Abstract
Background
The genetic contribution to complex diseases or traits, including cardio-metabolic traits, has been elucidated recently by large-scale genome-wide association studies. These genome-wide association studies have indicated that most pleiotropic loci contain genes associated with lipids. Clinically, lipid related abnormalities are strongly associated with other diseases such as type 2 diabetes, coronary artery disease and hypertension. The aim of this study was to evaluate the shared genetic background of lipids and other cardio-metabolic traits.
Methods
We conducted meta-analyses of the association between 157 published lipid-associated loci and 10 cardio-metabolic traits in 14,028 Korean individuals genotyped using the Exome chip (Illumina HumanExome BeadChip). We also examined whether the pleiotropic effects of such loci constituted independent (i.e., biological) pleiotropy or mediated pleiotropy in these metabolic pathways.
Results
Eighteen lipid-associated loci were significantly associated with one of six cardio-metabolic traits after correction for multiple testing (P < 3.70 × 10−4). Region 12q24.12 had pleiotropic effects on fasting plasma glucose, blood pressure and obesity-related traits (body mass index and waist-hip ratio) independent of its effects on the lipid profile. Lipid risk scores, calculated according to whether or not subjects carried the risk allele for lipid traits, were significantly associated with fasting plasma glucose, blood pressure and obesity-related traits.
Conclusions
The 12q24.12 region showed ethnic-specific genetic pleiotropy among cardio-metabolic traits in this study. Our findings may help to account for molecular mechanisms based on shared genetic background underlying not only dyslipidemia, but also cardiovascular disease and type 2 diabetes.
Electronic supplementary material
The online version of this article (doi:10.1186/s12933-016-0337-1) contains supplementary material, which is available to authorized users.
Keywords: Genome-wide association study, Lipids, Cardio-metabolic traits, Pleiotropy, 12q24.12
Background
The etiology of cardiovascular disease (CVD) is complex, as both genetic and environmental factors contribute [1]. Common risk factors for CVD are cardio-metabolic traits such as lipids, blood pressure, obesity and diabetes [2]. Of these, lipid-related abnormalities are strongly associated with type 2 diabetes (T2D)-related traits and hypertension [3]. For example, treatment of dyslipidemia can reduce the incidence of T2D [4], and circulating levels of triglycerides (TG) and high-density lipoprotein (HDL) cholesterol are predictors of T2D [3]. However, although there are strong correlations between lipid-related abnormalities and cardio-metabolic traits at the clinical level, the genetic and molecular mechanisms underlying the link between lipids and other cardio-metabolic traits remain unclear.
Genetic contributions to complex diseases or traits, including cardio-metabolic traits, have been analyzed by large-scale genome-wide association studies (GWASs) [5–7]. To date, more than 157 loci have been associated with lipids [5]. Some of these loci are pleiotropic, that is, they also showed significant association with other cardio-metabolic traits [8]. The forms of pleiotropy are important to distinguish because they have different implications for disease risk and pathogenesis. Independent pleiotropy is considered as genuine pleiotropy, and refers to a genetic variant that affects more than one trait independently [9]. In contrast, mediated pleiotropy exists when one genetic variant has an effect on a trait that in turn affects another trait [10].
In this study, we aimed to examine the pleiotropic effects of published lipid-associated loci on 10 cardio-metabolic traits in a Korean population using data from 14,028 individuals directly genotyped by the Exome chip (Illumina HumanExome BeadChip). We also explored whether or not the observed pleiotropic associations were independent or mediated, and tested the hypothesis that the effects of associations between lipid loci and other cardio-metabolic traits were controlled by lipids. By making lipid risk scores, we examined the cumulative genetic effect of lipid-associated loci on other cardio-metabolic traits. Our results suggest the presence of shared genetic backgrounds and biological pathways for lipids and other risk factors for CVD.
Methods
Study participants
We performed Exome chip genotyping of 14,616 study participants from three population-based cohorts that are comprised in the Korean Genome Epidemiology Study (KoGES). We selected 7981 participants from the Ansung and Ansan population-based cohort (the Korea Association REsource (KARE) project), 3448 participants from the Health Examinee (HEXA) cohort and 3187 participants from Cardio Vascular Disease Association Study (CAVAS). All participants were aged between 40 and 69 years. More detailed descriptions of these three cohorts are available in published articles [11–13]. All participants provided written informed consent, and this study was approved by the ethical committee of our institute (Korea Centers for Disease Control and Prevention Institutional Review Board).
Phenotype determination
Blood levels of glycemic traits [fasting plasma glucose (FPG), glycated hemoglobin (HbA1c)] and lipids [TG, total cholesterol (TC), and HDL] were measured in plasma (FPG), whole blood (HbA1c) and serum (lipids), respectively, using standard enzymatic methods from fasting-individuals. If the TG level was <400 mg/dl, the concentration of low-density lipoprotein (LDL) cholesterol was calculated using Friedewald’s formula [14]. Blood pressure (BP) was measured using a standard mercury sphygmomanometer after participants had been in a sitting position for at least 5 min. The mean of two measures (left and right arm) was taken as the BP. If participants were taking antihypertensive agents, BP values were adjusted for analyses by adding 10 mmHg to systolic BP (SBP) and 5 mmHg to diastolic BP (DBP) [7]. Body mass index (BMI) and waist-hip ratio (WHR) were defined as weight (in kilograms) divided by the square of height (in meters) and the ratio of waist to hip circumferences (in centimeters), respectively.
Individuals taking lipid lowering or anti-diabetes medication and individuals with FPG ≥ 126 mg/dl and HbA1c ≥ 6.5 % were excluded from the analyses to obtain only non-diabetic individuals.
Exome chip
The Illumina HumanExome BeadChip is a genotyping array that includes variants represented by ~250,000 single nucleotide polymorphisms (SNPs) selected from exome and whole-genome sequences in ~12,000 individuals [15]. This chip is focused on protein-altering variants (non-synonymous, stop and splice variants) and represents diverse populations and multiple complex traits [16, 17]. More detailed explanations about the contents of the Exome chip can be found at the Exome chip design web page (http://genome.sph.umich.edu/wiki/Exome_Chip_Design).
Genotyping and quality control
All 14,616 study participants were genotyped using the Illumina HumanExome BeadChip v1.1 (variants = 242,901 SNPs). For genotype calling, each genotype was automatically clustered by the Illumina GenomeStudio v2011.1 software [18]. To improve the accuracy of variant calling, manual re-clustering and visual inspection were performed for genotypes based on the CHARGE clustering method [15]. After genotype calling, variants with completely missing (n = 1431), monomorphic SNPs (n = 162,803), Hardy–Weinberg equilibrium values of P < 1.0 × 10−6 (n = 1184) or genotype call rates of < 0.95 (n = 11) were excluded for quality control. Samples with a genotype call rate of < 99 % (n = 43), sex discrepancy (n = 51) or cryptic relatedness (n = 487), and withdrawal of participation (n = 7) were also removed for sample quality control.
After genotype and sample quality control, 77,472 SNPs and 14,028 samples (7524 from KARE, 3436 from HEXA and 3068 from CAVAS) were available for further analyses. The number of SNPs with different minor allele frequencies is summarized in Additional file 1: Table S1.
Selection of lipid-associated SNPs for analyses
We focused on 157 loci that have been identified previously in large GWAS on lipids with genome-wide significance (P < 5 × 10−8) [5]. Different lipids can have different best-associated SNPs in the same locus. After selection of the most strongly associated SNPs for each locus, 157 regions were mapped to 129 genes and 135 SNPs [minor allele frequency (MAF) > 0.001, equivalent to a minor allele count ≥ 5 in each cohort].
Calculation of lipid risk score
To test the combined effects of lipid SNPs on other cardio-metabolic traits, we calculated two types of risk score, an unweighted risk score and a weighted risk score, for each SNP and each individual based on 49 lipid-related SNPs that were replicated in this study. The list of SNPs is provided in Additional file 1: Table S3. The unweighted risk score was determined as the total number of risk alleles per individual, and the weighted risk score was calculated by weighting the risk alleles by their estimated effect size based on our data in this study [19]. Associations between lipid risk scores and cardio-metabolic traits were evaluated based on a linear regression model using the R program (version 2.15.3; http://www.r-project.org/). In this program, a linear model (function “lm” in the “stats” package) is constructed that includes variables such as lipid risk scores and cardio-metabolic traits. For example, when we choose a model with three variables, the script is as follows; > fit <− lm (y ~ x1 + x2 + x3, data = riskscore), > summary (fit). Age, sex and lipids (TG, TC, LDL and HDL) were included in the analyses as covariates.
Statistical analyses
Single-variant association analysis was conducted between 135 SNPs and 10 cardio-metabolic traits (TG, TC, LDL, HDL, SBP, DBP, FPG, HbA1c, BMI and WHR) using a linear regression model via EPACTS v3.2.4 (http://genome.sph.umich.edu/wiki/EPACTS) and PLINK 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/) programs [20]. The main command of association analysis is “--test q.linear” and “--linear” to each program, respectively. To address the issue of multiple testing, a robust threshold of association significance (⍺ = 0.05/135 for each trait, P < 3.7 × 10−4) was used based on the Bonferroni correction. Phenotypes used in the analyses were approximately normally distributed, and age and sex were incorporated into the analyses as covariates. Meta-analyses of association results from each cohort were performed with the inverse variance method using the METAL program (http://genome.sph.umich.edu/wiki/METAL) [21].
To evaluate and clarify the type of pleiotropy (independent or mediated) for three pleiotropic SNPs located on region 12q24.12, additional analyses were performed including adjustment of lipids (TC, TG, LDL and HDL) for other cardio-metabolic traits to examine whether or not any association occurred independently of lipids effects.
Protein–protein interactions
To search for functional interactions among genes, protein–protein interactions were performed using Wiki-Pi (http://severus.dbmi.pitt.edu/wiki-pi/) [22]. This tool provides information about one or both proteins in the interaction based on automatically updated annotations of individual proteins from databases such as Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, REACTOME, Pubmed2ENSEMBL and DrugBank.
Results
One hundred and fifty-seven lipid-associated loci, mapping to 129 genes and 135 SNPs (MAF > 0.001, equivalent to a minor allele count ≥ 5) in our directly genotyped Exome chip data, were selected for analysis. All loci had a genome-wide significance P value (P < 5 × 10−8) according to the Global Lipids Genetics Consortium [5]. The associations between each of the 135 SNPs and each of the 10 cardio-metabolic traits (TG, TC, HDL, LDL, SBP, DBP, FPG, HbA1c, BMI and WHR) were examined in 14,028 Korean individuals from three cohorts based on Exome chip genotyping. Baseline characteristics of the study participants are summarized in Additional file 1: Table S2.
Genetic effects of previously implicated lipid-associated SNPs
In the meta-analyses, 49 loci with new lead SNPs were significantly associated with one of four lipid traits (10 loci associated with TC, 11 with TG, 14 with HDL and 14 with LDL) after correction for multiple testing (P < 3.70 × 10−4) (Additional file 1: Table S3). Most signals were associated with two or more lipid traits. The overlap of loci associated with different lipid traits is shown in Fig. 1. Among the 49 replicated SNPs, 22 (45 %) were located in exonic regions and, of these, 18 (82 %) were non-synonymous variants. Several non-synonymous variants, which were located on PCSK9, CETP and APOA1 associated with TC or HDL and were novel protein-coding variants that had not been revealed in the same loci in previous studies.
Fig. 1.
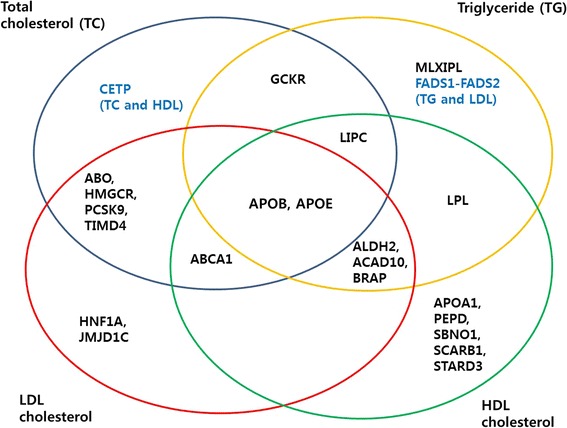
Overlap of loci associated with different lipids. This Venn diagram illustrates the name of lipid-associated loci that replicated in our study (P < 3.70 × 10−4)
Association between lipid-associated SNPs and other cardio-metabolic traits
Eighteen out of 135 SNPs were associated with other cardio-metabolic traits such as FPG, HbA1c, BMI, WHR, SBP and DBP after correction for multiple testing (P < 3.70 × 10−4) (Table 1; Fig. 2). Our findings replicated those of previous GWASs for five genes (GCKR, FADS1 and FADS2 for FPG, HFE for HbA1c and FTO for BMI) and also identified novel associations for 13 genes (three for FPG, four for WHR, three for SBP and three for DBP). Three associations were observed with one or more of four cardio-metabolic traits (FPG, WHR, SBP and DBP) within the genes BRAP, ACAD10 and ALDH2 in the 12q24.12 region (Table 1; Fig. 2). These associations had not been reported in previous GWASs. The three SNPs were composed of two non-synonymous and one intronic variant and had same direction of effect for each trait. All three SNPs showed monomorphic ethnic differences in allele frequency from European and South Asian descent in comparison with allele frequency from our data (Korean individuals, average MAF = 0.161) and East Asian descent (average MAF = 0.175; calculated using data from the 1000 Genomes Project, http://www.1000genomes.org/) (Additional file 1: Table S4).
Table 1.
Results of meta-analyses for associations between lipid loci and cardio-metabolic traits
| Category | Trait | CHR | SNP | Position | Gene | Function | A1/A2 | MAF | Effect size (beta ± s.e.) | P combined | P het (Q) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glycemic traits (n = 13,378) | FPG | 2 | rs780093 | 27742603 | GCKR | Intronic | G/A | 0.456 | 0.73 ± 0.13 | 6.26 × 10−9 | 0.72 (0.64) |
| 11 | rs174547 | 61570783 | FADS1 | Intronic | G/A | 0.319 | −0.70 ± 0.13 | 1.71 × 10−7 | 0.94 (0.12) | ||
| 11 | rs174570 | 61597212 | FADS2 | Intronic | A/G | 0.319 | −0.71 ± 0.13 | 1.09 × 10−7 | 0.97 (0.07) | ||
| 12 | rs3782886 | 112110489 | BRAP | Nonsynonymous | G/A | 0.167 | −0.90 ± 0.17 | 7.21 × 10−8 | 0.67 (0.79) | ||
| 12 | rs11066015 | 112168009 | ACAD10 | Intronic | A/G | 0.158 | −0.92 ± 0.17 | 5.93 × 10−8 | 0.74 (0.60) | ||
| 12 | rs671 | 112241766 | ALDH2 | Nonsynonymous | A/G | 0.157 | −0.96 ± 0.17 | 2.00 × 10−8 | 0.80 (0.44) | ||
| HbA1c | 6 | rs1799945 | 26091179 | HFE | Nonsynonymous | G/C | 0.045 | −0.06 ± 0.01 | 4.57 × 10−5 | – | |
| Blood pressure (n = 10,310) | SBP | 12 | rs3782886 | 112110489 | BRAP | Nonsynonymous | G/A | 0.167 | −1.34 ± 0.31 | 1.50 × 10−5 | 0.04 (4.15) |
| 12 | rs11066015 | 112168009 | ACAD10 | Intronic | A/G | 0.158 | −1.53 ± 0.32 | 1.33 × 10−6 | 0.03 (4.50) | ||
| 12 | rs671 | 112241766 | ALDH2 | Nonsynonymous | A/G | 0.157 | −1.49 ± 0.32 | 2.72 × 10−6 | 0.05 (4.01) | ||
| DBP | 12 | rs3782886 | 112110489 | BRAP | Nonsynonymous | G/A | 0.167 | −0.78 ± 0.20 | 1.02 × 10−4 | 0.49 (0.48) | |
| 12 | rs11066015 | 112168009 | ACAD10 | Intronic | A/G | 0.158 | −0.82 ± 0.21 | 7.12 × 10−5 | 0.41 (0.67) | ||
| 12 | rs671 | 112241766 | ALDH2 | Nonsynonymous | A/G | 0.157 | −0.80 ± 0.21 | 1.09 × 10−4 | 0.47 (0.52) | ||
| Obesity (n = 13,378) | BMI | 16 | rs1421085 | 53800954 | FTO | Intronic | G/A | 0.125 | 0.29 ± 0.06 | 1.84 × 10−7 | 0.70 (0.71) |
| WHR | 5 | rs3846662 | 74651084 | HMGCR | Intronic | A/G | 0.476 | 0.003 ± 0.001 | 3.69 × 10−4 | 0.29 (2.45) | |
| 12 | rs3782886 | 112110489 | BRAP | Nonsynonymous | G/A | 0.167 | −0.004 ± 0.001 | 3.45 × 10−5 | 0.12 (4.32) | ||
| 12 | rs11066015 | 112168009 | ACAD10 | Intronic | A/G | 0.158 | −0.005 ± 0.001 | 3.35 × 10−6 | 0.13 (4.14) | ||
| 12 | rs671 | 112241766 | ALDH2 | Nonsynonymous | A/G | 0.157 | −0.005 ± 0.001 | 2.87 × 10−6 | 0.13 (4.10) |
A threshold of association significance after correction for multiple testing is P < 3.70 × 10−4
CHR chromosome, A1/A2 minor allele/major allele, MAF minor allele frequency, n number of participants, SBP systolic blood pressure, DBP diastolic blood pressure, FPG fasting plasma glucose, HbA1c Glycated hemoglobin, BMI body mass index, WHR waist-hip ratio; All of analyses were adjusted by age and sex. s.e. standard error, P combined P value from meta-analyses using the results from three cohorts, P het P value from test of heterogeneity, Q Cochrane’s Q value based on Chi squared statistics
Fig. 2.
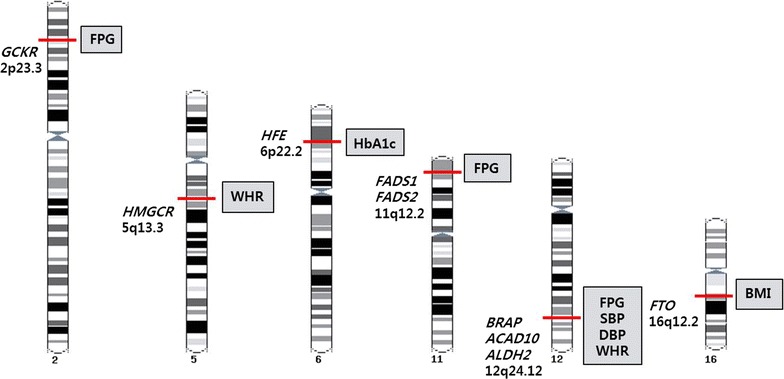
Summary of meta-analyses for associations between previously implicated lipid loci and cardio-metabolic traits. There are genes and genetic loci on chromosomes associated with cardio-metabolic traits that reached the statistical significance (P < 3.70 × 10−4) after correction for multiple testing in this study
The pleiotropic effect of SNPs in region 12q24.12
The pleiotropic effect of region 12q24.12 was identified for three SNPs (rs3782886 on BRAP, rs11066015 on ACAD10 and rs671 on ALDH2) on lipids and other cardio-metabolic traits (Table 2). These SNPs were associated with three lipid traits (TG, HDL and LDL), SBP, DBP, FPG, BMI and WHR (P < 0.05) and had the same direction of allelic effect for all cardio-metabolic traits (the linkage disequilibrium of these SNPs in our data, r2 = 0.988 between rs671 and rs11066015, r2 = 0.926 between rs671 and rs3782886) (Additional file 1: Figure S1). To identify the type of pleiotropy, adjusted association analyses were repeatedly performed across the eight cardio-metabolic traits. Mediated analyses showed that the association between rs671 and the three lipid traits (TG, HDL and LDL) remained significant after adjustments for all other cardio-metabolic traits (Table 3; Fig. 3; Additional file 1: Table S5). In addition, the association between rs671 and FPG, SBP, DBP, BMI and WHR remained significant after adjustments for the three lipid traits, suggesting that rs671 on ALDH2 is independently associated with each cardio-metabolic trait (Table 3; Fig. 3; Additional file 1: Table S5). The adjusted associations of rs3782886 and rs11066015 with each cardio-metabolic trait showed the same effects as rs671 (data not shown).
Table 2.
Pleiotropic effects of three SNPs located in the 12q24.12 region
| Traits | n | rs671, ALDH2 | rs11066015, ACAD10 | rs3782886, BRAP | |||
|---|---|---|---|---|---|---|---|
| Effect size (beta ± s.e.) | P | Effect size (beta ± s.e.) | P | Effect size (beta ± s.e.) | P | ||
| Lipids | |||||||
| TC | 13,378 | −0.03 ± 0.59 | 0.9547 | −0.03 ± 0.59 | 0.9633 | −0.26 ± 0.58 | 0.6502 |
| TG | 13,378 | −7.60 ± 1.72 | 9.47 × 10−6 | −7.28 ± 1.71 | 2.11 × 10−5 | −6.93 ± 1.68 | 3.53 × 10−5 |
| LDL | 13,378 | 2.55 ± 0.53 | 1.53 × 10−6 | 2.54 ± 0.53 | 1.50 × 10−6 | 2.13 ± 0.52 | 3.80 × 10−5 |
| HDL | 13,378 | −1.33 ± 0.18 | 1.20 × 10−13 | −1.33 ± 0.18 | 7.61 × 10−14 | −1.33 ± 0.17 | 2.97 × 10−14 |
| Blood pressure | |||||||
| SBP | 10,310 | −1.49 ± 0.32 | 2.72 × 10−6 | −1.53 ± 0.32 | 1.33 × 10−6 | −1.34 ± 0.31 | 1.50 × 10−5 |
| DBP | 10,310 | −0.80 ± 0.21 | 1.09 × 10−4 | −0.82 ± 0.21 | 7.12 × 10−5 | −0.78 ± 0.20 | 1.02 × 10−4 |
| Glycemic traits | |||||||
| FPG | 13,378 | −0.96 ± 0.17 | 2.00 × 10−8 | −0.92 ± 0.17 | 5.93 × 10−8 | −0.90 ± 0.17 | 7.21 × 10−8 |
| HbA1c | 7524 | 0.003 ± 0.008 | 0.6858 | 0.006 ± 0.008 | 0.4870 | 0.004 ± 0.008 | 0.5924 |
| Obesity | |||||||
| BMI | 13,378 | −0.16 ± 0.05 | 2.14 × 10−3 | −0.16 ± 0.05 | 2.10 × 10−3 | −0.13 ± 0.05 | 1.06 × 10−2 |
| WHR | 13,378 | −0.005 ± 0.001 | 2.87 × 10−6 | −0.005 ± 0.001 | 3.35 × 10−6 | −0.004 ± 0.001 | 3.45 × 10−5 |
n number of participants, s.e. standard error, TG Triglyceride, TC Total cholesterol, LDL Low density lipoprotein cholesterol, HDL high density lipoprotein cholesterol, SBP systolic blood pressure, DBP diastolic blood pressure, FPG fasting plasma glucose, HbA1c Glycated hemoglobin, BMI body mass index, WHR waist-hip ratio
All of analyses were adjusted by age and sex
Table 3.
Evaluation of pleiotropic effect of 12q24.12 (ALDH2, rs671) genotypes
| Trait | Effect size(beta ± s.e.) | P value after adjustment for age and sex | After adjustment for an additional covariate | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TG | LDL | HDL | FPG | |||||||
| Effect size | P value | Effect size | P value | Effect size | P value | Effect size | P value | |||
| TG | −7.60 ± 1.72 | 9.47 × 10−6 | – | – | −4.34 ± 1.18 | 2.44 × 10−4 | −12.24 ± 1.64 | 8.41 × 10−14 | −6.34 ± 1.68 | 1.58 × 10−4 |
| LDL | 2.55 ± 0.53 | 1.53 × 10−6 | 2.42 ± 0.54 | 7.47 × 10−6 | – | – | 2.76 ± 0.54 | 3.24 × 10−7 | 2.97 ± 0.56 | 9.78 × 10−8 |
| HDL | −1.33 ± 0.18 | 1.20 × 10−13 | −1.70 ± 0.18 | 1.80 × 10−21 | −1.53 ± 0.19 | 8.14 × 10−16 | – | – | −1.42 ± 0.19 | 8.88 × 10−14 |
| FPG | −0.96 ± 0.17 | 2.00 × 10−8 | −0.96 ± 0.19 | 2.00 × 10−7 | −1.11 ± 0.18 | 1.85 × 10−9 | −0.96 ± 0.19 | 2.38 × 10−7 | – | – |
| SBP | −1.49 ± 0.32 | 2.72 × 10−6 | −1.25 ± 0.33 | 1.37 × 10−4 | −1.45 ± 0.33 | 1.26 × 10−5 | −1.50 ± 0.33 | 5.68 × 10−6 | −1.09 ± 0.34 | 1.14 × 10−3 |
| DBP | −0.80 ± 0.21 | 1.09 × 10−4 | −0.67 ± 0.21 | 1.41 × 10−3 | −0.83 ± 0.21 | 1.03 × 10−4 | −0.90 ± 0.21 | 2.35 × 10−5 | −0.66 ± 0.22 | 2.35 × 10−3 |
| BMI | −0.16 ± 0.05 | 2.14 × 10−3 | −0.11 ± 0.05 | 2.90 × 10−2 | −0.20 ± 0.05 | 1.45 × 10−4 | −0.26 ± 0.05 | 3.86 × 10−7 | −0.14 ± 0.05 | 1.14 × 10−2 |
| WHR | −0.005 ± 0.001 | 2.87 × 10−6 | −0.004 ± 0.001 | 2.81 × 10−4 | −0.005 ± 0.001 | 1.92 × 10−5 | −0.007 ± 0.001 | 1.48 × 10−10 | −0.004 ± 0.001 | 3.54 × 10−4 |
| Trait | Effect size (beta ± s.e.) |
P value after adjustment for age and sex | After adjustment for an additional covariate | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SBP | DBP | BMI | WHR | |||||||
| Effect size | P value | Effect size | P value | Effect size | P value | Effect size | P value | |||
| TG | −7.60 ± 1.72 | 9.47 × 10−6 | −6.89 ± 1.94 | 3.80 × 10−4 | −6.92 ± 1.93 | 3.45 × 10−4 | −6.34 ± 1.71 | 2.14 × 10−4 | −5.71 ± 1.71 | 8.68 × 10−4 |
| LDL | 2.55 ± 0.53 | 1.53 × 10−6 | 2.66 ± 0.61 | 1.21 × 10−5 | 2.73 ± 0.61 | 7.29 × 10−6 | 2.72 ± 0.54 | 3.79 × 10−7 | 2.57 ± 0.54 | 2.09 × 10−6 |
| HDL | −1.33 ± 0.18 | 1.20 × 10−13 | −1.39 ± 0.19 | 3.16 × 10−13 | −1.40 ± 0.19 | 2.87 × 10−13 | −1.54 ± 0.19 | 1.45 × 10−16 | −1.47 ± 0.19 | 9.01 × 10−15 |
| FPG | −0.96 ± 0.17 | 2.00 × 10−8 | −0.84 ± 0.20 | 2.35 × 10−5 | −0.87 ± 0.20 | 1.20 × 10−5 | −0.91 ± 0.18 | 6.32 × 10−7 | −0.93 ± 0.19 | 4.85 × 10−7 |
| SBP | −1.49 ± 0.32 | 2.72 × 10−6 | – | – | −0.38 ± 0.19 | 4.78 × 10−2 | −1.16 ± 0.32 | 3.00 × 10−4 | −1.07 ± 0.32 | 9.21 × 10−4 |
| DBP | −0.80 ± 0.21 | 1.09 × 10−4 | −0.07 ± 0.12 | 0.5869 | – | – | −0.63 ± 0.20 | 2.08 × 10−3 | −0.57 ± 0.21 | 5.26 × 10−3 |
| BMI | −0.16 ± 0.05 | 2.14 × 10−3 | −0.10 ± 0.06 | 8.72 × 10−2 | −0.10 ± 0.06 | 8.47 × 10−2 | – | – | −0.05 ± 0.05 | 0.2964 |
| WHR | −0.005 ± 0.001 | 2.87 × 10−6 | −0.004 ± 0.001 | 8.28 × 10−4 | −0.004 ± 0.001 | 7.50 × 10−4 | −0.003 ± 0.001 | 5.59 × 10−4 | – | – |
Total cholesterol was excluded from this analysis. n = 13,378 for all traits except SBP and DBP, where n = 10,310
TG Triglyceride, LDL low density lipoprotein cholesterol, HDL high density lipoprotein cholesterol, FPG fasting plasma glucose, SBP systolic blood pressure, DBP diastolic blood pressure, BMI body mass index, WHR waist-hip ratio, s.e. standard error
Fig. 3.
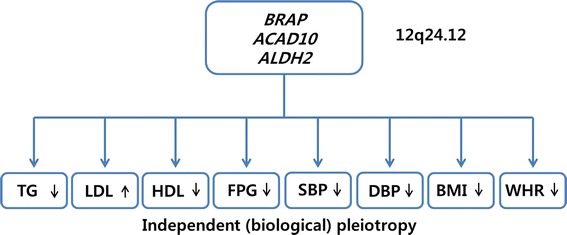
Pleiotropic effects on three genes (BRAP, ACAD10 and ALDH2) in the 12q24.12 region. This region was associated independently with each cardio-metabolic trait according to additional analyses including adjustments. The black arrows represented the direction of genetic effects on each trait
Protein–protein interactions
To assess the physical interactions among the three pleiotropic genes in the 12q24.12 region, protein–protein interactions were tested using the Wiki-Pi database (http://severus.dbmi.pitt.edu/wiki-pi/) [22]. There were numerous direct protein–protein interactions for the three genes: 12 interactions for ALDH2, one interaction for ACAD10 and 15 interactions for BRAP. According to this analysis, the three pleiotropic genes were functionally connected by one gene, APP, which encodes amyloid beta (A4) precursor protein (Additional file 1: Figure S2). This protein participated in several biological pathways and showed 1995 interactions with other proteins within Wiki-Pi data (Additional file 1: Table S6). In particular, APP has a role in the cholesterol metabolic process and in blood coagulation (Additional file 1: Table S6).
Associations between lipid risk scores and other cardio-metabolic traits
To examine the cumulative genetic effect of the 49 replicated lipid SNPs on other cardio-metabolic traits such as FPG, SBP, DBP, BMI and WHR, a lipid risk score was calculated for TG, TC, HDL and LDL from different sets of SNPs, and two types of lipid risk score were calculated for each lipid. The weighted risk score for LDL was associated with all five cardio-metabolic traits (P < 0.05) (Table 4). The unweighted risk score of the four lipids was associated with FPG (P < 0.05) (Table 4).
Table 4.
Pleiotropic association between lipid risk scores and cardio-metabolic traits
| Lipid RS | SBP | DBP | FPG | BMI | WHR | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect size | P value | Effect size | P value | Effect size | P value | Effect size | P value | Effect size | P value | |
| LDL_RS | −0.24 ± 0.07 | 2.87 × 10−4 | −0.12 ± 0.04 | 3.82 × 10−3 | −0.17 ± 0.04 | 2.43 × 10−6 | −0.02 ± 0.01 | 0.085 | −0.0006 ± 0.0002 | 8.55 × 10−3 |
| HDL_RS | −0.32 ± 0.07 | 9.05 × 10−6 | −0.15 ± 0.05 | 1.54 × 10−3 | −0.18 ± 0.04 | 7.17 × 10−6 | −0.02 ± 0.01 | 0.165 | −0.0004 ± 0.0002 | 0.113 |
| TG_RS | −0.20 ± 0.07 | 5.57 × 10−3 | −0.07 ± 0.05 | 0.103 | −0.19 ± 0.04 | 1.55 × 10−6 | −0.02 ± 0.01 | 0.160 | −0.0007 ± 0.0002 | 2.07 × 10−3 |
| TC_RS | 0.08 ± 0.10 | 0.457 | 0.03 ± 0.07 | 0.623 | 0.26 ± 0.06 | 3.27 × 10−6 | 0.04 ± 0.02 | 6.21 × 10−3 | 0.0010 ± 0.0003 | 2.52 × 10−3 |
| wLDL_RS | −0.04 ± 0.02 | 3.03 × 10−2 | −0.04 ± 0.01 | 4.95 × 10−3 | −0.06 ± 0.01 | 1.59 × 10−7 | −0.02 ± 0.00 | 5.77 × 10−9 | −0.0003 ± 0.0001 | 2.69 × 10−5 |
| wHDL_RS | 0.18 ± 0.05 | 4.42 × 10−4 | 0.17 ± 0.03 | 1.45 × 10−7 | 0.02 ± 0.03 | 0.396 | 0.07 ± 0.01 | 1.44 × 10−18 | 0.0017 ± 0.0002 | 8.11 × 10−25 |
| wTG_RS | 0.02 ± 0.01 | 0.070 | 0.01 ± 0.01 | 0.161 | −0.01 ± 0.005 | 0.237 | −0.003 ± 0.001 | 1.73 × 10−2 | −0.00001 ± 0.00003 | 0.798 |
| wTC_RS | −0.03 ± 0.02 | 0.183 | −0.02 ± 0.01 | 0.125 | −0.08 ± 0.01 | 2.32 × 10−10 | −0.02 ± 0.004 | 1.55 × 10−6 | −0.0002 ± 0.0001 | 8.87 × 10−3 |
Lipid RS lipid risk score, LDL_RS unweighted LDL risk score, HDL_RS unweighted HDL risk score, TG_RS unweighted TG risk score, TC_RS unweighted TC risk score, wLDL_RS weighted LDL risk score, wHDL_RS weighted HDL risk score, wTG_RS weighted TG risk score, wTC_RS weighted TC risk score, SBP systolic blood pressure, DBP diastolic blood pressure, FPG fasting plasma glucose, BMI body mass index, WHR waist-hip ratio
Effect size are shown as beta ± standard error. All of analyses were adjusted by age, sex and lipids
Discussion
In this study, we performed meta-analyses of associations between 135 SNPs and 10 cardio-metabolic traits using Exome chip data from 14,028 Korean individuals to assess whether lipid-associated loci reported by the Global Lipids Genetics Consortium are also applicable to cardio-metabolic traits in this population. The 49 SNPs out of 135 SNPs that located lipid-associated loci were significantly replicated in these Korean participants after correction for multiple testing. Eighteen SNPs showed substantial pleiotropic effects on other cardio-metabolic traits that are risk factors for cardiovascular disease, such as lipids. We also reported a major pleiotropic effect of three genes in the 12q24.12 region on cardio-metabolic traits and confirmed the relevance of independent pleiotropic effect. Independent pleiotropy can occur at the regional level, whereby multiple variants in the same region are associated with different traits [10]. The physiological interrelation among these three genes was revealed by protein–protein interactions. They were connected by a mediator gene, APP, that participates in the lipid metabolic process [23].
At the 12q24.12 pleiotropic region, the functional non-synonymous variant rs671 on ALDH2 which encodes the aldehyde dehydrogenase 2 family, has been reported to be associated with coronary artery diseases, blood pressure and alcohol consumption and is common in East-Asian populations in contrast to monomorphic in Europeans [7, 24]. One previous study provided evidence of positive selection at 12q24.12 by confirming the occurrence of a selective sweep in East-Asians by haplotype-based tests [7]. We predicted the functional effects, possibly damaging, potentially disease-causing biological effects, of the missense mutation at SNP rs671 using PolyPhen 2 and SIFT programs [25, 26]. In the current study, we established the genetic overlap of rs671 across eight cardio-metabolic traits, and first reported the association between this locus and FPG with genome-wide significance.
Two other SNPs in linkage disequilibrium (r2 > 0.9) with rs671 in Koreans, rs3782886 and rs11066015, showed the same genetic effects as rs671. Rs3782886 is a non-synonymous variant located on BRAP, which encodes BRCA1-associated protein, and rs11066015 is an intronic variant on ACAD10, which encodes acyl-CoA dehydrogenase family member 10. Previous studies have reported that ACAD10 is associated with coronary artery disease and T2D, and it has been suggested that the effect of ACAD10 on T2D might be mediated by disordered insulin resistance that is due to abnormal lipid oxidation [27, 28]. The two SNPs rs3782886 and rs11066015 were polymorphic only in individuals of East-Asian descent. This suggests that the pleiotropic associations described in our study may be specific to East-Asian individuals.
The three genes in the 12q24.12 region, ALDH2, BRAP and ACAD10, were functionally linked by a mediator gene, APP, that takes part in lipid-related metabolic processes (MIM104760). APP is a complex biological molecule that interacts with many types of receptors or proteins [29]. Mutations in APP have been implicated in Alzheimer’s disease and cerebroarterial amyloidosis [30]. Alzheimer’s disease is believed to be driven by the accumulation and deposition of the amyloid beta precursor protein that encoded by APP in brain region [23]. The abnormal accumulation or clearance of the amyloid beta precursor protein is regulated by cholesterol levels [23]. High LDL and low HDL is commonly found in patients with CVD and it has been shown to correlate with an increased risk of Alzheimer’s disease, with high risk being associated with advanced CVD [31]. As the three genes in 12q24.12 region is well known to association with CVD and lipid metabolism, the functional relation between APP and three genes in our study newly suggests that APP might play an important role in modulating CVD and dyslipidemia as well as Alzheimer’s disease.
An additional investigation was performed to reveal the combined genetic effect of lipid SNPs on other cardio-metabolic traits. This was conducted using lipid risk scores calculated by combination of the lipid SNPs implicated in this study. We were able to detect significant associations between unweighted or weighted lipid risk scores and cardio-metabolic traits. All four of the unweighted lipid risk scores were significantly associated with FPG. This provides compelling evidence that lipid risk SNPs have the potential to be used for identification of T2D risk. The finding that unweighted and weighted HDL risk scores were associated with SBP and DBP also suggests a shared genetic background between these traits. Our results showing an association between lipid risk scores and cardio-metabolic traits require replication in other ethnic groups.
Future directions
In this study, we have presented lipid-related genetic variants with pleiotropic effects on other cardio-metabolic traits. Recent studies suggested that pleiotropic effects on complex traits may be widespread. Genetic variants associated with two or more traits, such as rs7138803 on FAIM2 that related to obesity, T2D and myocardial infarction, rs8192673 on PGC-1α and rs1801282 on PPAR-γ associated with waist circumference and T2D, were revealed by previous studies [32–34]. The increased use of genetic information in clinical practice, has underscored the importance of understanding pleiotropy and its implications for genetic testing and personal genomics. A gene or multiple genes in a biological pathway that affect more than one trait present new chances and challenges for drug development. However, most studies have focused only on the detection of pleiotropic effects. Functionally characterizing identified variants through experiments and understanding the mechanisms of shared pathophysiology are necessary to establish causality. In addition, gene-environmental interactions (G × E) could provide insights the role of environmental factors in disease risk and hence to investigate their role as genetic factor modifiers [35]. For example, rs671 on ALDH2 was found to be associated with TG when the interaction between SNP and alcohol consumption was considered [36]. By integrating biological knowledge with genetic pleiotropy and environmental factors, G × E interactions may provide the potential to shed light on biological processes leading to diseases and could explain ‘missing heritability’ for phenotypic variance in GWASs. And also, it may be necessary to search for additional shared genetic patterns on multiple phenotypes in larger sample sizes across diverse ethnic cohorts as well as Korean population.
Conclusions
In summary, our results provide evidence that lipid-related loci implicated by previous GWASs also affect other cardio-metabolic traits in Korean individuals. We identified SNPs in the 12q24.12 region with ethnic-specific independent pleiotropic effects on both lipids and one or more metabolic traits. Our results may help to elucidate the East-Asian specific shared genetic background for cardio-metabolic traits that affect the risk of cardiovascular disease.
Authors’ contributions
YKK provided the study design and performed data analysis, writing and editing manuscript. MYH, YJK and SH participated in data analysis, SM and BJK participated in general discussion, reviewed data and editing manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by an intramural grant from the Korea National Institute of Health (2012-N73002-00, 2014-NI73001-00) and grants from the Korea Centers for Disease Control and Prevention (4845-301, 4851-302, 4851-307).
Competing interests
The authors declare that they have no competing interests.
Additional file
10.1186/s12933-016-0337-1 The direction of the allelic effect for each of three pleiotropic SNPs in the 12q24.12 region. Figure S2. Protein-protein interactions among three genes in the 12q24.12 region. Table S1. Frequency distribution of minor allele frequencies in Exome chip data. Table S2. Descriptive information of study subjects. Table S3. Replication results of known lipid-associated loci in Korean individuals. Table S4. Population diversity of three pleiotropic SNPs in the 12q24.12 region. Table S5. Evaluation of pleiotropic effect of 12q24.12 (ALDH2, rs671) genotypes. Table S6. Protein-protein interactions among three genes in 12q24.12 region.
Contributor Information
Yun Kyoung Kim, Email: bborumi@gmail.com.
Mi Yeong Hwang, Email: myhwang85@gmail.com.
Young Jin Kim, Email: anwltlarkr@gmail.com.
Sanghoon Moon, Email: moon.sanghoon@daum.net.
Sohee Han, Email: smallstar1999@hanmail.net.
Bong-Jo Kim, Phone: +82-43-719-8870, Email: kbj6181@cdc.go.kr.
References
- 1.Dauriz M, Meigs JB. Current insights into the joint genetic basis of type 2 diabetes and coronary heart disease. Cur Cardiovasc Risk Rep. 2014;8(1):368. doi: 10.1007/s12170-013-0368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donnell CJ, Elosua R. Cardiovascular risk factors. Insights from Framingham Heart Study. Rev Esp Cardiol. 2008;61(3):299–310. doi: 10.1157/13116658. [DOI] [PubMed] [Google Scholar]
- 3.Riserus U, Arnlov J, Berglund L. Long-term predictors of insulin resistance: role of lifestyle and metabolic factors in middle-aged men. Diabetes Care. 2007;30(11):2928–2933. doi: 10.2337/dc07-0360. [DOI] [PubMed] [Google Scholar]
- 4.Freeman DJ, Norrie J, Sattar N, Neely RD, Cobbe SM, Ford I, Isles C, Lorimer AR, Macfarlane PW, McKillop JH, et al. Pravastatin and the development of diabetes mellitus: evidence for a protective treatment effect in the West of Scotland Coronary Prevention Study. Circulation. 2001;103(3):357–362. doi: 10.1161/01.CIR.103.3.357. [DOI] [PubMed] [Google Scholar]
- 5.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43(6):531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rankinen T, Sarzynski MA, Ghosh S, Bouchard C. Are there genetic paths common to obesity, cardiovascular disease outcomes, and cardiovascular risk factors? Circ Res. 2015;116(5):909–922. doi: 10.1161/CIRCRESAHA.116.302888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ligthart S, de Vries PS, Uitterlinden AG, Hofman A, Franco OH, Chasman DI, Dehghan A. Pleiotropy among common genetic loci identified for cardiometabolic disorders and C-reactive protein. PLoS One. 2015;10(3):e0118859. doi: 10.1371/journal.pone.0118859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14(7):483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 12.Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, Hwang JY, Oh JH, Kim DJ, Kim NH, et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet. 2011;43(10):990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- 13.Hwang JY, Sim X, Wu Y, Liang J, Tabara Y, Hu C, Hara K, Tam CH, Cai Q, Zhao Q, et al. Genome-wide association meta-analysis identifies novel variants associated with fasting plasma glucose in East Asians. Diabetes. 2015;64(1):291–298. doi: 10.2337/db14-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 15.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One. 2013;8(7):e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, Brody JA, Khetarpal SA, Crosby JR, Fornage M, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94(2):223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, Stringham HM, Sim X, Yang L, Fuchsberger C, Cederberg H, et al. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45(2):197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, He J, Zhao S, Wu H, Zhong X, Sheng Q, Samuels DC, Shyr Y, Long J. Illumina human exome genotyping array clustering and quality control. Nat Protoc. 2014;9(11):2643–2662. doi: 10.1038/nprot.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li N, van der Sijde MR, Bakker SJ, Dullaart RP, van der Harst P, Gansevoort RT, Elbers CC, Wijmenga C, Snieder H, Hofker MH, et al. Pleiotropic effects of lipid genes on plasma glucose, HbA1c, and HOMA-IR levels. Diabetes. 2014;63(9):3149–3158. doi: 10.2337/db13-1800. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orii N, Ganapathiraju MK. Wiki-pi: a web-server of annotated human protein-protein interactions to aid in discovery of protein function. PLoS One. 2012;7(11):e49029. doi: 10.1371/journal.pone.0049029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puglielli L, Tanzi RE, Kovacs DM. Alzheimer’s disease: the cholesterol connection. Nat Neurosci. 2003;6(4):345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, Kobayashi S, Ogihara T, Yamori Y, Fujioka A, et al. Confirmation of ALDH2 as a Major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J. 2011;75(4):911–918. doi: 10.1253/circj.CJ-10-0774. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 27.Lee JY, Lee BS, Shin DJ, Woo Park K, Shin YA, Joong Kim K, Heo L, Young Lee J, Kyoung Kim Y, Jin Kim Y, et al. A genome-wide association study of a coronary artery disease risk variant. J Hum Genet. 2013;58(3):120–126. doi: 10.1038/jhg.2012.124. [DOI] [PubMed] [Google Scholar]
- 28.Bian L, Hanson RL, Muller YL, Ma L, Kobes S, Knowler WC, Bogardus C, Baier LJ. Variants in ACAD10 are associated with type 2 diabetes, insulin resistance and lipid oxidation in Pima Indians. Diabetologia. 2010;53(7):1349–1353. doi: 10.1007/s00125-010-1695-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadigh-Eteghad S, Sabermarouf B, Majdi A, Talebi M, Farhoudi M, Mahmoudi J. Amyloid-beta: a crucial factor in Alzheimer’s disease. Med Princ Pract. 2015;24(1):1–10. doi: 10.1159/000369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conidi ME, Bernardi L, Puccio G, Smirne N, Muraca MG, Curcio SA, Colao R, Piscopo P, Gallo M, Anfossi M, et al. Homozygous carriers of APP A713T mutation in an autosomal dominant Alzheimer disease family. Neurology. 2015;84(22):2266–2273. doi: 10.1212/WNL.0000000000001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofman A, Ott A, Breteler MM, Bots ML, Slooter AJ, van Harskamp F, van Duijn CN, Van Broeckhoven C, Grobbee DE. Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer’s disease in the Rotterdam Study. Lancet. 1997;349(9046):151–154. doi: 10.1016/S0140-6736(96)09328-2. [DOI] [PubMed] [Google Scholar]
- 32.Corella D, Sorli JV, Gonzalez JI, Ortega C, Fito M, Bullo M, Martinez-Gonzalez MA, Ros E, Aros F, Lapetra J, et al. Novel association of the obesity risk-allele near Fas Apoptotic Inhibitory Molecule 2 (FAIM2) gene with heart rate and study of its effects on myocardial infarction in diabetic participants of the PREDIMED trial. Cardiovasc Diabetol. 2014;13:5. doi: 10.1186/1475-2840-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruzliak P, Haley AP, Starcevic JN, Gaspar L, Petrovic D. Polymorphisms of the peroxisome proliferator-activated receptor-gamma (rs1801282) and its coactivator-1 (rs8192673) are associated with obesity indexes in subjects with type 2 diabetes mellitus. Cardiovasc Diabetol. 2015;14:42. doi: 10.1186/s12933-015-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams JN, Raffield LM, Freedman BI, Langefeld CD, Ng MC, Carr JJ, Cox AJ, Bowden DW. Analysis of common and coding variants with cardiovascular disease in the Diabetes Heart Study. Cardiovasc Diabetol. 2014;13:77. doi: 10.1186/1475-2840-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11(4):259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan A, Sun J, Xia N, Qin X, Hu Y, Zhang S, Tao S, Gao Y, Yang X, Zhang H, et al. A genome-wide association and gene-environment interaction study for serum triglycerides levels in a healthy Chinese male population. Hum Mol Genet. 2012;21(7):1658–1664. doi: 10.1093/hmg/ddr587. [DOI] [PubMed] [Google Scholar]


