categories (i.e., no agents tested as susceptible for the organism). Thus, a bacterial isolate that is characterized as XDR will also be categorized as MDR. Similarly, a bacterial isolate would have to be XDR in order for it to be further defined as PDR [5].
2.3. Phenotypic Tests for Detection of ESBLs
ESBL was detected by double-disk-diffusion test using cefotaxime (30 μg) and with cefotaxime/clavulanic acid (CTX 30 μg + CA 10 μg per disk) (MAST, Merseyside, UK). In accordance with the Clinical and Laboratory Standards Institute (CLSI) recommended guidelines, an increase in zone diameter ≥5 mm in the presence of clavulanic acid indicated presence of the extended spectrum-β-lactamases (ESBLs) in test organism [6].
2.4. Phenotypic Tests for the Detection of MBL
Imipenem-resistant isolates were screened for producing metallo-beta-lactamase (MBL). The double-disk synergy test (DDST) was performed for identification of MBL by imipenem (10 μg), alone and in combination with EDTA (750 μg/disk) (ROSCO, Denmark). An increase in the zone diameter of ≥7 mm around imipenem plus EDTA disk compared to that of imipenem disk alone was considered as positive for MBL production [6].
2.5. PCR Amplification of the Beta-Lactamase-Encoding Genes
A series of PCR reactions were performed to detect the different Ambler class bla genes. Primers were designed to amplify the following carbapenemase-encoding genes using a multiplex PCR: class D: bla OXA-23, bla OXA-24, bla OXA-51, and bla OXA-58; class A: bla PER-1 [7, 9]. IS elements (ISAba1) were amplified by PCR using previously described methods [8]. All isolates were subjected to the multiplex PCR to detect bla OXA-23, bla OXA-24, bla OXA-51, and bla OXA-58 genes. The primers used for the analysis are listed in Table 1.
Table 1.
Primers used in this study for amplification of genes from Acinetobacter baumannii isolates.
| Primer | Nucleotide sequence (5′ to 3′) | Amplicon size (bp) | Reference |
|---|---|---|---|
| OXA51LF | TAA TGC TTT GAT CGG CCT TG | 353 | Feizabadi et al. (2008) [7] |
| OXA51LR | TGG ATT GCA CTT CAT CTT GG | ||
|
| |||
| OXA23LF | GAT CGG ATT GGA GAA CCA GA | 501 | Feizabadi et al. (2008) [7] |
| OXA23LR | ATT TCT GAC CGC ATT TCC AT | ||
|
| |||
| OXA24LF | GGT TAG TTG GCC CCC TTA AA | 246 | Feizabadi et al. (2008) [7] |
| OXA24LR | AGT TGA GCG AAA AGG GGA TT | ||
|
| |||
| OXA58LF | AAGTATTGGGGCTTGTGCTG | 599 | Feizabadi et al. (2008) [7] |
| OXA58LR | CCCCTCTGCGCTCTACATAC | ||
|
| |||
| ISAba1F | CACGAATGCAGAAGTTG | 549 | Segal et al. (2005) [8] |
| ISAba1R | CGACGAATACTATGACAC | ||
|
| |||
| PER-1 F | ATGAATGTCATTATAAAAGC | 925 | Vahaboglu et al. (2001) [9] |
| PER-1 R | AATTTGGGCTTAGGGCAGAA | ||
PCR products run on 1.0% agarose gel, stained with ethidium bromide, and photographed by UV illumination (Ingenius, Syngene). The size of the PCR product was compared with 100 bp DNA ladder (Bioneer, Korea). A. baumannii ATCC 19606 was used as reference strain.
Sequencing Method. Sequencing was done by Sanger's method (Applied Biosystems 3730/3730xl DNA Analyzers Sequencing; Bioneer). The sequences were analyzed using ChromasPro version 1.7.5 Technelysium (http://www.technelysium.com.au/). GenBank accession numbers for bla OXA-23, bla OXA-24, bla OXA-51, and bla PER-1 are KP462887, KP462888, KP462889, and KP462892.
2.6. Statistical Analysis
Statistical data analysis was conducted using SPSS software version 19 (SPSS Inc., Chicago, IL). The chi-square test was used to compare antibiotic resistance rates in this study. P < 0.05 was considered statistically significant.
3. Results
The mean age of the studied patients was 54.2 ± 18.1 years, which ranged from 23 to 95 years. The majority of patients (61.3%) were male, 76 versus 48 female. The number and rates of isolates from different wards of the hospital were as follows: ICU, 69 (55.6%); Internal Medicine, 26 (21%); Emergency Room, 23 (18.5%); and Pediatrics, 6 (4.8%).
Antimicrobial Resistance Pattern. All strains were resistant to ceftazidime, ceftriaxone, and cefotaxime, while they were susceptible to colistin and polymyxin B. The resistance patterns of all A. baumannii isolates are shown in Figure 1. All of the A. baumannii isolates were considered as multidrug resistance (MDR), and 81 (96%) isolates were extensively drug resistant (XDR). Fortunately, none of the isolates were pan-drug resistant (PDR). Seven (5.6%) of isolates were ESBL positive and 68 (54.8%) isolates produced metallo-β-lactamases (MBLs). In our study, all of the A. baumannii isolates were positive for bla OXA-51 and ISAba1. Analysis of incidence for OXA encoding genes of isolates demonstrated that 79.8% of them were positive for bla OXA-23, 25% for bla OXA-24, and 3.2% for bla OXA-58. The coexistence of different bla OXA genes in isolates was observed, and their relative amounts were as follows: bla OXA-51 + bla OXA-23 in 75 (60.5%); bla OXA-51 + bla OXA-24 in 7 (5.6%); bla OXA-51 + bla OXA-58 in 3 (2.4%); bla OXA-51 + bla OXA-23 + bla OXA-24 in 23 (18.5%), and bla OXA-51 + bla OXA-23 + bla OXA-24 + bla OXA-58 in 1 isolate (0.8%). Amplification for the presence of oxacillinases genes is shown in Figure 2. bla PER-1 of Ambler class Aβ-lactamase-encoding gene was identified in 65 (52.4%) of the isolates. Table 2 presents genetic and phenotypic analyses of isolates. Figure 3 shows the variance in the size of amplicon products of the multiplex PCR assay for different OXA genes.
Figure 1.
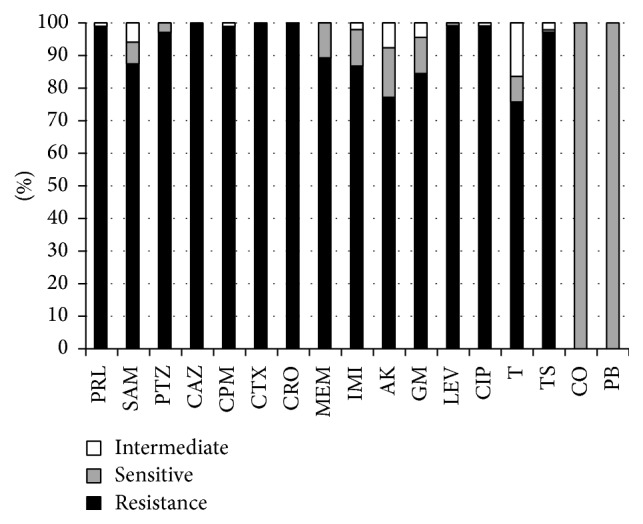
Antimicrobial susceptibility test of 124 A. baumannii isolates in Iran. PRL: piperacillin; SAM: ampicillin-sulbactam; PTZ: piperacillin-tazobactam; CAZ: ceftazidime; CPM: cefepime; CTX: cefotaxime; CRO: ceftriaxone; MEM: meropenem; IMI: imipenem; AK: amikacin; GM: gentamicin; LEV: levofloxacin; CIP: ciprofloxacin; T: tetracycline; TS: trimethoprim-sulfamethoxazole; CO: colistin; PB: polymyxin B.
Figure 2.
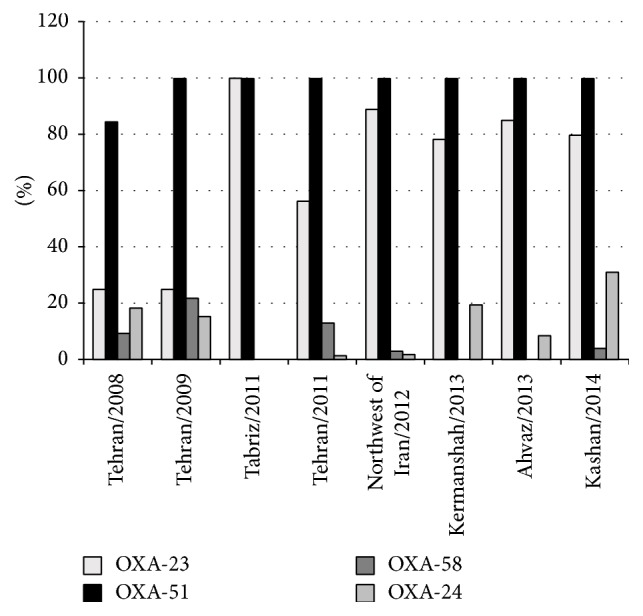
Frequency rates of bla OXA genes in A. baumannii isolates in Iran (2008–2014).
Table 2.
Summary of resistance: phenotypic and genetic characteristics of the isolatesa.
| Pattern | Resistance phenotypeb | bla genes | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | PRL | SAM | PTZ | CAZ | CPM | CTX | CRO | MEM | IMI | AK | GM | LEV | CIP | T | TS | CO | PB | OXA-23 | OXA-24 | OXA-51 | OXA-58 | PER-1 | ISAab1 | |
| 1 | 25 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | − | + | − | − | + |
| 2 | 16 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | − | + | − | + | + |
| 3 | 10 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | − | + | + |
| 4 | 9 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | − | − | + |
| 5 | 8 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | − | − | + |
| 6 | 5 | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | − | − | + | − | + | − | + | + |
| 7 | 4 | + | − | + | + | + | + | + | − | − | + | + | + | + | + | + | − | − | − | − | + | − | + | + |
| 8 | 3 | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | − | − | + | − | + | − | − | + |
| 9 | 3 | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | − | + | − | + | − | − | + |
| 10 | 2 | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | − | − | + | − | + | − | − | + |
| 11 | 2 | + | − | + | + | + | + | + | + | + | + | + | + | + | − | + | − | − | + | − | + | − | − | + |
| 12 | 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | − | − | − | + | − | + | + |
| 13 | 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | + | + | − | − | + |
| 14 | 1 | + | + | + | + | + | + | + | + | + | + | − | + | + | − | + | − | − | + | + | + | − | + | + |
| 15 | 1 | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | − | − | + | + | + | − | + | + |
| 16 | 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | − | + | + | |
| 17 | 1 | + | − | + | + | + | + | + | + | + | − | + | + | + | + | + | − | − | − | + | + | − | + | + |
| 18 | 1 | + | + | + | + | + | + | + | + | + | − | + | + | + | − | + | − | − | − | + | + | − | + | + |
| 19 | 1 | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | − | − | − | + | + | − | + | + |
| 20 | 1 | + | + | + | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | + | + | − | − | + |
| 21 | 1 | + | + | − | + | − | + | + | − | − | − | − | + | + | − | − | − | − | − | − | + | + | + | + |
| 22 | 1 | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | − | − | − | − | + | + | + | + |
| 23 | 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | − | − | − | + | + | − | + |
| 24 | 1 | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | − | − | + | + | + | − | − | + |
| 25 | 1 | + | − | + | + | + | + | + | + | + | − | + | + | + | + | + | − | − | + | + | + | − | − | + |
| 26 | 1 | + | − | + | + | + | + | + | + | + | − | − | + | + | + | + | − | − | + | + | + | − | − | + |
| 27 | 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | − | + |
| 28 | 1 | + | − | + | + | + | + | + | − | − | − | − | + | + | − | − | − | − | − | − | + | − | − | + |
| 29 | 1 | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + | − | − | − | − | + | − | − | + |
| 30 | 1 | + | − | − | + | + | + | + | − | − | − | − | + | + | − | − | − | − | − | − | + | − | − | + |
| 31 | 1 | + | − | + | + | + | + | + | − | − | + | − | + | + | − | + | − | − | − | − | + | − | − | + |
| 32 | 1 | + | + | + | + | + | + | + | − | − | + | + | + | + | + | + | − | − | − | − | + | − | + | + |
| 33 | 1 | − | + | + | + | + | + | + | − | − | − | + | + | + | + | + | − | − | − | − | + | − | + | + |
| 34 | 1 | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + | − | − | − | − | + | − | + | + |
| 35 | 1 | + | − | − | + | + | + | + | − | − | − | − | + | + | + | + | − | − | − | − | + | − | + | + |
| 36 | 1 | + | + | + | + | + | + | + | − | + | + | + | + | + | − | + | − | − | − | − | + | − | + | + |
| 37 | 1 | + | + | + | + | + | + | + | + | + | − | + | + | + | − | + | − | − | + | − | + | − | + | + |
| 38 | 1 | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | − | − | + | − | + | − | + | + |
| 39 | 1 | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | − | − | + | − | + | − | + | + |
| 40 | 1 | + | − | + | + | + | + | + | − | + | + | − | + | + | − | + | − | − | + | − | + | − | + | + |
| 41 | 1 | + | + | + | + | + | + | + | + | − | − | + | + | + | − | + | − | − | + | − | + | − | + | + |
| 42 | 1 | + | + | + | + | + | + | + | + | + | + | − | + | + | − | + | − | − | + | − | + | − | − | + |
| 43 | 1 | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | − | − | + | − | + | − | − | + |
| 44 | 1 | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | − | − | + | − | + | − | − | + |
| 45 | 1 | + | + | + | + | + | + | + | + | + | − | − | + | + | − | + | − | − | + | − | + | − | − | + |
| 46 | 1 | + | − | + | + | + | + | + | + | − | − | + | + | + | + | + | − | − | + | − | + | − | − | + |
a+Presence of indicated gene or resistance phenotype.
bPRL: piperacillin; SAM: ampicillin-sulbactam; PTZ: piperacillin-tazobactam; CAZ: ceftazidime; CPM: cefepime; CTX: cefotaxime; CRO: ceftriaxone; MEM: meropenem; IMI: imipenem; AK: amikacin; GM: gentamicin; LEV: levofloxacin; CIP: ciprofloxacin; T: tetracycline, TS: trimethoprim-sulfamethoxazole; CO: colistin; PB: polymyxin B.
Figure 3.
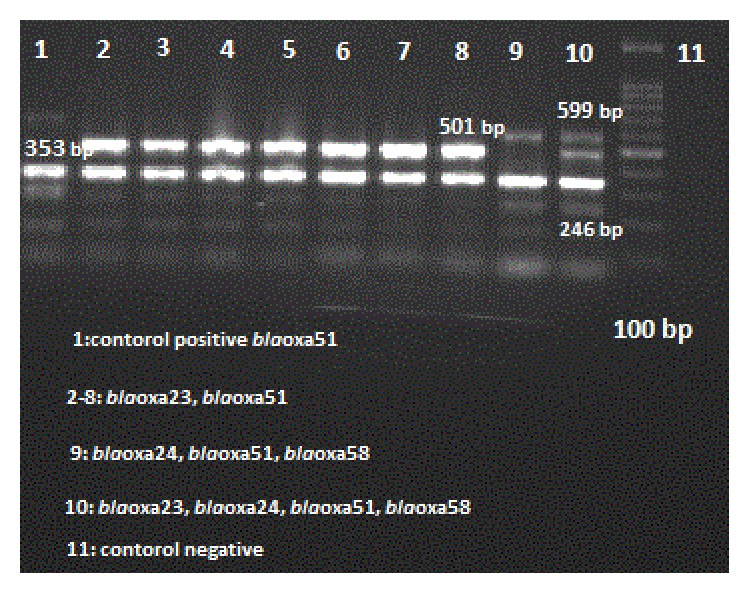
Amplification for presence of oxacillinases genes by multiplex PCR in A. baumannii isolates in Iran.
4. Discussion
The results of this study showed that the 124 A. baumannii isolates were extremely resistant to most common antibiotics, surprisingly, 100% to ceftazidime, ceftriaxone, and cefotaxime. In addition, 100% of isolates were MDR, 96% XDR, and 54.8% MBL producers. In comparison, the frequency rate of ESBL producing A. baumannii (5.6%) was not notable in the present study. According to the previous reports, high resistance rates to carbapenems have been reported in Iran, ranging from 25% to 96.1% [10–15]. Carbapenems have been the drugs of choice for the treatment of nosocomial infections caused by A. baumannii; however carbapenem-resistant strains of A. baumannii have been reported worldwide [16]. The extensive prescription of carbapenems in our hospitals in order to treat A. baumannii infections has led to occurrences of carbapenem-resistant isolates. Extreme use of third-generation cephalosporins and aztreonam has exacerbated the problem of carbapenem resistance rate. In hospitals with high rates of resistance to the broad-spectrum cephalosporin, a combination of beta-lactam/beta-lactamase inhibitors, or a carbapenem, selection of appropriate antibiotics is critical. Production of carbapenem-hydrolyzingβ-lactamases which is distributed worldwide is the major mechanism of resistance to carbapenems [17]. Polymyxin B and colistin are the most commonly used agents for Acinetobacter isolates which are resistant to first-line agents. The results of this study showed that 100% of the isolates were sensitive to polymyxins. It was found that 100% of A. baumannii were sensitive to colistin in Algeria, 70.9% in Saudi Arabia, 92.5% in Kuwait, and 95% in Egypt [18]. Epidemics of isolates harboring genes encoding OXA-type carbapenemase (bla OXA-23, bla OXA-24, bla OXA-51, and bla OXA-58 groups) have increasingly been described worldwide [19]. In the present study, 79.8% of A. baumannii isolates were positive for bla OXA-23 gene and more than 80% of them were resistant to both imipenem and meropenem. There are also studies on susceptibility test to carbapenems, in which an isolate is susceptible to meropenem but resistant to imipenem and vice versa; some studies have recently reported that bla OXA-23 is the most frequent type of carbapenemase identified among carbapenem-resistant A. baumannii [20]. The prevalence rate of bla OXA-23 has been detected from 0 to 98.4%; bla OXA-51 and ISAba1 were observed in all isolates. The prevalence rate of bla OXA-24/40 has been detected from 0 to 85.43%, while the rate for bla OXA-58 has been reported from 0% to 96.9% in Colombia, Greece, Korea, Poland, Taiwan, and Turkey [13, 18]. The comparative frequency rates for bla OXA genes in A. baumannii isolates in Iran are shown in Figure 2 [7, 12, 13, 21–23]. bla PER-1 was detected in 52.4% of the A. baumannii isolates. It was also found in Acinetobacter isolates from Argentina, Belgium, Egypt, France, India, Iran, Saudi Arabia, and South Korea [18, 24]. In addition, 48 isolates (48.5%) simultaneously were positive for both bla OXA-23 and bla PER-1 genes. Empiric antibiotic therapy for Acinetobacter isolates should be selected based on local susceptibility patterns, which consists of a broad-spectrum cephalosporin, a combination of beta-lactam/beta-lactamase inhibitors, or a carbapenem. In infectious cases with occurrence of highly antibacterial resistance rates, it is recommended to use a combination of the above agents with an antipseudomonal fluoroquinolone, an aminoglycoside, or colistin to impede the chance of treatment failure. With consideration of resistance rates to the first-line antibacterial agents, therapeutic choices are mandatory limited to polymyxins, minocycline, and tigecycline.
5. Conclusions
The results of the present study indicated the occurrence of high prevalence of MDR A. baumannii and signifies the alarming spreads of bla OXA-23 carbapenemase in our setting. On the whole, these findings recommend that screening can provide helpful information for comparisons of MDR genetic collection between different populations and countries and potentially support more infection control strategies and chemotherapeutic treatment programs.
Disclosure
The paper is based on the thesis of M.S. degree and financially was supported by Kashan University of Medical Sciences Research Fund (Grant no. 92149). The funder's had no role in study design, data collection and analysis, decision to publish, or preparation of the paper.
Conflict of Interests
The authors declare that there is no conflict of interests.
References
- 1.Zarrilli R., Pournaras S., Giannouli M., Tsakris A. Global evolution of multi-drug resistant Acinetobacter baumannii clonal lineages. International Journal of Antimicrobial Agents. 2013;41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Perez F., Hujer A. M., Hujer K. M., Decker B. K., Rather P. N., Bonomo R. A. Global challenge of multidrug-resistant Acinetobacter baumanni . Antimicrobial Agents and Chemotherapy. 2007;51(10):3471–3484. doi: 10.1128/aac.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vila J., Martí S., Sánchez-Céspedes J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii . Journal of Antimicrobial Chemotherapy. 2007;59(6):1210–1215. doi: 10.1093/jac/dkl509. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-First Informational Supplement. Wayne, Pa, USA: CLSI; 2011. [Google Scholar]
- 5.Falagas M. E., Karageorgopoulos D. E. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among gram-negative bacilli: need for international harmonization in terminology. Clinical Infectious Diseases. 2008;46(7):1121–1122. doi: 10.1086/528867. [DOI] [PubMed] [Google Scholar]
- 6.Owlia P., Azimi L., Gholami A., Asghari B., Lari A. R. ESBL- and MBL-mediated resistance in Acinetobacter baumannii: a global threat to burn patients. Infezioni in Medicina. 2012;20(3):182–187. [PubMed] [Google Scholar]
- 7.Feizabadi M. M., Fathollahzadeh B., Taherikalani M., et al. Antimicrobial susceptibility patterns and distribution of BlaAXA genes among Acinetobacter spp. isolated from patients at Tehran hospitals. Japanese Journal of Infectious Diseases. 2008;61(4):274–278. [PubMed] [Google Scholar]
- 8.Segal H., Garny S., Elisha B. G. Is ISABA-1 customized for Acinetobacter? FEMS Microbiology Letters. 2005;243:425–442. doi: 10.1016/j.femsle.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Vahaboglu H., Coskunkan F., Tansel O., et al. Clinical importance of extended-spectrum β-lactamase (PER-1-type)-producing Acinetobacter spp. and Pseudomonas aeruginosa strains. Journal of Medical Microbiology. 2001;50(7):642–645. doi: 10.1099/0022-1317-50-7-642. [DOI] [PubMed] [Google Scholar]
- 10.Safari M., Saidijam M., Bahador A., Jafari R., Alikhani M. Y. High prevalence of multi-drug resistance and metallo-beta-lactamase (MβL) producing Acinetobacter baumannii isolated from patients in ICU wards, Hamadan. Journal of Research in Health Sciences. 2013;13(2):162–167. [PubMed] [Google Scholar]
- 11.Moniri R., Farahani R. K., Shajari G., Shirazi M. H. N., Ghasemi A. Molecular epidemiology of aminoglycosides resistance in Acinetobacter spp. with emergence of multidrug-resistant strains. Iranian Journal of Public Health. 2010;39(2):63–68. [PMC free article] [PubMed] [Google Scholar]
- 12.Mohajeri P., Farahani A., Feizabadi MM., Ketabi H., Abiri R., Najafi F. Antimicrobial susceptibility profiling and genomic diversity of acinetobacter baumannii isolates: a study in western Iran. Iranian Journal of Microbiology. 2013;5(3):5195–5202. [PMC free article] [PubMed] [Google Scholar]
- 13.Shoja S., Moosavian M., Peymani A., Tabatabaiefar M. A., Rostami S., Ebrahimi N. Genotyping of carbapenem resistant Acinetobacter baumannii isolated from tracheal tube discharge of hospitalized patients in intensive care units, Ahvaz, Iran. Iranian Journal of Microbiology. 2013;5(4):315–322. [PMC free article] [PubMed] [Google Scholar]
- 14.Japoni S., Farshad S., Ali A. A., Japoni A. Antibacterial susceptibility patterns and cross-resistance of acinetobacter, isolated from hospitalized patients, Southern Iran. Iranian Red Crescent Medical Journal. 2011;13(11):832–836. [PMC free article] [PubMed] [Google Scholar]
- 15.Rahbar M., Mehrgan H., Aliakbari N. H. Prevalence of antibiotic-resistant Acinetobacter baumannii in a 1000-bed tertiary care hospital in Tehran, Iran. Indian Journal of Pathology and Microbiology. 2010;53(2):290–293. doi: 10.4103/0377-4929.64333. [DOI] [PubMed] [Google Scholar]
- 16.Hujer K. M., Hujer A. M., Hulten E. A., et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrobial Agents and Chemotherapy. 2006;50(12):4114–4123. doi: 10.1128/aac.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y.-C., Sheng W.-H., Chen Y.-C., Chang S.-C., Hsia K.-C., Li S.-Y. Differences in carbapenem resistance genes among Acinetobacter baumannii, Acinetobacter genospecies 3 and Acinetobacter genospecies 13TU in Taiwan. International Journal of Antimicrobial Agents. 2010;35(5):439–443. doi: 10.1016/j.ijantimicag.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 18.Al-Agamy M. H., Khalaf N. G., Tawfick M. M., Shibl A. M., Kholy A. E. Molecular characterization of carbapenem-insensitive Acinetobacter baumannii in Egypt. International Journal of Infectious Diseases. 2014;22:49–54. doi: 10.1016/j.ijid.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Evans B. A., Amyes S. G. B. OXA β-lactamases. Clinical Microbiology Reviews. 2014;27(2):241–263. doi: 10.1128/cmr.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mugnier P. D., Poirel L., Naas T., Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii . Emerging Infectious Diseases. 2010;16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taherikalani M., Fatolahzadeh B., Emaneini M., Soroush S., Feizabadi M. M. Distribution of different carbapenem resistant clones of Acinetobacter baumannii in Tehran Hospitals. New Microbiologica. 2009;32(3):265–271. [PubMed] [Google Scholar]
- 22.Sohrabi N., Farajnia S., Akhi M. T., et al. Prevalence of oxa-type β-lactamases among Acinetobacter baumannii isolates from northwest of Iran. Microbial Drug Resistance. 2012;18(4):385–389. doi: 10.1089/mdr.2011.0077. [DOI] [PubMed] [Google Scholar]
- 23.Peymani A., Higgins P. G., Nahaei M.-R., Farajnia S., Seifert H. Characterisation and clonal dissemination of OXA-23-producing Acinetobacter baumannii in Tabriz, Northwest Iran. International Journal of Antimicrobial Agents. 2012;39(6):526–528. doi: 10.1016/j.ijantimicag.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Yong D., Shin J. H., Kim S., et al. High prevalence of PER-1 extended-spectrum β-lactamase-producing Acinetobacter spp. in Korea. Antimicrobial Agents and Chemotherapy. 2003;47(5):1749–1751. doi: 10.1128/aac.47.5.1749-1751.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


