Abstract
Information‐bearing nucleic acids display universal 3′‐5′ linkages, but regioisomeric 2′‐5′ linkages occur sporadically in non‐enzymatic RNA synthesis and may have aided prebiotic RNA replication. Herein we report on the enzymatic synthesis of both DNA and RNA with site‐specific 2′‐5′ linkages by an engineered polymerase using 3′‐deoxy‐ or 3′‐O‐methyl‐NTPs as substrates. We also report the reverse transcription of the resulting modified nucleic acids back to 3′‐5′ linked DNA with good fidelity. This enables a fast and simple method for “structural mutagenesis” by the position‐selective incorporation of 2′‐5′ linkages, whereby nucleic acid structure and function may be probed through local distortion by regioisomeric linkages while maintaining the wild‐type base sequence as we demonstrate for the 10–23 RNA endonuclease DNAzyme.
Keywords: DNAzyme, nucleic acids, nucleotides, polymerase, regioselectivity
Genetic information storage and propagation in biology is based on nucleic acids with uniform 3′‐5′ phosphodiester linkages. In contrast, 2′‐5′ linked RNA oligoadenylates1 and mixed 2′‐5′/3′‐5′ dinucleotides (e.g. Gp(2′‐5′)Ap(3′‐5′)2) are involved in innate immune signaling but not genetic information transfer. Sporadic 2′‐5′ linkages occur in non‐enzymatic RNA synthesis3 and have been proposed to facilitate primordial RNA replication and evolution via transient duplex destabilization3d while retaining overall RNA folding and function.4 Duplex destabilization by sporadic 2′‐5′ linkages has been examined in detail5 and results from weakened Watson–Crick base pairing and base stacking caused by lateral displacement of the base and adoption of non‐canonical C2′ endo puckering.6 Nevertheless, 2′‐5′ linked nucleic acids specifically hybridize with complementary 3′‐5′ RNA and 2′‐5′ RNA and DNA, albeit more weakly.5a, d, 7 Thus, despite their destabilizing influence on duplex structures, we reasoned that 2′‐5′ linked nucleic acids should be able to both encode and transmit genetic information to the canonical 3′‐5′ linked nucleic acids. Furthermore, their non‐canonical backbone conformations might be harnessed to expand the structural and functional space of nucleic acid ligands and enzymes.
2′‐5′ linkages are accessible through solid‐phase synthesis, but this is time‐ and cost‐intensive. Herein we report a rapid, inexpensive and scalable strategy for synthesizing defined 2′‐5′ linked DNA and RNA regioisomers by an engineered DNA polymerase capable of forming both canonical 3′‐5′ and non‐canonical 2′‐5′ linkages in either DNA or RNA (Figure 1).
Figure 1.
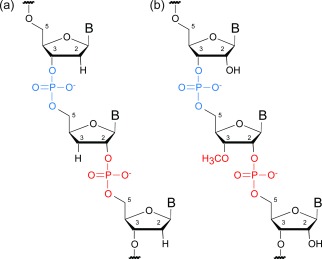
Structure of partially substituted a) 2′‐5′ DNA and b) 2′‐5′ RNA, with 3′O‐methyl groups as synthesized by polymerase TGLLK.
We first screened a panel of engineered8 and commercial polymerases for DNA synthesis in which dGTP was completely replaced by 3′deoxy‐GTP (3’dGTP). The engineered polymerases TgoT8b (a Tgo mutant comprising V93Q,9 D141A, E141A, A485L10) and TGK (TgoT: Y409G, E664K, previously described for primer‐dependent RNA synthesis8a) as well as the related commercial polymerase Vent(exo‐) were capable of full‐length DNA synthesis with mixed 3′‐5′/2′‐5′ linkages (Figure S1 in the Supporting Information). However, none of these efficiently synthesized RNA with mixed 3′‐5′/2′‐5′ linkages when GTP was replaced by 3’O‐methyl‐GTP (3′OMe‐GTP). To this end, we prepared a TGK variant with two further mutations known to expand the polymerase substrate spectrum (I521L,8b F545L11) yielding polymerase TGLLK (TgoT: Y409G, I521L, F545L, E664K; Figure S2), which proved effective at synthesizing both DNA and RNA with defined 2′‐5′ linkages (Figure 2).
Figure 2.
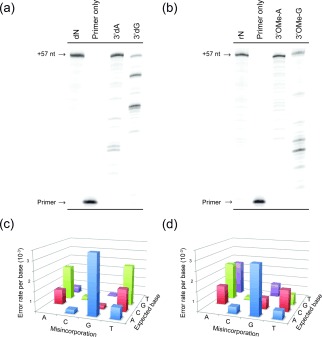
Enzymatic synthesis of partially substituted a) 2′‐5′ DNA b) 3′‐5′ RNA by TGLLK on a 57 nt template (TempN, see Supporting Information) encoding all possible dinucleotide combinations. Reactions in (a) involve dNTPs apart from 3′dA/G as indicated. Reactions in (b) involve NTPs apart from 3′OMeA/G as indicated. c),d) Error spectra of c) 3′dG/dHTP synthesis (aggregate misincorporation rate 5.08×10−4) and d) 3′OMe‐A/BTP synthesis (aggregate misincorporation rate 7.18×10−4). The columns show the misincorporation frequency for each incorrect nucleotide.
TGLLK is capable of fully replacing purine dNTPs and NTPs with their respective 3′deoxy (3′dATP, 3′dGTP) or 3′O‐methyl analogues (3′OMe‐ATP, 3′OMe‐GTP) during processive synthesis (Figure 2) even though these normally act as potent chain terminators (e.g. Cordycepin, 3′dATP12). Activity with 3′‐analogues of pyrimidine nucleotides or a combination of any two nucleotide analogues was poor, although some full‐length synthesis was possible with 3′dUTP or 3′OMe‐ATP/3′OMe‐GTP (Figure S3). Pausing is observed mostly at sequential 3′dNTP insertion sites, presumably because consecutive 2′‐5′ linkages weaken duplex stability and cause progressive conformational distortions relative to canonical 3′‐5′ helices.6c, 13
To ensure that full‐length synthesis is not the result of misincorporation utilizing only the dNTP substrates (as has been observed for some polymerases14), we sought to verify the presence of 2′‐5′ linkages using an HPLC assay, which distinguishes 2′‐5′ and 3′‐5′ phosphodiester linkages by mobility.15 This assay unambiguously confirmed the ability of TGLLK to insert specific 2′‐5′ linkages into DNA using 3′dATP (Figure 3).
Figure 3.
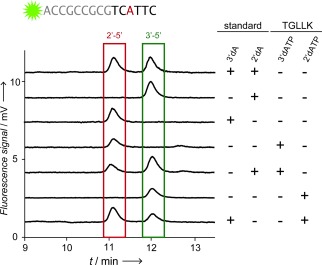
HPLC analysis of a fluorescent oligonucleotide that was synthesized by TGLLK either using 2′dATP or 3′dATP. Additional traces are corresponding chemically synthesized 2′‐5′ (3′dA)/3′‐5′ (2′dA) standards and an enzymatically synthesized 2′dA control. Standards and polymerase‐synthesized oligonucleotides were mixed in equimolar ratio and separated by HPLC under denaturing conditions.
Reverse transcription of 2′‐5′ substituted DNA and 3′OMe‐RNA back to all 3′‐5′ DNA using Taq DNA polymerase (Taq) and Avian Myleoblastosis Virus RT (AMV RT) respectively (as previously reported16) allowed us to determine aggregate fidelity of the information transfer through 2′‐5′ linkages in both DNA and RNA by deep sequencing, revealing misincorporation frequencies ranging from 8×10−3 for 3′dATP to 2×10−4 for 3′OMe‐GTP (Figure 2 c,d; Table S1).
Next, we set out to probe the impact of 2′‐5′ linkages on nucleic acid function. Limited, sporadic substitution (<25 %) of randomly distributed 2′‐5′ linkages is compatible with function in some ribozymes and aptamers as polyclonal populations.4 We reasoned that a better understanding of the functional impact of 2′‐5′ linkages might be gained from site‐specific insertion of regioisomeric linkages. Such a “structural mutagenesis” approach would not replace functional groups—like conventional mutagenesis—but instead alter their three‐dimensional positioning, potentially allowing a novel interrogation of the roles of different nucleotides in nucleic acid structure and function.
We chose the well‐studied 10–23 RNA‐endonuclease DNAzyme17 (Figure 4 a) to validate this “structural mutagenesis” approach and first synthesized 10–23 with progressively more 2′‐5′ linked purines (Figure 4 and Figure S4). This “primer scanning” approach allows a quick determination of obligatory 3′‐5′ linkages sensitive to distortion, neutral positions tolerating both 3′‐5′ and 2′‐5′ geometry, and any positions where 2′‐5′ introduction is beneficial. We identified nucleotide positions broadly falling into each category (Figure 4). For example, structural mutagenesis of the distal RNA‐binding arm revealed that introducing 2′‐5′ linkages after G18 and G22 results in a modest activity gain (Figure 4 b,c), presumably because of reduced product inhibition.18
Figure 4.
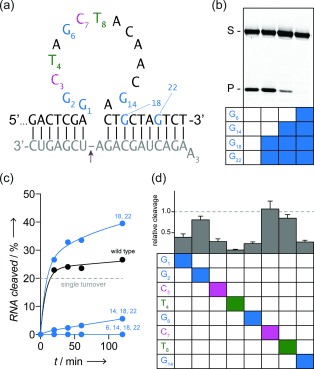
Structural mutagenesis of enzymatically synthesized 10–23 DNAzyme variants. a) 10–23 DNAzyme/substrate pair used in this study. b) Cleavage of the fluorescent RNA (200 nm) after 1 h at 37 °C by different DNAzyme variants (40 nm) containing 2′‐5′ linkages downstream of G6 or G14 shown in panel (a). Blue boxes indicate 2′‐5′ linkages after 3′dG. c) Cleavage reaction progress of DNAzymes with increasing amounts of 3′dG substitutions. d) Cleavage after 1 h by different DNAzyme variants containing single 2′‐5′ linkages produced by position‐selective enzymatic synthesis.
Previous analysis had shown that G14 is highly sensitive to mutation as changes to A, C, or T result in around a 20‐fold loss of activity and even conservative substitution of G with inosine or 2‐aminopurine results in a greater than 10‐fold drop in activity.19 In contrast, we find that structural mutagenesis of G14, whereby guanine functional groups are retained but a geometric perturbation of the active site is introduced,20 results in a comparatively mild reduction in 10–23 activity (around fivefold; Figure 4 b,c).
Primer scanning using 3′dA revealed modest activity gains when binding‐arm positions were mutated (A21); inserting 2′‐5′ linkages after all As at once resulted in complete loss of activity (Figure S4). Restoring the 3′‐5′ linkage after A5 rescued limited activity and restoring them after A9, A11, and A15 resulted in further progressive activity gains.
To obtain a higher resolution picture of the effects of 2′‐5′ linkages on 10–23 activity, we investigated the effects of discrete 2′‐5′ linkages using a “pulse‐chase” approach similar to the recently described position‐selective labeling of RNA (PLOR21) method, which we adapted to a simple one‐pot reaction using TGLLK polymerase. Briefly, primer, template, and TGLLK were incubated with a single 3′dNTP to introduce one defined 3′ deoxynucleotide (pulse) and then synthesis was completed with unmodified dNTPs (chase), generating a position‐selective 2′‐5′ linkage (Figure S5).
We inserted position‐selective 2′‐5′ linkages after several nucleotides in the catalytic core (G1, G2, C3, T4, G6, C7, T8, and G14, Figure 4 d) known to affect 10–23 function. Several positions (G2, C7, T8) were largely unaffected by insertion of a 2′‐5′ linkage (>75 % wild type activity), suggesting a high tolerance to conformational distortions, while others (G1, C3, G6, G14) displayed more substantial drops in activity (20–40 % wild type activity). One position (T4) proved highly sensitive to insertion of a 2′‐5′ linkage (<10 % wild type activity).
These data suggest that T4 and the region C3–G6 in general are sensitive to structural mutagenesis, as well as to base substitutions,19 base deletions,22 or replacement with abasic sites.23 This may be because the essential Mg2+ ion coordinated by the T4–A5 backbone linkage is displaced (Figure S6).24 We also find position G2, which is highly intolerant to base substitution,19 is largely insensitive to the backbone distorting effects of 2′‐5′ linkages. Thus, our results show that structural mutagenesis through site‐specific insertion of 2′‐5′ linkages can probe novel aspects of nucleic acid structure and function and may be particularly useful for the identification of backbone‐mediated metal‐ion coordination sites.
In summary, we have described a novel engineered polymerase capable of template‐dependent synthesis of DNA and RNA with regioisomeric 2′‐5′ linkages. In the case of RNA synthesis, these are currently associated with the concomitant insertion of a 3′O‐methyl group. Future approaches may allow the incorporation of NTPs with a cleavable 3′ group, whereby mild deprotection would yield “pure” 2′‐5′ RNA in a manner analogous to cyclic reversible termination sequencing technologies25 or deacetylation of 2′‐O‐acetylated RNA.15
We also describe a rapid method for position‐selective structural mutagenesis of nucleic acids, whereby structure, conformation, and activity may be altered through insertion of regioisomeric backbone distortions, or indeed any unnatural nucleotide. While currently limited by the inefficient incorporation of 3′deoxy and 3′O‐methyl pyrimidines as well as multiple substitutions, the described methodologies allow rapid, template‐directed synthesis and reverse transcription of nucleic acid polymers containing mixed 2′‐5′/3′‐5′ backbone linkages. This procedure forms the foundation for future in vitro evolution experiments and a new approach for expanding the structural and functional repertoire of nucleic acid enzymes, ligands, and sensors.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information
Acknowledgements
This work was supported by a FEBS long‐term fellowship (H.M.) and by the Medical Research Council (program no. U105178804).
References
- 1. Hartmann R., Justesen J., Sarkar S. N., Sen G. C., Yee V. C., Mol. Cell 2003, 12, 1173–1185. [DOI] [PubMed] [Google Scholar]
- 2. Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Rohl I., Hopfner K. P., Ludwig J., Hornung V., Nature 2013, 498, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. Lohrmann R., Orgel L. E., Tetrahedron 1978, 34, 853–855; [Google Scholar]
- 3b. Kanavarioti A., Lee L. F., Gangopadhyay S., Origins Life Evol. Biospheres 1999, 29, 473–487; [DOI] [PubMed] [Google Scholar]
- 3c. Monnard P. A., Kanavarioti A., Deamer D. W., J. Am. Chem. Soc. 2003, 125, 13734–13740; [DOI] [PubMed] [Google Scholar]
- 3d. Szostak J. W., J. Syst. Chem. 2012, 3, 2. [Google Scholar]
- 4. Engelhart A. E., Powner M. W., Szostak J. W., Nat. Chem. 2013, 5, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.
- 5a. Prakash T. P., Jung K. E., Switzer C., Chem. Commun. 1996, 1793–1794; [Google Scholar]
- 5b. Sheppard T. L., Breslow R. C., J. Am. Chem. Soc. 1996, 118, 9810–9811; [Google Scholar]
- 5c. Giannaris P. A., Damha M. J., Nucleic Acids Res. 1993, 21, 4742–4749; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5d. Wasner M., Arion D., Borkow G., Noronha A., Uddin A. H., Parniak M. A., Damha M. J., Biochemistry 1998, 37, 7478–7486. [DOI] [PubMed] [Google Scholar]
- 6.
- 6a. Premraj B. J., Yathindra N., J. Biomol. Struct. Dyn. 1998, 16, 313–328; [DOI] [PubMed] [Google Scholar]
- 6b. Li L., Szostak J. W., J. Am. Chem. Soc. 2014, 136, 2858–2865; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6c. Sheng J., Li L., Engelhart A. E., Gan J., Wang J., Szostak J. W., Proc. Natl. Acad. Sci. USA 2014, 111, 3050–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.
- 7a. Jung K. E., Switzer C., J. Am. Chem. Soc. 1994, 116, 6059–6061; [Google Scholar]
- 7b. Robinson H., Jung K. E., Switzer C., Wang A. H. J., J. Am. Chem. Soc. 1995, 117, 837–838. [Google Scholar]
- 8.
- 8a. Cozens C., Pinheiro V. B., Vaisman A., Woodgate R., Holliger P., Proc. Natl. Acad. Sci. USA 2012, 109, 8067–8072; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Pinheiro V. B., Taylor A. I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J. C., Wengel J., Peak‐Chew S.‐Y., McLaughlin S. H., Herdewijn P., Holliger P., Science 2012, 336, 341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fogg M. J., Pearl L. H., Connolly B. A., Nat. Struct. Biol. 2002, 9, 922–927. [DOI] [PubMed] [Google Scholar]
- 10. Gardner A. F., Jack W. E., Nucleic Acids Res. 1999, 27, 2545–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramsay N., Jemth A. S., Brown A., Crampton N., Dear P., Holliger P., J. Am. Chem. Soc. 2010, 132, 5096–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Axelrod V. D., Vartikyan R. M., Aivazashvili V. A., Beabealashvili R. S., Nucleic Acids Res. 1978, 5, 3549–3563; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b. Montefiori D. C., R. W. Sobol, Jr. , Li S. W., Reichenbach N. L., Suhadolnik R. J., Charubala R., Pfleiderer W., Modliszewski A., W. E. Robinson Jr. , Mitchell W. M., Proc. Natl. Acad. Sci. USA 1989, 86, 7191–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Premraj B. J., Patel P. K., Kandimalla E. R., Agrawal S., Hosur R. V., Yathindra N., Biochem. Biophys. Res. Commun. 2001, 283, 537–543. [DOI] [PubMed] [Google Scholar]
- 14. Gloeckner C., Sauter K. B. M., Marx A., Angew. Chem. Int. Ed. 2007, 46, 3115–3117; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2007, 119, 3175–3178. [Google Scholar]
- 15. Bowler F. R., Chan C. K., Duffy C. D., Gerland B., Islam S., Powner M. W., Sutherland J. D., Xu J., Nat. Chem. 2013, 5, 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.
- 16a. Lorsch J. R., Bartel D. P., Szostak J. W., Nucleic Acids Res. 1995, 23, 2811–2814; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b. Sinha S., Kim P. H., Switzer C., J. Am. Chem. Soc. 2004, 126, 40–41. [DOI] [PubMed] [Google Scholar]
- 17. Santoro S. W., Joyce G. F., Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cairns M. J., King A., Sun L. Q., Nucleic Acids Res. 2003, 31, 2883–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zaborowska Z., Furste J. P., Erdmann V. A., Kurreck J., J. Biol. Chem. 2002, 277, 40617–40622. [DOI] [PubMed] [Google Scholar]
- 20. Premraj B. J., Raja S., Yathindra N., Biophys. Chem. 2002, 95, 253–272. [DOI] [PubMed] [Google Scholar]
- 21. Liu Y., Holmstrom E., Zhang J., Yu P., Wang J., Dyba M. A., Chen D., Ying J., Lockett S., Nesbitt D. J., Ferre‐D′Amare A. R., Sousa R., Stagno J. R., Wang Y. X., Nature 2015, 522, 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaborowska Z., Schubert S., Kurreck J., Erdmann V. A., Febs Lett. 2005, 579, 554–558. [DOI] [PubMed] [Google Scholar]
- 23. Wang B., Cao L. Q., Chiuman W., Li Y. F., Xi Z., Biochemistry 2010, 49, 7553–7562. [DOI] [PubMed] [Google Scholar]
- 24. Nawrot B., Widera K., Wojcik M., Rebowska B., Nowak G., Stec W. J., FEBS J. 2007, 274, 1062–1072. [DOI] [PubMed] [Google Scholar]
- 25. Guo J., Xu N., Li Z., Zhang S., Wu J., Kim D. H., Sano Marma M., Meng Q., Cao H., Li X., Shi S., Yu L., Kalachikov S., Russo J. J., Turro N. J., Ju J., Proc. Natl. Acad. Sci. USA 2008, 105, 9145–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information


