Abstract
Background
Antibiotic resistance in bacteria leads to massive health problems. Incidence of carbapenem and multidrug resistance in Gram-negative bacteria are increasing globally and turn out to be a very urgent challenge in health care. Resistant bacteria play an important clinical role during hospital outbreaks as well as in sepsis. Rapid diagnostic tests are necessary to provide immediate information for antimicrobial treatment and infection control measures.
Methods
Our mass spectrometry-based assay was validated with 63 carbapenemase-producing Gram-negative bacterial isolates, and 35 carbapenem-resistant Gram-negative species with no carbapenemase production. These were analyzed from solid culture media and positive blood culture vials. After 4 h of incubation the carbapenemase products were analyzed with the MALDI-TOF MS. All the isolates were genotyped for carbapenemase genes by PCR and sequencing.
Results
For culture isolates the concordance of hydrolysis assay to genetic results was 98 % for OXA variants, KPC, VIM, IMP, GIM, and NDM. In contrast, only 14 of 29 Acinetobacter baumannii isolates carrying the OXA and NDM genes could be identified from blood culture. However, from blood culture vials our method allowed the detection of carbapenemases in 98 % of Pseudomonas and Enterobacteriaceae isolates harboring different genes.
Conclusions
This MALDI-TOF MS–based assay permitted the detection of carbapenemases either from solid culture media (98–100 %) or blood culture vials (96 %) for all non-A. baumannii isolates within 4 h. In case of A. baumannii isolates the assay was highly sensitive for the detection of carbapenemases directly from solid culture media.
Keywords: MALDI-TOF MS, Carbapenemases, Acinetobacter baumannii, MDR Gram-negative bacteria, Hydrolysis
Background
The resistance of bacteria to antibiotics has increased in recent decades. Resistant bacteria can significantly complicate the treatment of infections in critically ill patients; especially in surgery, hemato-oncology, and intensive care in general [1, 2]. Recently, bacterial isolates that are resistant to all available antibiotics have also been emerging [3, 4]. The identification and detection of specific resistance mechanisms in these bacteria are of paramount importance to clinical microbiologists for diagnosis and treatment of systemic infections, such as blood stream infections (BSIs), which allows for effective management of systemic infections, which allows de-escalation from broad spectrum to targeted antibiotics, thus reducing overuse of carbapenems and other broad spectrum antibiotics. The diagnostic methods for detection of carbapenemase producers include at first level the antibiotic susceptibility testing results obtained by disk diffusion or automated systems, E-test and the verification via modified Hodge test. However, the later method may lack specificity (high-level AmpC producers) and sensitivity, e.g. weak detection of NDM variants. Several technologies using molecular methods have been developed in recent years (i.e., targeted PCR assays [5, 6] and peptide nucleic acid fluorescent in situ hybridization [7, 8]). Nowadays, Matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) is increasingly utilized by clinical microbiology laboratories for the identification of bacteria and yeasts [9, 10]. MALDI-TOF MS has been performed for the detection of antimicrobial resistance, e.g. β-lactam resistance in Gram-negative bacteria [11, 12]. For instance, this method is effective for detection of carbapenemase-producing gram-negative bacteria [9, 12–15].
Methods
Bacterial isolates
Sixty three Gram-negative bacteria, comprised of Enterobacteriaceae and non-fermenters, were collected by the National Reference Center for Gram-negative bacteria at the Ruhr University Bochum and at the Institute in Wuppertal. All of the isolates had been previously characterized at the molecular level for the diverse types of carbapenemases (KPC, GIM, IMP, NDM, OXA, and VIM). As negative control, 35 non-carbapenemase-producing, but carbapenem-resistant isolates were included. Species identification was performed using a MALDI-TOF Biotyper system (Bruker Daltonics, Bremen, Germany) and phenotypic antimicrobial susceptibility testing by use of Phoenix™ automated system (Becton–Dickinson, Heidelberg, Germany), including ampicillin (± sulbactam), piperacillin (± tazobactam), cefuroxime, cefotaxime, cefepime, ceftazidime, imipenem, meropenem, gentamicin, ciprofloxacin, levoflocaxin, co-trimoxazole, amikacin, fosfomycin, tigecycline, and colistin.
Imipenem hydrolysis assay
A colony of an overnight bacterial culture on MacConkey agar (Becton–Dickinson, Heidelberg, Germany) was washed in 1 ml H2O dest. and centrifuged at 13,200 rpm for 1 min, the harvested pellet was resuspended in 20 mM Tris–HCl, 20 mM NaCl, pH 7.0, to an inoculum equivalent to 3.0 McFarland standard. A 1-ml aliquot of the suspension was centrifuged at 13,200 rpm for 1 min; the harvested pellet was resuspended in 50 µl of a reaction buffer (20 mM Tris–HCl), pH 7.0, supplemented with 0.1 mM imipenem (Fresenius Kabi, Bad Homburg, Germany). Four hours after incubation at 37 °C in an Eppendorf thermomixer [agitation of 950 rpm] (Eppendorf, Hamburg, Germany) the reaction mixture was centrifuged at 13,200 rpm for 1 min; 1 µl of the supernatant was mixed with 1 µl HCCA and allowed to dry on a target. Mass spectra were measured after drying between 160 and 700 m/z, using Microflex LT mass spectrometer. Spectra were analyzed using the software program FlexControl, version 3.0 (Bruker Daltonics, Bremen, Germany). For the analysis of the hydrolysis activity from blood culture vials containing the Gram-negative bacterial isolates, 8 ml of blood culture—after 10 min at 800 rpm—the supernatant was mixed with 200 μl of lysis buffer. After vortexing, the sample was centrifuged at 13,200 rpm for 2 min, the supernatant was discarded, and the pellet was washed with 1 ml of deionized H2O. The water was thoroughly removed, and the bacterial pellets (107–109 CFU) were resuspended in 10 μl of 0.1 mM imipenem solution and SDS (2 %). The suspension was incubated at 37 °C as mentioned above. Thereafter centrifugation at 13,200 rpm for 2 min followed and the cell-free supernatant was analyzed by MALDI-TOF–MS.
The absence of the imipenem + matrix component peak at 479 m/z (299 + 180 m/z) was considered as positive for the carbapenemase production, indicating the peaks of the degradation products with overlap of mass peaks. For non-carabapenemase-producing isolates, the complex peak of imipenem/matrix was recorded at 479 m/z. According to these findings the interpretation criteria were established.
Results
Identification of relevant peaks in the MS-spectra after hydrolysis assay
To generate and optimize our hydrolysis assay, we performed MALDI-TOF MS analysis following different periods of incubation with imipenem (1, 2, 3, and 4 h—data not shown). The assay was conducted with 3.0 of McFarland bacterial inoculum and 0.1 mM imipenem. Although after 2 h incubation time the KPC activity was measurable, for other enzyme variants 4 h of incubation was required to demonstrate significant signals of hydrolysis in MALDI-TOF MS.
As a negative control MALDI-TOF MS analysis was performed prior to the hydrolysis assay. Differences of peaks in the spectra generated were recorded and subsequently compared to the results gained after incubation. The MS signal of imipenem-matrix complex (Fig. 1) disappeared if the bacterial isolate produced carbapenemase. For non-carbapenemase-producing species we observed the signal of the complex at 479 m/z (Fig. 1).
Fig. 1.
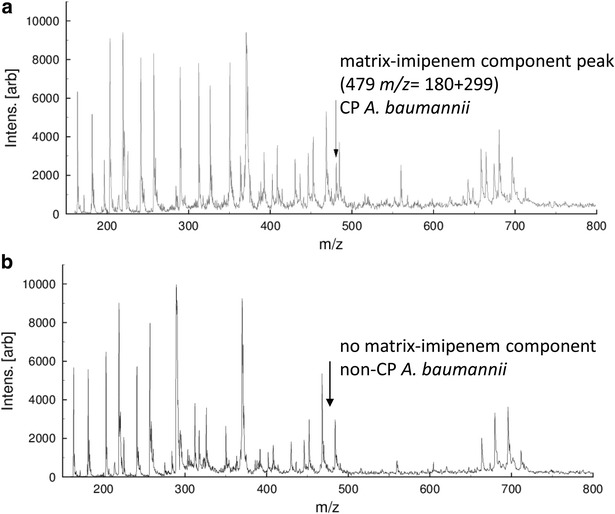
MALDI-TOF MS analysis showing the selected spectrum (range 160–800 m/z) after the performance of imipenem hydrolysis; OXA-23 carbapenemase-producing A. baumannii (a) and carbapenem-susceptible A. baumannii (b)
Hydrolysis assay for carbapenemase-producing Acinetobacter baumannii, Pseudomonas aeruginosa and different Enterobacteriaceae isolates directly from solid culture media
All investigated isolates were imipenem resistant as determined by automated susceptibility testing systems (BD Phoenix). In our study 38 A. baumannii isolates were investigated; all were positive for the genetic presence of carbapenemases. Twenty nine of these isolates carried the OXA gene variants OXA-23, -40, -58, -72, and OXA-164; the remaining isolates were positive for NDM-1, GIM-1 and VIP-2 genes. 25 P. aeruginosa and Enterobacteriaceae isolates were positive for different carbapenemase types. Seven of the later isolates produced the enzyme variant of OXA (Table 1). The IMP variants IMP-4 and IMP-14 were detected in single Enterobacter cloacae isolate whereas, the NDM variants, NDM-1 or NDM-6, were carried by Escherichia coli, Klebsiella pneumoniae and E. cloacae isolates. The OXA variants were detected in 6 different species, but one E. coli isolate with OXA-48 gene did not reveal significant hydrolysis of imipenem in the MALDI-TOF MS analysis. None of the control isolates (n = 35, Enterobacteriaceae and non-fermenters), which were resistant to imipenem and meropenem but negative for the presence of a carbapenemase-encoding genes, showed hydrolysis activity in the mass spectrometry, revealing 100 % specificity for the non-carbapenemase-producing bacteria. In brief, the sensitivity of the MALDI-TOF MS-based hydrolysis assay for A. baumannii isolates from solid culture was 97.4 % (n = 37 of 38), whereas the sensitivity of 100 % was reached for the different Enterobacteriaceae and P. aeruginosa isolates. All genotyping data for other enzyme variants of carbapenemases are detailed in Table 1.
Table 1.
Comparison of the genetic characteristics of the carbapenemase genes in A. baumannii, Pseudomonas aeruginosa and Enterobacteriaceae isolates and the activity of the respective enzymes and detection of imipenem hydrolysis by use of MALDI-TOF-MS directly from solid culture media and directly from positive blood culture vials
| Bacterial isolates | Enzyme variants Ambler classification |
Carbapenemase variants—genotyped | MALDI-TOF MS-based imipenem hydrolysis assay |
|---|---|---|---|
| Solid culture media | |||
| A. baumannii | B (n = 9) | NDM-1a (n = 7) | (n = 7) |
| GIM-1 (n = 1) | (n = 1) | ||
| VIM-2 (n = 1) | (n = 1) | ||
| A. baumannii | D (n = 29) | OXA-23 (n = 19) | (n = 19) |
| OXA-40 (n = 1) | (n = 1) | ||
| OXA-58 (n = 3) | (n = 3) | ||
| OXA-72 (n = 4) | (n = 3) | ||
| OXA-164 (n = 2) | (n = 2) | ||
| E. coli | A (n = 2) | KPC-2 (n = 2) | (n = 2) |
| K. pneumoniae | A (n = 3) | KPC-3 (n = 3) | (n = 3) |
| E. cloacae | B (n = 2) | IMP-4, IMP-14 (n = 2) | (n = 2) |
| S. marcescens | B (n = 1) | IMP-13 | (n = 1) |
| E. cloacae | B (n = 1) | NDM-1a (n = 1) | (n = 1) |
| E. coli | B (n = 2) | NDM-1a, -3 (n = 2) | (n = 2) |
| K. pneumoniae | B (n = 1) | NDM-1a (n = 1) | (n = 1) |
| K. pneumoniae | B (n = 2) | VIM-2 (n = 2) | (n = 2) |
| K. oxytoca | B (n = 1) | VIM-1, (n = 1) | (n = 1) |
| E. cloacae | B (n = 1) | VIM-4 (n = 1) | (n = 1) |
| P. aeruginosa | B (n = 2) | VIM-2 (n = 2) | (n = 2) |
| K. pneumoniae | D (n = 4) | OXA-48 (n = 4) | (n = 4) |
| C. freundii | D (n = 1) | OXA-162 (n = 1) | (n = 1) |
| E. coli | D (n = 1) | OXA-181 (n = 1) | (n = 1) |
| K. pneumoniae | D (n = 1) | OXA-204 (n = 1) | (n = 1) |
| Blood culture vials | |||
| A. baumannii | B (n = 9) | NDM-1a (n = 7) | (n = 2) |
| GIM-1 (n = 1) | (n = 1) | ||
| VIM-2 (n = 1) | None | ||
| A. baumannii | D (n = 29) | OXA-23 (n = 19) | (n = 15) |
| OXA-40 (n = 1) | None | ||
| OXA-58 (n = 3) | (n = 3) | ||
| OXA-72 (n = 4) | (n = 3) | ||
| OXA-164 (n = 2) | None | ||
| E. coli | A (n = 2) | KPC-2 (n = 2) | (n = 2) |
| K. pneumoniae | A (n = 3) | KPC-3 (n = 3) | (n = 3) |
| E. cloacae | B (n = 2) | IMP-4, IMP-14 (n = 2) | (n = 2) |
| S. marcescens | B (n = 1) | IMP-13 | (n = 1) |
| E. cloacae | B (n = 1) | NDM-1a (n = 1) | (n = 1) |
| E. coli | B (n = 2) | NDM-1a, −3 (n = 2) | (n = 2) |
| K. pneumoniae | B (n = 1) | NDM-1a (n = 1) | (n = 1) |
| K. pneumoniae | B (n = 2) | VIM-2 (n = 2) | (n = 2) |
| K. oxytoca | B (n = 1) | VIM-1, (n = 1) | (n = 1) |
| E. cloacae | B (n = 1) | VIM-4 (n = 1) | (n = 1) |
| P. aeruginosa | B (n = 2) | VIM-2 (n = 2) | (n = 2) |
| K. pneumoniae | D (n = 4) | OXA-48 (n = 4) | (n = 3) |
| C. freundii | D (n = 1) | OXA-162 (n = 1) | (n = 1) |
| E. coli | D (n = 1) | OXA-181 (n = 1) | (n = 1) |
| K. pneumoniae | D (n = 1) | OXA-204 (n = 1) | (n = 1) |
aNDM-1/-6
Hydrolysis assay directly from positive blood culture vials for the detection of carbapenemase-producing Acinetobacter baumannii, Pseudomonas aeruginosa and different Enterobacteriaceae isolates
To achieve faster results with the presented assay we set to run the hydrolysis assay on positive blood culture samples. Here, a total of 38 A. baumannii isolates were analyzed. Fourteen of the 38 isolates carried genes for a carabapenemase enzyme but did not show any activity in the MALDI-TOF MS hydrolysis assay (Table 1). Eight of the 29 OXA producing isolates (OXA-23, -40, -72, -164), five of the seven NDM-1 isolates and one VIM-2 A. baumannii isolate did not significantly show hydrolysis activity in the MALDI-TOF–MS (Table 1), although all the assays were performed at least in duplicate, revealing 63.2 % sensitivity.
Additionally, 25 multidrug-resistant Enterobacteriaceae and P. aeruginosa isolates spiked in blood culture vials were analyzed. Seven of 25 isolates produced a variant of OXA. The other enzyme variants are given in Table 1. All KPC-2/-3 producing isolates (n = 5) showed significant hydrolysis activity in the MALDI-TOF MS analysis. The hydrolysis activity of different class B enzymes was detected. Within the Ambler class D the OXA variants were detected in six different species, but one E. coli isolate with OXA-48 gene did not reveal significant hydrolysis activity in the MALDI-TOF MS hydrolysis analysis (Table 1). Taken together the present imipenem hydrolysis assay was able to detect carbapenemases in 24 of 25 (96 %) of blood cultures spiked with multidrug-resistant Enterobacteriaceae and P. aeruginosa. The 35 Gram-negative control isolates from positive blood culture vials showed no hydrolysis effects in the mass spectrometry.
Discussion
The rapid detection of clinically important beta-lactamases (e.g. extended-spectrum beta-lactamases and carbapenemases) in routine diagnostic laboratories is crucial for initial antibiotic therapy for patients as well as for the prevention of the spread of beta-lactamase-producing bacteria in health care settings. MALDI-TOF mass spectrometry allows clinical microbiologists to discern unique protein signatures, abundantly ribosomal proteins, in order to identify the pathogens [16]. In addition, there is potential for the detection of resistance mechanisms [10]. The most common mechanism of resistance to beta-lactams is the hydrolysis of the amide bond of the beta-lactam ring [10]. This last application includes identification of the antibiotic itself and its modified/degraded derivatives [9, 12–14, 17]; detection of the resistance proteins within the cell [11, 18], and discovery of mutations within the resistance genes through mini-sequencing [19, 20]. Almost 4 years ago two studies on direct carbapenemase detection by MALDI-TOF MS hydrolysis assay were reported [9, 13]. An analysis of antibiotics and their degradation products that are smaller than 1000 Da by MALDI-TOF MS is possible using a specific sample preparation [21]. Several studies have assessed the utility of MALDI-TOF mass spectrometry for the identification of beta-lactams and beta-lactam degradation products within four hours, either from the culture media or positive blood culture vials [9, 12–15, 17, 22].
Jung et al. reported the detection of ampicillin and cefotaxime resistances in hydrolysis assays directly from blood cultures [22]. However, these assays can only detect the presence of beta-lactamases as a resistance mechanism. Detection of the actual proteins conferring resistance in such resistant bacteria by MALDI-TOF–MS seems to be more challenging since the protein signatures of bacteria are very complex. Camara and Hays [11] established a proof of principle for this methodology in which they dissected the differences between wild-type and ampicillin-resistant E. coli strains. After the cultivation of resistant strains in broth supplemented with ampicillin, proteins were extracted by using a formic acid-isopropyl alcohol-water solution and spotted onto a MALDI-TOF MS target by a sandwich method using sinapinic acid as a matrix. They identified a 29.000 Da peak in resistant strains confirming that this mass/ion peak represented a beta-lactamase.
Kempf et al. detected the carbapenemase-production of Gram-negative bacteria by performing the imipenem hydrolysis and Ultraflex MS analysis [23]. They were able to detect the peaks of imipenem (299 m/z) and the degradation product (255 m/z) in the mass spectra. Furthermore, Sparbier et al. [17] reported that in their imipenem hydrolysis assay they observed a complex of imipenem and the matrix (190.05 m/z) at 489 m/z was observed. In our assay, we detected the imipenem-matrix-complex at 479 m/z and the peak of the matrix could be demonstrated at 180 m/z (Fig. 1). In cases of carbapenemase-producing isolates the peak of this complex disappeared, but neither degradation products nor sodium variants of the antibiotic of imipenem were detectable, as confirmed by Sparbier et al. [17]. Moreover, we were not able to detect the instable hydrolysis product of imipenem.
From culture isolates our mass spectrometry-based imipenem hydrolysis assay correlated with the genotypic detection in 98 % of the cases, i.e., one OXA-48 E. coli isolate did not demonstrate the hydrolysis of imipenem. The hydrolysis activity of such enzyme might be weak as described previously [24]. The other OXA-48 producing bacteria belonged to K. pneumoniae; these showed excellent hydrolysis activity in our MALDI-TOF–MS based hydrolysis assay. For the positive blood culture isolates, we were able to detect the carbapenemase activity in only 24 (63.2 %) of total 38 A. baumannii isolates. For the remaining 14 isolates no interpretation was possible concerning their carbapenemase activity; these included the enzyme groups OXA (OXA-23, -40, and -164), NDM-1 and VIM-2. Thus, eight of the 29 OXA producing isolates, five of the seven NDM-1 producing isolates and one VIM-2 positive isolate were not correctly detectable. Even the repeated incubation and measurements did not reveal better interpretable results.
In our hydrolysis assay we relied solely upon the disappearance of imipenem + matrix peak to consider an isolate as carbapenemase-producer. Kempf et al. distinguished carbapenemase producing isolates with regard to disappearance of imipenem peak or an intact/hydrolyzed imipenem ratio of <0.5 [23]. Recent studies consider Liquid Chromatography-Tandem Mass Spectrometry the gold standard method for small-molecule detection, and its performance is well suited to the detection of small amounts of compounds present in highly complex matrices, such as serum or bacterial growth broths [25]. As also observed by Kulkarni et al. [25], the effects of ion suppression on carbapenem detection have not been reported for MALDI-TOF, and this could be particularly important when test interpretation relies upon the lack of detection of a compound. In several studies, the matrix peak has been used as an internal standard, but the chemical differences between MALDI matrix and carbapenems are substantial, and differential ionization effects may not be effectively determined without an appropriate internal standard.
Authors’ contributions
BG and AH proposed, designed, carried out the study, BG analyzed the generated data and drafted the manuscript and MS participated in data analysis. MK provided with some of the strains from NRZ, MK and PAN participated in proofreading of the manuscript and in critical revision. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- MALDI-TOF MS
matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- MDR
multidrug resistant
- KPC
Klebsiellapneumoniae Carbapenemase
- NDM
New-Delhi-Metallo-betalactamase
- IMP
Imipenemase
- GIM
German Imipenemase
- OXA
Oxacillinase
Footnotes
B. Ghebremedhin and A. Halstenbach contributed equally to this work
Contributor Information
B. Ghebremedhin, Phone: +49-202-8962262, Email: bg.id@msn.com
A. Halstenbach, Email: alexander.halstenbach@uni-duesseldorf.de
M. Smiljanic, Email: manuela.smiljanic@uni-duesseldorf.de
M. Kaase, Email: martin.kaase@rub.de
P. Ahmad-Nejad, Email: parviz.ahmad-nejad@helioskliniken.de
References
- 1.Livermore DM. Fourteen years in resistance. Int J Antimicrob Agents. 2012;39:283–294. doi: 10.1016/j.ijantimicag.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Kaye KS, Pogue JM. Infections caused by resistant gram-negative bacteria: epidemiology and management. Pharmacotherapy. 2015;35(10):949–962. doi: 10.1002/phar.1636. [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20(9):821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 5.Raja S, Ching J, Xi L, Hughes SJ, Chang R, Wong W, et al. Technology for automated, rapid, and quantitative PCR or reverse transcription-PCR clinical testing. Clin Chem. 2005;51:882–890. doi: 10.1373/clinchem.2004.046474. [DOI] [PubMed] [Google Scholar]
- 6.Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother. 2012;67(4):906–909. doi: 10.1093/jac/dkr563. [DOI] [PubMed] [Google Scholar]
- 7.Forrest GN, Mankes K, Jabra-Rizk MA, Weekes E, Johnson JK, Lincalis DP, Venezia RA. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J Clin Microbiol. 2006;44:3381–3383. doi: 10.1128/JCM.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forrest GN, Mehta S, Weekes E, Lincalis DP, Johnson JK, Venezia RA. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother. 2006;58:154–158. doi: 10.1093/jac/dkl146. [DOI] [PubMed] [Google Scholar]
- 9.Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. Carbapenemase activity detection by matrix-assisted laser desorption–ionization time of flight mass spectrometry. J Clin Microbiol. 2011;49:3222–3227. doi: 10.1128/JCM.00984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrabák J, Chudácková E, Walková R. Matrix-assisted laser desorption ionization-time of flight (maldi-tof) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev. 2013;26(1):103–114. doi: 10.1128/CMR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camara JE, Hays FA. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Bioanal Chem. 2007;389:1633–1638. doi: 10.1007/s00216-007-1558-7. [DOI] [PubMed] [Google Scholar]
- 12.Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2012;50:2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burckhardt I, Zimmermann S. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol. 2011;49:3321–3324. doi: 10.1128/JCM.00287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooff GP, van Kampen JJ, Meesters RJ, van Belkum A, Goessens WH, Luider TM. Characterization of beta-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J Proteome Res. 2012;11:79–84. doi: 10.1021/pr200858r. [DOI] [PubMed] [Google Scholar]
- 15.Hoyos-Mallecot Y, Riazzo C, Miranda-Casas C, Rojo-Martín MD, Gutiérrez-Fernández J, Navarro-Marí JM. Rapid detection and identification of strains carrying carbapenemases directly from positive blood cultures using MALDI-TOF MS. J Microbiol Methods. 2014;105:98–101. doi: 10.1016/j.mimet.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Wieser A, Schneider L, Jung J, Schubert S. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review) Appl Microbiol Biotechnol. 2012;93(3):965–974. doi: 10.1007/s00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- 17.Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J Clin Microbiol. 2012;50:927–937. doi: 10.1128/JCM.05737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai JC, Hu YY, Zhang R, Zhou HW, Chen G-X. Detection of Ompk36 porin loss in Klebsiella spp. by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2012;50:2179–2182. doi: 10.1128/JCM.00503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikryannikova LN, Afanas’ev MV, Akopian TA, Il’ina EN, Kuz’min AV, Larionova EE, Smirnova TG, Chernousova LN, Govorun VM. Massspectrometry based minisequencing method for the rapid detection of drug resistance in Mycobacterium tuberculosis. J Microbiol Methods. 2007;70:395–405. doi: 10.1016/j.mimet.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Ikryannikova LN, Shitikov EA, Zhivankova DG, Il’ina EN, Edelstein MV, Govorun VM. A MALDI TOF MS-based minisequencing method for rapid detection of TEM-type extended-spectrum beta-lactamases in clinical strains of Enterobacteriaceae. J Microbiol Methods. 2008;75:385–391. doi: 10.1016/j.mimet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Pan C, Xu S, Zhou H, Fu Y, Ye M, Zou H. Recent developments in methods and technology for analysis of biological samples by MALDI-TOF-MS. Anal Bioanal Chem. 2007;387:193–204. doi: 10.1007/s00216-006-0905-4. [DOI] [PubMed] [Google Scholar]
- 22.Jung JS, Popp C, Sparbier K, Lange C, Kostrzewa M, Schubert S. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid detection of β-lactam resistance in Enterobacteriaceae derived from blood cultures. J Clin Microbiol. 2014;52(3):924–930. doi: 10.1128/JCM.02691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One. 2012;7:e31676. doi: 10.1371/journal.pone.0031676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67(7):1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 25.Kulkarni MV, Zurita AN, Pyka JS, Murray TS, Hodsdon ME, Peaper DR. Use of imipenem to detect KPC, NDM, OXA, IMP, and VIM carbapenemase activity from gram-negative rods in 75 minutes using liquid chromatography-tandem mass spectrometry. J Clin Microbiol. 2014;52(7):2500–2005. doi: 10.1128/JCM.00547-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


